Abstract
The COVID-19 pandemic caused by SARS-CoV-2 has emerged as a global catastrophe. The virus requires main protease for processing the viral polyproteins PP1A and PP1AB translated from the viral RNA. In search of a quick, safe and successful therapeutic agent; we screened various clinically approved drugs for the in-vitro inhibitory effect on 3CLPro which may be able to halt virus replication. The methods used includes protease activity assay, fluorescence quenching, surface plasmon resonance (SPR), Thermofluor® Assay, Size exclusion chromatography and in-silico docking studies. We found that Teicoplanin as most effective drug with IC50 ~ 1.5 μM. Additionally, through fluorescence quenching Stern–Volmer quenching constant (KSV) for Teicoplanin was estimated as 2.5 × 105 L·mol−1, which suggests a relatively high affinity between Teicoplanin and 3CLPro protease. The SPR shows good interaction between Teicoplanin and 3CLPro with KD ~ 1.6 μM. Our results provide critical insights into the mechanism of action of Teicoplanin as a potential therapeutic against COVID-19. We found that Teicoplanin is about 10–20 fold more potent in inhibiting protease activity than other drugs in use, such as lopinavir, hydroxychloroquine, chloroquine, azithromycin, atazanavir etc. Therefore, Teicoplanin emerged as the best inhibitor among all drug molecules we screened against 3CLPro of SARS-CoV-2.
Keywords: 3CL-protease, Activity inhibition, Drug repurposing
Highlights
-
•
3CLPro protease is an attractive drug target to halt the viral replications of SARS-CoV-2.
-
•
Screening of approved molecules against 3CLPro protease of SARS-CoV-2
-
•
Biochemical activity and biophysical analysis of molecules against 3CLPro protease
-
•
Teicoplanin found to be an effective inhibitor of 3CLPro protease of SARS-CoV-2
1. Introduction
COVID-19 is an infectious disease caused by a newly discovered positive-sense single-stranded RNA virus called the SARS-CoV-2 (severe acute respiratory syndrome coronavirus 2). The disease has emerged as a pandemic and causes respiratory complications in a majority of the cases and has caused lakhs of deaths worldwide owing to the lack of any treatment option. The family Coronaviridae under the order Nidovirales contains the members of Coronavirus (CoV) that are a potential health concern to human beings and possibly to other animals [1,2], having been responsible diseases such as severe acute respiratory syndrome (China) and Middle-East respiratory syndrome (MERS) [3]. Bats and other animals are natural reservoirs for CoVs, and SARS-CoV-2 [4]. However, the reported route of transmission till date is human to human that occurs by sneezing, coughing and spread of respiratory aerosols.
The symptoms manifested by infection of SARS-CoV-2 include the alteration in lung functioning, localized lesions, pneumonia, bronchiolitis and these are presented in a majority of the patients. The virus infects the lung endothelial cells and induces a pathological state like the lymphocytic endothelialitis and inflammatory cell invasion [5]. The SARS-CoV-2 infects multiple organs and recruits enormous numbers of immune cells and complexes in these organs [4]. It may also show central nervous system invasion can be presented in advanced stages of the CoV infection [6]. Other mild manifestations of the COVID-19 include fever, dry cough, dyspnoea, myalgia and fatigue. The hematological and serological examination reveals the augmentation in the levels of lactate dehydrogenase, serum amyloid A (SAA), and thrombocytopenia [7,8].
SARS-CoV-2 comprising of about 30,000 RNA, encodes for about 66% non-structural region. The nsP5 is chymotrypsin-like (CL) and possesses cysteine protease activity [9]. It is called the main protease or 3CLPro. This protease is essential for the processing of polyproteins PP1A and PP1B, translated from the RNA of the virus. The 3CLPro is very important for virus to replicate and propagate and its inhibitors may therefore be able to halt the replication of the virus. It recognizes and cleaves the virus non-structural polyprotein at 12 sites. It generally acts on the sequence Leu-Gln*(Ser, Ala, Gly) (* denotes the cleavage site). Due to its role in initiation events of viral replication, it is an attractive drug target. Besides 3CLPro, nucleocapsid protein (N), envelope protein (E), spike glycoprotein (S), membrane protein (M), and two isoforms of replicase polyprotein, namely 1a and 1ab are considered as potential drug/vaccine targets [10]. Its indispensable role in the initiation events of the replication cycle makes it an attractive drug target [11]. 3CLPro is an attractive and relatively safer drug target because its recognition sequence is dissimilar to any of the proteases in the human body.
A number of clinically approved drugs are being tested for their potential to ameliorate the effects of the SARS-CoV-2 infection. We tested the different classes of drugs - nucleoside analogues, antiretroviral agents, HIV protease inhibitors and neuraminidase inhibitors - for their potential antiviral effect. Teicoplanin is an effective glycopeptide antibiotic used in the prevention and treatment of various serious infections caused by gram-positive bacteria, including methicillin-resistant Staphylococcus aureus (MRSA) and Enterococcus faecalis. Some of the drugs under clinical trials for COVID-19 like hydroxychloroquine, chloroquine, nitazoxanide, oseltamivir, amoxicillin, famciclovir, aciclovir, lopinavir, atazanavir, and azithromycin [12] have been screened in this study. Here, we present Teicoplanin as a potent inhibitor of main protease and speculate its role as to halt the replication of the virus.
2. Materials and methods
2.1. Cloning and purification of 3CLPro
SARS-CoV-2 3CLPro gene was cloned between BamHI and XhoI sites in pET28a vector having N terminal 6×-His tag, FLAG tag and PreScission protease tag. Protein was transformed in BL-21DE3 Rosetta RIL cells and autoinduction method was used for expression and purification. The culture was grown at 37 °C for 2–3 h and 18 °C for overnight. The cell pellet after centrifugation at 10,000 ×g for 30 min, was resuspended in 50 mM Tris pH 7.5, 500 mM NaCl, 10% glycerol, 10 mM Imidazole, 1 mM DTT, 0.5 mM PMSF, 10 μg DNaseI and lysozyme (1 mg/ml). After 30 min incubation, it was sonicated at 50% duty cycles with 30 s ON and 30 s OFF for 20 min. The supernatant was applied on Ni-NTA columns in FPLC system (ÄKTA™ start, GE Healthcare). The bound proteins were eluted with 300 mM imidazole gradient. The protein fractions were applied on Q-FF ion-exchange chromatography, eluted with 500 mM NaCl gradient. The His tag was cleaved with PreScission protease and subjected to size exclusion chromatography (SEC) (Superdex™ 75 10/300 GL, GE Healthcare). Homogeneous fractions were pooled, dialyzed and kept at −80 °C for all biochemical and biophysical studies.
2.2. Activity inhibition assay
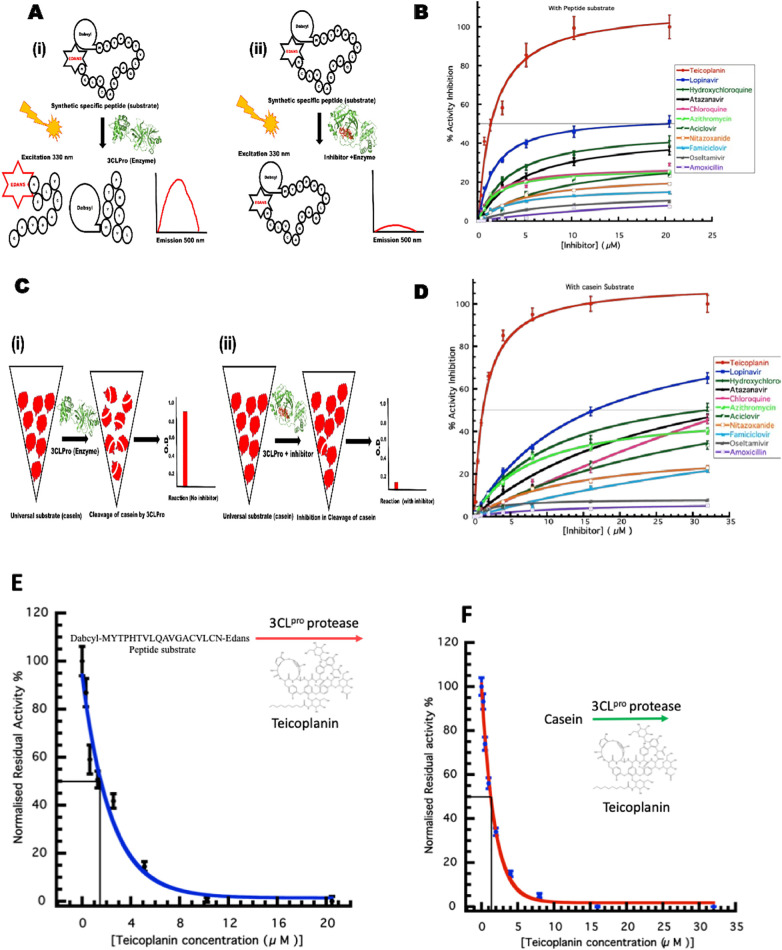
3CLPro amino acid sequence is highly conserved between SARS-CoV and SARS-CoV-2, it cleaves 11 sites in the polyproteins to result many functional proteins, including helicase, RNA-dependent RNA polymerase, protease, RNA-binding protein, exo-ribonuclease, endo-ribonuclease, and 2′-O-ribose methyltransferase etc. 3CLpro belongs to cysteine protease category and is excised from polyproteins by its own proteolytic activity, further exhibiting specificity for its own C-terminal auto processing. Various peptide cleavage sites have been reported in literature for the in-vitro 3CLPro protease activity measurement [13]. In this study cleavage sequence MYTPHTVLQ↓AVGACVLCN (cleavage at nsp12/nsp13) has been used for FRET based protease assay.
2.2.1. Protease activity using specific substrate
A well-established method for assessment of protease activity by 3CLPro of SARS-CoV has been described previously [15]. Thus, a custom synthesized fluorogenic peptide substrate was procured for the 3CLPro protease, DABCYL-MYTPHTVLQAVGACVLCN-EDANS {FRET pair as EDANS (Flourophore) and DABCYL (Quencher)} (Henan Tianfu Chemical Co., Ltd., China). This peptide substrate qualifies as a generic substrate for the main protease of various coronaviruses including the SARS-CoV, by the virtue of it possessing the nsp12/nsp13 cleavage sequence, MYTPHTVLQ↓AVGACVLCN. The peptide was dissolved in distilled water and used to measure the activity of the protease. Initial velocity for enzymatic activity using 1 μM 3CLPro protease and 0–10 μM of peptide substrate in 20 mM HEPES, pH 7.0 was plotted as a function of fluorescence intensity arbitrary unit and peptide concentration. The rate of proteolytic activity was determined by measuring the increase in fluorescence intensity of enzymatic reactions in 500 μl volume conducted in a fluorescence cuvette. The relative fluorescent unit (RFU) is linearly proportional to the amount of AVGACVLCN-EDANS generated by protease activity. The slope of the curve was used for enzymatic activity of enzyme for the FRET peptide substrate.
In the FRET assay for determination of kinetic parameters, 1 μM of 3CLPro enzyme was incubated with varied concentration (2.5–100 μM) of the peptide substrate DABCYL-MYTPHTVLQ↓AVGACVLCN-EDANS for an 1 h at 25 °C and the intensity of fluorescence was measured. The excitation and emission parameters for measurement were kept as 330 nm and 500 nm, respectively, using Cary Eclipse fluorescence spectrophotometer (Agilent Technologies). All reactions were performed at 25 °C in temperature-controlled conditions. The initial velocity slope was used for the fluorescence intensity rate calculation and plotted against varied substrate concentrations for the determination of the kinetic parameters including Km, Vmax, k cat and k cat/Km values.
For the inhibitor assay, the above experiment was conducted with 1 μM of 3CLPro in 20 mM HEPES, pH 7.0 buffer incubated with varying inhibitor concentrations and 50 μM of peptide substrate in 500 μl, incubated at 25 °C for an hour before the measurements. The enzyme inhibitory potential IC50 value was calculated by fitting the curve of normalised residual activity with inhibitor concentration. The reactions for each set were performed in triplicates.
2.2.2. Protease activity using casein substrate
Through the sequence analysis, we found that casein substrate has some cleavage junction peptides matching with the consensus cleavage sequences of 3CLPro of SARS-CoV-2. Hence, we have used the casein substrate also for the protease activity measurements. The protease activity of 3CLPro was previously established by using casein substrate [14,15] by the measurement of tyrosine liberated as a result of enzymatic activity. A calibration curve was prepared from absorbance measurements at 280 nm with standard L-tyrosine solutions and a slope was obtained. The activity of 3CLPro was estimated by using 1 μM of enzyme with different casein concentrations in 400 μl phosphate buffer pH 7.5 incubated at 37 °C for an hour before terminating the reaction with 200 μl of 1.7% trichloro-acetic acid. The reaction was centrifuged at 15,000 ×g for 10 min and tyrosine concentration was monitored in cuvette at 280 nm using Beckman Coulter DU 800 spectrophotometer. The amount of tyrosine liberated was calculated using tyrosine standard slope, plotted as a function of substrate concentration and fitted with Michaelis-Menten equation. Further, the Lineweaver-Burk plot was used to determine the kinetic parameters Km, Vmax, Kcat and catalytic efficiency Kcat/Km. In the compound inhibition study, 1 μM of 3CLPro was incubated with varying inhibitor concentration for an hour followed by addition of 135 μM of casein in 400 μl phosphate buffer at pH 7.5, incubated at 37 °C for an hour before terminating the reaction with 200 μl of 1.7% trichloro-acetic acid and absorbance measurements was done as above. The enzyme inhibitory potential IC50 value was calculated by fitting the curve of percentage residual activity with inhibitor concentration.
2.3. Studying protein-drug interaction studies using fluorescence spectroscopy
Fluorescence quenching is routinely used to ascertain the effect of a particular drug on the target protein. The impact of the drug on the protein can be dynamic due to collisions between the fluorophore (protein) and the quencher (drug), or it may be static, owing to the protein and the quencher forming a complex at ground-state [16,17]. An intense quenching of the fluorescence marked an unvarying local dielectric environment. The tertiary structure perturbations post-interaction to ligands were assessed using the principle of intrinsic fluorescence of 3CLPro which has three tryptophan residues which contribute to its hydrophobicity and fluorescence properties. In the fluorescence quenching experiment 1 μM of 3CLPro was allowed to interact with varying concentrations of drugs. Fluorescence signals were monitored at 295 nm excitation with emission scan between 310 and 400 nm using Cary Eclipse fluorescence spectrophotometer (Agilent technologies).
2.4. Bimolecular interaction by surface plasmon resonance spectroscopy
The binding between 3CLPro and Teicoplanin was measured using surface plasmon resonance spectroscopy (SPR) Biacore 3000 (GE Healthcare.). For our SPR experiments, 3CLPro protein (10 mg/ml) solution in PBS buffer pH 7.4 was first immobilized to a CM5 gold chip using EDC-NHS coupling and the binding of varying concentrations of Teicoplanin was tested. The resultant sensorgram was analysed using BIA evaluation software to determine the binding constants.
2.5. Thermofluor® assay
Bimolecular interactions of 3CLPro and Teicoplanin were measured by Thermofluor® assay [18]. 3CLPro enzyme was incubated with Teicoplanin and 20× SYPRO® Orange Protein Gel Stain (S5692) in buffer and thermal scanning was performed from 25 °C to 99 °C at an interval of 0.5 °C/min with hold of 0.5 min at each measurement. The shift in melting curve was plotted and analysed.
2.6. In-silico analysis of molecular interaction between Teicoplanin and 3CLPro
The backbone chain without water or other drugs of 3CLPro (PDB: 6LU7) was used for docking studies using ParDOCK+. The best prediction coordinates were in agreement with the reported active sites i.e. His41 and Cys145. The coordinate near the active site was fixed and subjected to site-specific docking by ParDOCK+. The 3-Dimensional structure of Teicoplanin (Pubchem ID: 16152170) was used as a ligand for docking. The partial charge model used was AM1BCC with minimization cycles of 2500. The four best poses were received and analysed.
3. Results
3.1. Purification and validation of 3CLPro
The protein 3CLPro was cloned in pET28a vector and purified using affinity, ion exchange and SEC (Fig. 1 ). The relative molecular weight of the protein was observed ~34 kDa on 12% SD-PAGE. The protein identity was confirmed with western blot using FLAG antibody (DYKDDDDK, # 2368 Cell Signaling Technology).
Fig. 1.

3CLPro purification and validation. 3CLPro showing a monomeric protein on SEC. In the inset (i) 12% denaturing gel showing the protein band ~34 kDa, (ii) western blot of the 3CLPro performed using anti-flag antibody.
3.2. Inhibition of 3CLPro enzymatic activity with inhibitors
FRET-based assay and casein assay was employed for the protease activity measurement. The relative fluorescence units were calculated for the correlation of fluorescence intensity and enzymatic activity. Initial velocity for enzymatic activity was plotted as a function of time which shows the typical fluorescence profile for the hydrolysis of the substrate. For calibration curve, 1 μM of enzyme was used with varied concentrations (from 0 to 10 μM) of substrate in a final volume of 500 μl reaction buffer, incubated for an hour. Initial velocities were determined from the linear section of the curve, slope obtained is 4.1 RFU per hour (or 0.00114 RFU per sec), and the corresponding relative fluorescence units per unit of time (RFU/s) was converted to the amount of the cleaved substrate by fitting to the calibration curve (Fig. S1A). The kinetic parameter was calculated using 1 μM enzyme in 500 μl volume by varying the peptide substrate concentrations (0–100 μM) and fitted with the Michaelis-Menten equation as well as Lineweaver-Burk plot (Fig. S1B and C). From the double reciprocal plot, ignoring the outlier points, parameters obtained are as: Km = (17.2 ± 1.7) × 10−6 M, k cat = (0.51 ± 0.05) s−1, k cat/Km = (29,316 ± 2990) M−1 s−1. The activity parameters are in consistent with recent findings where a different substrate Mca–AVLQ↓SGFRK(Dnp)K was used, the catalytic efficiency (k cat/Km) for SARS-CoV-2 Mpro was measured to be 28,500 M−1 s−1, which is slightly higher than that for SARS-CoV Mpro (k cat/Km = 26,500 M−1 s−1) [19].
Fig. S1.
(A) Plot of initial velocity at 5 μM of peptide substrate and 1 μM 3CLPro protease proteolytic activity for an hour for the release of fluorophore EDANS. The relative fluorescent unit (RFU) is linearly proportional to the amount of AVGACVLCN-EDANS. Data were fitted by linear regression. (B) Enzymatic activity by 1 μM 3CLPro enzyme and varying the peptide substrate (0.25–100 μM) fitted with Michaelis–Menten equation. (C) LB plot for peptide substrate to derive the kinetic parameters. (D) Standard plot for tyrosine absorbance at 280 nm. (E) Protease activity by keeping 1 μM 3CLPro enzyme and varying the casein substrate (0–270 μM) fitted with Michaelis–Menten equation. (F) LB plot for casein substrate for kinetic parameter calculations.
Further, we tried to find out and correlate with the previously reported kinetic parameters for 3CLpro of SARS-CoV-2 as well as SARS-CoV, as they share a high-level of sequence similarity, and have tabulated this information in Supplementary Table S1 [[20], [33], [34], [35], [36], [37], [38], [39], [40], [41], [43], [44], [45], [46], [47], [48], [49]]. The reason behind the huge variations in the kinetic parameter like k cat/Km values is unclear, as to whether it is because of the differences in experimental conditions, length of substrate, FRET pairs, peptide-substrate preparations, buffer compositions, or other assay methods. It is well documented that the protein preparations are very crucial for the enzymatically-active form of 3CLPro, compared in past, in the presence and absence of histidine tag, as it exists in monomer and dimer mixed states. The dimer of the 3C-like proteinase is the active form of the enzyme and predomination of the inactive monomeric form with low concentrations of the active dimers in the experiment might be a major reason for these observations.
We also used a non-specific casein substrate for determining the proteolytic activity of 3CLPro. The slope of the tyrosine standard plot (0.0011 Absorbance Unit 280 nm per μM) (Fig. S1D) was used for calculating the enzyme activity of 3CLPro, as a measure of tyrosine liberated from casein upon hydrolysis. The kinetic parameters for the enzyme were obtained by fitting the observed values in Michaelis-Menten equation and Lineweaver-Burk plot (Fig. S1E and F). From the double reciprocal plot, the values obtained were as follows: Km = (116 ± 12) × 10−6 M, k cat = (0.46 ± 0.05) s−1, k cat/Km = (3917 ± 470) M−1 s−1. The catalytic efficiency of 3CLPro towards specific peptide substrate DABCYL-MYTPHTVLQ↓AVGACVLCN-EDANS was found to be about 7.5 fold higher than broad specificity casein substrate possessing multiple cleavage sites.
From literature search and some computational studies, we have prepared a list of about 100 potential molecules. We found only 23 drugs in our initial screening which has some effect on protease activity and finalised 11 drugs for our further studies based on based on their inhibition profile under similar experimental conditions using 16 μM of inhibitors (Table 1 ). The effect of selected 11 inhibitors on the enzymatic activity of 3CLPro was monitored by peptide substrate and casein substrate (Fig. 2 A–D). Teicoplanin emerged as the best inhibitor in our studies with an IC50 value of 1.61 ± 0.09 μM and 1.46 ± 0.05 μM with peptide and casein substrate respectively (Fig. 2E, F). Teicoplanin was found to be more effective among the currently used/proposed anti-COVID19 drugs such as lopinavir, Hydroxychloroquine, Chloroquine, Atazanavir, Azithromycin.
Table 1.
Screening of drug molecules for 3ClPro protease activity inhibition.
| S. no. | Molecules | Activity inhibition at 16 μM conc of drug |
|---|---|---|
| 1 | Arbidol | N.D. |
| 2 | Aciclovir | + |
| 3 | Amoxicillin | + |
| 4 | Atazanavir | ++ |
| 5 | Azithromycin | ++ |
| 6 | Chloroquine | ++ |
| 7 | Digitoxin | N.D. |
| 8 | Dronendrone | N.D. |
| 9 | Ethylestradiol | N.D. |
| 10 | Famiciclovir | + |
| 11 | Febuxostat | N.D. |
| 12 | Halofantine | N.D. |
| 13 | Hydroxychloroquine | +++ |
| 14 | Imatinib | N.D. |
| 15 | Itraconazole | N.D. |
| 16 | Lapatinib | N.D. |
| 17 | Levonorgestrol | N.D. |
| 18 | Lopinavir | ++++ |
| 19 | Montelucast | N.D. |
| 20 | Nitazoxanide | + |
| 21 | Oseltamivir | + |
| 22 | Teicoplanin | ++++++++++ |
| 23 | Telmisartan | N.D. |
Fig. 2.
3CLPro protease activity using specific fluorogenic peptide and casein substrate (A) schematic representation of FRET based assay. (B) The enzyme kinetics using peptide substrate with different approved drugs at varying concentration. (C) Schematic representation of casein substrate assay. (D) The enzyme kinetics of 3CLPro with different approved drugs at varying concentration using casein substrate. (E) IC50 calculation for Teicoplanin using fluorogenic peptide as substrate. (F) IC50 calculation for Teicoplanin using casein as substrate.
3.3. Studying protein-ligand interactions by fluorescence spectroscopy
The fluorescence quenching spectra of 3CLPro protease titrated to increasing concentrations of various molecules are shown in Fig. 3 . The overall structural perturbations on the 3CLPro with ligands were assessed by fluorescence quenching. Upon titration with Teicoplanin there was a gradual decrease in fluorescence intensity with concentration (Fig. 3A). A significant quenching of the 3CLPro fluorescence emission was observed with Teicoplanin. This observation indicates the probable involvement of Trp residues of protein in the conformational dynamics. The concentration-dependent quenching was also observed in case of lopinavir, hydroxychloroquine and chloroquine (Fig. 3B–D). The interaction of 3CLPro with hydroxychloroquine and chloroquine led to protein conformation changes from native state to unfolded state (332 nm to 360 nm peak transitions). This depicts that three tryptophan residues in 3CLPro are exposed more in unfolded state with hydroxychloroquine and chloroquine. The other drugs like aciclovir, famciclovir, atazanavir, and azithromycin displayed some quenching (Fig. 3E–H). Rest of the drugs were not able to display any observable changes in the protein (Fig. 3I–K).
Fig. 3.
Fluorescence quenching study of 3CLPro with different inhibitors.
Fluorescence quenching measurement with different drugs (A) Teicoplanin (B) lopinavir (C) hydroxychloroquine (D) chloroquine (E) aciclovir (F) atazanavir (G) famiciclovir (H) azithromycin (I) oseltamivir (J) nitazoxanide (K) amoxicillin (L) Stern-Volmer plot for all the drugs titrated against 3CLPro.
The fluorescence quenching is usually described by the linear Stern–Volmer equations [19].
where F0 and F are the intrinsic fluorescence intensities of 3CLPro (fluorophore) in the absence or presence of the quencher respectively. [Q] is the concentration of the quencher, and KSV is the Stern–Volmer quenching constant.
The maximum fluorescence intensity at 333 nm was used for all KSV calculations. The Stern–Volmer plots for fluorescence quenching by all inhibitors are shown in Fig. 3L. The magnitude of KSV Stern–Volmer quenching constant for Teicoplanin was estimated 2.5 × 105 L·mol−1 (Table 2 ), which suggests a relatively high affinity between Teicoplanin and 3CLPro protease. The strongest effect was observed with Teicoplanin while the others show a diminished effect.
Table 2.
Stern Volmer constant calculation for drug protein interactions.
| Inhibitor | Stern-Volmer constant KSV ×105 L·mol−1 |
|---|---|
| Amoxicillin | 0.037 |
| Oseltamivir | 0.018 |
| Famiciclovir | 0.052 |
| Nitazoxanide | 0.035 |
| Aciclovir | 0.08 |
| Azithromycin | 0.08 |
| Chloroquine | 0.99 |
| Atazanavir | 0.13 |
| Hydroxychloroquine | 0.91 |
| Lopinavir | 0.11 |
| Teicoplanin | 2.58 |
3.4. Analysis of bimolecular interactions between 3CLPro and Teicoplanin
Surface plasmon resonance was employed to monitor the molecular interactions in real-time between 3CLPro and the identified ligand molecule. SPR is generally used to determine the binding specificity of molecules, including rates of association and dissociation between drug molecules and target proteins.
In this study, the SARS-CoV 3CLPro was immobilized on a sensor chip, and the identified molecule Teicoplanin was passed over the sensor's surface. The SPR sensorgram of the compound was recorded at varying concentrations of analyte molecule. The binding responses in resonance units (RUs) were continuously recorded and presented graphically as a function of time. The real-time binding kinetics and affinity of 3CLPro were measured for Teicoplanin. For 3CLPro, the molecule Teicoplanin increased the SPR sensorgram significantly and in a dose-dependent manner (Fig. 4A).
Fig. 4.
Analysis of 3CLPro and Teicoplanin interaction. (A) Binding kinetics and affinity using SPR (B) The melting curve of 3CLPro-Teicoplanin complex (blue curve) as compared to protein alone (red curve) by Thermofluor assay®. (C) The retention of 3CLPro monomer conformation post Teicoplanin binding was analysed by gel filtration chromatography. (D) The binding pose of Teicoplanin (cyan) in the three dimensional cleft of the 3CLPro. (E) The binding of the Teicoplanin (cyan) is in proximity to the hsitidine41 (red) and cysteine145 (magenta). (F) Two-dimensional interaction diagram of Teicoplanin interacting with the residues of active site of 3CLPro.
The association constant (k on) was recorded as 7.8 × 103 (1/Ms) and the dissociation rate constant (k off) was found to be 0.012 (1/s). The dissociation constant KD (k off/k on) was calculated by globally fitting the kinetic data at various concentrations of Teicoplanin according to fitting model “1:1 (Langmuir binding)”. The equilibrium rate constant (KD) was calculated by the formula
where, K is the equilibrium dissociation constant, k on is association rate constant, and k off is dissociation rate constant. The KD was recorded as 1.6 × 10−6 M defining a good interaction between the immobilized 3CLPro and Teicoplanin (Fig. 4A).
We tested various concentrations of Teicoplanin over 3CLPro and recorded the corresponding response units. We tried lower concentrations of Teicoplanin and found 1:1 binding at those concentrations. When we increased the concentrations of the Teicoplanin, the saturation level was not achieved, suggesting for multiple binding sites on 3CLPro with the analyte molecules. These promiscuous binding are seen when more molecule is available for binding to non-specific sites after the active sites are saturated with the molecule.
The effect of Teicoplanin on the 3CLPro protein was analysed using Thermofluor assay® which relies upon the binding of SYPRO® dye to exposed hydrophobic patches on heating. The melting temperature (TM) of native protein was recorded ~57 °C, whereas the same for the Teicoplanin-3CLPro complex was found ~49 °C (Fig. 4B). The interaction of inhibitors shifts the melting temperature which is a signature of protein stability. The binding doesn't alter the monomeric conformation of 3CLPro as depicted by SEC (Fig. 4C).
Our docking analysis using PARDock revealed that the ligand drug Teicoplanin fits perfectly in the active site pocket (Fig. 4D) and the binding energy obtained for this binding is about −8 kcal/mol. The cavity is lined by the active site amino acids i.e. histidine 41 and cysteine 145 (Fig. 4E). The residues His41 and Cys145 form the catalytic dyad, form the substrate binding region and are located at the cleft of domain I and II in which His acts as a proton acceptor while Cys behaves as a nucleophile. Previously, it is reported that Teicoplanin is well docked with SARS-CoV-2 main protease using AutoDock4, AutoDock Vina and Dock6 docking programs, which also revealed that docked Teicoplanin is well accommodated within the inhibitor binding cavity in a manner similar a Michael acceptor inhibitor—known as N3 which is a potent and irreversible inhibitor of SARS-CoV-2 [20].
Teicoplanin displays a bonding with the protein at the molecular level, via hydrophobic interactions, hydrogen bonding and halogen bonding. The hydrophobic interactions are seen with Asp187 and Glu166 (Fig. 4F). These two residues are also involved in the formation of hydrogen bonds. The involvement of H-bond donors and acceptors around hydrophobic sites compel the Teicoplanin molecule to interact within the inhibitor binding pocket of the 3CLPro protease. The formation of extensive hydrogen bonds with active site residues has been observed previously with Arg40 and Gly170 residues. In a recent MD simulation study, it was reported that Teicoplanin in complex with SARS-CoV-2 main protease has stable ligand-protein complex and intermolecular interactions during the simulated trajectory [21].
Further our careful analysis revealed that the Leu141 and Ser144 residues of 3CLPro form a halogen bond with the Teicoplanin molecule. It has been established that the halogen bond interactions are critical for the potential activity of the protease inhibitors. The halogen bond interaction characteristic of an inhibitor compound makes it more promising candidate as a novel anti-viral therapeutic [22].
We observed quite significant and different kinds of interactions between the Teicoplanin molecule and the 3CLPro protein suggesting that Teicoplanin can act as a good inhibitor of 3CLPro. These interactions around and at the active site amino acids most probably hinder the proton transfer and binding of substrate to the active site, leading to disruption in the protease activity.
3.5. Relative efficacy of drugs to inhibit 3CLPro
The relative efficacy of the drugs to inhibit the protease activity of main protease of SARS-CoV-2 was analysed using protease assay. Teicoplanin emerged as the most potent inhibitor in our involving 23 drugs (Table 1). As compared to Teicoplanin with IC50 value at 1.5 μM as 100%, lopinavir displayed only 9.12% activity inhibition and hydroxychloroquine showed 4.56% inhibition (Fig. 5 ). The other drugs showed less than 5% inhibition.
Fig. 5.
Relative efficacy of drugs. Activity Inhibition by Teicoplanin was considered 100% and the other drugs were compared for relative efficacy.
In this study, we have used a wide variety of drugs indicated in different infectious diseases such as retroviral HIV infections, bacterial infections, etc. and have used biochemical and biophysical analysis to compare these 23 drugs in terms of activity on the casein substrate, FRET peptide substrate as well as drug-protein interactions using fluorescence quenching. We have found that under similar laboratory conditions, Teicoplanin was found to be a much stronger inhibitor of 3CLPro as compared to the other drugs like hydroxychloroquine, lopinavir, chloroquine, azithromycin, atazanavir etc.
4. Discussion
The COVID-19 pandemic has become a major challenge for healthcare workers and governments of the world. The current treatment options are targeting symptoms of the disease which involve pulmonary complications including localized lesions, pneumonia, bronchiolitis. The virus infects the lung endothelial cells and promotes the development of a pathological state akin to lymphocytic endothelialitis and inflammatory cell invasion. The lack of a specific treatment has resulted in major loss of life and economy. Drug discovery usually takes a long time with proper validation to ensure the safety and efficacy of the drug. In these circumstances, drug-repositioning confers an edge along with 3CLPro as an attractive drug target of SARS-CoV-2. The sessile sequences of 3CLPro do not match with the protein sequences of human proteases and hence a number of different strategies are being used to inhibit the activity of this protease, to halt the viral replication with minimal toxicity.
There are many approved drugs for other viral infections such as Ebola, HIV and Influenza suggested for tackling COVID-19. The secondary infections resulting from the COVID-19 infection is also contributing to fatality [23]. Therefore, searching for drugs with antiviral properties with minimum side effects would be an ideal solution. Teicoplanin is an effective glycopeptide antibiotic with reported anti-MERS CoV activity [24,25]. We report Teicoplanin as an effective drug against 3CLPro which works at a micromolar concentration of 1.5 μM (Fig. 2) and acts by blocking the active site of the protease (Fig. 4F).
We studied protein-drug interactions in order to decipher the working of the drug and mechanism of inhibition at the molecular level. The protein-drug interaction shows fluorescence quenching by monitoring the intrinsic fluorescence, Teicoplanin interaction is gradual however the interaction of hydroxychloroquine and chloroquine is perturbing the conformational dynamics as depicted by wavelength shifts (Fig. 3). The SPR finding suggests that the good affinity of Teicoplanin with 3CLPro (Fig. 4A). Lopinavir is another anti-retroviral drug that acts by targeting HIV protease. It was under clinical trial for COVID-19 which concluded that its beneficial effects were minimal [26]. Hydroxychloroquine is in an ongoing drug trial and is currently included in the prime regimen of drugs used in the management of COVID-19. The proposed mechanism of action of the drug is the alteration in the pH of endosome thereby preventing the uncapping of capsid and genome release [27,28]. We found that the relative potency of lopinavir and hydroxychloroquine to inhibit the SARS-CoV-2 main protease was 9.12% and 4.56%, respectively (Fig. 5). The activity of other drugs was found negligible or undetectable. These drugs have already proven their potential in the treatment of different DNA and RNA virus diseases which further supports their anti-viral capabilities.
In our study, Teicoplanin showed significant reduction of the proteolytic activity of 3CLPro. As this protein is essential to the replication cycle of the virus, due to its irreplaceable role in the processing of viral polyproteins, which the virus needs to complete its life cycle, we propose that this reduction may contribute to its anti-SARS-CoV-2 effect via inhibition of the viral replication. We also observed some inhibitory activity in other drugs that we tested but as Teicoplanin was found most potent among them. Further experiments in higher validation systems are required to establish the efficacy of the molecule. We advocate the expedited systematic investigation in primate models and trials in terminally ill patients suffering from COVID-19.
Recently, the effect of Teicoplanin with respect to SARS-CoV-2 has been observed by other groups as well. For example, according to Zhou et al. [29], Teicoplanin acts on an early stage of the viral life cycle in coronaviruses by inhibiting the low-pH cleavage of the viral spike protein by cathepsin L in the late endosomes, thereby preventing the release of genomic viral RNA and continuation of the virus replication cycle. In a recent study, J. Zhang et al. [30] showed that Teicoplanin blocked virus entry by specifically inhibiting the activity of cathepsin L in SARS-CoV-2 virus. They identified that Teicoplanin inhibited the entry of SARS-CoV-2 pseudovirus, which provides a possible strategy for the prophylaxis and treatment for SARS-CoV-2 infection. They proposed that Teicoplanin may prevent virus infection and amplification at a very early stage. Their result indicates that the potential antiviral activity of Teicoplanin could be utilized for the treatment of SARS-CoV-2 virus infection [29,30].
Further, recently there has been a clinical study carried out with Teicoplanin as reported by Intensive Care COVID-19 Study Group of Sapienza University. They recruited a cohort of 21 patients affected by severe COVID-19 symptoms such as lung involvement, who were hospitalized in intensive care units (ICUs) of a hospital in Italy, Rome, and complementarily treated with Teicoplanin. On ICU admission, the patients received Teicoplanin in doses of 6 mg/kg every 24 h. The median duration of Teicoplanin therapy was 10 days (range 7–12 days). The ICU mortality rate was only 14.3% (3/21 patients). None of the patients had any adverse effects related to Teicoplanin [31].
Although this study has many obvious limitations, including its non-comparative retrospective observational nature, small sample size and short follow-up. But most importantly, their observations were investigated on critically ill patients. However, this is the first clinical report on the use of Teicoplanin in-vivo in subjects affected by COVID-19 and the results appear fairly acceptable when compared with other medications. However, a more detailed clinical investigation is required on large cohort, in different stages mild, moderate and critically ill patients to conclude the definite role of Teicoplanin against COVID-19.
In a nutshell, all these studies support each other that Teicoplanin might be a potential therapeutic option against COVID-19.
5. Conclusion
The approved drug Teicoplanin has already been tested for toxicity in human beings over a period of time. We propose Teicoplanin as an effective drug against SARS CoV-2. The study paves the path for an expedited research to an effective treatment of COVID-19. We also propose immediate attention for the randomized clinical trials for this drug to reduce infections and to help the society, as it has a proven mechanism of action and efficacy as compared to other recommended drugs for COVID-19.
The following are the supplementary data related to this article.
Kinetic parameters of 3CLPro constructs of SARS-CoV-2 and SARS-CoV.
CRediT authorship contribution statement
Ashok Kumar Patel: Conceptualization, Methodology, Software. Praveen Kumar Tripathi, Saurabh Upadhyay and Manju Singh: Data curation, Writing - original draft preparation. Praveen Kumar Tripathi, Saurabh Upadhyay and Manju Singh: Visualization, Investigation. Ashok Kumar Patel: Supervision.: Pradeep Sharma: Software, Validation. Praveen Kumar Tripathi, Saurabh Upadhyay, Manju Singh, Siva Raghvandhar, Mohit Bhardwaj: Writing - reviewing and editing.
Acknowledgments
Acknowledgment
Facilities of Kusuma School of Biological Sciences, IIT Delhi are acknowledged. Authors acknowledge ICMR and DBT for research scholarships. We acknowledge the funding support from the Dean, Research and Development, IIT Delhi, New Delhi, India through Grant No MI02217G. We acknowledge Dr. Likhesh Sharma (GE Healthcare) for helping in analysis and interpretation of SPR experiments.
Declaration of competing interest
Authors declare no conflict of interest.
References
- 1.Shi J., Wen Z., Zhong G., Yang H., Wang C., Huang B., Liu R., He X., Shuai L., Sun Z., Zhao Y., Liu P., Liang L., Cui P., Wang J., Zhang X., Guan Y., Tan W., Wu G., Chen H., Bu Z. Susceptibility of ferrets, cats, dogs, and other domesticated animals to SARS–coronavirus 2. Science. 2020 doi: 10.1126/science.abb7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.To K.K.W., Hung I.F.N., Chan J.F.W., Yuen K.-Y. From SARS coronavirus to novel animal and human coronaviruses. J. Thorac. Dis. 2013;5:S103–S108. doi: 10.3978/j.issn.2072-1439.2013.06.02. (S108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prompetchara E., Ketloy C., Palaga T. Immune responses in COVID-19 and potential vaccines: lessons learned from SARS and MERS epidemic. Asian Pac. J. Allergy Immunol. 2020;38:1–9. doi: 10.12932/AP-200220-0772. [DOI] [PubMed] [Google Scholar]
- 4.Liu J., Zheng X., Tong Q., Li W., Wang B., Sutter K., Trilling M., Lu M., Dittmer U., Yang D. Overlapping and discrete aspects of the pathology and pathogenesis of the emerging human pathogenic coronaviruses SARS-CoV, MERS-CoV, and 2019-nCoV. J. Med. Virol. 2020;92:491–494. doi: 10.1002/jmv.25709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Wit E., van Doremalen N., Falzarano D., Munster V.J. SARS and MERS: recent insights into emerging coronaviruses. Nat. Rev. Microbiol. 2016;14:523–534. doi: 10.1038/nrmicro.2016.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H., Xiang X., Ren H., Xu L., Zhao L., Chen X., Long H., Wang Q., Wu Q. SAA is a biomarker to distinguish the severity and prognosis of Coronavirus Disease 2019 (COVID-19) J. Inf. Secur. 2020 doi: 10.1016/j.jinf.2020.03.035. [DOI] [Google Scholar]
- 8.Lippi G., Plebani M., Henry B.M. Thrombocytopenia is associated with severe coronavirus disease 2019 (COVID-19) infections: a meta-analysis. Clin. Chim. Acta Int. J. Clin. Chem. 2020;506:145–148. doi: 10.1016/j.cca.2020.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astuti I., Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. 2020 doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naqvi A.A.T., Fatima K., Mohammad T., Fatima U., Singh I.K., Singh A., Atif S.M., Hariprasad G., Hasan G.M., Hassan Md.I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: structural genomics approach. Biochim. Biophys. Acta (BBA) - Mol. Basis Dis. 2020;1866 doi: 10.1016/j.bbadis.2020.165878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shamsi A., Mohammad T., Anwar S., AlAjmi M.F., Hussain A., Rehman M.T., Islam A., Hassan M.I. Glecaprevir and Maraviroc are high-affinity inhibitors of SARS-CoV-2 main protease: possible implication in COVID-19 therapy. Biosci. Rep. 2020;40 doi: 10.1042/BSR20201256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li G., Clercq E.D. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat. Rev. Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- 13.Muramatsu T., Takemoto C., Kim Y.-T., Wang H., Nishii W., Terada T., Shirouzu M., Yokoyama S. SARS-CoV 3CL protease cleaves its C-terminal autoprocessing site by novel subsite cooperativity. Proc. Natl. Acad. Sci. 2016;113:12997–13002. doi: 10.1073/pnas.1601327113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banik S., Biswas S., Karmakar S. Extraction, purification, and activity of protease from the leaves of Moringa oleifera. F1000Research. 2018;7:1151. doi: 10.12688/f1000research.15642.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cupp-Enyard C. Sigma’s non-specific protease activity assay - casein as a substrate. J. Vis. Exp. JoVE. 2008 doi: 10.3791/899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qashqoosh M.T.A., Manea Y.K., Alahdal F.A.M., Naqvi S. Investigation of conformational changes of bovine serum albumin upon binding with benzocaine drug: a spectral and computational analysis. BioNanoScience. 2019;9:848–858. doi: 10.1007/s12668-019-00663-7. [DOI] [Google Scholar]
- 17.Ghisaidoobe A.B.T., Chung S.J. Intrinsic tryptophan fluorescence in the detection and analysis of proteins: a focus on Förster resonance energy transfer techniques. Int. J. Mol. Sci. 2014;15:22518–22538. doi: 10.3390/ijms151222518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huynh K., Partch C.L. Analysis of protein stability and ligand interactions by thermal shift assay. Curr. Protoc. Protein Sci. 2015;79:28.9.1–28.9.14. doi: 10.1002/0471140864.ps2809s79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lakowicz J.R. 3rd ed. Springer; US: 2006. Principles of Fluorescence Spectroscopy. [DOI] [Google Scholar]
- 20.Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C., Duan Y., Yu J., Wang L., Yang K., Liu F., Jiang R., Yang X., You T., Liu X., Yang X., Bai F., Liu H., Liu X., Guddat L.W., Xu W., Xiao G., Qin C., Shi Z., Jiang H., Rao Z., Yang H. Structure of Mpro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- 21.Azam F. 2020. Targeting SARS-CoV-2 Main Protease by Teicoplanin: A Mechanistic Insight by In Silico Studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hattori S., Hayashi H., Bulut H., Rao K.V., Nyalapatla P.R., Hasegawa K., Aoki M., Ghosh A.K., Mitsuya H. Halogen bond interactions of novel HIV-1 protease inhibitors (PI) (GRL-001-15 and GRL-003-15) with the flap of protease are critical for their potent activity against wild-type HIV-1 and multi-PI-resistant variants. Antimicrob. Agents Chemother. 2019;63 doi: 10.1128/AAC.02635-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou P., Liu Z., Chen Y., Xiao Y., Huang X., Fan X.-G. Bacterial and fungal infections in COVID-19 patients: a matter of concern. Infect. Control Hosp. Epidemiol. 2020:1–2. doi: 10.1017/ice.2020.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramos-Martín V., Johnson A., McEntee L., Farrington N., Padmore K., Cojutti P., Pea F., Neely M.N., Hope W.W. Pharmacodynamics of teicoplanin against MRSA. J. Antimicrob. Chemother. 2017;72:3382–3389. doi: 10.1093/jac/dkx289. [DOI] [PubMed] [Google Scholar]
- 25.Baron S.A., Devaux C., Colson P., Raoult D., Rolain J.-M. Teicoplanin: an alternative drug for the treatment of COVID-19? Int. J. Antimicrob. Agents. 2020;55 doi: 10.1016/j.ijantimicag.2020.105944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li H., Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li H., Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang J., Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir-ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khan M., Santhosh S.R., Tiwari M., Lakshmana Rao P.V., Parida M. Assessment of in vitro prophylactic and therapeutic efficacy of chloroquine against Chikungunya virus in vero cells. J. Med. Virol. 2010;82:817–824. doi: 10.1002/jmv.21663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mauthe M., Orhon I., Rocchi C., Zhou X., Luhr M., Hijlkema K.-J., Coppes R.P., Engedal N., Mari M., Reggiori F. Chloroquine inhibits autophagic flux by decreasing autophagosome-lysosome fusion. Autophagy. 2018;14:1435–1455. doi: 10.1080/15548627.2018.1474314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou N., Pan T., Zhang J., Li Q., Zhang X., Bai C., Huang F., Peng T., Zhang J., Liu C., Tao L., Zhang H. Glycopeptide antibiotics potently inhibit cathepsin L in the late endosome/lysosome and block the entry of Ebola virus, Middle East respiratory syndrome coronavirus (MERS-CoV), and severe acute respiratory syndrome coronavirus (SARS-CoV) J. Biol. Chem. 2016;291:9218–9232. doi: 10.1074/jbc.M116.716100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J., Ma X., Yu F., Liu J., Zou F., Pan T., Zhang H. Teicoplanin potently blocks the cell entry of 2019-nCoV. BioRxiv. 2020 doi: 10.1101/2020.02.05.935387. 2020.02.05.935387. [DOI] [Google Scholar]
- 31.G C., F A., d’Ettorre G., C B., O S., A O., F R., Cm M., F P., M V. Is teicoplanin a complementary treatment option for COVID-19? The question remains. Int. J. Antimicrob. Agents. 2020:106029. doi: 10.1016/j.ijantimicag.2020.106029. (undefined) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rut W., Groborz K., Zhang L., Sun X., Zmudzinski M., Pawlik B., Młynarski W., Hilgenfeld R., Drag M. Substrate specificity profiling of SARS-CoV-2 main protease enables design of activity-based probes for patient-sample imaging. BioRxiv. 2020 doi: 10.1101/2020.03.07.981928. 2020.03.07.981928. [DOI] [Google Scholar]
- 35.Ma C., Sacco M.D., Hurst B., Townsend J.A., Hu Y., Szeto T., Zhang X., Tarbet B., Marty M.T., Chen Y., Wang J. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30:678–692. doi: 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muramatsu T., Kim Y.-T., Nishii W., Terada T., Shirouzu M., Yokoyama S. Autoprocessing mechanism of severe acute respiratory syndrome coronavirus 3C-like protease (SARS-CoV 3CLpro) from its polyproteins. FEBS J. 2013;280:2002–2013. doi: 10.1111/febs.12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang C., Wei P., Fan K., Liu Y., Lai L. 3C-like proteinase from SARS coronavirus catalyzes substrate hydrolysis by a general base mechanism. Biochemistry. 2004;43:4568–4574. doi: 10.1021/bi036022q. [DOI] [PubMed] [Google Scholar]
- 38.Blanchard J.E., Elowe N.H., Huitema C., Fortin P.D., Cechetto J.D., Eltis L.D., Brown E.D. High-throughput screening identifies inhibitors of the SARS coronavirus main proteinase. Chem. Biol. 2004;11:1445–1453. doi: 10.1016/j.chembiol.2004.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kao R.Y., To A.P.C., Ng L.W.Y., Tsui W.H.W., Lee T.S.W., Tsoi H.-W., Yuen K.-Y. Characterization of SARS-CoV main protease and identification of biologically active small molecule inhibitors using a continuous fluorescence-based assay. FEBS Lett. 2004;576:325–330. doi: 10.1016/j.febslet.2004.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bacha U., Barrila J., Velazquez-Campoy A., Leavitt S.A., Freire E. Identification of novel inhibitors of the SARS coronavirus main protease 3CLpro. Biochemistry. 2004;43:4906–4912. doi: 10.1021/bi0361766. [DOI] [PubMed] [Google Scholar]
- 41.Fan K., Wei P., Feng Q., Chen S., Huang C., Ma L., Lai B., Pei J., Liu Y., Chen J., Lai L. Biosynthesis, purification, and substrate specificity of severe acute respiratory syndrome coronavirus 3C-like proteinase. J. Biol. Chem. 2004;279:1637–1642. doi: 10.1074/jbc.M310875200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xue X., Yang H., Shen W., Zhao Q., Li J., Yang K., Chen C., Jin Y., Bartlam M., Rao Z. Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction. J. Mol. Biol. 2007;366:965–975. doi: 10.1016/j.jmb.2006.11.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li C., Qi Y., Teng X., Yang Z., Wei P., Zhang C., Tan L., Zhou L., Liu Y., Lai L. Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase. J. Biol. Chem. 2010;285:28134–28140. doi: 10.1074/jbc.M109.095851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen L., Chen S., Gui C., Shen J., Shen X., Jiang H. Discovering severe acute respiratory syndrome coronavirus 3CL protease inhibitors: virtual screening, surface plasmon resonance, and fluorescence resonance energy transfer assays. J. Biomol. Screen. 2006;11:915–921. doi: 10.1177/1087057106293295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Park J.-Y., Ko J.-A., Kim D.W., Kim Y.M., Kwon H.-J., Jeong H.J., Kim C.Y., Park K.H., Lee W.S., Ryu Y.B. Chalcones isolated from Angelica keiskei inhibit cysteine proteases of SARS-CoV. J. Enzyme Inhib. Med. Chem. 2016;31:23–30. doi: 10.3109/14756366.2014.1003215. [DOI] [PubMed] [Google Scholar]
- 47.Shi J., Han N., Lim L., Lua S., Sivaraman J., Wang L., Mu Y., Song J. Dynamically-driven inactivation of the catalytic machinery of the SARS 3C-like protease by the N214A mutation on the extra domain. PLoS Comput. Biol. 2011;7 doi: 10.1371/journal.pcbi.1001084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cheng S.-C., Chang G.-G., Chou C.-Y. Mutation of Glu-166 blocks the substrate-induced dimerization of SARS coronavirus main protease. Biophys. J. 2010;98:1327–1336. doi: 10.1016/j.bpj.2009.12.4272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krichel B., Falke S., Hilgenfeld R., Redecke L., Uetrecht C. Processing of the SARS-CoV pp1a/ab nsp7-10 region. Biochem. J. 2020;477:1009–1019. doi: 10.1042/BCJ20200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Kinetic parameters of 3CLPro constructs of SARS-CoV-2 and SARS-CoV.