Abstract
In French Guiana, the malaria, a parasitic infection transmitted by Anopheline mosquitoes, remains a disease of public health importance. To prevent malaria transmission, the main effective way remains Anopheles control. For an effective control, accurate Anopheles species identification is indispensable to distinguish malaria vectors from non-vectors. Although, morphological and molecular methods are largely used, an innovative tool, based on protein pattern comparisons, the Matrix Assisted Laser Desorption / Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS) profiling, emerged this last decade for arthropod identification. However, the limited mosquito fauna diversity of reference MS spectra remains one of the main drawback for its large usage. The aim of the present study was then to create and to share reference MS spectra for the identification of French Guiana Anopheline species. A total of eight distinct Anopheles species, among which four are malaria vectors, were collected in 6 areas. To improve Anopheles identification, two body parts, legs and thoraxes, were independently submitted to MS for the creation of respective reference MS spectra database (DB). This study underlined that double checking by MS enhanced the Anopheles identification confidence and rate of reliable classification. The sharing of this reference MS spectra DB should make easier Anopheles species monitoring in endemic malaria area to help malaria vector control or elimination programs.
Introduction
Since 2005, malaria cases declined significantly in French Guiana, an oversea territory of France located in South-America. The number of diagnosed cases has decreased from 4,479 cases in 2005 to 597 cases in 2017 [1]. However, the disease is still endemic in the inland forested areas, especially in illegal gold mining areas and so remains of public health importance [2–4]. Majority of the cases are caused by Plasmodium vivax (89%, p = 531/597), followed by P. falciparum (11%, p = 66/597).
Anopheles mosquitoes are known for their role in transmitting malaria. Historically, thirty-three mosquitoes from the genus Anopheles have been reported in French Guiana [5]. Members in the subgenus Anopheles and Nyssorhynchus have been implicated in malaria transmission in French Guiana. Anopheles darlingi is the recognized primary vector in the territory [6–9]. Recently, An. nuneztovari sl, An. oswaldoi sl, An. intermedius, An. marajoara and An. ininii were found naturally infected with Plasmodium sporozoites and were suspected to be secondary vectors [2,3,10]. Malaria transmission is further complicated as some of these secondary vectors belong to species complexes, characterized by similar morphological characteristics, such as An. oswaldoi [11], An. marajoara [12] and An. nuneztovari [13].
A rapid and accurate identification of Anopheles species is then critical when designing malaria vector control strategies which should be species-specific to be effective. The most common method for mosquito species identification remains the utilization of morphological criteria [14]. However, morphological identification is skill dependent requiring entomological expertise. The correct species assignation could also be compromised for damaged specimens with the loss of determinant characters. Moreover, the description of morphological characteristic variations between intact adult conspecific specimens underlined that correct mosquito species classification could be held only by experienced mosquito taxonomists [15]. Additionally, the availability of mosquito taxonomic keys is a cornerstone for identification success. Unfortunately, morphological keys are still missing for numerous Anopheles species, notably for closely related-species (cryptic and complex species).
The emergence of molecular biology approaches in the 2000’s has solved some long-standing taxonomic questions [16]. However, the choice of the target gene sequence for accurate mosquito identification could be complex. In South America, Anopheles species were classified by using different target genes: either 18S rRNA [17], either the second internal transcribed spacer (ITS2) [18] or cytochrome c oxidase I (COI) [19]. The absence of consensus for mosquito species identification complicate studies comparison. The development of the Barcode of Life Data (BOLD) system contributed to standardize gene sequencing for organism identification [20]. Nevertheless, for some cryptic mosquito species (eg, An. gambiae complex), the sequencing of COI could be insufficient to classify them unambiguously [21] and the sequencing of a second gene could be required [22]. For instance, some Anopheles and Culex sibling species could not be distinguished using uniquely the mitochondrial COI barcode [23,24]. Moreover, despite the large advances of this strategy this last decade, notably by shortening experiment duration and costs of reagents, gene sequencing remains time-consuming and expansive [25]. The development of a quick and low cost approach for mosquito monitoring with elevated rate of reliable identification is always in high demand.
The MALDI-TOF MS profiling has recently demonstrated its performance for reliable arthropod identification [25], including mosquitoes at adult [26] and immature stages [27,28]. At adult stage, for specimen identification by MS, different mosquito body parts were selected such as the cephalothorax [29,30] or legs, but this last compartment remains the more frequently used [26,31–33]. The recent successful identification of mosquitoes at immature stages by MALDI-TOF MS profiling validated the efficiency of this MS strategy for field monitoring of Culicidae [19]. The regent low costs, the rapidity and technical simplicity of protocols participated to the success of this approach. However, conversely to molecular analyses, MS protein profiles from conspecific specimens could vary according to several factors such as sample storing mode, developmental stage, homogenization mode or body part used [25,34]. To overcome these limitations, standardized protocols were established for some arthropod families [35] including mosquitoes [36]. The standardization of the protocols facilitated result comparisons and reference MS spectra sharing.
The main problem with legs is that they are breakable. The loss of one to all mosquito legs during trapping and/or storing is not infrequent, which could compromise specimen identification by MS. The selection of a second body part, the mosquito thorax, revealed that it could also generate mosquito species-specific MS but distinct from legs of conspecific specimens [37]. This last study underlined that the query of these two body parts against the reference MS spectra database (DB) improved mosquito species identification with accuracy and confidence. This pioneering study was assessed to distinct 7 mosquito species from 4 genera living in sympatry in Guadeloupe Island [37].
The aim of the present study was to assess whether the submission of two body parts could improve Anopheles identification from French Guiana. The creation of a reference MS spectra DB should make easier Anopheles species monitoring in endemic malaria area to help malaria vector control or elimination programs.
Methods
Mosquito collection and dissection
Anopheles adult female mosquitoes were selected from field mosquito collections done in 6 distinct sites from French Guiana, during entomological surveys, using different collection methods, over different sampling periods (Fig 1 and Table 1) [3,38–40]. No permits were required for the described study, which complied with all relevant regulations; the land accessed was public (not privately owned or protected) and no protected or sensitive animals or plants were sampled. After collections, mosquitoes were sorted by genera and Anopheles mosquitoes were morphologically identified under a binocular loupe at a magnification of ×56 (Leica M80, Leica, Nanterre, France) using standard taxonomic keys for the region (Floch and Abonnenc 1951, Forattini 1962, Faran and Linthicum 1981). Anopheles specimens were then stored individually at -20°C. According to their availability, one to 22 specimens per Anopheles species were selected for molecular and MS analyses (Table 1). Legs and thoraxes from mosquitoes were dissected for MALDI-TOF MS analysis [37]. The abdomens, wings and heads were kept for molecular analyses.
Fig 1. Map of mosquito collection sites in French Guiana.
The different sampling sites are indicated by circles.
Table 1. Overview of Anopheles mosquito origins and subgroup identification by COI molecular typing.
| Morphological identification | Catching site (Latitude/Longitude) | Catching period | Number of specimens (Ref. MS DB§) | Species identified by NCBi (Accession Number) | COI gene sequence coverage (%) / identity (%) | Species identified by BOLD | COI gene sequence similarity (%) |
|---|---|---|---|---|---|---|---|
| An. peryassui | Cacao (4°34'12.0"N 52°28'12.0"E) | (June-2015) | 3 (2) | NC_037790.1 | 99/99 | KF698875 | 97.84 |
| An. intermedius | Cacao (4°34'12.0"N 52°28'12.0"E) | (June-2015) | 10 (2) | MF381700.1 / NC_037789.1 | 99/98 | Early release | 98.62–99.39 |
| An. oswaldoi | Eau Claire (3°36'27.0"N 53°34'32.6"W) | (June-2014) | 1 (1) | MG241906.1 | 93/99 | 51917708 | 97.85 |
| An. aquasalis | Cayenne (4°55'21.5"N 52°18'47.9"W) | (Sept./Oct.-2014) | 17 (2) | KC354822.1 | 99/98 | KC354821 | 98.15–98.29 |
| An. braziliensis | Cacao (4°34'12.0"N 52°28'12.0"E) | (June-2015) | 18 (2) | NC_037791.1 / MF381732.1 | 99/99 | Private / DQ913839 / DQ913846 / DQ913825 | 98.91–99.73 |
| An. darlingi | Blondin (3°87’69.0” N 51°81’34.0” E) | (Sept.-2015) | 22 (2) | MF381596.1 / MF381713.1 | 99/99 | JF923694 / private | 99.85–100 |
| An. nuneztovari | Dorlin (3°45'12.9"N 53°33'21.3"W) | (March-2013) | 20 (2) | NC_037810.1 / MF381656.1 | 99/99 | KU865547 / KC167737 | 99.83–100 |
| An. triannulatus | Saint Georges (3°53'31.7"N 51°48'27.0"W) | (May/June-2014) | 20 (2) | MF381730.1 / JX205112.1 | 99/99 | Early-Release / KC167680 | 99.54–99.83 |
| Total | 111 (15) |
§Ref. MS DB: Number of specimens used to create the reference MS database.
*Mosquito species for whose COI gene sequences are not available (N.A.) in the database (27th November 2018). #No reliable ID: Identity with top match < 97%. BOLD: Barcode of Life Data Systems; COI: cytochrome oxidase one.
Molecular identification of mosquitoes
DNA was individually extracted from the head and abdomen of all mosquito specimens (n = 111) using the QIAamp DNA tissue extraction kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. Molecular identification of mosquito at the species level was performed by sequencing the PCR product of a fragment of the cytochrome c oxidase I gene (COI) (LCO1490 (forward): 5’-GGTCAACAAATCATAAAGATATTGG-3’; HC02198 (reverse): 5’-TAAACTTCAGGGTGACCAAAAAATCA-3’) as previously described [19,41]. The sequences were assembled and analyzed using the Molecular Evolutionary Genetics Analysis (MEGA) software version 7.0 and BioEdit Sequence alignment editor software version 7.2.6.0. All sequences were blasted against GenBank (http://blast.ncbi.nlm.nih.gov/Blast.cgi) and against the Barcode of Life Data Systems (BOLD; http://www.barcodinglife.org; [20]) to assign unknown COI sequences to mosquito species.
Sample homogenization and MALDI-TOF MS analysis
Each dissected compartment (legs and thoraxes) was homogenized individually 3 x 1 minute at 30 Hertz using TissueLyser (Qiagen) and glass beads (#11079110, BioSpec Products, Bartlesville, OK, US) in a homogenization buffer composed of a mix (50/50) of 70% (v/v) formic acid (Sigma) and 50% (v/v) acetonitrile (Fluka, Buchs, Switzerland), prepared with HPLC-grade water, according to the standardized automated setting [42]. After sample homogenization, a quick spin centrifugation at 200 g for 1 min was then performed and 1 μL of the supernatant of each sample was spotted on the MALDI-TOF steel target plate in quadruplicate (Bruker Daltonics, Wissembourg, France). After air-drying, 1 μL of matrix solution composed of saturated α-cyano-4-hydroxycinnamic acid (Sigma, Lyon, France), 50% (v/v) acetonitrile, 2.5% (v/v) trifluoroacetic acid (Aldrich, Dorset, UK) prepared with HPLC-grade water, was added. To control matrix quality (i.e. absence of MS peaks due to matrix buffer impurities) and MALDI-TOF apparatus performance, matrix solution was loaded in duplicate onto each MALDI-TOF plate alone and with a bacterial test standard (Bruker Bacterial Test Standard, ref: #8255343).
MALDI-TOF MS parameters
Protein mass profiles were obtained using a Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics, Germany), with detection in the linear positive-ion mode at a laser frequency of 50 Hz within a mass range of 2–20 kDa. The setting parameters of the MALDI-TOF MS apparatus were identical to those previously used [43]. Briefly, the acceleration voltage was 20 kV, and the extraction delay time was 200 ns. Each spectrum corresponds to ions obtained from 240 laser shots performed in six regions of the same spot and automatically acquired using the AutoXecute of the Flex Control v.2.4 software (Bruker Daltonics).
MS spectra analysis
MS spectra profiles were firstly controlled visually with flexAnalysis v3.3 software (Bruker Daltonics). MS spectra were then exported to ClinProTools v2.2 and MALDI-Biotyper v3.0. (Bruker Daltonics) for data processing (smoothing, baseline subtraction, peak picking). MS spectra reproducibility was assessed by the comparison of the average spectral profiles (MSP, Main Spectrum Profile) obtained from the four spots for each specimen according to body part with MALDI-Biotyper v3.0 software (Bruker Daltonics). MS spectra reproducibility and specificity taking into account mosquito body part were objectified using cluster analyses. Cluster analyses (MSP dendrogram) were performed based on comparison of the MSP given by MALDI-Biotyper v3.0. software and clustered according to protein mass profile (i.e. their mass signals and intensities). In addition, to visualize MS spectra distribution according to body part, principal component analysis (PCA) from ClinProTools v2.2 software were performed for each species.
The top-10 and top-5 of the most intense MS peaks per mosquito species and per body-part were analyzed with ClinProTools software to estimate their performance to discriminate the Anopheles species for each body-part. The parameter settings in ClinProTools software for spectrum preparation were as follows: a resolution of 300; a noise threshold of 2.00; a maximum peak shift of 800 ppm and a match to calibrating agent peaks of 10%. Peak calculation and selection were performed on individual spectra with a signal-to-noise threshold of 2.00 and an aggregation of 800 ppm. Based on the peak list obtained for each body part per species, the top-10 and top-5 of the most intense m/z peaks were selected to include them into the genetic algorithm (GA) model. The selected peaks by the operator gave a recognition capability (RC) value together with the highest cross-validation (CV) value. The presence or absence of all discriminating peak masses generated by the GA model was controlled by comparing the average spectra from each species per body-part.
Database creation and blind tests
The reference MS spectra were created using spectra from legs and thorax of two specimens per species when available using MALDI-Biotyper software v3.0. (Bruker Daltonics) [26]. MS spectra of legs and thoraxes from a total of 15 specimens, identified morphologically and molecularly, were added to our MS spectra database (DB), including already legs and thoraxes reference MS spectra from eight distinct mosquito species, but none from the Anopheles genus [37]. MS spectra were created with an unbiased algorithm using information on the peak position, intensity and frequency. MS spectra from legs and thoraxes of 96 mosquitoes were tested against the in-house MS reference spectra DB. The peak list from query samples were compared to reference peak lists of organisms in the MS reference spectra DB. The reliability of species identification was estimated using the log score values (LSVs). The calculation of this score, which ranged from 0 to 3, was done using a biostatistical algorithm from the MALDI Biotyper software v.3.0. According to previous studies [26,43], LSVs greater than 1.8 were considered reliable for species identification. Data were analyzed by using GraphPad Prism software version 5.01 (GraphPad, San Diego, CA, USA).
Phylogenetic analyses
After gene sequences alignment with the Clustal ω2 algorithm in the MEGA 7.0 software, a maximum likelihood tree based on the COI gene were constructed using the MEGA 7.0 software [44]. The tree with the highest log likelihood was kept. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Support for internal nodes was estimated using the nonparametric bootstrap method with 1000 replications.
Results
Morphological identification and molecular validation
Among the mosquitoes captured in the 6 distinct sites from French Guiana (Fig 1), uniquely anopheline specimens were selected. These mosquitoes were classified morphologically into eight distinct species, six from the Nyssorhynchus subgenus (An. aquasalis, An. braziliensis, An. darlingi, An. nuneztovari sl, An. triannulatus sl, An. oswaldoi sl) and two from the Anopheles subgenus (An. intermedius, An. peryassui) (Table 1). According to their availability, one to 22 specimens per species were included in the present study. A total of 111 Anopheles specimens were selected. The COI gene sequencing of all specimens was done to valid morphological identification. COI gene sequences were queried against GenBank (NCBI) and the Barcode of Life Data (BOLD) Systems. The query of COI gene sequences allowed to obtain reliable mosquito species identification for all samples with identity ranges of 98–99% against GenBank and 98.15–100% against BOLD databases (Table 1). Concordant mosquito species identification were obtained between the two molecular DBs. The COI gene sequencing corroborated morphological classifications, at the exception of one mosquito. It was morphologically classified as An. peryassui but molecular analysis revealed that it was identified as An. intermedius. The phylogenetic analysis was done with the COI gene sequences of the 15 mosquito specimens selected for MS reference creation (S1 Fig). Mosquitoes belonging to the same subgenus clustered together.
Reproducible and specific MS spectra from both mosquito body parts
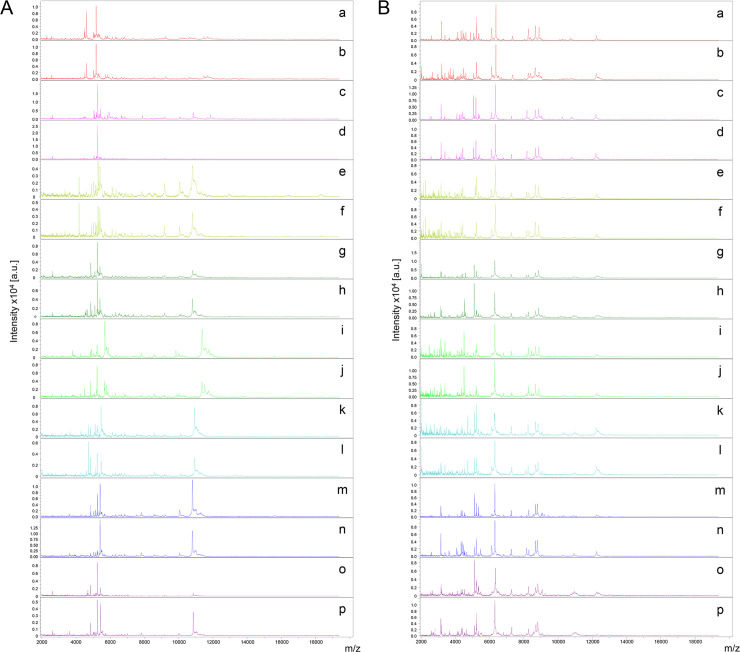
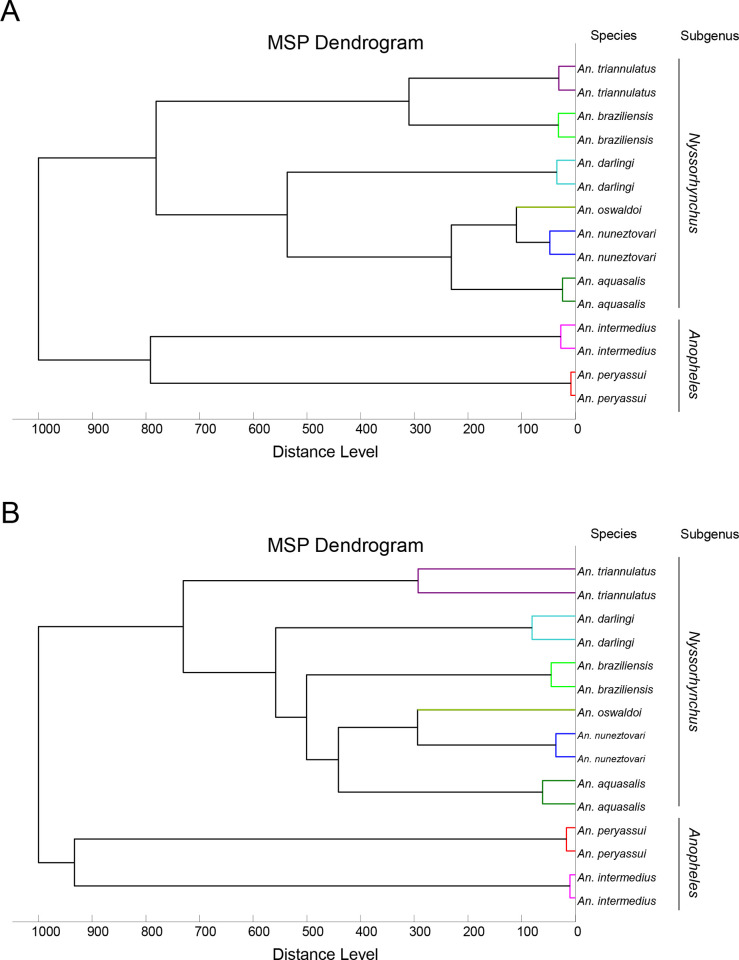
MS profiles of high intensity (>2000 a.u.) were obtained for legs (Fig 2A) and thoraxes (Fig 2B) from each of the 111 mosquitoes submitted to MALDI-TOF MS. Visual reproducible MS spectra were obtained for specimens of the same species according to body part (Fig 2). To evaluate the reproducibility and specificity of MS spectra from legs and thoraxes according to species, cluster analyses were performed. Two specimens per species were used for MSP dendrogram creation, at the exception of An. oswaldoi for which only one specimen was available. The clustering of specimens from the same species on the same branch and the absence of species intertwining underlined the reproducibility and specificity of the protein profiles for each Anopheles species for legs (Fig 3A) and thoraxes (Fig 3B). Interestingly, Anopheles species ordination from MSP dendrograms was not similar between legs and thoraxes of paired species (Fig 3). Nevertheless, Anopheles species from the same subgenus clustered on the same branch on both MSP dendrograms. To visualize specificity of MS spectra according body part per Anopheles species, PCAs were performed. PCAs revealed a clear separation of the dots corresponding to MS spectra from the legs and thoraxes, confirming a specificity of MS profiles between these two body parts for the seven Anopheles species tested (S2 Fig).
Fig 2.
Comparison of MALDI-TOF MS spectra from legs (A) and thoraxes (B) of Anopheles mosquitoes. Representative MS spectra of An. peryassui (a, b), An. intermedius (c, d), An. oswaldoi (e, f), An. aquasalis (g, h), An. braziliensis (i, j), An. darlingi (k, l), An. nuneztovari (m, n), and An. triannulatus (o, p) are shown. MS spectra from two distinct specimens per species were selected, excepted for An. oswaldoi. As only one specimen was available for this species, MS spectra from biological replicates were presented. a.u., arbitrary units; m/z, mass-to-charge ratio.
Fig 3.
MSP dendrogram of MALDI-TOF MS spectra from legs (A) and thoraxes (B) of Anopheles mosquitoes. Two specimens per species were used to construct the dendrogram, at the exception of An. oswaldoi, for which only one specimen was available. The dendrogram was created using Biotyper v3.0 software and distance units correspond to the relative similarity of MS spectra. The Anopheles and Nyssorhynchus subgenus were indicated at the right part.
As correct specimen species classification relies mainly on the intensity of resulting MS spectra, we assessed whether the most intense mass peaks from legs and thoraxes per mosquito species could be enough to distinct these Anopheles species, at the exception of An. oswaldoi for which only one specimen was available. The selection of the top-ten and top-five mass peak lists per species conducted to a total of 41 and 27 MS peaks for legs and 39 and 24 for thoraxes, respectively (S1 and S2 Tables). These MS peak lists were included in the genetic algorithm (GA) model from ClinProTools 2.2 software. The combination of the presence/absence of these top-ten and top-five mass peak lists per Anopheles species displayed, respectively, RC values of 99.6% and 97.0% and CV values of 97.9% and 98.3% for MS spectra from legs. For MS spectra from thoraxes, RC values of 99.4% and 97.5% and CV values of 99.8% and 99.6% were obtained for the top-ten and top-five selected mass peak lists, respectively.
MS reference spectra database creation and validation step
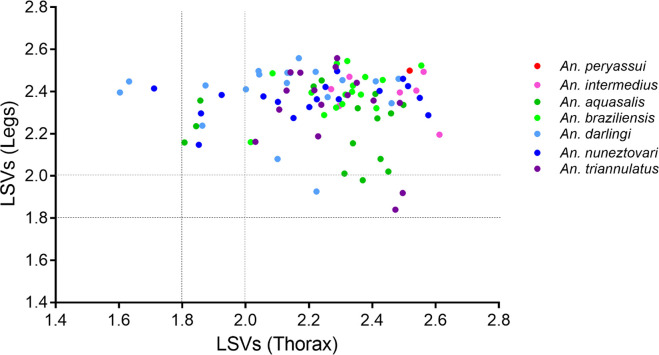
MS spectra of legs and thoraxes from the 15 specimens used for MSP dendrogram analysis (Table 1 and S1 File), validated morphologically and molecularly, were added to our MS spectra database (DB) including already legs and thoraxes reference MS spectra from eight distinct mosquito species [37]. The legs and thoraxes MS spectra from the 96 remaining specimens were queried against this upgraded DB. Interestingly, 100% of the identification results were concordant between paired MS spectra from legs and thoraxes. Among them, the MS identification of 95 specimens corroborated morphological results. One specimen, classified as An. peryassui based on morphological criteria was identified as An. intermedius based on MS tool with LSVs of 2.40 and 2.49 for legs and thorax, respectively, confirming the results of COI gene sequencing. The LSVs ranged from 1.84 to 2.56 for legs and from 1.60 to 2.61 for thoraxes (S3 Fig). As a threshold LSV upper than 1.8 is require for reliable identification [26,42], correct classification could be considered for 100% (96/96) of MS spectra from legs and 96.9% (3/96) from thoraxes. However, if we considered the LSV results from paired-samples per specimen, 100% of the mosquitoes tested, succeeded to obtain a LSV upper than 1.8 for at least one body-part (Fig 4). Interestingly, an increase of the LSV cut-off at 2.0, which improves identification confidence, revealed that 95.8% (92/96) and 88.5% (85/96) of the specimens reached this threshold, based on their legs and thoraxes MS spectra, respectively. However, the rate of at least one body-part from paired-samples per specimen achieving this threshold (LSVs>2.0) remained at 100%.
Fig 4. Comparison of paired body parts LSVs from MS spectra of Anopheles species.
Dashed lines represent the threshold values (black and grey for LSV threshold of 1.8 and 2.0, respectively), for relevant identification. LSV, log score value.
Discussion
The correct identification of mosquito species is essential for adapted control management, allowing to index circulating species in a given region and, consequently, to estimate vector borne disease transmission risks. Border regions such as French Guiana may be influenced by their neighboring countries. The legal and illegal flow of human and merchandises through this frontier could induce mosquito vectors migration and conducting to colonization of new areas. Solely, a rapid, accurate and low cost surveillance method of mosquitoes will succeed to improve control measures.
In French Guiana, An. darlingi is the main malaria vector transmitting several Plasmodium species, such as P. falciparum and P. vivax [45], this last one representing about 75% of human cases [46]. In addition to An. darlingi, other Anopheles species were reported to transmit Plasmodium pathogens in this area during their blood feeding [2,3,10,40]. Moreover, these malaria vectors live in sympatry with other anopheline species non-malaria vectors [39]. Indeed, in an effective program aiming to prevent or to eliminate malaria transmission, an accurate identification of Anopheles species remains a key factor. The recent repetitive success of the use of MALDI-TOF MS profiling for arthropod identification, including mosquitoes, was applied in the present work to implement our home-made MS reference spectra DB with anopheline specimens from French Guiana and to palliate of the limitations of morphological and molecular analyses [25].
In the present study, we confirmed that legs and thoraxes MS spectra from paired-specimens of the same species were distinct at least for the seven species for which more than one specimen was available. The species-specificity demonstrated for each body-part, underlined that these two compartments could be used independently for mosquito identification. The independent MS submission of several body-part to MALDI-TOF MS for specimen identification, was recently reported for mosquitoes [37] but also for ticks [47,48]. The advantages to test two distinct body parts are the possibility to cross-validate the results and to improve rate of identification confidence and reliability. Effectively, the combination of the results obtained independently by the query against the MS reference DB conducted to concordant results for each body part tested. Here, based on morphological criteria, a specimen was classified as An. peryassui. The MS submission of its legs and thorax indicated a matching with An. intermedius MS reference spectra for both body parts with high LSVs (>2.40), which was concordant with COI gene sequencing. Moreover, if the cut-off threshold of LSV to consider identification as reliable was raised to 2.0, 100% of the specimens succeeded to reach this cut-off with MS spectra from at least one body part. It is interesting to note that, for bacteria identification, it was assumed that species were assigned for LSVs upper or equal than 2.0, and genera were assigned for scores of upper or equal to 1.7 but lower than 2.0 [49,50]. Then, the combination of the results from both body-parts allowed to enhance ratio of LSVs upper or equal than 2.0 for each specimen compared to the use of only one body-part.
Finally, the concordance of the MS identification results between the two body-parts more the reaching of the LSV threshold of 2.0 for at least one body-part, are complementary data which improve identification confidence. The previous study using two body parts for mosquito identification was done on mosquitoes from distinct genera [37]. Here, the eight mosquito species came all from the Anopheles genus including two subgenera, Nyssorhynchus and Anopheles. The correct identification of mosquito specimens by MALDI-TOF MS profiling for these close-related Anopheles species comfirms the accuracy of this innovative tool.
Among the mosquito species included in the MS reference spectra DB, 4 species are malaria vectors (An. darlingi, An. nuneztovari sl, An. intermedius, An. oswaldoi sl), for the 4 remaining species their malaria vector competence was not yet demonstrated (An. aquasalis, An. braziliensis, An. triannulatus sl, An. peryassui) [5]. Interestingly, malaria vectors and non-vectors are present in each of these two subgenera, underlining the importance to classify them correctly for disease prevention and vector control of species living in sympatric.
The main risks of misidentification or to fail identification by MALDI-TOF MS profiling are generally attributed to either the comprehensiveness of the species included in the MS reference database, or more frequently the low intensity of MS spectra. The problem of incomplete reference MS database could be easily solved by performing COI gene sequencing of un-matched good quality MS spectra, using the remaining body part of the specimen (ie, head, abdomen or wings). In case of new mosquito species, the addition of respective MS spectra, not yet included in the MS reference DB, could be done. The application of this strategy could resolved step by step MS spectra of good quality failing to find correspondence in the reference MS DB. Concerning the MS spectra un-matched, attributed to the low intensity of protein profiles, this phenomenon is frequently observed for legs MS spectra due to the low protein quantity contained in this compartment [26]. Moreover, as the legs are highly breakable, it is frequent that the loose of one to five legs occurred during specimen collection, transport or storing. This decrease of leg number reduces the success rate of specimen identification [42]. Moreover, for specimens which have lost all legs, their identification become not possible if the identification was based uniquely on this compartment by MS. The creation of reference MS spectra from two distinct mosquito body parts allows to succeed specimen identification by this rapid proteomic approach.
Moreover, in the present work, it was highlighted that the classification of Anopheles species could be correctly done using the most intense MS peaks. Effectively, the selection of the top-10 or also the top-5 of the MS peaks possessing the higher intensity appeared sufficiently discriminants to classify correctly these mosquito species. This correct classification is valid for both body-parts. These results underline that a correct identification remain possible with MS spectra of low intensity for which uniquely the most intense MS peaks could be detected.
The comparison of MSP dendrograms between legs and thoraxes revealed that, despite species from the same subgenus were clustered in the same branch, the ordination of the species inside these branches was not similar. This unreproducible classification objectifies that the MS profiles proximity were different between the Anopheles species for legs and thorax. These distinct ordination of species based on their MS profiles, should reduce the risk of misidentification by the submission of both body parts. Interestingly, none of these MSP dendrograms proposed a classification comparable with those obtained using COI gene sequences for phylogenetic tree construction. The MSP dendrograms are not adapted for phylogenetic analyses as previously reported [51,52].
Conclusion
Mosquito monitoring with fast, highly reproducible and reliable tools such as MALDI-TOF appears essential in today's globalization scenario. The specificity of MS protein profiles for mosquito legs and thoraxes confirmed that these two body parts are relevant for specimen identification. Moreover, as the most intense MS peaks were demonstrated to be sufficient for correct classification, the sample which will generate MS spectra of low quality, could anyway be identified. The sharing of reference MS spectra is primordial to accelerate the dissemination of this innovative tool for a routine use in mosquito identification contributing to adapt control of vectors.
Supporting information
(TIF)
PCA dimensional image from MS spectra of legs (red dots) and thoraxes (green dots) from An. intermedius (A), An. aquasalis (B), An. braziliensis (C), An. darlingi (D), An. nuneztovari (E), An. triannulatus (F) and An. peryassui (G). Respectively, 10, 22, 18, 22, 20, 20 and 3 specimens per species were included. Quadruplicate of each sample per body part were presented.
(TIF)
LSVs obtained following homemade MS reference database query with MS spectra of legs (A) and thoraxes (B) from Anopheles mosquitoes. Horizontal dashed lines represent the threshold value for reliable identification (black and grey for LSV threshold of 1.8 and 2.0, respectively). LSVs, log score values; a.u., arbitrary units.
(TIF)
(DOCX)
(DOCX)
MS spectra were obtained using Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics).
(7Z)
Acknowledgments
We would like to acknowledge Samuel Vezenegho and Antoine Adde, from Unite d’Entomologie Médicale, Institut Pasteur de la Guyane, Cayenne, French Guiana, for their involvement in sample management. We also acknowledge Albin Fontaine (UPE, IRBA, Marseille) for his help in map building.
List of abbreviations
- sl
sensu lato
- ss
sensu stricto
- MALDI-TOF MS
Matrix Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry
- PCR
Polymerase Chain Reaction
- CCI
Composite Correlation Index
- LSV
Log Score Value
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been supported by the Délégation Générale pour l’Armement (DGA, MoSIS project, Grant no PDH-2-NRBC-2-B-2113).
References
- 1.Ardillon V, Carvalho L, Prince C, Abboud P, Djossou F. Bilans 2013 et 2014 de la situation du paludisme en Guyane. Bull Veille Sanit. 2015;1: 16–20. [Google Scholar]
- 2.Pommier de Santi V, Dia A, Adde A, Hyvert G, Galant J, Mazevet M, et al. Malaria in French Guiana Linked to Illegal Gold Mining. Emerg Infect Dis. 2016;22: 344–346. 10.3201/eid2202.151292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pommier de Santi V, Girod R, Mura M, Dia A, Briolant S, Djossou F, et al. Epidemiological and entomological studies of a malaria outbreak among French armed forces deployed at illegal gold mining sites reveal new aspects of the disease’s transmission in French Guiana. Malar J. 2016;15: 35 10.1186/s12936-016-1088-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Douine M, Sanna A, Hiwat H, Briolant S, Nacher M, Belleoud D, et al. Investigation of a possible malaria epidemic in an illegal gold mine in French Guiana: an original approach in the remote Amazonian forest. Malar J. 2019;18: 91 10.1186/s12936-019-2721-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Talaga S, Dejean A, Carinci R, Gaborit P, Dusfour I, Girod R. Updated Checklist of the Mosquitoes (Diptera: Culicidae) of French Guiana. J Med Entomol. 2015;52: 770–782. 10.1093/jme/tjv109 [DOI] [PubMed] [Google Scholar]
- 6.Girod R, Gaborit P, Carinci R, Issaly J, Fouque F. Anopheles darlingi bionomics and transmission of Plasmodium falciparum, Plasmodium vivax and Plasmodium malariae in Amerindian villages of the Upper-Maroni Amazonian forest, French Guiana. Mem Inst Oswaldo Cruz. 2008;103: 702–710. 10.1590/s0074-02762008000700013 [DOI] [PubMed] [Google Scholar]
- 7.Girod R, Roux E, Berger F, Stefani A, Gaborit P, Carinci R, et al. Unravelling the relationships between Anopheles darlingi (Diptera: Culicidae) densities, environmental factors and malaria incidence: understanding the variable patterns of malarial transmission in French Guiana (South America). Ann Trop Med Parasitol. 2011;105: 107–122. 10.1179/136485911X12899838683322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fouque F, Gaborit P, Carinci R, Issaly J, Girod R. Annual variations in the number of malaria cases related to two different patterns of Anopheles darlingi transmission potential in the Maroni area of French Guiana. Malar J. 2010;9: 80 10.1186/1475-2875-9-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiwat H, Issaly J, Gaborit P, Somai A, Samjhawan A, Sardjoe P, et al. Behavioral heterogeneity of Anopheles darlingi (Diptera: Culicidae) and malaria transmission dynamics along the Maroni River, Suriname, French Guiana. Trans R Soc Trop Med Hyg. 2010;104: 207–213. 10.1016/j.trstmh.2009.07.007 [DOI] [PubMed] [Google Scholar]
- 10.Dusfour I, Issaly J, Carinci R, Gaborit P, Girod R. Incrimination of Anopheles (Anopheles) intermedius Peryassú, An. (Nyssorhynchus) nuneztovari Gabaldón, An. (Nys.) oswaldoi Peryassú as natural vectors of Plasmodium falciparum in French Guiana. Mem Inst Oswaldo Cruz. 2012;107: 429–432. 10.1590/s0074-02762012000300021 [DOI] [PubMed] [Google Scholar]
- 11.Ruiz-Lopez F, Wilkerson RC, Ponsonby DJ, Herrera M, Sallum MAM, Velez ID, et al. Systematics of the oswaldoi complex (Anopheles, Nyssorhynchus) in South America. Parasit Vectors. 2013;6: 324 10.1186/1756-3305-6-324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conn JE, Wilkerson RC, Segura MNO, de Souza RTL, Schlichting CD, Wirtz RA, et al. Emergence of a new neotropical malaria vector facilitated by human migration and changes in land use. Am J Trop Med Hyg. 2002;66: 18–22. 10.4269/ajtmh.2002.66.18 [DOI] [PubMed] [Google Scholar]
- 13.Montoya-Lerma J, Solarte YA, Giraldo-Calderón GI, Quiñones ML, Ruiz-López F, Wilkerson RC, et al. Malaria vector species in Colombia: a review. Mem Inst Oswaldo Cruz. 2011;106 Suppl 1: 223–238. 10.1590/s0074-02762011000900028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beebe NW, Cooper RD. Systematics of malaria vectors with particular reference to the Anopheles punctulatus group. Int J Parasitol. 2000;30: 1–17. 10.1016/s0020-7519(99)00171-x [DOI] [PubMed] [Google Scholar]
- 15.Munstermann LE, Conn JE. Systematics of mosquito disease vectors (Diptera, Culicidae): impact of molecular biology and cladistic analysis. Annu Rev Entomol. 1997;42: 351–369. 10.1146/annurev.ento.42.1.351 [DOI] [PubMed] [Google Scholar]
- 16.Van Rensburg AJ, Hunt RH, Koekemoer LL, Coetzee M, Shiff CJ, Minjas J. The polymerase chain reaction method as a tool for identifying members of the Anopheles gambiae complex (Diptera:Culicidae) in northeastern Tanzania. J Am Mosq Control Assoc. 1996;12: 271–274. [PubMed] [Google Scholar]
- 17.Figueiredo MAP, Di Santi SM, Manrique WG, Gonçalves LR, André MR, Machado RZ. Molecular identification of Plasmodium spp. and blood meal sources of anophelines in environmental reserves on São Luís Island, state of Maranhão, Brazil. Parasit Vectors. 2017;10: 203 10.1186/s13071-017-2133-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrelli MT, Floeter-Winter LM, Malafronte RS, Tadei WP, Lourenço-de-Oliveira R, Flores-Mendoza C, et al. Amazonian malaria vector anopheline relationships interpreted from ITS2 rDNA sequences. Med Vet Entomol. 2005;19: 208–218. 10.1111/j.0269-283X.2005.00558.x [DOI] [PubMed] [Google Scholar]
- 19.Nebbak A, Koumare S, Willcox AC, Berenger J-M, Raoult D, Almeras L, et al. Field application of MALDI-TOF MS on mosquito larvae identification. Parasitology. 2018;145: 677–687. 10.1017/S0031182017001354 [DOI] [PubMed] [Google Scholar]
- 20.Ratnasingham S, Hebert PDN. bold: The Barcode of Life Data System (http://www.barcodinglife.org). Mol Ecol Notes. 2007;7: 355–364. 10.1111/j.1471-8286.2007.01678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Versteirt V, Nagy ZT, Roelants P, Denis L, Breman FC, Damiens D, et al. Identification of Belgian mosquito species (Diptera: Culicidae) by DNA barcoding. Mol Ecol Resour. 2015;15: 449–457. 10.1111/1755-0998.12318 [DOI] [PubMed] [Google Scholar]
- 22.Norris LC, Norris DE. Phylogeny of anopheline (Diptera: Culicidae) species in southern Africa, based on nuclear and mitochondrial genes. J Vector Ecol J Soc Vector Ecol. 2015;40: 16–27. 10.1111/jvec.12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Laurito M, Oliveira TMP de, Almirón WR, Sallum MAM. COI barcode versus morphological identification of Culex (Culex) (Diptera: Culicidae) species: a case study using samples from Argentina and Brazil. Mem Inst Oswaldo Cruz. 2013;108 Suppl 1: 110–122. 10.1590/0074-0276130457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang G, Li C, Guo X, Xing D, Dong Y, Wang Z, et al. Identifying the main mosquito species in China based on DNA barcoding. PloS One. 2012;7: e47051 10.1371/journal.pone.0047051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yssouf A, Almeras L, Raoult D, Parola P. Emerging tools for identification of arthropod vectors. Future Microbiol. 2016;11: 549–566. 10.2217/fmb.16.5 [DOI] [PubMed] [Google Scholar]
- 26.Yssouf A, Parola P, Lindström A, Lilja T, L’Ambert G, Bondesson U, et al. Identification of European mosquito species by MALDI-TOF MS. Parasitol Res. 2014;113: 2375–2378. 10.1007/s00436-014-3876-y [DOI] [PubMed] [Google Scholar]
- 27.Dieme C, Yssouf A, Vega-Rúa A, Berenger J-M, Failloux A-B, Raoult D, et al. Accurate identification of Culicidae at aquatic developmental stages by MALDI-TOF MS profiling. Parasit Vectors. 2014;7: 544 10.1186/s13071-014-0544-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaffner F, Kaufmann C, Pflüger V, Mathis A. Rapid protein profiling facilitates surveillance of invasive mosquito species. Parasit Vectors. 2014;7: 142 10.1186/1756-3305-7-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mewara A, Sharma M, Kaura T, Zaman K, Yadav R, Sehgal R. Rapid identification of medically important mosquitoes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Parasit Vectors. 2018;11: 281 10.1186/s13071-018-2854-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Müller P, Pflüger V, Wittwer M, Ziegler D, Chandre F, Simard F, et al. Identification of cryptic Anopheles mosquito species by molecular protein profiling. PloS One. 2013;8: e57486 10.1371/journal.pone.0057486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yssouf A, Socolovschi C, Flaudrops C, Ndiath MO, Sougoufara S, Dehecq J-S, et al. Matrix-assisted laser desorption ionization—time of flight mass spectrometry: an emerging tool for the rapid identification of mosquito vectors. PloS One. 2013;8: e72380 10.1371/journal.pone.0072380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Raharimalala FN, Andrianinarivomanana TM, Rakotondrasoa A, Collard JM, Boyer S. Usefulness and accuracy of MALDI-TOF mass spectrometry as a supplementary tool to identify mosquito vector species and to invest in development of international database. Med Vet Entomol. 2017;31: 289–298. 10.1111/mve.12230 [DOI] [PubMed] [Google Scholar]
- 33.Tandina F, Almeras L, Koné AK, Doumbo OK, Raoult D, Parola P. Use of MALDI-TOF MS and culturomics to identify mosquitoes and their midgut microbiota. Parasit Vectors. 2016;9: 495 10.1186/s13071-016-1776-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nebbak A, Almeras L. Identification of Aedes mosquitoes by MALDI-TOF MS biotyping using protein signatures from larval and pupal exuviae. Parasit Vectors. 2020;13: 161 10.1186/s13071-020-04029-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nebbak A, El Hamzaoui B, Berenger J-M, Bitam I, Raoult D, Almeras L, et al. Comparative analysis of storage conditions and homogenization methods for tick and flea species for identification by MALDI-TOF MS. Med Vet Entomol. 2017;31: 438–448. 10.1111/mve.12250 [DOI] [PubMed] [Google Scholar]
- 36.Nebbak A, Willcox AC, Bitam I, Raoult D, Parola P, Almeras L. Standardization of sample homogenization for mosquito identification using an innovative proteomic tool based on protein profiling. Proteomics. 2016;16: 3148–3160. 10.1002/pmic.201600287 [DOI] [PubMed] [Google Scholar]
- 37.Vega-Rúa A, Pagès N, Fontaine A, Nuccio C, Hery L, Goindin D, et al. Improvement of mosquito identification by MALDI-TOF MS biotyping using protein signatures from two body parts. Parasit Vectors. 2018;11: 574 10.1186/s13071-018-3157-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Adde A, Roux E, Mangeas M, Dessay N, Nacher M, Dusfour I, et al. Dynamical Mapping of Anopheles darlingi Densities in a Residual Malaria Transmission Area of French Guiana by Using Remote Sensing and Meteorological Data. PloS One. 2016;11: e0164685 10.1371/journal.pone.0164685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adde A, Dusfour I, Roux E, Girod R, Briolant S. Anopheles fauna of coastal Cayenne, French Guiana: modelling and mapping of species presence using remotely sensed land cover data. Mem Inst Oswaldo Cruz. 2016;111: 750–756. 10.1590/0074-02760160272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vezenegho SB, Adde A, Pommier de Santi V, Issaly J, Carinci R, Gaborit P, et al. High malaria transmission in a forested malaria focus in French Guiana: How can exophagic Anopheles darlingi thwart vector control and prevention measures? Mem Inst Oswaldo Cruz. 2016;111: 561–569. 10.1590/0074-02760160150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3: 294–299. [PubMed] [Google Scholar]
- 42.Nebbak A, Willcox AC, Bitam I, Raoult D, Parola P, Almeras L. Standardization of sample homogenization for mosquito identification using an innovative proteomic tool based on protein profiling. Proteomics. 2016;16: 3148–3160. 10.1002/pmic.201600287 [DOI] [PubMed] [Google Scholar]
- 43.Lafri I, Almeras L, Bitam I, Caputo A, Yssouf A, Forestier C-L, et al. Identification of Algerian Field-Caught Phlebotomine Sand Fly Vectors by MALDI-TOF MS. PLoS Negl Trop Dis. 2016;10: e0004351 10.1371/journal.pntd.0004351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10: 512–526. 10.1093/oxfordjournals.molbev.a040023 [DOI] [PubMed] [Google Scholar]
- 45.Sinka ME, Bangs MJ, Manguin S, Rubio-Palis Y, Chareonviriyaphap T, Coetzee M, et al. A global map of dominant malaria vectors. Parasit Vectors. 2012;5: 69 10.1186/1756-3305-5-69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Musset L, Pelleau S, Girod R, Ardillon V, Carvalho L, Dusfour I, et al. Malaria on the Guiana Shield: a review of the situation in French Guiana. Mem Inst Oswaldo Cruz. 2014;109: 525–533. 10.1590/0074-0276140031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyer PH, Boulanger N, Nebbak A, Collin E, Jaulhac B, Almeras L. Assessment of MALDI-TOF MS biotyping for Borrelia burgdorferi sl detection in Ixodes ricinus. PloS One. 2017;12: e0185430 10.1371/journal.pone.0185430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boyer PH, Almeras L, Plantard O, Grillon A, Talagrand-Reboul É, McCoy K, et al. Identification of closely related Ixodes species by protein profiling with MALDI-TOF mass spectrometry. PloS One. 2019;14: e0223735 10.1371/journal.pone.0223735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murugaiyan J, Lewin A, Kamal E, Bakuła Z, van Ingen J, Ulmann V, et al. MALDI Spectra Database for Rapid Discrimination and Subtyping of Mycobacterium kansasii. Front Microbiol. 2018;9: 587 10.3389/fmicb.2018.00587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Piamsomboon P, Jaresitthikunchai J, Hung TQ, Roytrakul S, Wongtavatchai J. Identification of bacterial pathogens in cultured fish with a custom peptide database constructed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS). BMC Vet Res. 2020;16: 52 10.1186/s12917-020-2274-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kumsa B, Laroche M, Almeras L, Mediannikov O, Raoult D, Parola P. Morphological, molecular and MALDI-TOF mass spectrometry identification of ixodid tick species collected in Oromia, Ethiopia. Parasitol Res. 2016;115: 4199–4210. 10.1007/s00436-016-5197-9 [DOI] [PubMed] [Google Scholar]
- 52.Karger A, Kampen H, Bettin B, Dautel H, Ziller M, Hoffmann B, et al. Species determination and characterization of developmental stages of ticks by whole-animal matrix-assisted laser desorption/ionization mass spectrometry. Ticks Tick-Borne Dis. 2012;3: 78–89. 10.1016/j.ttbdis.2011.11.002 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
PCA dimensional image from MS spectra of legs (red dots) and thoraxes (green dots) from An. intermedius (A), An. aquasalis (B), An. braziliensis (C), An. darlingi (D), An. nuneztovari (E), An. triannulatus (F) and An. peryassui (G). Respectively, 10, 22, 18, 22, 20, 20 and 3 specimens per species were included. Quadruplicate of each sample per body part were presented.
(TIF)
LSVs obtained following homemade MS reference database query with MS spectra of legs (A) and thoraxes (B) from Anopheles mosquitoes. Horizontal dashed lines represent the threshold value for reliable identification (black and grey for LSV threshold of 1.8 and 2.0, respectively). LSVs, log score values; a.u., arbitrary units.
(TIF)
(DOCX)
(DOCX)
MS spectra were obtained using Microflex LT MALDI-TOF Mass Spectrometer (Bruker Daltonics).
(7Z)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.