Abstract
Current geographic spread of documented severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections shows heterogeneity. This study explores the role of age in potentially driving differentials in infection spread, epidemic potential, and rates of disease severity and mortality across countries. An age-stratified deterministic mathematical model that describes SARS-CoV-2 transmission dynamics was applied to 159 countries and territories with a population ≥1 million. Assuming worst-case scenario for the pandemic, the results indicate that there could be stark regional differences in epidemic trajectories driven by differences in the distribution of the population by age. In the African Region (median age: 18.9 years), the median R0 was 1.05 versus 2.05 in the European Region (median age: 41.7 years), and the median (per 100 persons) for the final cumulative infection incidence was 22.5 (versus 69.0), for severe and/or critical disease cases rate was 3.3 (versus 13.0), and for death rate was 0.5 (versus 3.9). Age could be a driver of variable SARS-CoV-2 epidemic trajectories worldwide. Countries with sizable adult and/or elderly populations and smaller children populations may experience large and rapid epidemics in absence of interventions. Meanwhile, countries with predominantly younger age cohorts may experience smaller and slower epidemics. These predictions, however, should not lead to complacency, as the pandemic could still have a heavy toll nearly everywhere.
Introduction
The current geographic spread of the documented severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) [1] infections and associated Coronavirus Disease 2019 (COVID-2019) [1] shows heterogeneity [2]. This remains unexplained but may possibly reflect delays in virus introduction into the population, differentials in testing and/or reporting, differentials in the implementation, scale, adherence, and timing of public health interventions, or other epidemiological factors. In this study, we explore the role of age in explaining the differential spread of the infection and its future epidemic potential.
As our understanding of the SARS-CoV-2 transmission dynamics is rapidly evolving [3], the role of age in the epidemiology of this infection is becoming increasingly apparent [4–7]. In a recent study examining SARS-CoV-2 epidemiology in China [4], we quantified the effect of age on the biological susceptibility to infection acquisition. We found that relative to individuals aged 60–69 years, susceptibility was 0.06 in those aged 0–19 years, 0.34 in those aged 20–29 years, 0.57 in those aged 30–39 years, 0.69 in those aged 40–49 years, 0.79 in those aged 50–59 years, 0.94 in those aged 70–79 years, and 0.88 in those aged ≥80 years (S1 Fig in S1 File). Notably, this age-dependence was estimated after accounting for the assortativeness in population mixing by age [4].
Age also affects disease progression [7–10] and mortality risk [11–13] among those infected. The proportion of infections that eventually progress to severe disease, critical disease, or death, increases rapidly with age, especially among those ≥50 years of age (S2 Fig in S1 File) [8–12]. Since the demographic structure of the population (that is the distribution of the population across the different age groups) varies by country and region, this poses a question as to the extent to which age effects can drive geographic differentials in the basic reproduction number (R0), epidemic potential, and rates of disease severity and mortality. R0 is defined here as the number of infections caused by an index case in a fully susceptible population [14].
We aimed here to provide an answer to this question and to estimate for each country (with a population ≥1 million), region, and globally, R0, and the rate per 100 persons (out of the total population by the end of the epidemic cycle) of each of the cumulative number of incident infections, mild infections, severe and/or critical disease cases, and deaths, in addition to the number of days needed for the national epidemic to reach its incidence peak (a measure of how fast the epidemic will grow).
Materials and methods
We adapted and applied our recently developed deterministic mathematical model [4] describing SARS-CoV-2 transmission dynamics in China (S3 Fig in S1 File), to 159 countries and territories, virtually covering the world population [15]. Since our focus was on investigating the natural course of the SARS-CoV-2 epidemic and on assessing its full epidemic potential, the model was applied assuming the worst-case scenario for the epidemic in each country, that is in absence of any intervention.
The model stratified the population into compartments according to age (0–9, 10–19, …, ≥80 years), infection status (uninfected, infected), infection stage (mild, severe, critical), and disease stage (severe, critical), using a system of coupled nonlinear differential equations (Section 1 in S1 File). Susceptible individuals in each age group were assumed at risk of acquiring the infection at a hazard rate that varies based on the age-specific susceptibility to the infection (S1 Fig in S1 File), the infectious contact rate per day, and the transmission mixing matrix between the different age groups (Section 1 in S1 File). Following a latency period of 3.69 days [16–19], infected individuals develop mild, severe, or critical infection, as informed by the observed age-specific distribution of cases across these infection stages in China [8–10]. The duration of infectiousness was assumed to last for 3.48 days [8, 16, 18, 19] after which individuals with mild infection recover, while those with severe and critical infection develop, respectively, severe and critical disease over a period of 28 days [8] prior to recovery. Individuals with critical disease have the additional risk of disease mortality, as informed by the age-stratified disease mortality rate in China [11, 12].
Model parameters were based on current data for SARS-CoV-2 natural history and epidemiology, or through model fitting to the China outbreak data [4] (S1, S2 Tables in S1 File). Namely, the overall infectious contact rate, age-specific biological susceptibility profile, and age-specific mortality rate (assuming the observed case fatality rate) were assumed as those in China [4]. The population size, demographic structure, and life expectancy in countries and territories with a population of ≥1 million, as of 2020, were extracted from the United Nations World Population Prospects database [15]. Whenever available data for a parameter covered a wider age bracket, we assumed that this parameter has the same value for all age groups within that bracket. Model parameters, definitions, values, and justifications are in Section B and S1, S2 Tables in S1 File.
Ranges of uncertainty around model-predicted outcomes were determined using five-hundred simulation runs that applied Latin Hypercube sampling [20, 21] from a multidimensional distribution of the model parameters, including both the natural history parameters and the age-specific susceptibility profile. At each run, input parameter values were selected from ranges specified by assuming ±30% uncertainty around parameters’ point estimates [22–26]. The resulting distribution for each model-predicted outcome was then used to derive the most probable estimate (based on maximum likelihood) and 95% uncertainty interval. Mathematical modelling analyses were performed in MATLAB R2019a [27], while statistical analyses were performed in STATA/SE 16.1 [28].
Results
In what follows, we highlight results for select (mostly populous) countries that are of broad geographic representation and characterized by diverse demographic structures. These include Brazil, China, Egypt, India, Indonesia, Italy, Niger, Pakistan, and USA. Estimates and uncertainty ranges for these countries are illustrated in Figs 1, 3 and 4. Detailed results for all 159 countries and territories are in S3-S8 Tables in S1 File.
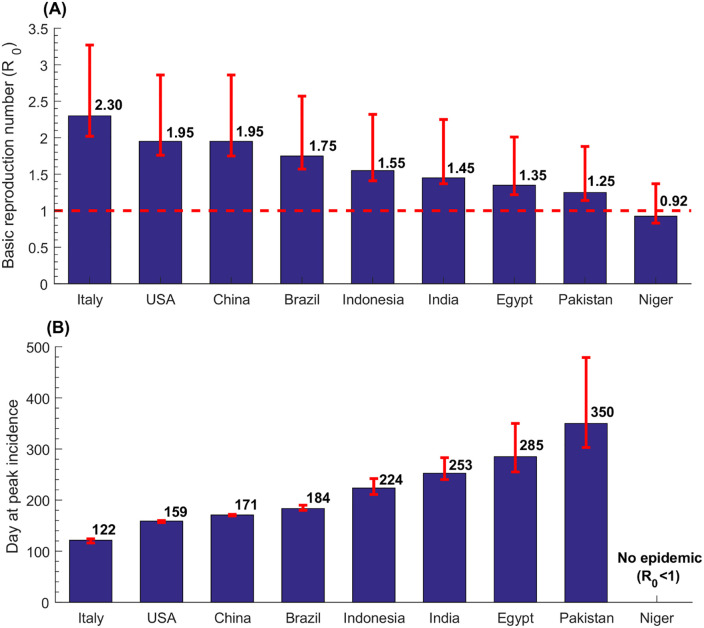
Fig 1. Estimates for the basic reproduction number, R0, and the number of days needed for the national epidemic to reach its incidence peak, in select countries.
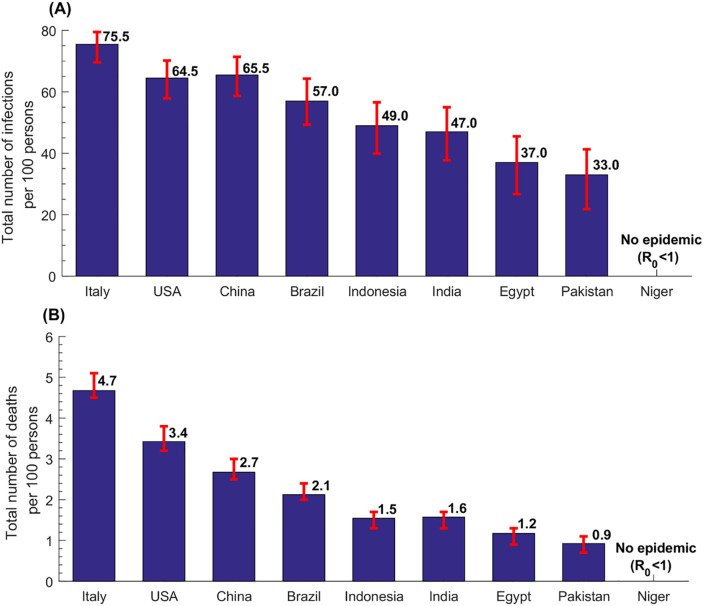
Fig 3. Estimates for the total number of infections and the total number of deaths per 100 persons in select countries.
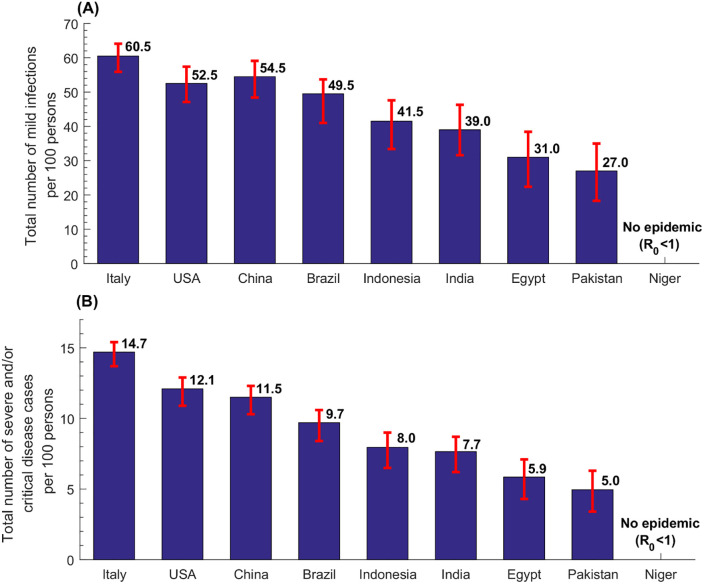
Fig 4. Estimates for the total number of mild infections and the total number of severe and/or critical diseases cases per 100 persons in select countries.
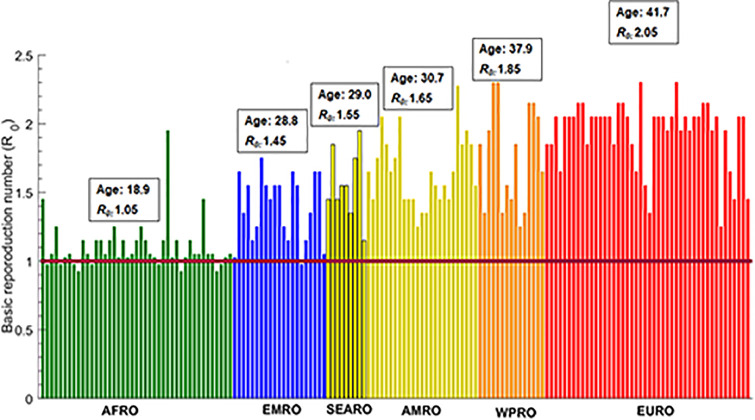
Fig 1A shows the estimated R0 in these select countries, which was highest in Italy at 2.30, followed by USA and China at 1.95, Brazil at 1.75, Indonesia at 1.55, India at 1.45, Egypt at 1.35, Pakistan at 1.25, and lowest in Niger at 0.93. Country-specific estimates of R0 grouped by World Health Organization (WHO) region are illustrated in Fig 2. The median R0 was 1.05 (range: 0.93–1.95) in African Region (AFRO), 1.45 (range: 0.98–1.75) in Eastern Mediterranean Region (EMRO), 1.55 (range: 1.15–1.95) in South-East Asia Region (SEARO), 1.65 (range: 1.25–2.28) in Region of the Americas (AMRO), 1.85 (range: 1.25–2.30) in Western pacific Region (WPRO), and 2.05 (range: 1.25–2.30) in European Region (EURO). Globally, the median R0 was 1.55 with a range of 0.93–2.30.
Fig 2. Estimates for the basic reproduction number R0 in 159 countries and territories with a population of at least one million, across World Health Organization regions.
These are the African Region (AFRO), Eastern Mediterranean Region (EMRO), South-East Asia Region (SEARO), Region of the Americas (AMRO), Western Pacific Region (WPRO), and European Region (EURO). The figure shows also the median age in years and the median R0 for each world region.
Fig 1B shows the estimated number of days needed for the national epidemic to reach its incidence peak in the select countries. In Italy, the peak was reached after 122 days (~4 months), whereas in Pakistan it was reached after 350 days (nearly a year). Meanwhile, since R0<1, no epidemic would emerge in Niger. Of note that the number of days needed for the national epidemic to reach its incidence peak depends on the population size in addition to R0—it takes more time for the epidemic to reach its peak in larger nations (S3-S8 Tables in S1 File).
The estimated incidence rate per 100 persons in the nine select countries, defined as the cumulative number of infections by the end of the epidemic cycle out of the total population, was highest in Italy at 75.5 and lowest in Pakistan at 33.0 (Fig 3A). Across world regions, the median incidence rate per 100 persons was 22.5 (range: 0.25–63.5) in AFRO, 45.0 (range: 1.5–65.0) in EMRO, 49.0 (range: 25.0–66.5) in SEARO, 53.0 in AMRO (range: 35.0–70.5), 63.5 (range: 31.0–76.5) in WPRO, and 69.0 (range: 31.0–75.5) in EURO, and 49.0 (range: 0.25–76.5) globally.
The estimated death rate per 100 persons in the nine select countries followed the same pattern, where it was highest in Italy at 4.7 and lowest in Pakistan at 0.9 (Fig 3B). Across world regions, the median death rate per 100 persons was 0.5 (range: 0.0–2.8) in AFRO, 0.9 (range: 0.0–2.0) in EMRO, 1.6 (range: 0.8–2.9) in SEARO, 2.0 in AMRO (range: 1.0–4.2), 2.7 (range: 0.8–5.3) in WPRO, and 3.9 (range: 0.8–4.7) in EURO, and 1.6 (range: 0.0–5.3) globally.
The estimated rate of mild infections per 100 persons in the nine select countries was highest in Italy at 60.5 and lowest in Pakistan at 27.0 (Fig 4A). Across world regions, the median per 100 persons was 17.0 (range: 0.25–52.5) in AFRO, 39.0 (range: 1.5–55.0) in EMRO, 41.0 (range: 23.0–54.5) in SEARO, 43.5 in AMRO (range: 29.0–56.5), 51.5 (range: 27.0–61.5) in WPRO, and 56.5 (range: 25.0–65.0) in EURO, and 41.0 (range: 0.25–61.5) globally.
Meanwhile, the estimated rate of severe and/or critical disease cases per 100 persons in the nine select countries was highest in Italy at 14.7 and lowest in Pakistan at 5.0 (Fig 4B). Across world regions, the median per 100 persons was 3.3 (range: 0.0–11.3) in AFRO, 7.1 (range: 0.3–9.5) in EMRO, 7.7 (range: 4.1–11.9) in SEARO, 8.7 in AMRO (range: 5.0–13.5), 11.5 (range: 5.0–15.5) in WPRO, and 13.0 (range: 4.7–14.7) in EURO, and 8.0 (range: 0.0–15.5) globally.
Discussion
The above results suggest that although this pandemic is a formidable challenge globally, its intensity and toll in terms of morbidity and mortality may vary substantially by country and regionally. While some countries, particularly in resource-rich countries, may experience large and rapid epidemics, other countries, particularly in resource-poor countries, may experience smaller and slower epidemics. While the scale of the epidemic may be smallest in sub-Saharan Africa with its young population, it could be intense in countries with a relatively small proportion of children and sizable proportions of adults and/or elderly.
The key finding of this study is the potential role of age in driving divergent SARS-CoV-2 epidemic trajectories and disease burdens. The effect of age reflects the interplay of three main factors: the variation in susceptibility by age (S1 Fig in S1 File), variation in disease severity and mortality by age (S2 Fig in S1 File), and variation in demographic structure across countries (S3-S8 Tables in S1 File). The combined effect of these factors can result in stark variations in R0 (Fig 2), and consequently epidemic potential, disease severity, and disease mortality—there was strong correlation between R0 and median age across countries (S4 Fig in S1 File). Incidentally, the globally diagnosed cases as of July 16, 2020 show a significant correlation with median age across countries (Spearman correlation coefficient: 0.34 (95% CI: 0.19–0.47), p<0.001). Although this may be explained by more diagnosed infections being reported by countries with advanced testing infrastructure, this may support the role of age presented here. Importantly, as infection transmission dynamics is a non-linear phenomenon, even small changes in R0, when R0 is in the range of 1–2, can drive considerable differences in epidemic size and trajectory. Variability by age, though of different magnitude, was also noted in a recent other research work [7].
An illustration of the role of age can be seen in Niger, a nation with a median age of only 15 years, where the exclusion of children from the population, while maintaining the same susceptibility profile for the other age groups, would have increased R0 from 0.93 to 2.6. This demonstrates how the lower susceptibility among younger persons, particularly children, acts as a “herd immunity” impeding the ferocious strength of the force of infection of an otherwise very infectious virus. This epidemiological feature contrasts that of other respiratory infections, such as the 2009 influenza A (H1N1) pandemic (H1N1pdm) infection (S5 Fig in S1 File) [29], where the cumulative incidence was found to be highest among children and young adults, and much smaller among older adults.
One possible inference to be drawn from the above results is that epidemic size and associated disease burden could be highest in settings with sizable mid-age and/or elderly populations, as currently exemplified by Italy. This may also possibly explain sub-national patterns, such as the large number of diagnosed cases in the city of New York, although additional fine-grained analyses are needed to delineate within-country heterogeneities in transmission dynamics.
Another possible inference of relevance to containment efforts relates to the likelihood of the infection establishing itself in a population. In addition to the role of travel patterns/restrictions and multiple introductions for seeding the infection [30], the probability of a major outbreak upon introduction of one infection is given approximately by 1−1/R0 [31, 32]. Accordingly, in countries where R0 is just above the epidemic threshold of R0 = 1, the virus will need to be introduced multiple times before it can generate sustainable chains of transmission. In such countries, less disruptive social distancing measures may be sufficient to contain the epidemic compared to countries with larger R0.
Our study explores the potential effect of age on the epidemiology of SARS-CoV-2, but other factors that remain poorly understood, may also contribute to driving different epidemic trajectories. Transmission of the virus may be affected by seasonality, environmental and genetic factors, differences in social network structure and cultural norms (such as shaking hands, kissing, and other person-to-person contacts), age-dependent transmissibility effects, and underlying co-morbidities which also impact disease severity and mortality [33–36]. These factors may have contributed to a slowly growing epidemic in Japan, despite its demographic structure, as opposed to fast growing epidemics in the European Region and the USA.
This study has limitations. Model estimations are contingent on the validity and generalizability of input data. Our estimates were based on SARS-CoV-2 natural history and disease progression data from China, but these may not be applicable to other countries. The key factor driving the heterogeneity in transmission dynamics is the age-specific biological susceptibility profile. We used the susceptibility profile as derived from the China outbreak data [4], but this may not be generalizable to other countries. More estimates from other countries are needed to investigate the potential variation in this profile across countries, and different analytical approaches may also arrive at different estimates [7]. Some of the effects of biological susceptibility may simply reflect differences in social behavior or the probability of infection ascertainment. The presence and contribution of such factors are, however, difficult to disentangle.
Model 10-year age-bands were informed by available empirical evidence, which hindered analysis using narrower age intervals [37]. While current data on the attack rate from different countries supports age heterogeneity and lower exposure among children, different countries show still variation in the age-stratified attack rate (S6, S7 Figs in S1 File). Of note that the observed lower susceptibility to the infection at younger age [4] does not necessarily imply absence of infection, but may reflect rapid infection clearance or subclinical infection. This may impact our estimates depending on these infections’ degree of infectiousness. Two sensitivity analyses assuming a 50% increase in the susceptibility of younger age cohorts or equal susceptibility among those aged <20 to those aged 20–29 (to somewhat reflect the effect of subclinical infections) resulted in smaller differences in R0 across countries, but still supported our findings of wide heterogeneity in R0 (S8 and S9 Figs in S1 File, respectively). It is conceivable that such subclinical infections may have lower viral load, and therefore rapid infection clearance and limited transmission potential. Data on infection clusters from China and other countries suggested that children do not appear to play a significant role in the transmission of this infection [7, 8, 38].
Increasingly, evidence suggests that R0 is higher than assumed here based on fitting the China epidemic [4, 39, 40], which would increase the R0 values reported here, but will not affect the relative differences across countries. We assumed a constant overall contact rate across countries, but contact rate patterns, and therefore R0, can vary across and within countries [34, 35], and also over time with the implementation of interventions and associated behavioral changes [41]. There could be also transient effects affecting transmission patterns and epidemic dynamics [41, 42]. We assumed low degree of assortativeness in age group mixing based on fitting the China epidemic [4], however, a sensitivity analysis assuming high assortativeness, though yielded higher estimates for R0, still preserved the relative differences in these estimates across countries (S10 Fig in S1 File).
Our study may have overestimated disease mortality by basing mortality rates on estimates of the crude case fatality rate in China, as suggested by a recent study [13]. Our results are based on the most probable value for R0 as estimated through 500 runs of uncertainty analysis. The latter may have biased our reported results towards lower R0. For instance, the most probable value for R0 in China was 1.95, but the point estimate assuming the baseline values of the input parameters was higher at 2.10 [4]. Despite these limitations, our parsimonious model, tailored to the nature of available data, was able to reproduce the epidemic as observed in China [4], and generated results that are valid to a wide range of model assumptions.
Age appears to be a driver of variable SARS-CoV-2 epidemic trajectories worldwide. Countries with sizable adult and/or elderly populations and smaller children populations may experience large and rapid epidemics in absence of interventions, necessitating disruptive social distancing measures to contain the epidemic. Meanwhile, countries with predominantly younger age cohorts may experience smaller and slower epidemics and may require less disruptive social distancing measures to contain the epidemic. These predictions, however, should not lead to complacency, and should not affect national response in strengthening preparedness plans and in implementing prevention interventions, as although many countries are predicted to have lower R0, these also often coincide with those having limited healthcare capacity and testing. Once established, and even if smaller in scale due to their predominantly younger population, the SARS-CoV-2 epidemic would still cause a heavy toll on developing countries with resource-poor healthcare infrastructure.
Supporting information
(DOCX)
Data Availability
All relevant data are within the manuscript and its Supporting Information files. The MATLAB code is available at: https://github.com/HousseinAyoub/COVID-19-Code.git.
Funding Statement
This publication was made possible by NPRP grant number 9-040-3-008 and NPRP grant number 12S-0216-190094 from the Qatar National Research Fund (a member of Qatar Foundation). The authors are also grateful for support provided by the Biomedical Research Program and the Biostatistics, Epidemiology, and Biomathematics Research Core, both at Weill Cornell Medicine-Qatar. GM acknowledges support by UK Research and Innovation as part of the Global Challenges Research Fund, grant number ES/P010873/1. The statements made herein are solely the responsibility of the authors. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. The publication of this article was funded by the Qatar National Library.
References
- 1.World Health Organization (WHO). Naming the coronavirus disease (COVID-19) and the virus that causes it. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/technical-guidance/naming-the-coronavirus-disease-(covid-2019)-and-the-virus-that-causes-it. Accessed on March 11, 2020. 2020.
- 2.COVID-19 Outbreak Live Update. https://www.worldometers.info/coronavirus/. Accessed on March 27, 2020. 2020.
- 3.Callaway E, Cyranoski D, Mallapaty S, Stoye E, Tollefson J. The coronavirus pandemic in five powerful charts. Nature. 2020;579(7800):482–3. 10.1038/d41586-020-00758-2 . [DOI] [PubMed] [Google Scholar]
- 4.Ayoub HH, Chemaitelly H, Mumtaz GR, Seedat S, Awad SF, Makhoul M, et al. Characterizing key attributes of the epidemiology of COVID-19 in China: Model-based estimations. medRxiv. 2020:2020.04.08.20058214. 10.1101/2020.04.08.20058214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang J, Litvinova M, Liang Y, Wang Y, Wang W, Zhao S, et al. Changes in contact patterns shape the dynamics of the COVID-19 outbreak in China. Science. 2020;368(6498):1481–6. 10.1126/science.abb8001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizumoto K, Omori R, Nishiura H. Age specificity of cases and attack rate of novel coronavirus disease (COVID-19). medRxiv. 2020:2020.03.09.20033142. 10.1101/2020.03.09.20033142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davies NG, Klepac P, Liu Y, Prem K, Jit M, group CC-w, et al. Age-dependent effects in the transmission and control of COVID-19 epidemics. Nat Med. 2020. 10.1038/s41591-020-0962-9 . [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization. Report of the WHO-China Joint Mission on Coronavirus Disease 2019 (COVID-19). https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf. Accessed on March 10, 2020. 2020.
- 9.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N Engl J Med. 2020. 10.1056/NEJMoa2002032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395(10223):497–506. 10.1016/S0140-6736(20)30183-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Novel Coronavirus Pneumonia Emergency Response Epidemiology Team. [The epidemiological characteristics of an outbreak of 2019 novel coronavirus diseases (COVID-19) in China]. Zhonghua Liu Xing Bing Xue Za Zhi. 2020;41(2):145–51. 10.3760/cma.j.issn.0254-6450.2020.02.003 . [DOI] [PubMed] [Google Scholar]
- 12.Wu Z, McGoogan JM. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention. JAMA. 2020. 10.1001/jama.2020.2648 . [DOI] [PubMed] [Google Scholar]
- 13.Verity R., Okell L.C., Dorigatti I., Winskill P., Whittaker C., Imai N., et al. Estimates of the severity of coronavirus disease 2019: a model-based analysis. The Lancet. 2020;20:30243–7. 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delamater PL, Street EJ, Leslie TF, Yang YT, Jacobsen KH. Complexity of the Basic Reproduction Number (R0). Emerg Infect Dis. 2019;25(1):1–4. 10.3201/eid2501.171901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.United Nations Department of Economic and Social Affairs Population Dynamics. The 2019 Revision of World Population Prospects. https://population.un.org/wpp/. Accessed on March 1st, 2020. 2020.
- 16.Li R, Pei S, Chen B, Song Y, Zhang T, Yang W, et al. Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2). Science. 2020;368(6490):489–93. 10.1126/science.abb3221 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lauer SA, Grantz KH, Bi Q, Jones FK, Zheng Q, Meredith HR, et al. The Incubation Period of Coronavirus Disease 2019 (COVID-19) From Publicly Reported Confirmed Cases: Estimation and Application. Ann Intern Med. 2020;172(9):577–82. 10.7326/M20-0504 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, et al. SARS-CoV-2 Viral Load in Upper Respiratory Specimens of Infected Patients. N Engl J Med. 2020;382(12):1177–9. 10.1056/NEJMc2001737 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382(10):970–1. 10.1056/NEJMc2001468 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mckay MD, Beckman RJ, Conover WJ. A Comparison of Three Methods for Selecting Values of Input Variables in the Analysis of Output from a Computer Code. Technometrics. 1979;21(2):239–45. 10.2307/1268522 [DOI] [Google Scholar]
- 21.Sanchez MA, Blower SM. Uncertainty and sensitivity analysis of the basic reproductive rate—Tuberculosis as an example. Am J Epidemiol. 1997;145(12):1127–37. 10.1093/oxfordjournals.aje.a009076 [DOI] [PubMed] [Google Scholar]
- 22.Ayoub HH, Chemaitelly H, Abu-Raddad LJ. Characterizing the transitioning epidemiology of herpes simplex virus type 1 in the USA: model-based predictions. BMC Med. 2019;17(1):57 10.1186/s12916-019-1285-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awad SF, Sgaier SK, Tambatamba BC, Mohamoud YA, Lau FK, Reed JB, et al. Investigating Voluntary Medical Male Circumcision Program Efficiency Gains through Subpopulation Prioritization: Insights from Application to Zambia. PLoS One. 2015;10(12):e0145729 10.1371/journal.pone.0145729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chemaitelly H, Awad SF, Abu-Raddad LJ. The risk of HIV transmission within HIV-1 sero-discordant couples appears to vary across sub-Saharan Africa. Epidemics. 2014;6:1–9. 10.1016/j.epidem.2013.11.001 . [DOI] [PubMed] [Google Scholar]
- 25.Chemaitelly H, Awad SF, Shelton JD, Abu-Raddad LJ. Sources of HIV incidence among stable couples in sub-Saharan Africa. J Int AIDS Soc. 2014;17:18765 10.7448/IAS.17.1.18765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ayoub HH, Abu-Raddad LJ. Impact of treatment on hepatitis C virus transmission and incidence in Egypt: A case for treatment as prevention. J Viral Hepat. 2017;24(6):486–95. 10.1111/jvh.12671 . [DOI] [PubMed] [Google Scholar]
- 27.MATLAB®. The Language of Technical Computing. The MathWorks, Inc. 2019.
- 28.StataCorp. Statistical Software: Release 16.1. College Station, TX: Stata Corporation. 2019.
- 29.Van Kerkhove MD, Hirve S, Koukounari A, Mounts AW, group HNpsw. Estimating age-specific cumulative incidence for the 2009 influenza pandemic: a meta-analysis of A(H1N1)pdm09 serological studies from 19 countries. Influenza Other Respir Viruses. 2013;7(5):872–86. 10.1111/irv.12074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gilbert M, Pullano G, Pinotti F, Valdano E, Poletto C, Boelle PY, et al. Preparedness and vulnerability of African countries against importations of COVID-19: a modelling study. Lancet. 2020;395(10227):871–7. 10.1016/S0140-6736(20)30411-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Whittle P. The Outcome of a Stochastic Epidemic. Biometrika. 1955;42(1–2):116–22. [Google Scholar]
- 32.Allen LJS, Lahodny GE. Extinction thresholds in deterministic and stochastic epidemic models. J Biol Dynam. 2012;6(2):590–611. 10.1080/17513758.2012.665502 [DOI] [PubMed] [Google Scholar]
- 33.Kissler SM, Tedijanto C, Goldstein E, Grad YH, Lipsitch M. Projecting the transmission dynamics of SARS-CoV-2 through the postpandemic period. Science. 2020. 10.1126/science.abb5793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mossong J, Hens N, Jit M, Beutels P, Auranen K, Mikolajczyk R, et al. Social contacts and mixing patterns relevant to the spread of infectious diseases. Plos Med. 2008;5(3):381–91. 10.1371/journal.pmed.0050074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Prem K, Cook AR, Jit M. Projecting social contact matrices in 152 countries using contact surveys and demographic data. Plos Computational Biology. 2017;13(9). 10.1371/journal.pcbi.1005697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Viner RM, Mytton OT, Bonell C, Melendez-Torres GJ, Ward JL, Hudson L, et al. Susceptibility to and transmission of COVID-19 amongst children and adolescents compared with adults: a systematic review and meta-analysis. medRxiv. 2020:2020.05.20.20108126. 10.1101/2020.05.20.20108126 [DOI] [Google Scholar]
- 37.Goldstein E, Lipsitch M. Temporal rise in the proportion of younger adults and older adolescents among coronavirus disease (COVID-19) cases following the introduction of physical distancing measures, Germany, March to April 2020. Eurosurveillance. 2020;25(17):2–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Bloxham CJ, Hulme KD, Sinclair JE, Tong ZWM, Steele LE, et al. Children are unlikely to have been the primary source of household SARS-CoV-2 infections. medRxiv. 2020:2020.03.26.20044826. 10.1101/2020.03.26.20044826 [DOI] [Google Scholar]
- 39.He W, Yi GY, Zhu Y. Estimation of the basic reproduction number, average incubation time, asymptomatic infection rate, and case fatality rate for COVID-19: Meta-analysis and sensitivity analysis. J Med Virol. 2020. 10.1002/jmv.26041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MIDAS Online COVID-19 Portal. COVID-19 parameter estimates: basic reproduction number. https://github.com/midas-network/COVID-19/tree/master/parameter_estimates/2019_novel_coronavirus. Accessed on: MAy 19, 2020. 2020.
- 41.Riccardo F, Ajelli M, Andrianou X, Bella A, Del Manso M, Fabiani M, et al. Epidemiological characteristics of COVID-19 cases in Italy and estimates of the reproductive numbers one month into the epidemic. medRxiv. 2020:2020.04.08.20056861. 10.1101/2020.04.08.20056861 [DOI] [Google Scholar]
- 42.Adhikari R, Bolitho A, Caballero F, Cates ME, Dolezal J, Ekeh T, et al. Inference, prediction and optimization of non-pharmaceutical interventions using compartment models: the PyRoss library. arXiv: Populations and Evolution. 2020. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files. The MATLAB code is available at: https://github.com/HousseinAyoub/COVID-19-Code.git.