Abstract
Bone is a composite material consisting primarily of cells, extracellular matrices, accessory proteins and the complex calcium phosphate salt hydroxyapatite. Collectively, the extracellular network of proteins and accessory molecules that provide the organic component of bone tissue is referred to as the osteogenic extracellular matrix (OECM). OECM provides tensile strength and increases the durability of bone, but the OECM also serves as an attachment site and regulatory substrate for attached cells and a repository for growth factors and cytokines.
Increasingly, purified OECM generated by osteogenic cells in culture has attracted interest because it has the capacity to improve the growth and viability of attached cells, enhances the osteogenic program in vitro and in vivo, and shows great promise as a therapeutic tool for orthopedic tissue engineering.
This chapter will describe fundamental protocols for the selection and culture of osteogenic cells and conditions for their osteogenic differentiation, and the synthesis, purification and characterization of OECM. Some examples of immobilization to surfaces for the purpose of 2 and 3 dimensional culture will also be described.
Keywords: Osteogenesis, osteogenic, bone, extracellular matrix, tissue culture, purification
1. Introduction.
Bone tissue is a composite material consisting primarily of cells, extracellular matrices, accessory proteins and the complex calcium phosphate salt hydroxyapatite (HAP) (Moreira, Dempster, & Baron, 2000; Sommerfeldt & Rubin, 2001; Zimmermann & Ritchie, 2015). The hydroxyapatite gives bone its strength, resilience, and stiffness while providing an attachment substrate for cells and a repository for calcium, phosphate and other inorganic ions (Moreira et al., 2000; Murshed, 2018; Zimmermann & Ritchie, 2015). Embedded within, and also coating the surface of HAP, are heterotypic fibrils consisting primarily of collagens, non-collagenous matrix proteins, polymeric saccharides, and immobilized soluble factors (Murshed, 2018; Sommerfeldt & Rubin, 2001; Weiner & Traub, 1992). Collectively, the extracellular network of proteins and accessory molecules that provide the organic component of bone tissue is referred to as the osteogenic extracellular matrix (ECM). The heterotypic fibrils of the osteogenic ECM (OECM) provide additional tensile strength and increase durability of the HAP, but the OECM also serves as an attachment site and regulatory substrate for attached cells and a repository for growth factors and cytokines (Marastoni, Ligresti, Lorenzon, Colombatti, & Mongiat, 2008; Martino, Briquez, Maruyama, & Hubbell, 2015; Sedlmeier & Sleeman, 2017).
Osteoblasts, the cells that synthesize bone tissue, initially deposit a network of collagen-rich ECM on the surface of the bone, and the unique composition of the OECM catalyzes the growth of HAP crystals resulting in the formation of mature, hardened bone (Murshed, 2018). The process of bone formation is termed ossification, and the ECM that serves as the precursor of bone tissue is known as osteoid. For decades, the composition, assembly, and function of OECM has been the subject of intense research for the purposes of our understanding of bone development and disease, but also as a foundation for the generation of materials with osteoinductive properties.
A manufacturable OECM with the regenerative properties of osteoid would be of significant value to the orthopedic and tissue engineering industries and as a result of this research, a broad array of synthetic bone mimics, osteoinductive biological factors, and promising cell-based technologies have arisen (Fernandez de Grado et al., 2018; Lobb, DeGeorge, & Chhabra, 2019). Despite efforts, the gold standard for bone tissue engineering remains as it was in the 1970s, the autologous bone graft (autograft), where bone is explanted from an autologous donor site in the patient and implanted at the site of the defect. This procedure is often very successful with the donor bone tissue readily engrafting into the defect site, becoming contiguous with the surrounding bone within several weeks (Jakoi, Iorio, & Cahill, 2015). Unfortunately, there are obvious limitations to this technique, primarily related to availability of the donor tissue and how this relates to the size of the bone deficit. An additional drawback of this approach is donor site morbidity and associated pain, which can become more problematic than the injury itself (Costa Mendes, Sauvigne, & Guiol, 2016). To address the problem of volumetric availability, bone marrow cells or morselized autograft has been combined with chemically synthesized bone mimics such as synthetic HAP, tricalcium phosphate or even forms of glass (Fernandez de Grado et al., 2018). Unfortunately these “graft extenders” as they are so-called can reduce the overall effectiveness of the bone graft because they often exhibit poor biocompatibility with osteoblasts and other cells reducing capacity to support osteogenesis. In some cases, synthetic bone mimics are also highly toxic to cells (Murphy, Suzuki, Sand, Chaput, & Gregory, 2013). An alternative approach involves the use of demineralized cadaveric bone matrix (DBM). On the whole, DBM exhibits superior biocompatibility to synthetic bone mimics, but harsh processing to remove cells and antigens can reduce the osteogenic efficacy of the matrix, and in spite of harsh treatments, there is always risk of pathogen transfer (H. Zhang et al., 2019). To stimulate bone formation by the host, recombinant bone morphogens such as bone morphogenic protein 2 (BMP2) can be utilized, but these factors, often administered in supraphysiological doses, can leak from the site causing dangerous levels of ectopic bone formation and inflammation (Tannoury & An, 2014). Limitations of current bone repair technologies, and the continued use of autograft indicate that a greater understanding of bone biology is needed if we are to improve our ability to repair it.
Our understanding of bone development and repair originates from the study of small animal models such as rodents, chickens, amphibians, fish and rabbits, but these approaches, while informative, tend to lack direct experimental accessibility and cannot be readily simplified. Optimization of the culture of osteoprogenitors (OPs) and mesenchymal stem cells (MSCs), the presumptive progenitors of osteoblasts, permit the execution of simple in vitro experiments (C. A. Gregory, Prockop, D. J., 2007; McNeill et al., 2018), but these tend to be performed on plastic monolayers under conditions that fail to recapitulate the complex composition and topology of the osteoid. Given the complexity of interactions between the OECM and the osteoblast during bone formation, a faithful in vitro recapitulation of the osteogenic niche involves, at minimum, culture of appropriate isolates of osteoprogenitors on purified extracts of functional OECM (McNeill et al., 2018). This type of experiment permits simultaneous study (and manipulation) of osteoblast and matrix, facilitating a better understanding of the osteogenic process, especially when human cells and tissues are employed. Indeed, recent work has demonstrated that ECM generated from monolayer-cultured OPs and MSCs can enhance proliferation, viability and osteogenic differentiation of attached cells (Mao et al., 2019; McNeill et al., 2018; Zeitouni et al., 2012). Some studies have also demonstrated that ECM composition can be conditioned by controllable factors such as the age or source of the donor cell or by media composition to alter bioactive properties (Mao et al., 2019; Zeitouni et al., 2012).
This chapter will describe fundamental protocols for the selection and culture of osteogenic cells and conditions for their osteogenic differentiation, and the synthesis, purification and characterization of OECM. Some examples of immobilization to surfaces for the purpose of 2D and 3D culture will also be described.
2. Osteogenic Tissue Culture and Relevant Assays.
OECM is readily purified from monolayers of osteogenic cells attached to tissue culture plastic (W. Zhang et al., 2016). The detailed protocols described herein refer primarily to marrow derived human MSCs, but a broad range of tissue-derived or immortalized cell lines can be employed to generate OECM-containing monolayers and a non-exhaustive selection of cell sources and conditions are presented in Table 1.
Table 1:
A selection of plastic adherent osteogenic cell lines from various sources.
| Cell type and species | Osteogenic conditions | Ref. |
|---|---|---|
| Human bone marrow derived MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (C. A. Gregory, Gunn, Peister, & Prockop, 2004; Wei et al., 2017) |
| Human bone marrow derived MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, GW9662 | (Krause et al., 2010; Zeitouni et al., 2012) |
| Chicken bone marrow derived MSCs | DMEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Bai et al., 2013) |
| Rabbit bone marrow derived MSCs | DMEM-LG, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Solchaga, Johnstone, Yoo, Goldberg, & Caplan, 1999) |
| Canine various tissues MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone, BMP2 | (Bearden et al., 2017) |
| Rat bone marrow derived MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Javazon, Colter, Schwarz, & Prockop, 2001) |
| Mouse bone marrow derived MSCs | IMDM, FBS/HS, β-glycerophosphate, ascorbate, dexamethasone, thyroxine | (Peister et al., 2004) |
| Human adipose derived MSCs | DMEM, FBS, β-glycerophosphate, ascorbate, dexamethasone or DHVD3 | (Zuk et al., 2002) |
| Murine adipose derived MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Xie et al., 2018) |
| Human iPSC-derived MSCs | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Zhao et al., 2015) |
| Human MG63 osteosarcoma (OS) cells | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (C. A. Gregory et al., 2004) |
| Human SAOS OS cells | McCoy’s 5A, β-glycerophosphate, ascorbate | (Jiang et al., 2013) |
| Human MNNG/HOS OS cells | StemPro (ThermoFisher, Waltham, MA) Osteogenesis Kit | (Martins-Neves et al., 2012) |
| Murine MC3T3 OP line | DMEM-LG, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Liu et al., 2014) |
| Murine MOSJ osteosarcoma cells | α-MEM, FBS, β-glycerophosphate, ascorbate, dexamethasone | (Krause, Ryan, Clough, & Gregory, 2014) |
α-MEM: alpha minimal essential medium, DMEM: Dulbecco’s minimal essential medium, LG: low glucose, FBS: fetal bovine serum, BMP2: bone morphogenic protein 2, IMDM: Iscove’s modified Dulbecco media. DHVD3: dihydroxy vitamin D3, iPSC: induced pluripotent cells
To prepare monolayers for osteogenic differentiation, the cells are plated at low density and allowed to establish a 70-80% confluent monolayer under standard expansion conditions. This approach allows cells to gradually adapt to nutritional and chemical changes associated with a decreasing availability of resources before the additional challenge of media containing osteogenic supplementation. Osteogenic differentiation typically takes place in the presence of rich media containing a source of organic phosphate, ascorbic acid, serum, and in some cases, dexamethasone. In specific cases, additional osteoinductive agents are also added to the media. The differentiation process can take between 5-21 days, with media changes usually every 48 hr. The yield of ECM usually correlates with the degree of osteogenic differentiation in the culture, and a variety of assays can be performed to measure progress. Measurements of activity of the osteogenic biomarker alkaline phosphatase (ALP) (Buchet, Millan, & Magne, 2013), secretion of the ligand osteoprotegerin (OPG) (Lacey et al., 1998), and measurement of monolayer mineralization (C. A. Gregory et al., 2004) are readily performed in parallel with the use of quantitative real-time RT-PCR (qRT-PCR) to measure transcription of stage-specific osteogenic biomarkers.
2.1. Human MSC expansion and osteogenic differentiation.
Materials.
5 mL - 50 mL assorted serological pipettes
10 cm and 15 cm diameter plastic tissue culture dish (about 50 cm2 and 150 cm2 surface area respectively)
6-well or 12-well tissue culture plates (about 4 and 9 cm2 surface area respectively)
15 mL and 50 mL sterile polypropylene centrifuge tubes
37°C water bath.
70 % (v/v) reagent ethanol
500 mL - 1000 mL low protein binding vacuum filter units with 0.22 μm filtration capacity
Alpha minimal essential medium (αMEM) without ribonucleosides or deoxy ribonucleosides.
Antibiotic 10,000 units mL−1 penicillin G sodium, 10,000 μg mL−1 streptomycin sulphate (100x).
Bench-top swinging bucket centrifuge with 1000 G capacity.
Class IIA/B3 biological safety cabinet (BSC) set to 37°C and 5% (v/v) CO2.
Complete Culture Media (CCM): α-MEM containing 20% (v/v) FBS, containing 1x antibiotic, 2 mM additional glutamine.
Dexamethasone dissolved at 1 μM in dimethyl sulphoxide.
Fetal bovine serum (FBS) hybridoma grade and not heat-inactivated.
Peroxisome proliferator-activating receptor-γ (PPARγ) inhibitor GW9662 dissolved at 10 mM in dimethyl sulphoxide.
Inverted phase contrast microscope or equivalent fitted with 4x, 10x and 20x objectives
L-glutamine solution (200 mM)
Neubauer hemacytometer
Porcine trypsin 0.05% (w/v) and 0.02% (w/v) EDTA solution (trypsin/EDTA)
Sterile phosphate buffered saline (PBS) without magnesium and calcium
Trypan blue solution in 0.85% saline for cell enumeration
Procedure.
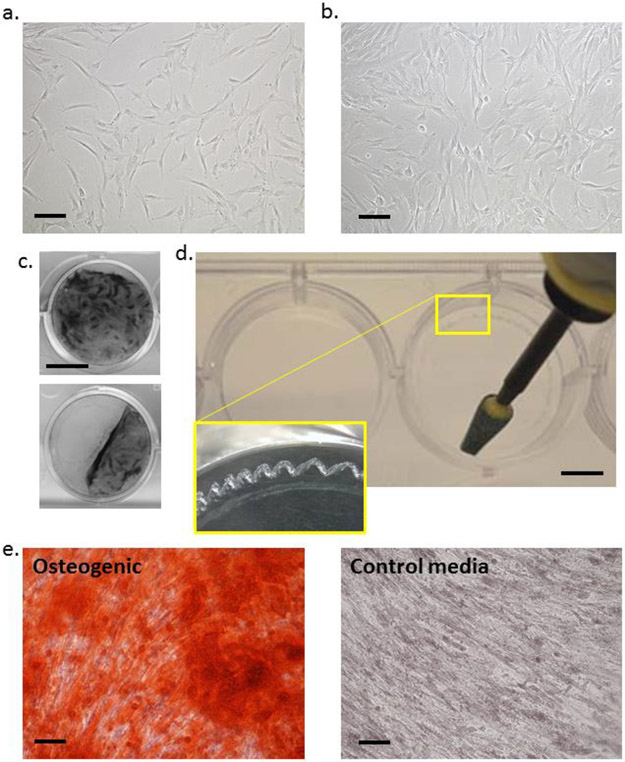
This protocol assumes that hMSCs have been plated onto 15 cm plates and expanded in accordance with standard protocols and the hMSCs are ready to seed into osteogenic cultures. Inspect the monolayers with the phase contrast microscope, if adherent cells are fibroblastoid and are at approximately 60% confluency (Fig 1a), proceed. If the monolayer is sparse, wash, replenish medium and culture for a further 1-2 days.
In the BSC, wash the monolayer with 20 mL of pre-warmed (37°C) PBS, then recover the cells by addition of 7 mL of pre-warmed (37°C) trypsin/EDTA. Place the plate in the incubator for 2 min, then inspect for cell dissociation. Repeat every 2 min until monolayer is 95% dissociated then add 7 mL of CCM. Transfer suspended cells to a 15 mL conical tube and centrifuge for 10 min at 500 g.
Carefully remove the supernatant from the cell pellet and suspend 1 mL warm PBS. Add 10 μL of the cell solution to 10 μL of trypan blue and count with a hemacytometer. A suitable cell suspension should be between 200,000 to 500,000 cells per mL with a viability of above 80%.
Prepare plates necessary for OECM purification and osteogenic assays. OECM recovery is performed on multiple 15 cm dishes (the yield is cell-specific, but generally 10-20 mm3 per plate(Zeitouni et al., 2012)). A 10 cm dish is typically sufficient for mRNA generation, and 12-well plates are suitable for assays of ALP activity, OPG secretion and mineralization. In each case, plate hMSCs at a density of 500 cells per cm2 in CCM. For 15 cm dishes, 10 cm dishes and 12-well plates use 20 mL, 10 mL and 1 mL per well media respectively.
Culture cells in CCM changes every 2 days at 37°C in a humidified incubator in the presence of 5% (v/v) CO2 until reaching 80% confluency (≈20,000 cells per cm2, Fig 1b).
Change media to OBM, with optional supplementation with dexamethasone (1-10 nM) to increase the rate of mineralization, or 1-10 μM GW9662 to increase the yield and calcium content of OECM. Culture in the incubator for 7-21 days, dependent on the maturity of the OECM required. In some cases, the confluent monolayer can detach from the plastic (Fig 1c). The probability of this happening can be reduced by gentle handling of the monolayers and pipetting of media. If detachment still occurs, score the perimeter of the plates with a sterile abrasive burr to provide a more durable attachment site for the cells and associate matrix (Fig 1d).
Figure 1: Osteogenic monolayer culture of MSCs for generation of OECM:
Panel a: hMSCs at the appropriate confluency for reseeding into osteogenic cultures. Panel b: hMSCs at the appropriate confluency for initiation of osteogenic assays. Panel c: Crystal violet-stained monolayers of osteogenic hMSCs detaching form the tissue culture plastic. Panel d: Use of rotary burr to generate enhanced attachment sites (inset) at the edges of wells prior to osteogenic culture. Panel e: hMSCs stained with ARS cultured in osteogenic differentiation media (left) or control CCM media (right). For panel a,b,e: bar = 200 mm. For panel c: bar = 10 mm. For panel d: bar = 5 mm.
2.2. Assays of OPG secretion by enzyme linked immunosorbent assay (ELISA).
OPG, also referred to as tumor necrosis factor receptor superfamily, member 11b or osteoclast inhibitory factor is a secreted glycoprotein with affinity for the osteoclast activating factor, receptor activator of NF kappaB ligand (RANKL). OPG acts as a soluble decoy receptor for RANKL, thereby serving as its competitive inhibitor. In MSCs and OPs, OPG is an early indicator of osteogenic differentiation and serves as a useful and minimally invasive osteogenic biomarker (Glass et al., 2005).
Materials.
3, 3’, 5, 5’ tetramethyl benzidine substrate (TMB)
Analytical grade bovine serum albumin (BSA).
Block buffer:
PBS containing 0.1 % (v/v) Tween 20 and 4 % (w/v) BSA.
Colorless, flat bottomed ELISA microtiter plates.
Media dilution buffer:
PBS containing 0.1 % (v/v) Tween 20 and 1 % (w/v) BSA.
Microplate reader with 450 nm absorbance capability.
OPG ELISA DuoSet (R&D Systems, Minneapolis, MN)
Stop buffer:
Sulphuric acid (2M)
Tween 20 detergent
Wash buffer:
PBS containing 0.1 % (v/v) Tween 20.
Procedure.
After 2-8 days of osteogenic stimulus in 12-well format, recover 100 μL of 2-day conditioned media.
Prepare antibody solutions, biotin streptavidin, and OPG standards as directed by the DuoSet manufacturer.
Coat microtiter plates for 15 hr at 4°C with 100 μL of capture antibody (DuoSet) per well diluted in PBS at the recommended concentration.
Wash the wells twice with 100 μL wash buffer and block for 2 hr with 150 μL block buffer.
While blocking the plate, prepare a standard solution of OPG (DuoSet) at 4000 pg mL−1 in media dilution buffer. In the microtiter plate, prepare a triplicate 7-point standard curve (4000 pg mL−1 to 62.5 pg mL−1) by doubling dilution of 100 μL aliquots.
Dilute media samples 5-20 fold in media dilution buffer and plate 100 μL in a fresh well, in triplicate.
After 15 hr of incubation at 4°C, remove samples and standards. Wash wells 3 times with 100 μL wash buffer.
Add to each well 100 μL of the appropriate concentration of biotinylated detection antibody (DuoSet) and incubate at room temperature for 2 hr. Wash wells 3 times with 100 μL wash buffer.
Add to each well 100 μL diluted peroxidase conjugated streptavidin (DuoSet) and incubate at room temperature for 30 min. Wash wells 3 times with 100 μL wash buffer.
Add to each well 100 μL of TMB and incubate at room temperature, with manual agitation, for 2-10 min until the blue color develops.
Add to each well 50 μL stop solution to develop the yellow color.
Read at 450 nm on a microtiter plate reader. Plot standards and calculate unknowns.
2.3. Assays of ALP activity.
ALP is expressed on the membranes osteoblasts to generate phosphate for incorporation into HAP (Anderson, 2003) and to degrade inhibitory pyrophosphatases(Balcerzak et al., 2003). The activity of ALP can be determined by measuring the rate of p-nitrophenyl phosphate (PNPP) conversion to nitrophenolate which results in a color change from colorless to yellow, with absorbance at 405 nm(Bessey, Lowry, & Brock, 1946). ALP can be measured by direct addition of PNPP solution to the monolayer and results can then be normalized by cell enumeration. In this protocol, cells are enumerated with the DNA-chelating fluorescent CyQuant dye incorporation assay.
Materials.
Automated microplate reader with 12-well format compatibility, preferably with kinetic capability.
ALP reaction buffer (ARB):
Tris HCl pH 9.0 (100 mM) containing 1 mM MgCl2 and 100 mM NaCl.
PBS without calcium or magnesium.
Stabilized PNPP solution or a freshly prepared 1 mg mL−1 solution of PNPP in 1 M diethanolamine.
For cell enumeration:
Black, opaque, flat bottomed microtiter plates
CyQuant cell GR DNA dye (Invitrogen, Carlsbad, CA) or equivalent.
Extraction and lysis buffer (ELB):
PBS containing 0.1 % (v/v) Triton X100, 100 mM NaCl, 1 mM MgCl2, 5 μg mL−1 RNAse A and 10 U mL−1 EcoRI and HindIII restriction endonucleases.
Frozen (−80 °C) pellets of human cells (120,000 per pellet) for reference material.
Microplate reader with 480/520 nm (fluorescein equivalent) fluorescence capability.
Procedure.
Perform osteogenic assays in 12 well plates.
Wash monolayers twice with 1 mL PBS, then once with 1 mL room temperature ALP reaction buffer.
Set plate reader path length to 0.25 cm and activate orbital averaging if possible. For automated kinetic measurements, set plate reader to detect absorbance at 405 nm for 15 minutes, with measurements every 30 seconds.
Quickly add 1 mL room temperature ARB to the wells of the plate. Then add an equal volume of PNPP equilibrated to 4 °C, mix by gentle orbital agitation, and immediately initiate absorbance readings at 405 nm. For manual measurements, repeat readings every 30 s until 15 min has passed.
Plot the rate of accumulation of p-nitrophenolate (proportional to the absorbance at 405 nm) and derive the rate by calculating the slope at the linear phase of the reaction. If you choose to calculate ALP activity based on known standards, be aware that in some cases, the specific activity of commercially acquired ALP reference material may not be comparable to the specific activity of the membrane bound ALP.
After the reaction, wash the monolayers in PBS and store at −20 °C. The monolayers can be stored indefinitely prior to cell number measurements.
To the frozen monolayers, add the 1 mL of ELB and incubate for 4 −15 hr in a humidified incubator at 37 °C with orbital agitation at 60 rpm. Process a pellet of reference cells with the samples by suspending an aliquot in 200 μL of extraction buffer.
Transfer 800 μL of each digestion to a microcentrifuge tube and centrifuge at 15,000 g for 15 min then transfer supernatant to a fresh tube. This can be stored at −20°C for 1 month.
Prepare 1.5 x CyQuant GR working solution in ELB and add 200 μL of the extract to 400 μL of the CyQuant GR solution in a fresh microcentrifuge tube and incubate at room temperature in the dark for 5 min.
Prepare standard curve by transferring 200 μL of the standard cell extract into wells A1-3 of an opaque black microtiter plate. Using ELB, perform doubling dilutions from wells A1-3 to wells G1-3 to generate a 7 point standard curve from 20,000 to 625 cell equivalents per well. Add 200 μL ELB to wells H1-3 as blanks.
Load 100 μL of sample in remaining wells in triplicate.
Read fluorescence at 480/520 nm. Plot standards and calculate unknowns.
If necessary, dilute samples in additional ELB to attain values that are within the linear range of the assay.
2.4. Mineralization assays.
Biomineralization of cultured cell monolayers can occur in response to osteogenic stimuli. The calcium phosphate-rich monolayers can be readily visualized by the red calcium binding dye, alizarin red S (ARS). ARS can be easily extracted and quantified spectrophotometrically as a surrogate assay for calcium deposition and the following is a simplified version of the original protocol(C. A. Gregory et al., 2004).
Materials.
Alizarin red S solution (ARS):
2 % (w/v) in water carefully adjusted to pH 4.2 with 0.5 M ammonium hydroxide.
ARS extraction solution:
10% (v/v) acetic acid
Colorless flat-bottomed 96 well microtiter plates.
Microtiter plate reader
PBS without magnesium and calcium.
Phase contrast inverted microscope, preferably with camera attachment.
Phosphate buffered formalin.
Procedure.
Aspirate the media from monolayers in 12 well plates and wash wells with 2 mL of PBS. Fix with formalin at room temperature for 15 min. Wash monolayers twice with 2 m dH2O.
Add 1mL of ARS per well and incubate the plates at room temperature for 20 min with orbital shaking (45 rpm).
Aspirate off the unincorporated ARS and carefully wash wells four times with 2 mL dH2O. For each wash, shake for 3 min. While washing, care should be taken not to scrape or dislodge the calcium phosphate matrix.
Visualize and photograph the degree of differentiation of stained monolayers by microscopy using a phase-contrast inverted microscope. Mineralization is indicated by a deep red stain (Fig 1C). Plates can be stored at −20 °C at this point prior to dye extraction. Freeze/thawing will adversely affect the quality of micrographs.
For quantification of staining, add 500 mL ARS extraction solution pre-warmed to 50°C and incubate for 30 min at room temperature. The ARS should solubilize into the extraction buffer, resulting in a color change from red to yellow.
Read 150 μL aliquots of the ARS extraction solution in a 96 well plate reader at 405 nm in triplicates using transparent-bottomed plates. The results can be plotted against a “standard curve” of known ARS concentrations, but note that complete extraction of the dye cannot be guaranteed. The results should therefore be regarded as comparative or semi-quantitative.
2.5. Quantitative real-time RT-PCR (qRT-PCR) assays of biomarker transcription.
The progression of an osteogenic culture can be followed by measurement of the transcription of stage-specific osteogenic biomarkers by qRT-PCR assays on mRNA extracted from the monolayers. While RNA extraction, reverse transcription and quantitative PCR are established techniques and a range of products and kits are available for this purpose, some specific considerations are provided below.
Standard miniature spin columns are generally adequate for mRNA purification, but in samples associated with high levels of ECM, larger extraction columns designed for tissue specimens are often more effective, providing flexibility with respect to the addition of lysis buffer and the reduced danger of blocking the column with particulates. Typically, confluent monolayers generated on 10 cm diameter plates yield sufficient mRNA (100 - 500 ng per culture) for several assays, but it is important to note that the yields of mRNA can be reduced as the monolayers deposit more ECM. Monolayers associated with a high level of ECM can be recovered from the plate by a combination of trypsinization and scraping, and lysed by addition of 2-3 fold excess lysis buffer and incubation with orbital shaking (60 rpm) at room temperature for 2-10 min. It is advisable to perform a centrifugation step (19,000 g for 5 min) to remove excess matrix from the supernatant before applying to purification columns. Copy DNA can be generated using standard commercial products, but it is advisable to perform the priming with a mixture of oligo(dT) and random hexamers to maximize yields. The qRT-PCR reactions can be performed using an intercalating dye such as SYBR-Green, or by primer-probe technologies such as the TaqMan system. A list of commonly employed osteogenic biomarkers are provided in Table 2.
Table 2:
A selection of commonly employed osteogenic biomarkers for qRTPCR
| Transcript | Stagea | SYBR primersb | Ref. | TaqMan assay |
|---|---|---|---|---|
| GAPDH | Normalization | GAGCCACATCGCTCAGACA CCTCTTCAAGGGGTCTAC |
(Carraro, Albertin, Forneris, & Nussdorfer, 2005) | Hs01548420_m1 |
| Alkaline phosphatase | Day 8-14 | GACCCTTGACCCCCACAAT GCTCGTACTGCATGTCCCCT |
(Zhao et al., 2015) | Hs01029144_m1 |
| BMP2 | Day 2-5 | CCCAGCGTGAAAAGAGAG GAGACCGCAGTCCGTCTA |
(McNeill et al., 2018) | Hs00272516_m1 |
| Collagen I | Day 2-21 | GAACGCGTGTCACCTTGT GTCAACGCGTGTCATCCCTTGT |
(Zhao et al., 2015) | Hs00164004_m1 |
| Collagen X | Day 8-21 | AATGCCTGTGTCTGCTTTTAC ACAAGTAAAGATTCCAGTCCT |
(Alaseem et al., 2015) | Hs00166657_m1 |
| Osteocalcin | Day 14-21 | TGAGAGCCCTCACACTCC CGCCTGGGTCTCTTCACTAC |
(Salem et al., 2014) | Hs01587814_g1 |
| Osterix | Day 2-5 | GTGGGCAGCTAGAAGGGAGT AATTAGGGCAGTCGCAGGA |
(Schaap-Oziemlak et al., 2010) | Hs00541729_m1 |
| Runx2 | Day 2-5 | GCAAGGTTCAACGATCTGAGA TCCCCGAGGTCCATCTACTG |
(Schaap-Oziemlak et al., 2010) | Hs01047973_m1 |
approximate time-window when maximally activated, but dependent on cells assayed.
primers provided for human cells.
3. Purification and Characterization of OECM.
The following section describes a protocol for the decellularization and purification of OECM generated by monolayers of OPs or osteogenic hMSCs. Depending on the cell type employed, and the differentiation conditions, it is possible to generate approximately 10-20 mm3 of purified OECM per plate, which is equivalent to roughly 1.5 - 3 mg protein. Protocols for quantification of protein and calcium, visualization by scanning electron microscopy and guidelines for proteomic analyses are also provided.
3.1. Purification of matrix.
Materials.
1 mL or 5 mL polypropylene pipettes with filters.
5 mL – 25 mL sterile serological pipettes.
50 mL and 15 mL standard conical polypropylene “Falcon” tubes
Acetone, analytical grade
Chemical fume hood
Chloroform, analytical grade
Matrix extraction buffer (MEB):
PBS supplemented with 0.1% (v/v) Triton X-100, 1 mM MgCl2, and 1 U.mL−1 DNAse I, filtered through a 0.2 μm pore-diameter membrane.
Sterile PBS
Sterile plastic cell scrapers
Sterile plastic spatulas
Trypsin, tissue culture grade.
Procedure.
The volumes below refer to ECM purified from a single 150cm2 monolayer culture generated in a 15 cm diameter plate. ECM from up to four 150cm2 cultures can be combined and purified simultaneously in the 50 mL Falcon tube.
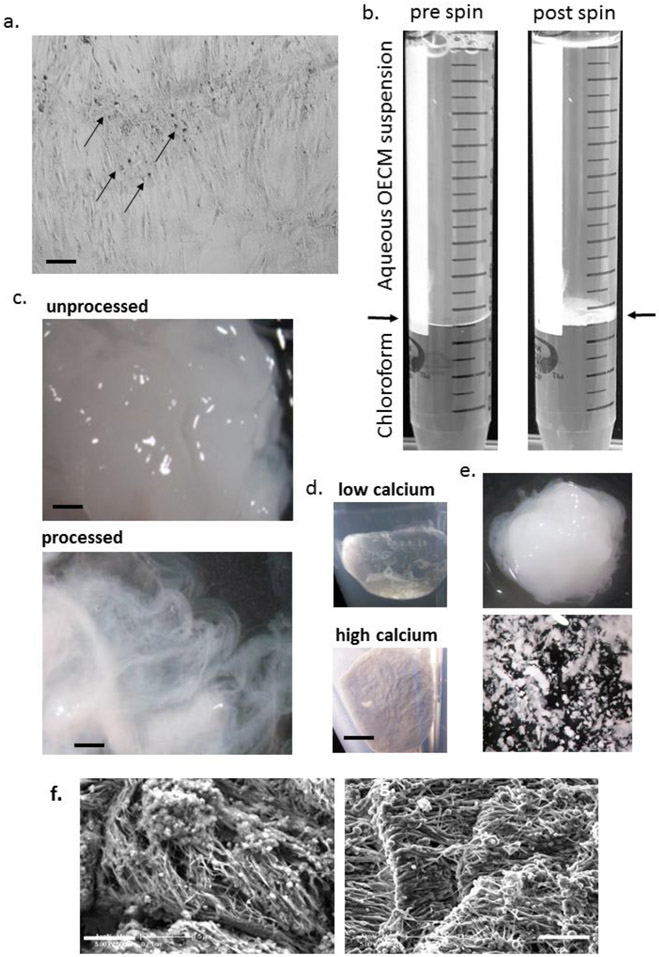
Wash the osteogenic monolayers (Fig 2a) twice in PBS and freeze at - 80°C for 15 hr to disrupt the cell membranes.
Thaw the monolayers by adding 5 mL room temperature PBS followed by incubation at room temperature for 20 min. Scrape the thawed monolayers into a 50 mL Falcon tube. Wash the plate with 5 mL PBS and transfer to the Falcon tube to ensure all material is collected.
Pellet the OECM/cell suspension by centrifugation at 1,800 g for 10 min.
Suspend the OECM/cell suspension in 3 mL of freshly made MEB equilibrated to 37°C and incubate for 2 hours at 37°C with rocking at 60 rpm. After 2 hours add a 10x solution of trypsin (suspended in MEB) to the cell/ECM mixture to attain a final concentration of 0.001 - 0.1% (v/v) and allow the lysis to proceed for a further 15 hours. The aim of this step is to degrade contaminating non-OECM associated proteins, and should be optimized to maximize yields of OECM while minimizing levels of abundant marker proteins such as glyceraldehyde phosphate dehydrogenase and β-actin. These proteins can be assayed by immunoblotting(Zeitouni et al., 2012).
Pellet the OECM by centrifugation at 1,800 g for 10 minutes, wash the OECM twice in 10 mL ice-cold dH2O, suspend pellet in 8 mL dH2O, and transfer suspension to a 15 mL Falcon tube.
In a fume hood, add 5 mL chloroform to the suspension, close the tube tightly and shake (Fig 2b). Centrifuge the suspension at 1,800 g for 10 minutes at room temperature. The OECM should accumulate as a tight white disc at the interface of the chloroform and aqueous phase (Fig 2b). First, carefully aspirate the upper aqueous phase with a serological pipette, then aspirate the lower chloroform phase with a 1 mL or 5 mL polypropylene pipette. Add another 8 mL dH2O, then 5 mL chloroform and repeat step 6. This step removes hydrophobic contaminants such as lipids.
Remove the aqueous and chloroform layers, then wash the OECM twice more with 10 mL dH2O, with centrifugation steps in between. After washes, aspirate as much dH2O as possible without disrupting the OECM pellet.
In a fume hood, rinse the OECM with 5 mL of acetone and allow to air dry for at least 4 hours. This step facilitates drying and destroys microbial contamination. The ECM should consist of a white-yellow putty-like material with a fibrous appearance when inspected by microscope (Fig 2c,d). Malleability is related to calcium and water content, and high calcium content combined with desiccation can make the material brittle (Fig 2d,e).
The OECM yield can be determined by weighing with an analytical balance after transferal to a sterile tube in a BSC (be sure to tare the balance with the empty tube prior to transferal). The volumetric yield can also be measured by micro-computed tomography (μCT) scanning with a very low energy x-ray beam (Zeitouni et al., 2012).
Figure 2: Purification and morphology of OECM:
Panel a: phase-contrast micrograph of osteogenic culture with dense nodules of OECM (arrowed). Panel b: processing of OECM with chloroform before and after centrifugation. The disk of OECM is present at the interface (arrowed) after spinning. Panel c: Appearance of cell/OECM mixture before processing (above) and OECM after processing (below) dispersed in water illustrating fibrous appearance (adapted with permission from ref. 18). Panel d: Appearance of acetone-dried pellets generated under conditions that deposit low calcium (above) or high calcium (below). Panel e: Low calcium OECM (above) generates a putty-like matrix upon partial drying, OECM with high calcium content fragment upon manipulation (below). Panel f: Scanning electron microscopy of OECM before (left) and after (right) processing. For panel a: bar = 200 μm. For panel c: bar = 500 μm. For panel d, e: bar = 2 mm. For panel f: bar = 5μm.
3.2. Assays of protein and calcium.
Measurements of calcium content in OECM can provide valuable insights into downstream biological activity and handling properties. For example, calcium levels above 100 μM per mg protein predict excellent osteogenic activity in vivo(Zeitouni et al., 2012), but calcium levels above this threshold can result in a brittle matrix with diminished handling properties (Fig 2d,e). Calcium can be readily measured chemically using standard laboratory equipment, but OECM is inherently insoluble, and mineral is challenging to extract and dissolve. A protocol for the accurate determination of protein and calcium levels in OECM is described below.
Materials.
Quantification of protein content.
ECM protein extraction buffer (EPEB):
2 M urea in 100 mM Tris-HCl pH 8.0
Bicinchoninic acid (BCA) protein assay kit (Pierce Chemical, or equivalent)
Colorless flat-bottomed 96-well microtiter plate
Visible range microtiter plate reader
Quantification of calcium content.
50 mL standard conical polypropylene “Falcon” tubes
Analytical grade hydrochloric acid 6 M
Analytical grade Trizma base 2.5 M
Analytical grade calcium chloride
Analytical grade arsenazo III colorimetric calcium chelator.
Analytical grade dH2O
Microprobe-equipped pH meter for small volumes (5 mL)
Colorless flat-bottomed 96-well plate
Visible range spectrophotometer with 96-well capability
Procedures.
Quantification of protein content.
Suspend the OECM pellet in 5 mL EPEB at 60°C until the pellet is completely dissolved.
Determine OECM protein content with a BCA protein assay kit following the manufacturer’s microplate procedure.
Quantification of calcium content.
Suspend the OECM pellet in 10 mL of 6M HCl in a 50 mL screw top Falcon tube and heat at 80°C until the pellet has completely dissolved. For this procedure, use appropriate personal protective equipment and perform in a chemical fume hood. Never uncap the sample until properly cooled to room temperature.
Cool the solution to room temperature and carefully neutralize to pH 5.0 with the addition of 2.5 M Trizma base. Perform a mock titration with 10 mL 6M HCl to determine an approximate volume of 2.5 M Trizma base to add. Retain the resultant solution for dilution of the standards.
Prepare a 100 μM stock of arsenazo III in dH20 and 10-100 μM CaCl2 standards in the neutralized Tris-HCl solution.
Transfer 10 μL of sample or standard followed by 90 μL of Arsenazo III solution onto a flat-bottomed 96-well colorless plate.
Incubate the plate for 5 minutes at room temperature. A transition from pink-purple-blue should be observed with increasing concentration of calcium.
Measure the absorbance at 595 nm and plot a standard curve to determine the concentration of calcium in the samples.
A calcium level of above 150 μM per mg protein indicates that the resultant OECM will probably adopt brittle handling properties whereas those OECM specimens with lower calcium levels will be more malleable (Fig 2e).
3.3. Morphology of OECM.
Prior to trypsinization, but after lysis with MEB, OECM adopts a white-yellow gelatinous appearance (Fig 2c). When non-trypsinized OECM is subjected to standard SEM procedures(Zeitouni et al., 2012), heterotypic fibrils are visible but these are frequently associated with particulate structures that often generate a calcium signal when probed by electron diffraction spectrometry. After trypsinization, the OECM adopts a fibrous appearance when suspended in water and visualized by low power light microscopy (Fig 2c, below), and a distinct fibrous structure in the absence of particulates is evident when analyzed by SEM (Fig 2f, right). Fully processed OECM should have a fibrous, cotton-like appearance when suspended in aqueous buffers, with opacity ranging from a translucent gel with OECM of low calcium content (Fig 2b, above), to an opaque, chalky substance when calcium content is relatively high (Fig 2b, below).
3.4. Composition of OECM.
The composition of OECM is most comprehensively assayed by tryptic fingerprinting followed by liquid chromatography and mass spectrometry (LC/MS) in accordance with standard proteomic approaches. Due to the insoluble and complex nature of OECM, we have found in-gel tryptic approaches to be most successful because the matrix of the gel retains insoluble particulates and partially digested proteins after digestion. The sample can be electrophoresed completely and individual clusters of bands with well-defined molecular masses can be analyzed. To reduce cost and workload, it is also possible to process a limited number of gel slabs generated by partially electrophoresed samples. OECM is a complex material consisting of many proteins, but analysis of multiple forms of OECM in independent laboratories(Mao et al., 2019; Ragelle et al., 2017) and our own, has revealed the robust presence of collagens I, III, V, IV, VI, IX, XI, XII and ECM factors such as fibronectin, laminin, fibrillin and biglycan. Traces of membrane proteins such as integrins (β1) and ECM-associated proteins such as periostin and matrix metalloproteinases have also been reproducibly detected.
Materials.
At least 1 mg of OECM
Analalytical grade ammonium bicarbonate
Analytical grade acetonitrile
Analytical grade formic acid
Colloidal Coomassie blue stain
Dionex UltiMate 3000 ultra-high performance liquid chromatography system attached to Orbitrap Fusion Tribrid Mass Spectrometer or equivalent LC/MS system
Dithiothrietol
Iodoacetamide
Mapping grade trypsin
Novex Mini-Cell (Invitrogen) or equivalent
NuPage 4-12% bis-Tris gel and running buffer (Invitrogen) or equivalent
NuPage lithium dodecyl sulphate (LDS) sample buffer (Invitrogen, Carlsbad, CA) or equivalent
PowerEase 500 Power Supply (Invitrogen) or equivalent
Ultra-pure urea (8 M)
Ziptip C18 column or equivalent
Procedure.
Solubilize the OECM in 2X NuPage LDS sample buffer at 60°C. If the sample fails to solubilize, add 8 M urea to a final concentration of 2 M and continue incubation.
Separate the OECM proteins by gel electrophoresis on NuPage 4-12% bis-Tris gel.
Stain the gel with colloidal Coomassie blue for 1 hour at room temperature and wash off excess stain with dH2O.
Excise the bands and de-stain with 2 cycles of 2:1 acetonitrile in 50 mM ammonium bicarbonate for 30 min, followed with 25 mM ammonium bicarbonate for 30 min.
Reduce the proteins for 1 hr in 5 mM dithiothrietol, followed by alkylation in 5.5 mM iodoacetamide.
Digest the proteins with 25 μg.mL−1 trypsin for 16 hrs at 36°C.
Desalt and purify the peptides with a C18 column according to the manufacturer’s directions.
Suspend the digested samples in 0.1% (v/v) formic acid before carrying out separation analysis by LC/MS. The LC elution is optimizable, but initially set to 4μL.min−1 with a 2 - 90% gradient of 0.1% (v/v) formic acid in acetonitrile against 0.1% (v/v) formic acid in dH2O. Settings for detection are optimizable, but initially set to: spray voltage 2.3 kV, orbitrap resolution 120 K, scan range 400 - 1600, higher-energy C-trap dissociation energy (HCD) 28%.
4. Coating of OECM onto growth surfaces.
Cell attachment to OECM has been demonstrated to improve cell survival, proliferation and enhance progenitor cell functionality in vitro and in vivo (Clough, McCarley, Krause, et al., 2015; Clough et al., 2017; Harvestine, Orbay, Chen, Sahar, & Leach, 2018; Mao et al., 2019; McNeill et al., 2018; Ragelle et al., 2017; Sun et al., 2011; Zeitouni et al., 2012). This can be demonstrated experimentally in several ways, but most approaches involve immobilization of OECM components onto supporting substrates. This section describes several approaches for the generation of OECM coated surfaces for assays with attached MSCs or OPs.
4.1. Solubilization and coating of OECM onto tissue culture plastic.
Materials.
At least 10 mg OECM (from approximately 5 × 15 cm plates)
15 mL standard conical polypropylene “Falcon” tubes
Analytical grade dH2O
Glacial acetic acid
Rat tail collagen solution or equivalent
UV visible spectrophotometer compatible with 50 μL volume or lower
Spectroscopy cuvettes compatible with 280 nm and 50 μL volume or lower
Sterile PBS without calcium or magnesium
Tissue culture treated plates or dishes for coating
Procedure.
Make a fresh 0.1 M acetic acid solution by adding 0.6 mL of glacial acetic acid to 100 mL dH2O. Note that glacial acetic acid is approximately 17.5 M.
Add 1.0 mL of 0.1 M acetic acid per 1 mg of OECM in a 15 mL Falcon tube and incubate with stirring or inversion for up to 2 days.
Prepare a 4 mg.mL−1 solution of rat tail collagen in 0.1 M acetic acid and prepare a standard curve by a 7-point doubling dilution with 0.1 M acetic acid. Prepare a standard curve of absorbance at 280 nm using the spectrophotometer.
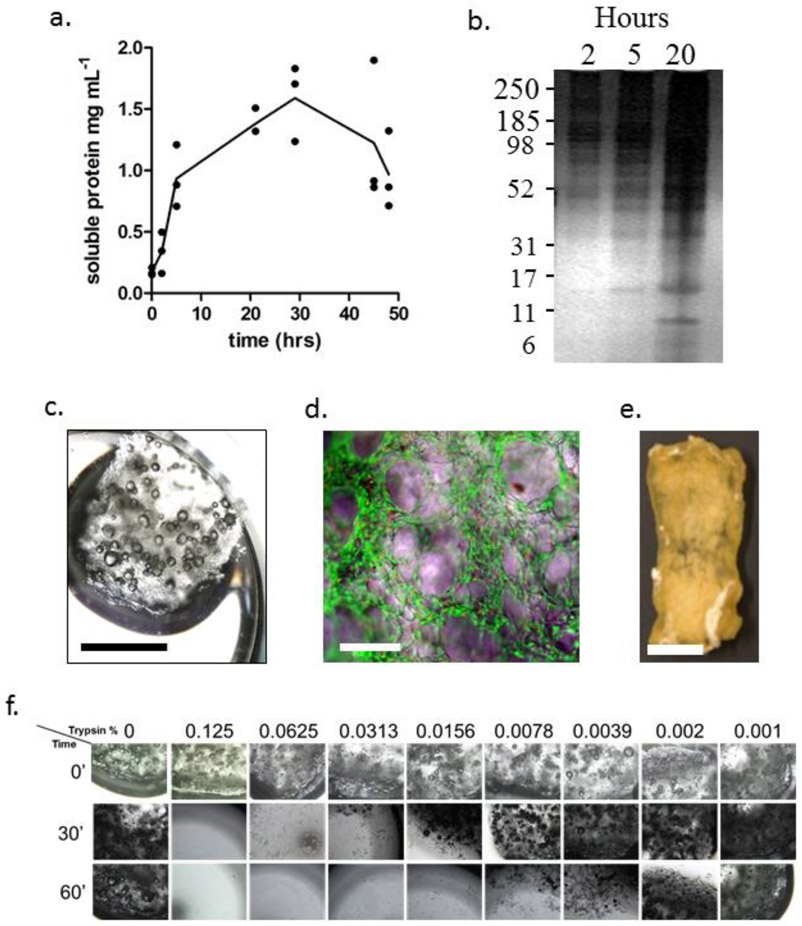
Every 6 - 12 h take a 50 μL sample from the dissolving OECM sample, centrifuge at 8,000 g for 5 min and take the absorbance at 280 nm. Calculate concentration using the standard curve generated in Step 3. The concentration should reach approximately 1 - 1.5 mg.mL−1 after 24 - 30 hr, but beware that this concentration can diminish with extended incubation if the sample contains high levels of acid proteases (e.g. Fig 3a). Ten μL of the sample can then be analyzed by standard SDS-PAGE (see Section 3.4) after neutralization by addition of 5 μL of 10x PBS (e.g. Fig 3b).
Use the solution immediately for downstream applications.
For coating of tissue culture-treated plastic, make a 30 μg mL−1 solution of solubilized OECM in PBS and add sufficient solution to cover the growth area of the desired tissue culture dishes. Incubate covered in a BSC at room temperature for at least 3 hr, then wash once in excess PBS. Store covered and aspirated at 4°C for up to 1 week.
Figure 3: Solubilization and coating of OECM:
Panel a: Plot of solubilization of OECM over time in acetic acid. Panel b: Silver stained SDS-polyacrylamide electrophoresis of solubilized OECM. Numbers of left represent approximate molecular mass in kilodaltons. Panel c: GF fragment in 12-well culture dish for purposes of MSC culture and OECM coating. Panel d: Composite phase-fluorescence micrograph of live-dead stained fragment of GF illustrating attached live MSCs (green) and occasional dead MSC nuclei (red). Panel e: Bioconditioned GF construct for the purposes of a bone repair assay in mice. OECM is incorporated throughout the thickness of the bioconditioned construct, and has the handling properties of decellularized bone (adapted with permission from ref.48). Panel f: Dose response experiment demonstrating the sensitivity of unmodified GF to trypsin. Note that the construct in Panel e was incubated in 0.125 (v/v) trypsin for 15 hr. For panel c,e: bar = 5 mm. For panel d: bar = 100 μm.
4.2. In situ coating of OECM by culture and decellularization.
A rapid approach for coating of OECM onto tissue culture plastic or other substrates can be achieved by simply culturing a monolayer of MCSs or OPs on the desired surface followed by a light decellularization step. This method is rapid and cost-effective, but depending on the attachment substrate selected, it may be possible to include a trypsinization step, or the downstream wash steps with chloroform and acetone. Omission of these steps will, of course, increase the complexity of the OECM and increase levels of cellular debris. Coating of tissue culture plastic by culture of MSCs or OPs on followed by decellularization has been achieved by several groups using similar, but distinct protocols (Clough, McCarley, Krause, et al., 2015; Harvestine et al., 2018; Mao et al., 2019; Ragelle et al., 2017; Sun et al., 2011). Herein, we describe a protocol for culture of MSCs on gelatin foam (GF) followed by decellularization, resulting in OECM-coated GF (bioconditioned GF or BGF) for 3D culture and in vivo applications and a protocol for the generation of OECM-coated tissue culture plastic.
Materials.
For coating of gelatin foam.
Acetone, analytical grade
Chemical fume hood
Chloroform, analytical grade
Inverted phase contrast microscope or equivalent fitted with 4x, 10x and 20x objectives and fluorescent capability (495/519 for green, 535/617 for red) if Live/Dead Assay utilized
Live dead cell viability assay (e.g. Live/Dead Assay, Invitrogen)
Matrix extraction buffer (MEB):
PBS supplemented with 0.1% (v/v) Triton X-100, 1 mM MgCl2, and 1 U.mL−1 DNAse I, filtered through a 0.2 μm pore-diameter membrane.
Porcine gelatin foam (e.g. Gelfoam, Baxter or equivalent) in 5 mm thick sheets
Polypropylene or glass dishes large enough to accommodate GF fragments
Sterile dH2O
Sterile PBS
Sterile scalpel and forceps
Tissue culture materials from Protocol 2.1
Trypsin, tissue culture grade
Low binding tissue culture plates or dishes.
For coating of tissue culture plastic.
Cell culture treated tissue culture plates, dishes or micro-carriers.
Inverted phase contrast microscope or equivalent fitted with 4x, 10x and 20x objectives.
Matrix extraction buffer (MEB): PBS supplemented with 0.1% (v/v) Triton X-100, 1 mM MgCl2, and 1 U.mL−1 DNAse I, filtered through a 0.2 μm pore-diameter membrane.
Sterile dH2O
Sterile PBS
Tissue culture materials from Protocol 2.1
Procedure.
For coating of gelatin foam.
In a BSC, using a sterile scalpel and forceps, cut GF into appropriate sizes and place in an appropriately sized low binding tissue culture plate or dish. Overlay with CCM and allow to hydrate for at least 15 min.
Grow MSCs or OPs to the desired density for reseeding as directed in Protocol 2.1 and recover a single-celled suspension by trypsinization, adjusted to 100,000 cells per mL in CCM.
Aspirate the CCM from the hydrated GF and pipette cell suspension directly into the GF by placing the tip of the pipette gently on the surface of the GF and releasing approximately 1 mL every 10 × 10 × 5 mm. Perform the seeding on saturated, but not immersed GF (Fig 3c). Prepare duplicate cultures with sufficient material to perform live/dead assays (see Step 5).
Add CCM to just cover the GF and culture for 5 days. At day 5, replace media with OBM and culture for a further 8 - 21 days with additional supplements such as dexamethasone or GW9662 as required. Flip GF and replace media every 2 days.
Every 2-5 days, recover a 5 × 5 × 5 mm fragment using sterile tools for live/dead visualization. Wash the GF in excess PBS, then perform the live/dead incubation in accordance with the manufacturer’s directions. Visualize live and dead cells attached to the GF using the fluorescent green and red channels respectively. Expect fewer than 10% red nuclei as compared to green cells spread over the surface of the GF (Fig 3e).
After at least 8 days of incubation under osteogenic and high-density conditions, wash the GF-cell composite 3 times with PBS by incubation in excess PBS for 3 min. Store at −80°C for at least 15 hr.
Thaw the GF-cell composite in excess MEB and incubate for 4 hr at 37°C, then add trypsin to the required concentration if necessary. Note that GF is highly sensitive to trypsin, but combination of the GF matrix with OECM will increase resilience. The example given in Fig 3e was incubated with 0.125 (w/v) trypsin for 15 hr(Clough, McCarley, Krause, et al., 2015), a condition that would degrade unmodified GF rapidly. It is therefore advisable to perform a dose response experiment to determine the highest dose of trypsin that can be used without destruction of the construct (Fig 3e).
In a polypropylene or glass dish wash the construct in at least 100 construct volumes of dH2O 3 times for 5 min with orbital shaking at 80 rpm, then once for 15 min to ensure trypsin is removed.
In a chemical fume hood, wash once for 5 min in chloroform sufficient to cover the construct, then repeat process with acetone.
Allow acetone to evaporate from BGF in chemical fume hood while covered to prevent contamination.
For coating of tissue culture plastic.
Note that an alternate protocol for this purpose is provided in Sun et al. (Sun et al., 2011).
Generate confluent monolayer of osteogenic MSCs or OPs (Fig 2a) using Protocol 2.1 herein or equivalent using the required plates, dishes or micro-carriers. A protocol for growing MSCs on polystyrene micro-carriers is provided in McNeill et al. (McNeill et al., 2018).
Aspirate media, and wash monolayers 3 times with PBS. Freeze aspirated monolayers for at least 15 hr at −80°C.
Cover monolayers with MEB and incubate for 15 hr at 37°C. Was plates in excess dH2O 3 times, aspirate and store dry at 4°C for up to 7 days.
5. Discussion, future prospects.
This chapter provides a selection of protocols to facilitate the generation of OECM from osteogenic progenitors, characterize its morphology and composition, and facilitate utilization in downstream experiments. The protocols for the generation of OECM from hMSCs are used routinely in our laboratory and the in vitro activity and osteogenic efficacy of the pure OECM and BGF has been published several times (Clough, McCarley, Krause, et al., 2015; Clough et al., 2017; McNeill et al., 2018; Zeitouni et al., 2012). In this chapter we have focused primarily on hMSCs, but there are many types of OP cells and a large body of published methodology, such that it is impossible to provide a comprehensive treatise of the field (W. Zhang et al., 2016). Therefore, the information provided herein is intended to represent an initial series of optimizable procedures that can be adapted for specific purposes. While beyond the scope of this chapter, it is useful to note that our group has developed in vivo assays for the study of OECM or BGF in calvarial healing (Zeitouni et al., 2012), femoral healing (Clough, McCarley, & Gregory, 2015; Clough, McCarley, Krause, et al., 2015) and spine fusion (Clough et al., 2017; Clough et al., 2018).
OECM can be coated onto surfaces through its natural affinity to certain materials, as described in Section 4, but because the OECM can be solubilized, it is amenable to covalent crosslinking approaches such as click chemistry(Azagarsamy & Anseth, 2013; Tang & Becker, 2014) and more classical approaches such as use of EDAC/NHS coupling for attachment to polystyrene and similar plastics(McNeill et al., 2018). Solubilized OECM may also be utilized in bioinks for 3D printing purposes and future work is focused in this area.
ECM manufactured from cultured cells has several potential uses; it can more accurately recapitulate tissue conditions in vitro and inform on mechanism, enhance useful properties of cells, increase cell yields, and in the case of musculoskeletal tissue engineering, it could serve as a potent regenerative tool (W. Zhang et al., 2016). The protocols described herein are hoped to facilitate progress in this growing field.
Acknowledgements.
The authors wish to acknowledge Dr Ulf Krause and Daryl Blalock for contributions of images. This work was funded by a research grant from the National Institute of Arthritis and Musculoskeletal and Skin Diseases R01AR066033, a research grant from the Center for Advancement of Science in Space, an Investigator Initiated Award from the Cancer Prevention Research Institute of Texas and an X-grant from the Texas A&M President’s Excellence Fund.
References.
- Alaseem AM, Madiraju P, Aldebeyan SA, Noorwali H, Antoniou J, & Mwale F (2015). Naproxen induces type X collagen expression in human bone-marrow-derived mesenchymal stem cells through the upregulation of 5-lipoxygenase. Tissue Eng Part A, 21(1-2), 234–245. doi: 10.1089/ten.TEA.2014.0148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson HC (2003). Matrix vesicles and calcification. Curr Rheumatol Rep, 5(3), 222–226. [DOI] [PubMed] [Google Scholar]
- Azagarsamy MA, & Anseth KS (2013). Bioorthogonal Click Chemistry: An Indispensable Tool to Create Multifaceted Cell Culture Scaffolds. ACS Macro Lett, 2(1), 5–9. doi: 10.1021/mz300585q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Hou L, Ma Y, Chen L, Zhang M, & Guan W (2013). Isolation and characterization of mesenchymal stem cells from chicken bone marrow. Cell Tissue Bank, 14(3), 437–451. doi: 10.1007/s10561-012-9347-8 [DOI] [PubMed] [Google Scholar]
- Balcerzak M, Hamade E, Zhang L, Pikula S, Azzar G, Radisson J, . . . Buchet R (2003). The roles of annexins and alkaline phosphatase in mineralization process. Acta Biochim Pol, 50(4), 1019–1038. doi:0350041019 [PubMed] [Google Scholar]
- Bearden RN, Huggins SS, Cummings KJ, Smith R, Gregory CA, & Saunders WB (2017). In-vitro characterization of canine multipotent stromal cells isolated from synovium, bone marrow, and adipose tissue: a donor-matched comparative study. Stem Cell Res Ther, 8(1), 218. doi: 10.1186/s13287-017-0639-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessey OA, Lowry OH, & Brock MJ (1946). A method for the rapid determination of alkaline phosphates with five cubic millimeters of serum. J Biol Chem, 164, 321–329. [PubMed] [Google Scholar]
- Buchet R, Millan JL, & Magne D (2013). Multisystemic functions of alkaline phosphatases. Methods Mol Biol, 1053, 27–51. doi: 10.1007/978-1-62703-562-0_3 [DOI] [PubMed] [Google Scholar]
- Carraro G, Albertin G, Forneris M, & Nussdorfer GG (2005). Similar sequence-free amplification of human glyceraldehyde-3-phosphate dehydrogenase for real time RT-PCR applications. Mol Cell Probes, 19(3), 181–186. doi:S0890–8508(04)00106–9 [pii] 10.1016/j.mcp.2004.11.004 [DOI] [PubMed] [Google Scholar]
- Clough BH, McCarley MR, & Gregory CA (2015). A Simple Critical-sized Femoral Defect Model in Mice. J Vis Exp (97). doi: 10.3791/52368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough BH, McCarley MR, Krause U, Zeitouni S, Froese JJ, McNeill EP, . . . Gregory CA (2015). Bone regeneration with osteogenically enhanced mesenchymal stem cells and their extracellular matrix proteins. J Bone Miner Res, 30(1), 83–94. doi: 10.1002/jbmr.2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough BH, McNeill EP, Palmer D, Krause U, Bartosh TJ, Chaput CD, & Gregory CA (2017). An allograft generated from adult stem cells and their secreted products efficiently fuses vertebrae in immunocompromised athymic rats and inhibits local immune responses. Spine J, 17(3), 418–430. doi: 10.1016/j.spinee.2016.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough BH, Zeitouni S, Krause U, Chaput CD, Cross LM, Gaharwar AK, & Gregory CA (2018). Rapid Osteogenic Enhancement of Stem Cells in Human Bone Marrow Using a Glycogen-Synthease-Kinase-3-Beta Inhibitor Improves Osteogenic Efficacy In Vitro and In Vivo. Stem Cells Transl Med. doi: 10.1002/sctm.17-0229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Mendes L, Sauvigne T, & Guiol J (2016). [Morbidity of autologous bone harvesting in implantology: Literature review from 1990 to 2015]. Rev Stomatol Chir Maxillofac Chir Orale, 117(6), 388–402. doi: 10.1016/j.revsto.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Fernandez de Grado G, Keller L, Idoux-Gillet Y, Wagner Q, Musset AM, Benkirane-Jessel N, . . . Offner D (2018). Bone substitutes: a review of their characteristics, clinical use, and perspectives for large bone defects management. J Tissue Eng, 9, 2041731418776819. doi: 10.1177/2041731418776819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DA 2nd, Bialek P, Ahn JD, Starbuck M, Patel MS, Clevers H, . . . Karsenty G (2005). Canonical Wnt signaling in differentiated osteoblasts controls osteoclast differentiation. Dev Cell, 8(5), 751–764. doi:S1534–5807(05)00097–3 [pii] 10.1016/j.devcel.2005.02.017 [DOI] [PubMed] [Google Scholar]
- Gregory CA, Gunn WG, Peister A, & Prockop DJ (2004). An Alizarin red-based assay of mineralization by adherent cells in culture: comparison with cetylpyridinium chloride extraction. Anal Biochem, 329(1), 77–84. doi: 10.1016/j.ab.2004.02.002S0003269704001332 [pii] [DOI] [PubMed] [Google Scholar]
- Gregory CA, Prockop DJ (2007). Fundementals of Culture and Characterization of Mesenchymal Stem/Progenitor Cells from Bone Marrow Stroma In R. I. S. Freshney GN and Auerbach JM (Ed.), Culture of Human Stem Cells (pp. 208). Hoboken NJ: Wiley-Liss. [Google Scholar]
- Harvestine JN, Orbay H, Chen JY, Sahar DE, & Leach JK (2018). Cell-secreted extracellular matrix, independent of cell source, promotes the osteogenic differentiation of human stromal vascular fraction. J Mater Chem B, 6(24), 4104–4115. doi: 10.1039/C7TB02787G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoi AM, Iorio JA, & Cahill PJ (2015). Autologous bone graft harvesting: a review of grafts and surgical techniques. Musculoskelet Surg, 99(3), 171–178. doi: 10.1007/s12306-015-0351-6 [DOI] [PubMed] [Google Scholar]
- Javazon EH, Colter DC, Schwarz EJ, & Prockop DJ (2001). Rat marrow stromal cells are more sensitive to plating density and expand more rapidly from single-cell-derived colonies than human marrow stromal cells. Stem Cells, 19(3), 219–225. doi: 10.1634/stemcells.19-3-219 [DOI] [PubMed] [Google Scholar]
- Jiang L, Cui Y, Luan J, Zhou X, Zhou X, & Han J (2013). A comparative proteomics study on matrix vesicles of osteoblast-like Saos-2 and U2-OS cells. Intractable Rare Dis Res, 2(2), 59–62. doi: 10.5582/irdr.2013.v2.2.59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause U, Harris S, Green A, Ylostalo J, Zeitouni S, Lee N, & Gregory CA (2010). Pharmaceutical modulation of canonical Wnt signaling in multipotent stromal cells for improved osteoinductive therapy. Proc Natl Acad Sci U S A, 107(9), 4147–4152. doi:0914360107 [pii] 10.1073/pnas.0914360107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause U, Ryan DM, Clough BH, & Gregory CA (2014). An unexpected role for a Wnt-inhibitor: Dickkopf-1 triggers a novel cancer survival mechanism through modulation of aldehyde-dehydrogenase-1 activity. Cell Death Dis, 5, e1093. doi: 10.1038/cddis.2014.67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey DL, Timms E, Tan HL, Kelley MJ, Dunstan CR, Burgess T, . . . Boyle WJ (1998). Osteoprotegerin ligand is a cytokine that regulates osteoclast differentiation and activation. Cell, 93(2), 165–176. doi:S0092–8674(00)81569-X [pii] [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang B, Song S, Ma M, Si S, Wang Y, . . . Guo Y (2014). Bovine collagen peptides compounds promote the proliferation and differentiation of MC3T3-E1 pre-osteoblasts. PLoS One, 9(6), e99920. doi: 10.1371/journal.pone.0099920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobb DC, DeGeorge BR Jr., & Chhabra AB (2019). Bone Graft Substitutes: Current Concepts and Future Expectations. J Hand Surg Am. doi: 10.1016/j.jhsa.2018.10.032 [DOI] [PubMed] [Google Scholar]
- Mao Y, Block T, Singh-Varma A, Sheldrake A, Leeth R, Griffey S, & Kohn J (2019). Extracellular matrix derived from chondrocytes promotes rapid expansion of human primary chondrocytes in vitro with reduced dedifferentiation. Acta Biomater, 85, 75–83. doi: 10.1016/j.actbio.2018.12.006 [DOI] [PubMed] [Google Scholar]
- Marastoni S, Ligresti G, Lorenzon E, Colombatti A, & Mongiat M (2008). Extracellular matrix: a matter of life and death. Connect Tissue Res, 49(3), 203–206. doi: 10.1080/03008200802143190 [DOI] [PubMed] [Google Scholar]
- Martino MM, Briquez PS, Maruyama K, & Hubbell JA (2015). Extracellular matrix-inspired growth factor delivery systems for bone regeneration. Adv Drug Deliv Rev, 94, 41–52. doi: 10.1016/j.addr.2015.04.007 [DOI] [PubMed] [Google Scholar]
- Martins-Neves SR, Lopes AO, do Carmo A, Paiva AA, Simoes PC, Abrunhosa AJ, & Gomes CM (2012). Therapeutic implications of an enriched cancer stem-like cell population in a human osteosarcoma cell line. BMC Cancer, 12, 139. doi: 10.1186/1471-2407-12-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNeill EP, Reese RW, Tondon A, Clough BH, Pan S, Froese J, . . . Gregory CA (2018). Three-dimensional in vitro modeling of malignant bone disease recapitulates experimentally accessible mechanisms of osteoinhibition. Cell Death Dis, 9(12), 1161. doi: 10.1038/s41419-018-1203-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira CA, Dempster DW, & Baron R (2000). Anatomy and Ultrastructure of Bone - Histogenesis, Growth and Remodeling In Feingold KR, Anawalt B, Boyce A, Chrousos G, Dungan K, Grossman A, Hershman JM, Kaltsas G, Koch C, Kopp P, Korbonits M, McLachlan R, Morley JE, New M, Perreault L, Purnell J, Rebar R, Singer F, Trence DL, Vinik A, & Wilson DP (Eds.), Endotext. South Dartmouth (MA). [Google Scholar]
- Murphy MB, Suzuki RK, Sand TT, Chaput CD, & Gregory CA (2013). Short Term Culture of Human Mesenchymal Stem Cells with Commercial Osteoconductive Carriers Provides Unique Insights into Biocompatibility. Journal of Clinical Medicine, 2(3), 49–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshed M (2018). Mechanism of Bone Mineralization. Cold Spring Harb Perspect Med, 8(12). doi: 10.1101/cshperspect.a031229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF, & Prockop DJ (2004). Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood, 103(5), 1662–1668. doi: 10.1182/blood-2003-09-3070 [DOI] [PubMed] [Google Scholar]
- Ragelle H, Naba A, Larson BL, Zhou F, Prijic M, Whittaker CA, . . . Anderson DG (2017). Comprehensive proteomic characterization of stem cell-derived extracellular matrices. Biomaterials, 128, 147–159. doi: 10.1016/j.biomaterials.2017.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salem O, Wang HT, Alaseem AM, Ciobanu O, Hadjab I, Gawri R, . . . Mwale F (2014). Naproxen affects osteogenesis of human mesenchymal stem cells via regulation of Indian hedgehog signaling molecules. Arthritis Res Ther, 16(4), R152. doi: 10.1186/ar4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaap-Oziemlak AM, Raymakers RA, Bergevoet SM, Gilissen C, Jansen BJ, Adema GJ, . . . Jansen JH (2010). MicroRNA hsa-miR-135b regulates mineralization in osteogenic differentiation of human unrestricted somatic stem cells. Stem Cells Dev, 19(6), 877–885. doi: 10.1089/scd.2009.0112 [DOI] [PubMed] [Google Scholar]
- Sedlmeier G, & Sleeman JP (2017). Extracellular regulation of BMP signaling: welcome to the matrix. Biochem Soc Trans, 45(1), 173–181. doi: 10.1042/BST20160263 [DOI] [PubMed] [Google Scholar]
- Solchaga LA, Johnstone B, Yoo JU, Goldberg VM, & Caplan AI (1999). High variability in rabbit bone marrow-derived mesenchymal cell preparations. Cell Transplant, 8(5), 511–519. [DOI] [PubMed] [Google Scholar]
- Sommerfeldt DW, & Rubin CT (2001). Biology of bone and how it orchestrates the form and function of the skeleton. Eur Spine J, 10 Suppl 2, S86–95. doi: 10.1007/s005860100283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Li W, Lu Z, Chen R, Ling J, Ran Q, . . . Chen XD (2011). Rescuing replication and osteogenesis of aged mesenchymal stem cells by exposure to a young extracellular matrix. FASEB J, 25(5), 1474–1485. doi:fj.10–161497 [pii] 10.1096/fj.10-161497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, & Becker ML (2014). “Click” reactions: a versatile toolbox for the synthesis of peptide-conjugates. Chem Soc Rev, 43(20), 7013–7039. doi: 10.1039/c4cs00139g [DOI] [PubMed] [Google Scholar]
- Tannoury CA, & An HS (2014). Complications with the use of bone morphogenetic protein 2 (BMP-2) in spine surgery. Spine J, 14(3), 552–559. doi: 10.1016/j.spinee.2013.08.060 [DOI] [PubMed] [Google Scholar]
- Wei F, Yang S, Guo Q, Zhang X, Ren D, Lv T, & Xu X (2017). MicroRNA-21 regulates Osteogenic Differentiation of Periodontal Ligament Stem Cells by targeting Smad5. Sci Rep, 7(1), 16608. doi: 10.1038/s41598-017-16720-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner S, & Traub W (1992). Bone structure: from angstroms to microns. FASEB J, 6(3), 879–885. [PubMed] [Google Scholar]
- Xie J, Zhang D, Zhou C, Yuan Q, Ye L, & Zhou X (2018). Substrate elasticity regulates adipose-derived stromal cell differentiation towards osteogenesis and adipogenesis through beta-catenin transduction. Acta Biomater, 79, 83–95. doi: 10.1016/j.actbio.2018.08.018 [DOI] [PubMed] [Google Scholar]
- Zeitouni S, Krause U, Clough BH, Halderman H, Falster A, Blalock DT, . . . Gregory CA (2012). Human mesenchymal stem cell-derived matrices for enhanced osteoregeneration. Sci Transl Med, 4(132), 132ra155. doi:4/132/132ra55 [pii] 10.1126/scitranslmed.3003396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Yang L, Yang XG, Wang F, Feng JT, Hua KC, . . . Hu YC (2019). Demineralized Bone Matrix Carriers and their Clinical Applications: An Overview. Orthop Surg. doi: 10.1111/os.12509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Zhu Y, Li J, Guo Q, Peng J, Liu S, . . . Wang Y (2016). Cell-Derived Extracellular Matrix: Basic Characteristics and Current Applications in Orthopedic Tissue Engineering. Tissue Eng Part B Rev, 22(3), 193–207. doi: 10.1089/ten.TEB.2015.0290 [DOI] [PubMed] [Google Scholar]
- Zhao Q, Gregory CA, Lee RH, Reger RL, Qin L, Hai B, . . . Liu F (2015). MSCs derived from iPSCs with a modified protocol are tumor-tropic but have much less potential to promote tumors than bone marrow MSCs. Proc Natl Acad Sci U S A, 112(2), 530–535. doi: 10.1073/pnas.1423008112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann EA, & Ritchie RO (2015). Bone as a Structural Material. Adv Healthc Mater, 4(9), 1287–1304. doi: 10.1002/adhm.201500070 [DOI] [PubMed] [Google Scholar]
- Zuk PA, Zhu M, Ashjian P, De Ugarte DA, Huang JI, Mizuno H, . . . Hedrick MH (2002). Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell, 13(12), 4279–4295. doi: 10.1091/mbc.E02-02-0105 [DOI] [PMC free article] [PubMed] [Google Scholar]