Abstract
Background:
Methadone, a synthetic opioid with longer duration of action and lower abuse potential compared with morphine, is used to prevent opioid withdrawal, as well as to manage chronic and acute surgical pain. The variability in response to methadone has been widely recognized. The purpose of this article is to review the literature on the pharmacogenetic factors underlying this variability.
Materials & methods:
This is a narrative overview of the literature on the genetic variants affecting pharmacodynamics and pharmacokinetics of methadone, retrieved from searches of databases such as PubMed and google scholar.
Discussion:
Clinical responses to methadone may be affected by genetic variants in the opioidergic, dopaminergic and neurotrophic pathways. Polymorphisms in genes related to disposition and elimination of methadone alter the pharmacokinetics, and possibly pharmacodynamics of methadone. Cytochrome P450 enzymes and P-glycoprotein variants contribute to the interindividual variability in methadone pharmacokinetics. Evidence for single gene variants affecting methadone response remains weak. Multiple genetic variants must be considered in conjunction to improve predictive ability.
Conclusion:
Evidence remains scarce at this time, to recommend pharmacogenetic testing before methadone administration. Well-powered clinical studies are needed with population pharmacokinetic-pharmacodynamic modeling and multigenetic signature-based predictions to enable tailored use of methadone in clinical practice.
Keywords: : gene variants, methadone, opioid, opioid maintenance therapy, personalized analgesia, pharmacodynamics, pharmacogenetics, pharmacogenomics, pharmacokinetics, polymorphism
Methadone is a long-acting, synthetic opioid analgesic, which acts as a MOR agonist, NMDA antagonist and central serotonin–norepinephrine reuptake inhibitor [1–3]. It was first synthesized in 1938 as a congener to morphine but soon fell into disrepute due to the associated adverse effects [4,5]. It gained importance again in the early 1960s as the treatment of opioid dependence and late 1970s as an analgesic. Presently, it is used for maintenance therapy to prevent opioid withdrawal, management of chronic pain and acute surgical pain [4].
Methadone has lower abuse potential, is longer acting and less sedating compared with morphine [6]. Due to these properties, problems such as tolerance, opioid induced-hyperalgesia and withdrawal are uncommon with methadone. This makes it an ideal drug for maintenance therapy in opioid abuse and in long-term management of chronic and cancer pain. There has been a growing body of literature on using methadone perioperatively, and its benefits in preventing persistent postoperative pain [7,8].
Despite this wide spectrum of clinical use, response to methadone varies greatly among patients. A number of studies have been performed to characterize the genetic variants causing this inter-person variability. A number of SNPs impacting methadone pharmacodynamics as well as pharmacokinetics have been described. The purpose of this review is to provide a narrative overview of literature related to pharmacogenomics of methadone.
Materials & methods
Articles were searched for on PubMed and Google Scholar. MeSH terms such as ‘methadone’, ‘pharmacogenetics’, ‘pharmacodynamics', ‘pharmacokinetics', ‘gene variants', ‘genetic polymorphism’, ‘SNP’ along with other additional specific keywords relevant to the topic were used to build a search. All studies in which genotyping was done in patients on methadone with clinical data such as efficacy, adverse effects and concentrations of drug, were chosen. Additional articles were found using hand searches of references in the retrieved literature. All citations were added to the citation manager EndNote X9 and duplicates were removed. Full-text papers of all the articles were accessed and articles not fitting the scope of the review were deleted manually. In the studies describing genetics of pharmacokinetics, there were studies that have associated CYP genotype with daily doses of methadone without concentration data, with varying results. These studies contribute to the overall scientific knowledge, but they were not included in this review as the effects of pharmacodynamic variability and pharmacokinetic variability cannot be disentangled in these studies.
Discussion
Pharmacodynamics of methadone
Methadone acts as an agonist to MOR, DOR and KOR [5]. Most of the therapeutic and adverse effects are primarily mediated through MOR agonism [6]. This is responsible for the analgesic, euphoric, miotic, respiratory depressant, sedative and gastrointestinal effects of methadone [6]. Methadone has greater affinity toward DOR compared with morphine [9]. This may contribute to the reduction of opioid tolerance associated with the use of methadone [10].
The methadone commercially available in the USA is a 50:50 mixture of R- and the S-methadone. The R-enantiomer is responsible for most of its MOR-related analgesic, therapeutic activity and related adverse events like respiratory depression and sedation. The S-enantiomer is responsible for most of methadone’s adverse events including QT prolongation [5]. The receptor activity of R- and S-methadone are quite specific reflecting their roles in different pharmacological activities. R-methadone has ten-times greater affinity for the MOR compared with the S isomer, resulting in the greater analgesic potency of R-methadone [5]. S-methadone, on the other hand, is a noncompetitive antagonist at the NMDA receptor [11,12]. NMDA activation has been linked to opioid tolerance and opioid induced hyperalgesia [13]. MOR agonist and NMDA antagonist properties result in an additive analgesic response with racemic methadone [14]. S-methadone also has serotonin and norepinephrine reuptake inhibitory properties in the central nervous system (CNS) [3,6].
Methadone can prolong QTC interval, with the potential risk of life-threatening torsades de pointes [15,16]. Particularly, the S isomer of methadone is an inhibitor of the potassium voltage-gated channel subfamily H member 2 in a dose-dependent manner [17]. This affects the delayed rectifier potassium current (IK) in Phase III of cardiac action potential, resulting in an increased time to repolarization and a corresponding increase in QT interval [18]. Patients with a baseline increased QTC due to congenital long QT syndromes, subclinical long QT syndromes, and those on other medications causing QT prolongation are at increased risk of torsades de pointes with methadone administration [18,19]. Methadone can also cause bradycardia due to its anticholinesterase and calcium channel blocking properties [20,21].
Genetic factors affecting the pharmacodynamics of methadone
Most of the studies on genetic variants affecting pharmacodynamics of methadone have looked into methadone dose requirements or response in patients on methadone maintenance therapy (MMT) for treatment of opioid use disorders.
Opioidergic pathway
OPRM1 coding for the MOR has been the most widely studied gene. Others include OPRD1 coding for DOR, POMC which encodes a precursor of endogenous opiates, and ARRB2 which encodes β-arrestin-2 (a downstream regulator in opioid signaling) [22].
OPRM1 polymorphism is an important factor influencing pharmacodynamics of opioids. The 118A>G (rs1799971) polymorphism has been associated with increased opiate dose requirements and decreased effects, including adverse effects and susceptibility to opioid dependence [23]. This was verified for methadone by Lotsch et al. in a pharmacogenetic study of CNS effects of methadone on 51 healthy volunteers [24]. They studied variants of CYP genes, ABCB1 and OPRM1 along with blood concentration of methadone and CNS effects quantified in terms of methadone-induced miosis. A significant association was found between OPRM1 118A>G carriers and methadone effect; however, there was no association with ABCB1 or CYP genes. Carriers of the 118G allele had 1.74-times (95% CI: 1.4–2.2) lower miotic potency of levomethadone compared with noncarriers. The maximum percent decrease in pupil diameter from baseline was lowest for homozygous carriers (118GG).
However, the association between OPRM1 118A>G and methadone dose for MMT was not conclusive as described in the systematic review by Oueslati et al. [25] In a study on 238 patients on MMT, Crettol et al. were not able to demonstrate an association between OPRM1 118A>G and methadone requirements [26]. Barratt et al. in their study on 119 subjects on MMT were able to demonstrate that an interaction between ABCB1 haplotype and OPRM1 variant influenced methadone dose requirements [27]. But an isolated effect of OPRM1 on methadone requirements was not found [27]. Hung et al. studied the association of genetic variants with methadone dose requirements in a sample of Han Chinese population on methadone maintenance therapy [28]. In a pair-wise comparison and genotyping of 321 opioid-dependent patients on MMT and 202 healthy controls, no association of 118A>G SNP and methadone maintenance dose was found. However, in a proportional odds regression model, 118A>G variant along with variants in CYP2B6, ANKK1 and DRD2 genes showed association with maximum methadone maintenance doses.
Levran et al. studied 227 former heroin users on MMT and found that OPRM1 rs558025 SNP carriers required lower methadone doses compared with noncarriers [22]. OPRM1 has also been studied as a risk factor for methadone-related deaths [29]. The 118A>G variant was associated with higher postmortem blood methadone concentration, although it did not reach a statistical significance. OPRM1 SNPs have also been associated with methadone adverse effects [30].
Luo et al. studied 257 patients on MMT for genetic factors affecting methadone dose [31]. They selected SNPs of OPRD1, ABCB1, ARRB2 and DRD1 for analysis. Carriers of OPRD1 rs529520TG variant had significantly greater methadone dose requirements compared with noncarriers. A significant SNP–SNP interaction of ABCB1–ARRB2–OPRD1 was also found to be associated with dose requirement. Levran et al. also evaluated POMC and ARRB2 variants in this study, along with OPRM1, but found no association with methadone dose [22].
Dopaminergic pathway
The mesolimbic dopaminergic system has been implicated in the rewarding effects of opioid use, dependence and withdrawal [22,32]. The genes DRD1 and DRD2 encode dopamine receptors, and ANKK1 (located near DRD2 on chromosome 11) may influence DRD2 expression [22,28]. Many variants of ANKK1 and DRD2 have been found to be in strong linkage disequilibrium [22]. In a study of MMT in Chinese patients, Hung et al. analyzed ANKK1–DRD2 haplotypes as part of their genetic panel [28]. They identified DRD2 variants 214A>G and 939C>T which lower methadone dose requirement. Patients with ANKK1–DRD2 haplotype CTACC or TCAAT were at greater risk of opioid addiction and needed larger methadone maintenance doses. A combined effect of variants in ABCB1, CYP2B6, OPRM1 and ANKK1–DRD2 could explain 53% of variation in methadone maintenance dose [28].
Levran et al. also reported that ANKK1–DRD2 variants influence methadone dose requirements [22]. Carriers of variant A allele of ANKK1 rs7118900 or variant T allele of DRD2 rs2283265 required lower methadone doses. Doehring et al. studied ANKK1–DRD2 polymorphism in Caucasian patients on MMT, evaluating 85 patients and 99 healthy controls [32]. The carrier frequency of a minor allele variant was greater in the patient group. The average and maximum daily doses of methadone were significantly greater in carriers of DRD2 rs6275C>T SNP. Similarly, Duan et al. showed a significant association between methadone dose requirement and DRD2 rs6275C>T variant in 257 Chinese patients on MMT [33]. One other DRD1 variant (rs686) was not found to be associated with methadone dose requirements for MMT [33].
Glutaminergic pathway
The GRIN1 and GRIN2A genes encode glutamate ionotropic receptor NMDA type subunits. Polymorphisms have been explored in the context of methadone pharmacodynamics; however, no significant associations have been discovered [22].
Neurotrophins
Neurotrophins are molecules involved in neuron growth, synaptic plasticity, learning, memory and, behavior, and these play a significant role in opioid-induced plasticity and reward effect of opioids [22,34]. The most notable neurotrophins include NGF and BDNF with their receptors, NTRK1 and NTRK2. Genetic variants in BDNF have been linked to variability in response to MMT [35]. Levran et al. showed an association between methadone dose and three BDNF intronic variants [22]. Similarly, six SNPs in NTRK2 were linked to methadone dose requirement [22]. The NGFβ intronic variant rs2239622 has been associated with variations in methadone dose [34].
Pharmacokinetics of methadone
Absorption
Methadone is a basic liposoluble drug with a pKa of 9.2. The drug is available as a hydrochloride salt (tablet for oral administration) and as an injectable formulation for intramuscular, subcutaneous and intravenous use. Other routes such as rectal, transmucosal, sublingual and epidural have been evaluated in the past. The R- and S-enantiomers of methadone are similar in absorption parameters such as the lag time and bioavailability [36]. When given orally, bioavailability is approximately 70% (ranging from 36 to 100% between individuals) [37]. This variability is attributed mainly to differences in first-pass metabolism by CYP enzymes and efflux transport by P-glycoprotein in the gut – governed by underlying pharmacogenetics [37]. The time to peak concentration is about 1–5 h, although therapeutic activity is initiated by 30–60 min following administration and lasts for about 4–6 h after a single dose [38]. The duration of analgesia increases to 8–12 h with repeated dosing, as seen in chronic therapy [39]. Methadone undergoes a minimal amount of enterohepatic recirculation with small secondary peaks observed in the concentration–time curve [40]. P-glycoprotein has been shown to be an efflux protein for methadone both in the gut and the brain in in vitro studies. However, inhibition of P-glycoprotein does not affect the bioavailability of the drug in vivo [41].
Distribution
Methadone is highly distributed in the body with apparent volume of distribution reported between 496 and 896 L for R-methadone, and 289 and 360 L for S-methadone [36,42,43]. The tissue protein binding of methadone is more than the plasma protein binding leading to a substantial peripheral reservoir [44]. In the plasma, methadone is predominantly protein bound (80–90%) but the amount of binding is dependent on the enantiomer. Methadone is mainly bound to α-1 AGP and to a lesser extent to other lipoproteins. Albumin appears to play a very minor role in plasma protein binding [45]. The amount of unbound drug observed is approximately 9–10 and 12–14% for R-methadone and the S-methadone, respectively [45,46]. About 56% of the variability in the unbound drug fraction of the racemates can be predicted by the total AGP concentration and by one of its major subtypes, ORM 2A [46]. The concentration of AGP is affected by physiological factors such as age, gender, menstrual cycle and pregnancy as well as regular diurnal variation [47]. Moreover, AGP, as an acute phase protein, increases in concentration from two to fivefold during stresses such as inflammation, burns, infections and surgeries [48]. The free concentration of methadone can fall drastically following such stressors, and this may reduce its efficacy.
Metabolism & elimination
Methadone is mainly eliminated by hepatic metabolism by CYP enzymes. These enzymes show stereoselectivity in preference for metabolism of the R- and S-enantiomers. About 10–20% of systemic drug is eliminated through urinary excretion, although this is dependent on urine pH. As methadone is a basic drug, urinary excretion decreases with increasing urinary pH [49]. The metabolic pathway of methadone is shown in detail in Figure 1. In the liver, methadone undergoes N-demethylation and spontaneous cyclization to form its major metabolite, 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine (EDDP), and an inactive metabolite, 2-ethyl-5-methyl-3,3-diphenylpyrroline (EMDP). Neither of the metabolites have any analgesic activity, although EDDP may have a role in QT interval prolongation reported with methadone [50].
Figure 1. . Methadone metabolism.
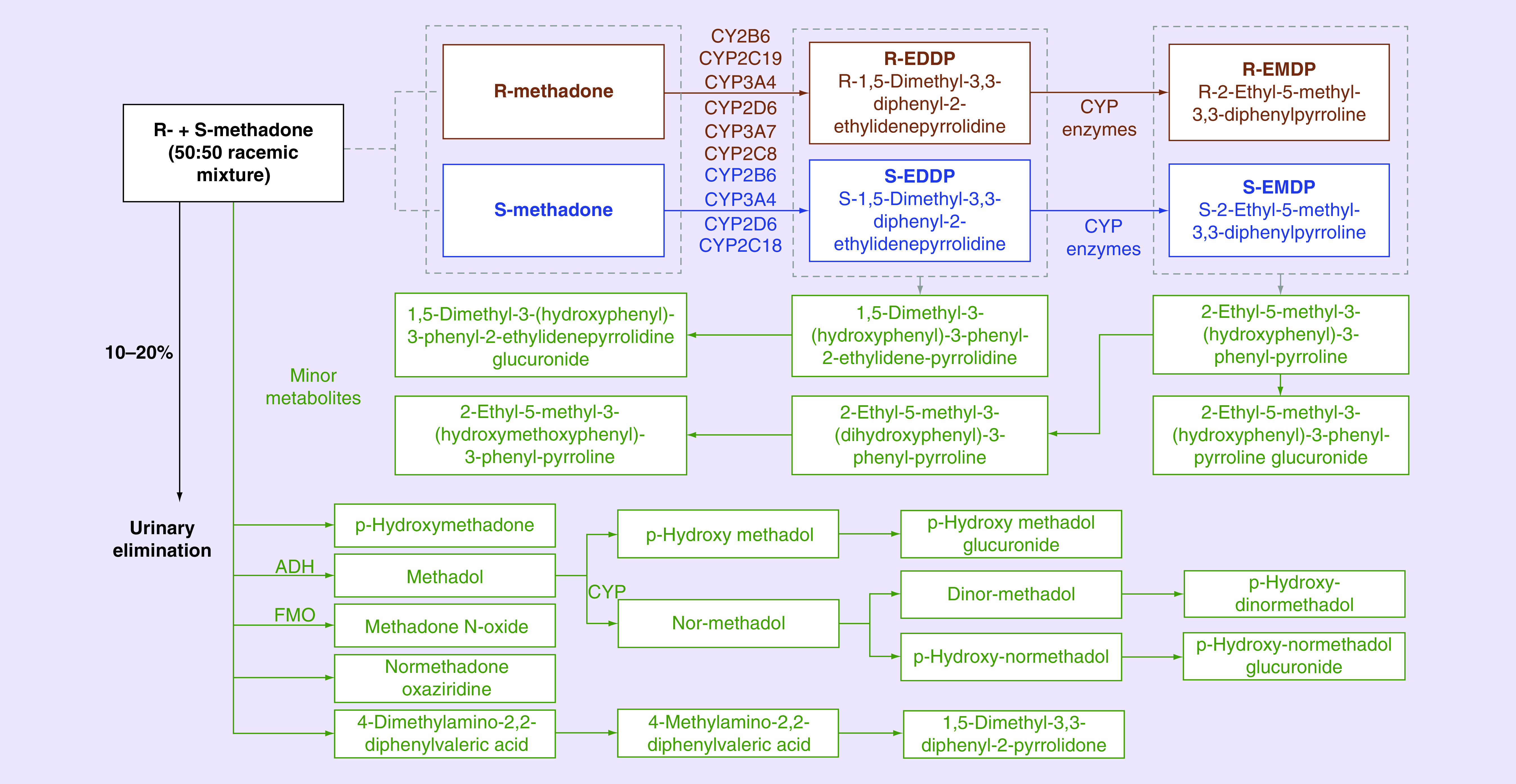
Methadone, a racemic mixture of R-methadone and S-methadone (50:50 mixture), is metabolized predominantly in the liver by N-demethylation in to a major metabolite, 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine, which is further N-methylated to an inactive metabolite, EMDP. The major metabolites of methadone are 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine and EMDP. The pathway of the R- and S-enantiomers are in brown and blue, respectively. Different liver CYP enzymes, as depicted, are involved in the metabolism of methadone’s each of the enantiomers as shown above. The other metabolites are methadol and p-hydroxy methadone. There are many minor metabolites of methadone, which are colored green in the pathway. 10–20% of methadone is excreted in the urine unchanged.
EMDP: 2-ethyl-5-methyl-3,3-diphenyl-1-pyrroline.
The understanding of the metabolic pathway for methadone has improved considerably in recent years. Initial human liver microsomal studies using specific enzyme inhibitors revealed a potential role of CYP3A4 in the metabolism of methadone without any preference to either of the enantiomers [51,52]. It is now evident that CYP3A4 is only one of several metabolizing enzymes of R- and S-methadone.
Gerber et al. assessed the role of several CYPs in methadone metabolism through experiments of recombinantly expressed CYP enzymes in Supersomes™ (Corning Inc., NY, USA). Their results showed the following order of importance: CYP2B6 > CYP2C19 ≥ CYP3A4 > CYP2C9 ≥ CYP2D6 [53]. The study also showed that CYP2B6 metabolizes S-methadone faster than R-methadone at low concentrations, and CYP2C19 metabolizes R-methadone better than S-methadone at all concentrations. However, it should be noted that this study in Supersomes™ could not account for the relative abundance of these enzymes expressed in the liver. For example, CYP3A4 is present in higher quantities in the liver than the other CYP enzymes tested. A follow-up study with relative abundance scaling demonstrated the ability of CYP enzymes to metabolize methadone in the following order of importance: CYP3A4 > CYP2B6 > CYP2C19 > CYP2D6 > CYP2C18, CYP3A7 > CYP2C8, CYP2C9, CYP3A5. Out of these CYPs, CYP2B6, 2D6 and 2C18 demonstrated a preference for (S)-EDDP formation and CYP2C19, 3A7 and 2C8 for (R)-EDDP formation. CYP3A4 showed no preference to either [54].
To follow-up on the in vitro characterization of methadone metabolism, Kharasch et al. conducted a series of drug interaction trials in humans focusing on CYP enzymes involved in metabolism of methadone. In one of these studies, ritonavir unexpectedly increased the clearance of both R- and S-methadone by about 1.5-fold and twofold, respectively, despite known CYP3A inhibitory activity [55]. This finding was reproduced in studies where indinavir, nelfinavir or ritonavir/indinavir were used to inhibit CYP3A4 enzymes [41,56–58]. Totah et al. reported that rifampicin increased the metabolism of R- and S-methadone while the CYP3A selective inhibitor troleandamycin did not have any effect on the enantiomers. The increase in ratio of R-/S-methadone with rifampicin also demonstrated the importance of CYP2B6 enzyme in metabolism of methadone in vivo [59]. Therefore, the CYP3A4 mediated metabolism of methadone may not be as significant as inferred from previous in vitro work.
The R- and S-enantiomers of methadone are metabolized by the same enzymes, albeit at different rates. Therefore, there is a possibility of interaction between the enantiomers. Indeed, Totah et al. found that the metabolism of R-methadone by CYP2B6 could be inhibited by S-methadone in vitro. However, similar interactions could not be elicited for CYP2C19 and CYP3A4 [60]. Methadone could also be involved in auto-induction. Activation on pregnane X receptor and the constitutive androstane receptor can increase the expression of CYP3A4 and CYP2B6 in the liver. Methadone has been shown to activate these receptors, thus potentially inducing its own metabolism [61,62].
Genetic factors affecting the pharmacokinetics of methadone
The high variability in methadone pharmacokinetics is principally caused by genetic variations in factors related to disposition and elimination [63]. These genetic factors have varying effects on the R- and S-enantiomers. Several in vivo studies have reported on this topic with varying results; however, there are limited clinical studies (Table 1) demonstrating an association of genetic polymorphisms.
Table 1. . Genetic polymorphisms with significant in vivo effect on methadone pharmacokinetics.
| Gene | Allele/haplotype | Effect | Study (year) | Ref. |
|---|---|---|---|---|
| CYP2B6 | *6 (*4 [rs2279343] and *9 [rs3745274]) | Decreased clearance and increased concentrations of both R- and S-methadone (S > R) | Csajka (2016) Crettol (2006) Bunten (2010) Dennis (2014) Kharasch (2015) Kringen (2017) |
[42,64–68] |
| *6 (*4 [rs2279343] and *9 [rs3745274]) | Decreased clearance and increased concentration of S-methadone | Wang (2011) Crettol (2005) Fonseca (2011) Bart (2014) |
[69–72] | |
| *5 (rs3211371) | Decreased clearance of racemic methadone in univariate analysis | Ahmad (2017) | [73] | |
| *5 (rs3211371) | Improved the clearance estimate in population PK model of R- and S-methadone, when used as a part of activity score | Csajka (2016) | [42] | |
| *5 (rs3211371) | Increase in clearance of S-methadone | Dobrinas (2013) | [74] | |
| *11 (rs35303484) | Improved the clearance estimate in population PK model of R- and S-methadone, when used as a part of activity score | Csajka (2016) | [42] | |
| *11 (rs35303484) | Trend for decreased S-methadone clearance | Dobrinas (2013) | [74] | |
| Intronic (rs2279344) | Trend for decreased S-methadone clearance | Csajka (2016) Dobrinas (2013) |
[42,74] | |
| Intronic (rs8192719) | Trend for decreased S-methadone clearance | Csajka (2016) Dobrinas (2013) |
[42,74] | |
| rs1038376 | Decreased S-methadone clearance and increased S-methadone dose corrected concentrations | Wang (2011) | [70] | |
| Intronic (rs10403955, rs2279345, rs707265) |
Increased S-methadone clearance and decreased S-methadone dose corrected concentrations | Wang (2011) | [70] | |
| Intronic (rs10500282) | Decreased S-methadone dose corrected concentrations | Wang (2011) | [70] | |
| Haplotypes TTT (rs8100458, rs10500282, rs10403955) and AGATAA hexanucleotide haplotypes (rs2279342–rs3745274–rs2279343–rs2279345–rs1038376–rs707265) | Highest concentrations including dose corrected concentrations and lowest clearance of S-methadone, while TCG showed opposite trend | Wang (2011) | [70] | |
| AGATAA (rs2279342, rs3745274, rs2279343, rs2279345, rs1038376 and rs707265) | Lower concentrations and dose corrected concentrations of S-methadone than the ATGCAG and ATGCTG combinations | Wang (2011) | [70] | |
| Haplotypes TAATCG and TCCTTT (rs8100458, rs7250601, rs7250991, rs11882424, rs8192719 and rs10853744) | Increased S-methadone clearance in sliding-window haplotype-based association analysis | Yang (2016) | [75] | |
| CYP2C19 |
CYP2C19 *2 rs4244285 CYP2C19 *3 rs4986893 PM (*2/*2, *2/*3 and *3/*3) IM (*1/*2 and *1/*3) EM (*1/*1) |
Decreased R-methadone clearance; EM > IM > PM | Wang (2013) | [76] |
| CYP2C19*2 or *3 | Decreased total methadone clearance with *2 and *3 | Kringen (2017) | [67] | |
| CYP2C19*2 | Increased total (S) and (R) EDDP concentrations | Carlquist (2015) | [50] | |
| CYP3A4 | CYP3A4*22 (rs35599367) | Decreased clearance of R-methadone and trend toward decreased clearance of S-methadone | Csajka (2016) | [42] |
| CYP3A4*1b (rs2740574) | Trend toward decreased clearance of R- and S-methadone | Crettol (2006) | [64] | |
| Intronic (rs2242480) and *1b (rs2740574) | Trend toward increased concentrations | Richards (2014) | [77] | |
| CYP2D6† | EM (homo): *1/*1 EM (hetero): *1/*4, *1/*5, *1/*3, *1/*6 PM: *4/*4, *4/*3, *4/*6 |
Increased clearance of both R- and S-methadone; UM > EM > PM | Eap (2001) | [78] |
| UM (*1×N, *2×N) EM (*1, *2, *3, *6, *35) IM (*9, *10, *41) PM (*4/*4) |
Concentrations of both R- and S-methadone were paradoxically higher in UM compared with EM, but UM patients had received much higher doses | Fonseca (2011) | [72] | |
| EM (*1/*1) IM (*1/*3, *1/*4, *1/*5, and *1/*6) PM (*3/*4, *4/*4, *4/*5, and *4/*6) UM (*1/*xN) |
UM had significantly lower trough S-methadone plasma concentrations compared with EM/IM. A similar trend was seen with R-methadone plasma concentrations. | Crettol (2006) | [64] | |
| CYP2C9‡ |
CYP2C9 *2 (rs1799853) CYP2C9 *3 (rs1057910) PM (*2/*2, *2/*3 and *3/*3) IM (*1/*2 and *1/*3) EM (*1/*1) |
Decreased total methadone clearance with *2 and *3 | Kringen (2017) | [67] |
| CYP3A5 | CYP3A5*3 (rs776746) | Decreased total methadone clearance | Kringen (2017) | [67] |
| ABCB1 | rs1045642 (3435C>T) | Increased clearance of S-methadone in univariate analysis and population PK modeling | Csajka (2016) | [42] |
| rs1045642 (3435C>T) | Increased clearance of R- and S-methadone | Crettol (2006) | [64] | |
| rs1045642 (3435C>T) | Trend toward increased clearance of R- and S-methadone | Dennis (2014) | [65] | |
| rs2032582 (2677G>T/A) | TT genotype was associated with lower concentrations of R- and S-methadone | Crettol (2006) | [64] | |
| rs2032582 (2677G>T/A) | GG genotype was associated with a reduction in CL/F of R- and S-methadone in PK modeling | Bart (2014) | [71] | |
| rs2032582 (2677G>T/A) | TT genotype was associated with higher concentrations of both R- and S-methadone | Lee (2013) | [79] | |
| rs9282564 | Increased clearance of R- and S-methadone | Crettol (2006) | [64] | |
| CGC/TTT diplotype rs1128503 (1236C>T) rs2032582 (2677G>T/A) rs1045642 (3435C>T) |
Increased dose-adjusted serum methadone concentration | Zahari (2016) | [80] | |
| Haplotype AGCTT rs9282564 61A>G rs2229109 1199G>A rs1128503 1236C>T rs2032582 2677G>T rs1045642 3435C>T |
Decreased trough methadone concentrations (both in homozygous and heterozygous forms) | Barratt (2012) | [27] | |
| POR | POR*28 (rs1057868) | Increased clearance of R- and S-methadone | Csajka (2016) | [42] |
| rs17180299 | rs17180299 | Increased concentrations of R-methadone | Yang (2016) | [75] |
CYPD6; UM: duplications (Ex: CYP2D6*1×N and CYP2D6*2×N [2–13 copies]) or alleles with promoter mutation; EM: 1 null/reduced + 1 normal activity (hetero) or 2 normal activity (homo); IM: 1 null + 1 reduced activity or 2 reduced activity allele; PM: 2 null allele.
CYP2C9; EM: homozygous normal activity allele; IM: heterozygous with one normal activity allele; PM: both reduced activity allele-homozygous or heterozygous.
CL/F: Apparent clearance; EDDP: 2-Ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine; EM: Extensive metabolizer; IM: Intermediate metabolizer; PK: Pharmacokinetic; PM: Poor metabolizer; UM: Ultrarapid metabolizer.
Genetics affecting absorption
Methadone, as a lipid-soluble drug, is absorbed primarily by passive diffusion before entering the jejunum. Human intestinal microsomes containing CYP enzymes have been found to be involved in metabolism of the drug [81]. Also, P-glycoprotein was shown to affect the absorption of methadone in an in vivo study employing quinidine as an inhibitor of the efflux transporter [82]. This P-glycoprotein efflux transporter is also known as MDR1 or ABCB1, and the encoding gene is highly polymorphic with multiple SNPs. However, there is no current evidence to support genetic polymorphisms affecting the absorption of the drug [83]. The likely explanation for this is the high bioavailability of methadone, such that the effects of these polymorphisms become negligible.
Genetics affecting disposition
As discussed earlier, methadone is primarily bound to AGP in plasma. The orosomucoid genes ORM1 and ORM2 encode this protein. Methadone binds selectively to the ORM2 [84]. Few studies regarding polymorphisms in ORM2 have been conducted. However, to our knowledge, no studies have evaluated ORM2 variants with methadone.
Genetics affecting metabolism
The CYP enzymes contribute to about 90% of the metabolism of methadone; therefore, their genetics remain the cornerstone of the pharmacogenetics of methadone [37]. The enzymes involved in metabolism for which studies with genotyping information available are CYP2B6, CYP2C19, CYP3A4, CYP2D6, CYP2C9 and CYP3A5. The importance of genetic polymorphisms for CYP enzymes has been determined either through univariate analysis, multivariate analysis or population pharmacokinetic modeling (Tables 1 & 2).
Table 2. . Characteristics of pharmacokinetics studies of methadone with genotyping available.
| Study (year) | n | Male | Female | Methadone samples | Genotype | Analysis† | Ref. |
|---|---|---|---|---|---|---|---|
| Csajka (2016) | 251 | 190 | 61 | PK modeling 244 peak (C4) and/or trough (C0), seven rich samples (11 each) in steady state |
CYP2B6: *6 [*4 and *9], *5, *11, intronic (rs2279344), intronic (rs8192719) CYP2C9 *2, *3 CYP2C19 *2, *3 CYP2D6 *3, *4, *5, *6 CYP3A4*1B, *22 CYP3A5*3 CYP3QA7*1C ABCB1 61A>G, 1199G>A, 1236C>T, 2677G>T, 3435C>T |
Population PK analysis with genotypes and supposed activity scores as covariates on clearance parameter | [42] |
| Crettol (2005) | 209 | NA | NA | 192 patients with trough (C0) and/or peak (C4) samples in steady state; 17 with trough only |
CYP2B6 *4, *5, *6, *7, *9 CYP2C9 *2, *3 CYP2C19 *2, *3 |
Univariate analysis between genotype and concentrations | [69] |
| Crettol (2006) | 245 | 185 | 60 | 245 trough (C0) and 203 (C4) peak samples under steady state |
CYP1A2*1F CYP2B6*4, *5, *9, CYP2C9*2, *3 CYP2C19*2, *3, CYP2D6*3, *4, *5, *6, xN CYP3A4*1B, *3 ABCB1 61A>G, 2677G>T, 3435C>T UGT2B7*2a |
Univariate analysis between genotype and concentrations | [64] |
| Dennis (2014) Meta-analysis |
NA | NA | NA | Data from four PK studies out of total seven studies selected for review. Crettol et al., Fonseca et al. and Uehlinger et al. |
CYP2B6*6 ABCB1 3435C>T |
Univariate analysis between genotype and concentrations | [64,65,69,72,85] |
| Kharasch (2015) | 64 | NA | NA | 489 genotyped out of which three groups of 20 subjects each were created with CYP2B6*1/*1, CYP2B6*1/*6 and CYP2B6*6/*6. Multiple PK samples through 96 h after single dose in healthy volunteers. | CYP2B6 *4, *5, *6, *7, *9, *16 and *18 | Univariate analysis between plasma EDDP/methadone, AUC ratio and genotype | [66] |
| Bunten (2010) | 67 | NA | NA | Concentration measured during autopsy | CYP2B6 *6 (*4 and *9) | Univariate analysis between genotype and concentrations | [68] |
| Kringen (2017) | 64 | NA | NA | At steady state (≥12 h after last intake of methadone) |
CYP2B6 *6 (*4 and *9) CYP3A5 *3 CYP2C9 *2, *3 CYP2D6 *3, *4, *5, *6 CYP2C19 *2, *3 |
Linear mixed model analysis along with age, gender and time with repeated measurements of concentration | [67] |
| Wang (2011) | 366 | 297 | 69 | Trough samples at steady state | rs8100458, rs10500282, rs10403955, rs2279342, rs3745274 (*9), rs2279343 (*4) rs2279345, rs1038376, rs707265, rs1042389 |
Univariate analysis between identified genotype and concentrations after multiple testing correction | [70] |
| Fonseca (2017) | 105 | 74 | 31 | 76 responders and 29 nonresponders. Trough samples at steady state |
CYP2B6 *1, *4, *6 CYP2C19 *1, *2, *3 CYP2D6 *1, *2, *3, *4, *5, *6, *9, *10, *17, *35, *41 CYP3A5 *1*3 CYP2C9 *1*2*3 ABCB1 3435C>T |
Univariate analysis between genotype and concentrations after multiple testing correction | [72] |
| Bart (2014) | 206 | NA | NA | PK modeling with 441 samples at steady state |
CYP3A4 *1b, rs28371759, rs4986909 CYP2B6*4, *5, *9, intronic (rs8192709) CYP2D6 rs1065852, rs5030656 CYP2C19 rs3758581 ABCB1 61A>G, 1236C>T, 2677G>T, 3435C>T, rs6949448, rs2235067, rs1922242, rs1128503, rs2520464, rs3789243 CYP1A2 rs762551 |
Population PK analysis with genotypes as covariates on clearance parameter | [71] |
| Ahmad (2017) | 228 | NA | NA | Concentration measured during autopsy compared with 297 controls |
CYP2B6 *5, *9, *2, *8, *15 intronic (rs2279344, rs4803419, rs8192719) |
Univariate analysis between genotype and concentrations after multiple testing correction | [73] |
| Dobrinas (2013) | 276 | NA | NA | Trough samples at steady state | CYP2B6 sequencing of 3917 bp including all nine exons and exon–intron boundaries | Univariate analysis between identified genotype and concentrations after multiple testing correction | [74] |
| Yang (2016) | 344 + 76 | 281 + 59 | 63 + 17 | At steady state. Sampling schedule unknown. Genotyping in 344 patients to identify susceptibility loci. Additional 76 patients used for validation. |
Whole-genome pharmacogenomic study using Axiom Genome-Wide CHB 1 Array (Affymetrix, CA, usa) | Genome-wide singe-locus association analysis after multiple test correction and sliding-window haplotype-based association analysis with concentration | [75] |
| Wang (2013) | 366 | NA | NA | Samples at 24 ± 2 h after the last methadone dose at steady state | CYP2C19 *2, *3 | Univariate analysis between genotype and concentrations | [76] |
| Carlquist (2015) | 31 | NA | NA | Day 1 and day 21 peak and trough concentrations |
CYP2B6 *9 CYP3A4 *1b, *22 CYP2C19 *2 ABCB1 3435C>T NOS1AP (rs12143842) |
Univariate analysis between genotype and concentrations | [50] |
| Richards (2014) | 228 | NA | NA | Concentration measured during autopsy |
CYP3A4 *1B rs4987161, rs4986910 intronic (rs2246709, rs3735451, rs4646437 and rs2242480) |
Univariate analysis between genotype and concentrations | [77] |
| Eap (2001) | 256 | 202 | 54 | Plasma trough at steady state. 15 patients with bd dosing were removed from analysis | CYP2D6*3, *4, *5, *6, *xN | Univariate analysis between genotype and concentrations after multiple testing correction | [78] |
| Lee (2013) | 178 | 156 | 22 | Plasma trough at steady state | CYP2B6 *4, *9, and *5, CYP2C19 *2, *3, *17 and ABCB1 1236C>T, 2677G>T/A and 3435C>T | Univariate analysis between genotype and concentrations | [79] |
| Barratt (2012) | 119 | NA | NA | Plasma trough at steady state | ABCB1 61A>G, 1199G>A, 1236C>T and 3435C>T | Univariate analysis between genotype and concentrations without multiple testing correction | [27] |
| Zahari (2016) | 148 | 148 | NA | Trough, 0.5, 1, 2, 4, 8, 12 and 24 h after morning dose at steady state | ABCB1 1236C>T, 2677G>T/A and 3435C>T | Univariate analysis between genotype and concentrations without multiple testing correction | [80] |
| Uehlinger (2007) | 14 | NA | NA | Trough samples at steady state before and after >1 week of quetiapine |
CYP2B6 *4, *5, *6, *7, *9 CYP3A5 *3 CYP2D6 *3, *4, *5, *6, *xN ABCB1 3435C>T |
Univariate analysis between before and after concentrations in each of the genotypes | [85] |
As mentioned in the published literature.
AUC: Area under the curve; bp: Base pair; EDDP: 2-ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidine; NA: Not available; PK: Pharmacokinetic.
CYP2B6
CYP2B6 is the most important CYP enzyme involved in the metabolism of S-methadone and to an extent, R-methadone. The expression of CYP2B6 is highly variable among individuals (20–250-fold) [86] due to genetic polymorphisms and differences in transcriptional regulation [87]. Several polymorphisms and various linkages with multiple haplotypes are known for CY2B6 (https://www.pharmvar.org/htdocs/archive/cyp2b6.htm), most of which result in reduced enzyme activity. The most commonly implicated allele is the *6 haplotype which is a combination of *4 and *9 allelic variants. The frequency of *6 haplotype varies greatly among different ethnicities (10–21% in Asians, 14–27% in Caucasians, 33–50% in Africans and African–Americans and ~62% in Papua New Guineans) [88]. Crettol et al. demonstrated a relationship between the haplotype and concentrations of S-methadone. There was a 2.1- and 1.7-fold increase in trough and peak concentrations, respectively, of S-methadone in homozygous *6 carriers compared with the noncarriers [64,69]. Other studies have shown a decrease in clearance of 19 to 39% with the homozygous *6 allele [42,65,66,70,71]. This allele has also been implicated in the metabolism of R-methadone (albeit to a lesser extent), as a 1.3-fold increase in both trough and peak plasma concentrations were seen [64,69]. As noted previously, the homozygous *6 slow metabolizer variant, which decreases clearance of S-methadone, has been shown to increase risk of QT prolongation with methadone [89].
The effect of the CYP2B6*5 allele on methadone clearance is unclear. An increase in activity was suggested by Csajka et al. when this allele used as a part of the activity score for both R-and S-methadone metabolism [42]. Dobrinas et al. observed a high *5 allele frequency in the group with lower S-methadone concentrations, also suggesting an increase in CYP2B6 activity [74]. However, contradictory results were reported by Ahmad et al. in a study of methadone fatalities. The racemic methadone concentrations in the patients with homozygous *5 allele were found to be about 2.5-fold higher than the others, suggesting a lower clearance. This contradictory result could possibly be related to the study design, as patient samples were collected at autopsy [73].
The presence of CYP2B6*11, an infrequent allele with reduced activity has been used to predict interindividual variability in clearance as part of an activity score [42]. Also Dobrinas et al. observed that the frequency of this allele was higher in patients with high concentrations of methadone [74]. Other variants like rs2279344, rs8192719, rs1038376 and rs10500282 decrease S-methadone clearance, while rs10403955, rs2279345 and rs707265 increase S-methadone clearance [42,74]. In an extensive genome-wide association study using sliding-window haplotype-based association analysis, Yang et al. identified novel CYP2B6 haplotypes which could affect the clearance of methadone. The haplotypes TAATCG and TCCTTT (rs8100458, rs7250601, rs7250991, rs11882424, rs8192719 and rs10853744) were associated with lower concentrations of S-methadone [75].
Transcription of CYP2B6 is mainly regulated by two nuclear receptors – pregnane X receptor and constitutive androstane receptor – encoded by NR1I2 and NR1I3, respectively [61]. However, variants in these genes have not yet been explored in relation to methadone metabolism.
Methadone may induce QT prolongation in the absence of any known risk factors [15]. The CYP2B6 (*6/*6) slow metabolizer variant, associated with decreased clearance of S-methadone, has been shown to increase risk of QT prolongation related to methadone [89].
A recent study identified the importance of CYP2B6 LOF alleles, sex and BMI as determinants of methadone metabolism and suggested including sex, BMI and CYP2B6 genotype as predictors in multivariate models to create methadone-dosing algorithms [90]. It is also crucial to include African-origin individuals in genetic studies as some CYP2B6 alleles are race specific [91].
CYP2C
CYP2C19 is involved mainly in the metabolism of R-methadone. Lan et al. demonstrated that the CYP2C19 *2, *29, *30, *31, *32 and *33 variants were associated with decreased in vitro clearance of racemic methadone. The *3 and *35 alleles show no detectable enzyme activity [92]. The *2 and *3 alleles are known to occur frequently. The *2 allele is found in approximately 18, 12, 28 and 61% in African, Caucasian, Asian and Oceanian populations, respectively. The *3 allele is mainly found in Asians and Oceanians, with a frequency of about 7 and 15%, respectively. The *17 allele, found commonly in the world population (10–20%), is known to increase activity of CYP2C19 in metabolism of most drugs. No specific information is known about this allele related to methadone.
Depending on the combination of alleles in an individual, the phenotype is classified as poor metabolizer (PM; homozygous for reduced-function variant allele), intermediate metabolizer (IM; heterozygous for nonfunctional allele with a normal function allele) and extensive or normal metabolizer (EM/NM; homozygous wild-type alleles). In one of the clinical studies with *2 and *3 alleles genotyping, Wang et al. showed a significant increase in dose adjusted trough R-methadone concentration from 3.62 ng/ml (EM) to 4.31 ng/ml (IM) to 4.36 ng/ml (PM) [76]. Similar results were observed in a smaller study where the total methadone concentration was found to be significantly increased with *2 and *3 genotypes [67]. Conflicting results were observed in a study by Carlquist et al. where both the metabolites R- and S-EDDP concentrations were significantly increased with these polymorphisms. However, this study did not find any association of CYP2C19 polymorphisms with the drug concentrations themselves [50].
CYP2C9 *2 and *3 are found at a frequency of 8–14 and 4–16%, respectively, in the general population and have a small role in the metabolism of methadone. These alleles have been reported to increase the dose-corrected concentration of racemic methadone by about 30% compared with the homozygous wild-type *1 [67]. The metabolic activity of CYP2C8 and CYP2C18 though documented in in vitro studies have not been studied in clinical studies with genotype associations.
CYP3A
CYP3A4 is the most abundantly expressed CYP enzyme that is involved in the metabolism of methadone. Although it has many variants, very few are clinically significant. The most commonly studied alleles are *1b, *2, *3 and *22 [93]. A population genetic-based pharmacokinetic modeling study by Csajka et al. showed that *22 allele reduced the clearance of R-methadone by 23% [42]. Crettol et al. observed a trend toward increased peak and trough concentrations of S-methadone in patients having the *1b allele, although statistical significance could not be demonstrated [64]. The intronic rs2242480 and *1b alleles were found more commonly in fatal methadone toxicity cases compared with other groups of fatal toxicity [77].
CYP3A5*3 is the most common nonfunctional variant of CYP3A5, with a frequency of 85% in Caucasians and 65% in Asians. It is usually found in linkage with CYP3A4*1b. In homozygous *3 carriers, the dose corrected trough racemic methadone concentration was 13.7 nmol/l/mg compared with 9.5 nmol/l/mg in others (p = 0.009) [67]. This association needs further study to accumulate more evidence before considering it as an important predictor of methadone clearance in vivo.
CYP2D6
CYP2D6 demonstrates a complex genetic variation with gene duplications, tandem arrangements, gene deletion and extensive regular allelic variants. This complexity makes the prediction of phenotype difficult. Duplications result in increased expression and contribute to an ultrarapid metabolizer (UM) phenotype (Table 1). The *1 and *2 alleles demonstrate normal activity while the *10 allele accounts for the majority of reduced activity. The CYP2D6 *3, *4, *5 and *6 alleles account for most of the null activity alleles. The alleles that most commonly influence clearance of drugs are CYP2D6*2, *3, *4, *5, *10, *17 and *41 [94]. Crettol et al. showed that the EM/IM group had significantly lower trough concentrations of both R- and S-methadone compared with the UM group (0.8- and 0.7-fold, respectively). This difference in concentration was not seen between EM/IM and the PM groups [64]. Eap et al. demonstrated that the dose-adjusted trough concentrations of both R- and S-methadone increased from UM to PM, with an increase of 46 and 32% for R- and S-methadone, respectively, from UM to PM [78].
ABCB1
Polymorphisms in P-glycoprotein possibly affect the hepatic and renal clearance of methadone, as there are studies describing the role of P-glycoprotein in intestine and brain in transporting methadone. The most frequently studied SNPs in ABCB1 are rs1128503 (1236C1), rs2032582 (2677G>T/A) and rs1045642 (3435C>T). Csajka et al. reported that the 3435C>T polymorphism decreased the clearance of S-methadone by 17.8% and used it as a covariate to predict the clearance of S-methadone along with CYP2B6*6 [42]. By univariate analysis, the 3435C>T genotype was associated with increased trough methadone concentrations (but not peak concentrations of either R- or S-methadone) [64]. The 2677G>T/A genotype is a triallelic polymorphism, where A is a rare variant. The 2677G>T polymorphism decreases trough concentrations of R- and S-methadone [64,71]. The infrequent allele rs9282564 (61A>G, 0.2–8% prevalence) is known to increase the Vmax of P-glycoprotein in vitro. It decreased the trough concentration of R-methadone by 0.7-fold and S-methadone by 0.6-fold [64]. Csajksa et al. also observed that 61A > G (rs9282564) increased the clearance of both R- and S-methadone [42].
The relationship of P-glycoprotein diplotypes to trough methadone concentrations was evaluated by Zahari et al. [80] They reported that the CGC/TTT diplotype (1236C>T, 2677G>T/A and 3435C>T) was associated with a greater dose-adjusted serum methadone concentration. Another haplotype AGCTT (61A>G, 1199G>A, 1236C>T, 2677G>T/A and 3435C>T) was found to be associated with lower trough concentration either in its homozygous or heterozygous form by Barratt et al. [27].
POR
Cytochrome POR is the sole electron donor in the oxidoreductive metabolism of all substrates by the cytochrome enzymes. Polymorphisms of POR have been reported to decrease the activity of the CYP enzymes. The POR genotype enhances the prediction of activity of CYP2B6 and CYP3A4 [95]. The POR*28 allele was a covariate to predict clearance along with other factors for both R- and S-methadone [42].
Special considerations in children
The pharmacokinetics of children (including neonates and infants) surprisingly does not differ much from adults both for racemic methadone and R- and S-methadone [96,97]. Ward et al. showed that clearance could be predicted allometrically in neonates without any influence of age (although age did affect the volume of distribution) [96]. In a study by Horst et al., the dose corrected area under the curve (AUC) and other pharmacokinetic parameters between adolescents (aged 15 ± 2 years) and adults (aged 25 ± 6 years) were not different [98]. Similar results have been reported in other studies [96,99]. The CYP3A4, CYP2B6 and CYP2D6 enzymes are immature at birth; therefore, it is possible that CYP3A7 abundantly expressed in neonates could be involved in the clearance of methadone in this population [96]. Pharmacogenomic studies of methadone in children and adolescents are generally lacking and may involve genes coding different key metabolizing CYP450 enzymes.
Conclusion & future perspective
Methadone is a remarkable opioid analgesic in clinical practice with large interindividual variations in response. Individualizing methadone therapy to patients based on underlying genetic factors would improve its efficacy while mitigating adverse effects. There is an abundance of literature related to specific aspects of methadone pharmacogenomics; however, evidence remains weak till date to guide clinical therapy according to genotype. Well-powered clinical studies are needed with population pharmacokinetic/pharmacodynamic modeling, analyzing the effect of methadone treatment on pharmacodynamic markers like pain, respiratory depression and QT prolongation in relation to various genetic variants. Genome-wide association studies in a large population with robust clinical phenotypes are needed to identify novel genetic variants regulating pain, analgesic activity, QT prolongation and pharmacokinetics of methadone to gain insight into unexplained variability in the drug’s activity and targeted dosing. Epigenetic changes may also alter methadone activity during chronic treatment and could be an important factor contributing to the interindividual variability. The CYP enzymes involved in the metabolism of methadone in neonates and children are probably different from that of adults; therefore, pediatric patients may have unique clinically relevant genotypes that warrant further investigation.
Executive summary.
Methadone is a MOR agonist and N-methyl-D-aspartate receptor antagonist.
Racemic methadone used in clinical practice comprises the R- and S-enantiomers which have distinct pharmacodynamic and pharmacokinetic properties. R-methadone is a MOR agonist, with greater receptor affinity compared with S-methadone and is responsible for most of the opioid-receptor related analgesic as well as adverse effects. S-methadone has inhibitory action on serotonin and norepinephrine reuptake.
OPRM1 118A>G variant (G-allele) has been largely associated with greater opioid dose requirements. Though some studies have replicated this finding with methadone, it remains largely inconclusive.
Variants in ANKK1–DRD2, BDNF, NGFB, NTRK1 and 2, OPRD1, and ARRB2 have also shown promise in relation to methadone pharmacodynamics.
No strong single gene associations have been established, although multiple genetic variants (e.g., OPRM1, CYP2B6, ANKK1 and DRD2) together have been found to strongly influence response to methadone and dose requirements.
The factors which reliably contribute to interindividual variability in pharmacokinetics are the genotypes of CYP enzymes and P-glycoprotein.
-
The major CYP enzymes involved in the metabolism of methadone are CYP2B6, CYP2C19 and CYP3A4:
The CYP2B6*6 haplotype (*4 and *9 alleles) is a reliable predictor of the clearance of the S-enantiomer and somewhat for the R-enantiomer;
The CYP2C19 phenotype based on the *2 and *3 allelic variants could be a predictor of the clearance of the R-enantiomer and needs to be studied more;
CYP3A4 was found to be the most important CYP in the metabolism of methadone in vitro; however, drug interaction studies in humans showed conflicting results. Pharmacokinetic studies demonstrate that CYP3A4*22 and *1b allelic variants have a role in the prediction of clearance of R- and S-methadone;
CYP2D6 activity, as predicted by its complex genotypic variants, is associated with the trough concentrations of both R- and S-methadone. More evidence is required to confirm these findings;
CYP2C9 and CYP3A5 contribute to the metabolism of methadone to a lesser extent; limited genotype information is available.
P-glycoprotein polymorphisms like rs1128503, rs2032582, rs1045642 and rs9282564 have been found to predict the clearance of methadone in few pharmacokinetic studies and needs to be studied further.
Other variants coding for the α-1 acid glycoprotein/orosomucoid, pregnane X receptor, constitutive androstane receptor, cytochrome P450 oxidoreductase should be further studied in relation to pharmacokinetics of methadone.
Footnotes
Author contributions
All authors contributed to the drafting of the work or revising it critically for content. All authors have reviewed and approved of this manuscript for submission. All authors agree to be accountable for all aspects of the work.
Financial & competing interests disclosure
Research reported in this publication was supported by the Eunice Kennedy Shriver National Institute of Child Health & Human Development of the National Institutes of Health under award no. R01HD089458 (PI: S Sadhasivam), R21HD094311 (PI: S Sadhasivam) and R01HD096800 (PI: S Sadhasivam). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The departments of Anesthesia and Clinical Pharmacology at Indiana University School of Medicine (IUSM) provided salary support for the authors. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest
- 1.Gottschalk A, Durieux ME, Nemergut EC. Intraoperative methadone improves postoperative pain control in patients undergoing complex spine surgery. Anesth. Analg. 112(1), 218–223 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Murphy GS, Szokol JW, Avram MJ. et al. Clinical effectiveness and safety of intraoperative methadone in patients undergoing posterior spinal fusion surgery: a randomized, double-blinded, controlled trial. Anesthesiology 126(5), 822–833 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Codd EE, Shank RP, Schupsky JJ, Raffa RB. Serotonin and norepinephrine uptake inhibiting activity of centrally acting analgesics: structural determinants and role in antinociception. J. Pharmacol. Exp. Ther. 274(3), 1263–1270 (1995). [PubMed] [Google Scholar]
- 4.Leppert W. The role of methadone in cancer pain treatment – a review. Int. J. Clin. Pract. 63(7), 1095–1109 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Barbosa Neto JO, Garcia MA, Garcia JBS. Revisiting methadone: pharmacokinetics, pharmacodynamics and clinical indication. Revista Dor. 16, 60–66 (2015). [Google Scholar]
- 6.Davis MP, Walsh D. Methadone for relief of cancer pain: a review of pharmacokinetics, pharmacodynamics, drug interactions and protocols of administration. Support. Care Cancer 9(2), 73–83 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Murphy GS, Avram MJ, Greenberg SB. et al. Postoperative pain and analgesic requirements in the first year after intraoperative methadone for complex spine and cardiac surgery. Anesthesiology 132(2), 330–342 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Murphy GS, Szokol JW. Intraoperative methadone in surgical patients: a review of clinical investigations. Anesthesiology 131(3), 678–692 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Liu JG, Liao XP, Gong ZH, Qin BY. The difference between methadone and morphine in regulation of delta-opioid receptors underlies the antagonistic effect of methadone on morphine-mediated cellular actions. Eur. J. Pharmacol. 373(2–3), 233–239 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Liu JG, Liao XP, Gong ZH, Qin BY. Methadone-induced desensitization of the delta-opioid receptor is mediated by uncoupling of receptor from G protein. Eur. J. Pharmacol. 374(2), 301–308 (1999). [DOI] [PubMed] [Google Scholar]
- 11.Callahan RJ, Au JD, Paul M, Liu C, Yost CS. Functional inhibition by methadone of N-methyl-D-aspartate receptors expressed in xenopus oocytes: stereospecific and subunit effects. Anesth. Analg. 98(3), 653–659 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Gorman AL, Elliott KJ, Inturrisi CE. The d- and l-isomers of methadone bind to the non-competitive site on the N-methyl-D-aspartate (NMDA) receptor in rat forebrain and spinal cord. Neurosci. Lett. 223(1), 5–8 (1997). [DOI] [PubMed] [Google Scholar]
- 13.Silverman SM. Opioid induced hyperalgesia: clinical implications for the pain practitioner. Pain Physician. 12(3), 679–684 (2009). [PubMed] [Google Scholar]
- 14.Davis AM, Inturrisi CE. D-methadone blocks morphine tolerance and N-methyl-D-aspartate-induced hyperalgesia. J. Pharmacol. Exp. Ther. 289(2), 1048–1053 (1999). [PubMed] [Google Scholar]
- 15.Mujtaba S, Romero J, Taub CC. Methadone, QTc prolongation and torsades de pointes: current concepts, management and a hidden twist in the tale? J. Cardiovasc. Dis. Res. 4(4), 229–35 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treece J, Al Madani M, El Khoury G. et al. Comprehensive review on methadone-induced QT prolongation and torsades. J. Pharmacol. Pharmacothera. 9(2), 66–75 (2018). [Google Scholar]
- 17.Katchman AN, McGroary KA, Kilborn MJ. et al. Influence of opioid agonists on cardiac human ether-a-go-go-related gene K+ currents. J. Pharmacol. Exp. Ther. 303(2), 688–694 (2002). [DOI] [PubMed] [Google Scholar]
- 18.Gupta A, Lawrence AT, Krishnan K, Kavinsky CJ, Trohman RG. Current concepts in the mechanisms and management of drug-induced QT prolongation and torsade de pointes. Am. Heart J. 153(6), 891–899 (2007). [DOI] [PubMed] [Google Scholar]
- 19.George S, Moreira K, Fapohunda M. Methadone and the heart: what the clinician needs to know. Curr. Drug Abuse Rev. 1(3), 297–302 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Čolović MB, Krstić DZ, Lazarević-Pašti TD, Bondžić AM, Vasić VM. Acetylcholinesterase inhibitors: pharmacology and toxicology. Curr. Neuropharmacol. 11, 315–335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seyler DE, Borowitz JL, Maickel RP. Calcium channel blockade by certain opioids. Fundam. Appl. Toxicol. 3(6), 536–542 (1983). [DOI] [PubMed] [Google Scholar]
- 22.Levran O, Peles E, Randesi M. et al. Association of genetic variation in pharmacodynamic factors with methadone dose required for effective treatment of opioid addiction. Pharmacogenomics 14(7), 755–768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walter C, Lotsch J. Meta-analysis of the relevance of the OPRM1 118A>G genetic variant for pain treatment. Pain 146(3), 270–275 (2009). [DOI] [PubMed] [Google Scholar]
- 24.Lotsch J, Skarke C, Wieting J, J et al. Modulation of the central nervous effects of levomethadone by genetic polymorphisms potentially affecting its metabolism, distribution, and drug action. Clin. Pharmacol. Ther. 79(1), 72–89 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Oueslati B, Moula O, Ghachem R. The impact of OPRM1‘s genetic polymorphisms on methadone maintenance treatment in opioid addicts: a systematic review. Pharmacogenomics 19(8), 741–747 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Crettol S, Besson J, Croquette-Krokar M. et al. Association of dopamine and opioid receptor genetic polymorphisms with response to methadone maintenance treatment. Prog. Neuropsychopharmacol. Biol. Psychiatry 32(7), 1722–1727 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Barratt DT, Coller JK, Hallinan R. et al. ABCB1 haplotype and OPRM1 118A>G genotype interaction in methadone maintenance treatment pharmacogenetics. Pharmgenomics Pers. Med. 5, 53–62 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hung CC, Chiou MH, Huang BH. et al. Impact of genetic polymorphisms in ABCB1, CYP2B6, OPRM1, ANKK1 and DRD2 genes on methadone therapy in Han Chinese patients. Pharmacogenomics 12(11), 1525–1533 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Bunten H, Liang WJ, Pounder DJ, Seneviratne C, Osselton D. OPRM1 and CYP2B6 gene variants as risk factors in methadone-related deaths. Clin. Pharmacol. Ther. 88(3), 383–389 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Wang SC, Tsou HH, Chen CH. et al. Genetic polymorphisms in the opioid receptor mu1 gene are associated with changes in libido and insomnia in methadone maintenance patients. Eur. Neuropsychopharmacol. 22(10), 695–703 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Luo R, Li X, Qin S. et al. Impact of SNP–SNP interaction among ABCB1, ARRB2, DRD1 and OPRD1 on methadone dosage requirement in Han Chinese patients. Pharmacogenomics 18(18), 1659–1670 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Doehring A, Hentig N, Graff J. et al. Genetic variants altering dopamine D2 receptor expression or function modulate the risk of opiate addiction and the dosage requirements of methadone substitution. Pharmacogenet. Genomics 19(6), 407–414 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Duan LX, Li XL, Hu PW, Luo R, Luo X, Chen YY. Association between DRD2 gene polymorphisms and the dosage used on methadone maintenance treatment program. Zhonghua Liu Xing Bing Xue Za Zhi. 39(2), 194–198 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Levran O, Peles E, Hamon S. et al. Nerve growth factor beta polypeptide (NGFB) genetic variability: association with the methadone dose required for effective maintenance treatment. Pharmacogenomics J. 12(4), 319–327 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Cid R, Fonseca F, Gratacos M. et al. BDNF variability in opioid addicts and response to methadone treatment: preliminary findings. Genes Brain Behav. 7(5), 515–522 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Kristensen K, Blemmer T, Angelo HR. et al. Stereoselective pharmacokinetics of methadone in chronic pain patients. Ther. Drug Monit. 18(3), 221–227 (1996). [DOI] [PubMed] [Google Scholar]
- 37.Eap CB, Buclin T, Baumann P. Interindividual variability of the clinical pharmacokinetics of methadone. Clin. Pharmacokinet. 41(14), 1153–1193 (2002). [DOI] [PubMed] [Google Scholar]
- 38.Ayonrinde OT, Bridge DT. The rediscovery of methadone for cancer pain management. Med. J. Australia 173(10), 536–540 (2000). [PubMed] [Google Scholar]
- 39.Weschules DJ, Bain KT, Richeimer S. Actual and potential drug interactions associated with methadone. Pain Med. 9(3), 315–344 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Wolff K, Hay AW, Raistrick D, Calvert R. Steady-state pharmacokinetics of methadone in opioid addicts. Eur. J. Clin. Pharmacol. 44(2), 189–194 (1993). [DOI] [PubMed] [Google Scholar]
- 41.Kharasch ED, Hoffer C, Whittington D, Walker A, Bedynek PS. Methadone pharmacokinetics are independent of cytochrome P4503A (CYP3A) activity and gastrointestinal drug transport: insights from methadone interactions with ritonavir/indinavir. Anesthesiology 110(3), 660–672 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Csajka C, Crettol S, Guidi M, Eap CB. Population genetic-based pharmacokinetic modeling of methadone and its relationship with the QTc interval in opioid-dependent patients. Clin. Pharmacokinet. 55(12), 1521–1533 (2016). [DOI] [PubMed] [Google Scholar]; • A population pharmacokinetic modeling study of methadone which had included the genotypes of most of the relevant factors including CYP2B6, CYP2C9, CYP2C19, CYP2D6, CYP3A4, CYP3A5, CYP3A7, ABCB1 and POR involved in the clearance of R- and S-methadone individually. This is the most comprehensive approach to date.
- 43.Foster DJ, Somogyi AA, White JM, Bochner F. Population pharmacokinetics of (R)-, (S)- and rac-methadone in methadone maintenance patients. Br. J. Clin. Pharmacol. 57(6), 742–755 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Toombs JD, Kral LA. Methadone treatment for pain states. Am. Fam. Physician 71(7), 1353–1358 (2005). [PubMed] [Google Scholar]
- 45.Romach MK, Piafsky KM, Abel JG, Khouw V, Sellers EM. Methadone binding to orosomucoid (alpha 1-acid glycoprotein): determinant of free fraction in plasma. Clin. Pharmacol. Ther. 29(2), 211–217 (1981). [DOI] [PubMed] [Google Scholar]
- 46.Eap CB, Cuendet C, Baumann P. Binding of d-methadone, l-methadone, and dl-methadone to proteins in plasma of healthy volunteers: role of the variants of alpha 1-acid glycoprotein. Clin. Pharmacol. Ther. 47(3), 338–346 (1990). [DOI] [PubMed] [Google Scholar]
- 47.Yost RL, de Vane CL. Diurnal variation of alpha-acid glycoprotein concentration in normal volunteers. J. Pharm. Sci. 74(7), 777–779 (1985). [DOI] [PubMed] [Google Scholar]
- 48.Israili ZH, Dayton PG. Human alpha-1-glycoprotein and its interactions with drugs. Drug Metab. Rev. 33(2), 161–235 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Brodin K, Lybeck A, Beck O. et al. 175 monitoring of methadone dose intake: importance of urinary pH. Ther. Drug Monit. 19(5), 591 (1997). [Google Scholar]
- 50.Carlquist JF, Moody DE, Knight S. et al. A possible mechanistic link between the CYP2C19 genotype, the methadone metabolite ethylidene-1,5-dimethyl-3,3-diphenylpyrrolidene (EDDP), and methadone-induced corrected QT interval prolongation in a pilot study. Mol. Diagn. Ther. 19(2), 131–138 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Iribarne C, Berthou F, Baird S. et al. Involvement of cytochrome P450 3A4 enzyme in the N-demethylation of methadone in human liver microsomes. Chem. Res. Toxicol. 9(2), 365–373 (1996). [DOI] [PubMed] [Google Scholar]
- 52.Foster DJ, Somogyi AA, Bochner F. Methadone N-demethylation in human liver microsomes: lack of stereoselectivity and involvement of CYP3A4. Br. J. Clin. Pharmacol. 47(4), 403–412 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gerber JG, Rhodes RJ, Gal J. Stereoselective metabolism of methadone N-demethylation by cytochrome P4502B6 and 2C19. Chirality 16(1), 36–44 (2004). [DOI] [PubMed] [Google Scholar]; • Landmark paper which first claimed the importance of CYP2B6 in the metabolism of methadone. This changed the direction of research in this field which was previously mainly directed at CYP3A4 only.
- 54.Chang Y, Fang WB, Lin SN, Moody DE. Stereo-selective metabolism of methadone by human liver microsomes and cDNA-expressed cytochrome P450s: a reconciliation. Basic Clin. Pharmacol. Toxicol. 108(1), 55–62 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This study expanded on the Gerber et al.'s work, with relative abundance of hepatic enzymes introduced which was a more realistic representation.
- 55.Kharasch ED, Bedynek PS, Park S, Whittington D, Walker A, Hoffer C. Mechanism of ritonavir changes in methadone pharmacokinetics and pharmacodynamics: I. Evidence against CYP3A mediation of methadone clearance. Clin. Pharmacol. Ther. 84(4), 497–505 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]; • This was the first of many human drug interaction studies done by the group which proved the importance of CYP2B6 in the metabolism of methadone over CYP3A4.
- 56.Kharasch ED, Whittington D, Ensign D. et al. Mechanism of efavirenz influence on methadone pharmacokinetics and pharmacodynamics. Clin. Pharmacol. Ther. 91(4), 673–684 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kharasch ED, Bedynek PS, Hoffer C, Walker A, Whittington D. Lack of indinavir effects on methadone disposition despite inhibition of hepatic and intestinal cytochrome P4503A (CYP3A). Anesthesiology 116(2), 432–447 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kharasch ED, Walker A, Whittington D, Hoffer C, Bedynek PS. Methadone metabolism and clearance are induced by nelfinavir despite inhibition of cytochrome P4503A (CYP3A) activity. Drug Alcohol Depend. 101(3), 158–168 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Totah RA, Sheffels P, Roberts T, Whittington D, Thummel K, Kharasch ED. Role of CYP2B6 in stereoselective human methadone metabolism. Anesthesiology 108(3), 363–374 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Totah RA, Allen KE, Sheffels P, Whittington D, Kharasch ED. Enantiomeric metabolic interactions and stereoselective human methadone metabolism. J. Pharmacol. Exp. Ther. 321(1), 389–399 (2007). [DOI] [PubMed] [Google Scholar]
- 61.Tolson AH, Li H, Eddington ND, Wang H. Methadone induces the expression of hepatic drug-metabolizing enzymes through the activation of pregnane X receptor and constitutive androstane receptor. Drug Metab. Dispos. 37(9), 1887–1894 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Campbell SD, Crafford A, Williamson BL, Kharasch ED. Mechanism of autoinduction of methadone N-demethylation in human hepatocytes. Anesth. Analg. 117(1), 52–60 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Kantelip JP, Gerritsen-van Schieveen P, Davani S. Interindividual variability of methadone response: impact of genetic polymorphism. Mol. Diagn. Ther. 12(2), 109–124 (2008). [DOI] [PubMed] [Google Scholar]
- 64.Crettol S, Déglon J-J, Besson J. et al. ABCB1 and cytochrome P450 genotypes and phenotypes: influence on methadone plasma levels and response to treatment. Clin. Pharmacol. Ther. 80(6), 668–681 (2006). [DOI] [PubMed] [Google Scholar]; • This pharmacokinetic study of methadone evaluates the effect of genotypes of all relevant CYP enzymes and P-glycoprotein on plasma trough and peak levels of R- and S-methadone.
- 65.Dennis BB, Bawor M, Thabane L, Sohani Z, Samaan Z. Impact of ABCB1 and CYP2B6 genetic polymorphisms on methadone metabolism, dose and treatment response in patients with opioid addiction: a systematic review and meta-analysis. PLoS ONE 9(1), e86114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kharasch ED, Regina KJ, Blood J, Friedel C. Methadone pharmacogenetics: CYP2B6 polymorphisms determine plasma concentrations, clearance, and metabolism. Anesthesiology 123(5), 1142–1153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kringen MK, Chalabianloo F, Bernard J-P, Bramness JG, Molden E, Høiseth G. Combined effect of CYP2B6 genotype and other candidate genes on a steady-state serum concentration of methadone in opioid maintenance treatment. Ther. Drug Monit. 39(5), 550–555 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Bunten H, Liang W-J, Pounder D, Seneviratne C, Osselton MD. CYP2B6 and OPRM1 gene variations predict methadone-related deaths. Addiction Biology. 16(1), 142–144 (2011). [DOI] [PubMed] [Google Scholar]
- 69.Crettol S, Déglon J-J, Besson J. et al. Methadone enantiomer plasma levels, CYP2B6, CYP2C19, and CYP2C9 genotypes, and response to treatment. Clin. Pharmacol. Ther. 78(6), 593–604 (2005). [DOI] [PubMed] [Google Scholar]
- 70.Wang SC, Ho IK, Tsou HH. et al. CYP2B6 polymorphisms influence the plasma concentration and clearance of the methadone S-enantiomer. J. Clin. Psychopharmacol. 31(4), 463–469 (2011). [DOI] [PubMed] [Google Scholar]; • The most comprehensive study of the genotypes of CYP2B6 on the clearance of S-methadone specifically using a gene sequencing-resequensing approach.
- 71.Bart G, Lenz S, Straka RJ, Brundage RC. Ethnic and genetic factors in methadone pharmacokinetics: a population pharmacokinetic study. Drug Alcohol Depend. 145, 185–193 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]; • A comprehensive population pharmacokinetic modeling study of methadone, which included the genotypes of ABCB1, CYP3A4, CYP2B6, CYP2D6, CYP2C19 and CYP1A2 as covariates in the clearance of R- and S-methadone.
- 72.Fonseca Casals F, Fornell T, Díaz L. et al. Contribution of cytochrome P450 and ABCB1 genetic variability on methadone pharmacokinetics, dose requirements, and response. PLoS ONE 6(5), e19527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ahmad T, Sabet S, Primerano DA, Richards-Waugh LL, Rankin GO. Tell-tale SNPs: the role of CYP2B6 in methadone fatalities. J. Anal. Toxicol. 41(4), 325–333 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dobrinas M, Crettol S, Oneda B. et al. Contribution of CYP2B6 alleles in explaining extreme (S)-methadone plasma levels: a CYP2B6 gene resequencing study. Pharmacogenet. Genomics 23(2), 84–93 (2013). [DOI] [PubMed] [Google Scholar]
- 75.Yang HC, Chu SK, Huang CL. et al. Genome-wide pharmacogenomic study on methadone maintenance treatment identifies SNP rs17180299 and multiple haplotypes on CYP2B6, SPON1, and GSG1L associated with plasma concentrations of methadone R- and S-enantiomers in heroin-dependent patients. PLoS Genet. 12(3), e1005910 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang SC, Ho IK, Tsou HH. et al. Functional genetic polymorphisms in CYP2C19 gene in relation to cardiac side effects and treatment dose in a methadone maintenance cohort. Omics 17(10), 519–526 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Richards-Waugh LL, Primerano DA, Dementieva Y, Kraner JC, Rankin GO. Fatal methadone toxicity: potential role of CYP3A4 genetic polymorphism. J. Anal. Toxicol. 38(8), 541–547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Eap CB, Broly F, Mino A. et al. Cytochrome P450 2D6 genotype and methadone steady-state concentrations. J. Clin. Psychopharmacol. 21(2), 229–234 (2001). [DOI] [PubMed] [Google Scholar]
- 79.Lee HY, Li JH, Sheu YL. et al. Moving toward personalized medicine in the methadone maintenance treatment program: a pilot study on the evaluation of treatment responses in Taiwan. Biomed. Res. Int. 2013, 741403 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zahari Z, Lee CS, Ibrahim MA. et al. Relationship between ABCB1 polymorphisms and serum methadone concentration in patients undergoing methadone maintenance therapy (MMT). Am. J. Drug Alcohol Abuse 42(5), 587–596 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Oda Y, Kharasch ED. Metabolism of methadone and levo-alpha-acetylmethadol (LAAM) by human intestinal cytochrome P450 3A4 (CYP3A4): potential contribution of intestinal metabolism to presystemic clearance and bioactivation. J. Pharmacol. Exp. Ther. 298(3), 1021–1032 (2001). [PubMed] [Google Scholar]
- 82.Kharasch ED, Hoffer C, Whittington D. The effect of quinidine, used as a probe for the involvement of P-glycoprotein, on the intestinal absorption and pharmacodynamics of methadone. Br. J. Clin. Pharmacol. 57(5), 600–610 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Meissner K, Blood J, Francis AM, Yermolenka V, Kharasch ED. Cyclosporine-inhibitable cerebral drug transport does not influence clinical methadone pharmacodynamics. Anesthesiology 121(6), 1281–1291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Boulton DW, Arnaud P, DeVane CL. Pharmacokinetics and pharmacodynamics of methadone enantiomers after a single oral dose of racemate. Clin. Pharmacol. Ther. 70(1), 48–57 (2001). [DOI] [PubMed] [Google Scholar]
- 85.Uehlinger C, Crettol S, Chassot P. et al. Increased (R)-methadone plasma concentrations by quetiapine in cytochrome P450s and ABCB1 genotyped patients. J. Clin. Psychopharmacol. 27(3), 273–278 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Thorn CF, Lamba JK, Lamba V, Klein TE, Altman RB. PharmGKB summary: very important pharmacogene information for CYP2B6. Pharmacogenet. Genomics 20(8), 520–523 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lamba V, Lamba J, Yasuda K. et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J. Pharmacol. Exp. Ther. 307(3), 906–922 (2003). [DOI] [PubMed] [Google Scholar]
- 88.Zanger UM, Klein K. Pharmacogenetics of cytochrome P450 2B6 (CYP2B6): advances on polymorphisms, mechanisms, and clinical relevance. Front. Genet. 4, 24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eap CB, Crettol S, Rougier JS. et al. Stereoselective block of hERG channel by (S)-methadone and QT interval prolongation in CYP2B6 slow metabolizers. Clin. Pharmacol. Ther. 81(5), 719–728 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Talal AH, Ding Y, Venuto CS. et al. Toward precision prescribing for methadone: determinants of methadone deposition. PLoS ONE 15(4), e0231467 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Y, Ingelman-Sundberg M, Lauschke VM. Worldwide distribution of cytochrome P450 alleles: a meta-analysis of population-scale sequencing projects. Clin. Pharmacol. Ther. 102(4), 688–700 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lan T, Yuan L-J, Hu X-X. et al. Effects of CYP2C19 variants on methadone metabolism in vitro. Drug Test. Anal. 9(4), 634–639 (2017). [DOI] [PubMed] [Google Scholar]
- 93.Klein K, Zanger UM. Pharmacogenomics of cytochrome P450 3A4: recent progress toward the “missing heritability” problem. Front. Genet. 4, 12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: part I. Clin. Pharmacokinet. 48(11), 689–723 (2009). [DOI] [PubMed] [Google Scholar]
- 95.Oneda B, Crettol S, Jaquenoud Sirot E, Bochud M, Ansermot N, Eap CB. The P450 oxidoreductase genotype is associated with CYP3A activity in vivo as measured by the midazolam phenotyping test. Pharmacogenet. Genomics 19(11), 877–883 (2009). [DOI] [PubMed] [Google Scholar]
- 96.Ward RM, Drover DR, Hammer GB. et al. The pharmacokinetics of methadone and its metabolites in neonates, infants, and children. Paediatr. Anaesth. 24(6), 591–601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stemland CJ, Witte J, Colquhoun DA. et al. The pharmacokinetics of methadone in adolescents undergoing posterior spinal fusion. Paediatr. Anaesth. 23(1), 51–57 (2013). [DOI] [PubMed] [Google Scholar]
- 98.Horst J, Frei-Jones M, Deych E, Shannon W, Kharasch ED. Pharmacokinetics and analgesic effects of methadone in children and adults with sickle cell disease. Pediatr. Blood Cancer 63(12), 2123–2130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Sharma A, Tallchief D, Blood J, Kim T, London A, Kharasch ED. Perioperative pharmacokinetics of methadone in adolescents. Anesthesiology 115(6), 1153–1161 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]


