Abstract
Resistance has developed in Plasmodium malaria parasites to every antimalarial drug in clinical use, prompting the need to characterize the pathways mediating resistance. Here, we report a framework for assessing development of resistance of Plasmodium falciparum to new antimalarial therapeutics. We investigated development of resistance by P. falciparum to the dihydroorotate dehydrogenase (DHODH) inhibitors DSM265 and DSM267 in tissue culture and in a mouse model of P. falciparum infection. We found that resistance to these drugs arose rapidly both in vitro and in vivo. We identified 13 point mutations mediating resistance in the parasite DHODH in vitro that overlapped with the DHODH mutations that arose in the mouse infection model. Mutations in DHODH conferred increased resistance (ranging from 2- to ~400-fold) to DHODH inhibitors in P. falciparum in vitro and in vivo. We further demonstrated that the drug-resistant parasites carrying the C276Y mutation had mitochondrial energetics comparable to the wild-type parasite and also retained their fitness in competitive growth experiments. Our data suggest that in vitro selection of drug-resistant P. falciparum can predict development of resistance in a mouse model of malaria infection.
INTRODUCTION
One of the biggest threats to malaria eradication efforts is the possibility of widespread resistance to current antimalarial drugs. Resistance to current frontline artemisinin-based therapies has been detected in several countries in the Greater Mekong subregion of Southeast Asia. Artemisinin resistance is defined as delayed parasite clearance in individuals infected with the malaria parasite Plasmodium falciparum (1, 2), requiring extended treatment periods to maintain drug efficacy (3, 4). With no alternative therapeutic options currently available, there is an urgent need to develop new antimalarial drugs that target different aspects of parasite biology.
Unfortunately, even for drugs that have never been introduced to parasite populations, resistance can emerge and spread rapidly, limiting their useful lifetime. For example, resistance to the dihydrofolate reductase inhibitor pyrimethamine emerged shortly after clinical introduction (5-7). Even when pyrimethamine was later combined with sulfadoxine, parasites resistant to the sulfadoxine-pyrimethamine combination were identified less than 1 year after its adoption as a frontline therapy (8). Because of this, use of the sulfadoxine-pyrimethamine combination is primarily limited to intermittent preventive treatment during pregnancy [reviewed in (9, 10)]. Resistance to the cytochrome b inhibitor atovaquone was detected in clinical trials before the drug was widely in use (11). This rapid emergence of drug resistance is thought to be due to selection of de novo mutations in malaria parasites that arose during the treatment of P. falciparum–infected individuals (10, 12). These examples illustrate the importance of identifying de novo mutations in P. falciparum and understanding their contributions to development of resistance to drug candidates early in the drug development pipeline.
A powerful tool to study drug resistance is experimental selection of resistance in vitro followed by whole-genome sequencing of resistant parasites (13). By exposing malaria parasites to antimalarial drugs in vitro and in vivo, we have been able to identify or confirm the mechanism of drug resistance in some cases [reviewed in (14)]. For example, treatment of cultured P. falciparum parasites with atovaquone in vitro led to the development of resistant parasites with single point mutations in the atovaquone-binding pocket of cytochrome b (15). It was subsequently found that failure of atovaquone-combination therapy in infected individuals was associated with point mutations in the same binding pocket (14, 16, 17). More recently, in vitro selection with a sensitive field isolate of P. falciparum provided the first evidence that mutations in the kelch13 malaria parasite gene are a major genetic determinant of artemisinin resistance (1, 18). These studies illustrate that in vitro selection can reflect the resistance pathways that arise in natural parasite populations.
Our work and that of others have demonstrated the usefulness of in vitro selection for identifying both targets and mechanisms of resistance in P. falciparum against new antimalarial drugs (19, 20). However, there are aspects of the in vivo environment, such as pharmacokinetics/pharmacodynamics (PK/PD) and pathophysiology, that are not captured in tissue culture. To further explore the translational relevance of experimental in vitro selection to the emergence of resistance in vivo, we developed a proof-of-concept study to directly compare the development of resistance of P. falciparum in tissue culture to that in a mouse model of P. falciparum infection. We focused on antimalarial drugs that inhibit Plasmodium dihydroorotate dehydrogenase (DHODH), an essential enzyme that catalyzes the rate-limiting step of pyrimidine biosynthesis (21). Multiple high-throughput screens have identified a range of structurally diverse molecules that target this protein (22-26). We and others have previously demonstrated that resistance to DHODH inhibitors can be acquired through point mutations in the inhibitor-binding pocket or copy number variations (CNVs) at the dhodh locus (22, 27-32). We chose the triazolopyrimidine inhibitor of DHODH, DSM265, for drug resistance studies in vitro and in vivo because it is a next-generation antimalarial drug candidate with demonstrated efficacy in recent phase 2 clinical trials (33).
RESULTS
Drug resistance emerges rapidly in P. falciparum during in vitro selection
We performed four in vitro selection experiments with two DHODH inhibitors, DSM265 (Fig. 1A) and DSM267 (Fig. 1D), using two independent P. falciparum 3D7 clones. The malaria parasite isolates used were Pf3D7 A10, used previously in large-scale in vitro drug selection studies (19), and Pf3D70087/N9 (34), which was the same clone used for in vivo selection in a mouse model of P. falciparum infection. We used two in vitro selection procedures: an intermittent pulse of DHODH inhibitor treatment and continuous exposure of parasite to drug (figs. S1 and S2). For pulse selections, P. falciparum cultures were treated with the 99% effective concentration (EC99) of the compound DSM265 for 6 to 8 days (selection 1 and selection 3). In addition to DSM265, we also used the chemical analog DSM267 as a tool compound to explore the landscape of resistance development to triazolopyrimidine-based inhibitors of DHODH (selection 2) (35).
Fig. 1. Emergence of resistance to DSM267 and DSM265 in P. falciparum in vitro.
(A) Structure of the DHODH inhibitor DSM265 used in in vitro resistance selection experiments [selection 1 (S1)]. (B) Representative dose-response curves for bulk-selected parasite populations. Parasite populations in three independent culture flasks containing human blood were exposed to DSM265 at the EC99 and allowed to recrudesce. Cultures recovered after selection were tested in a dose-response assay to determine the resistance phenotype (flask 1, yellow; flask 2, blue; Flask 3, red). (C) Shown is the dose-response phenotype of clones isolated from in vitro resistance selection experiments using pulsed treatments with DSM265. Bulk resistant populations were cloned in the absence of compound pressure. Clonal parasite lines from flasks 1 and 3 exhibited heterogeneous resistance phenotypes. Each clonal line is labeled on the x axis with a unique identifier that includes the selection number (S), flask number (F), and clone number (C), for example, S1-F1-C1. Individual replicate EC50 values are shown as a scatter plot, with error bars depicting mean and SD. Statistically significant differences in specific clonal phenotypes relative to the wild-type (WT) 3D7 A10 parental parasite line are indicated on the graph. Significance was calculated using a nonparametric one-way ANOVA (Kruskal-Wallis) with post hoc multiple comparisons (Dunn’s test): **P < 0.01, ****P < 0.0001; ns, not significant. (D to F) Increasing continuous drug selection pressure yielded parasites with highly drug-resistant phenotypes. (D) Structure of the DHODH inhibitor DSM267 used in in vitro drug resistance selection experiments (selection 2). (E) Parasites were exposed to two steps of selective pressure (steps 1 and 2). In step 1, parasites were exposed to a single pulse of 25 nM DSM267, leading to ~10-fold resistance in two of three flasks (flask 1, yellow; flask 2, blue; flask 3, green). In step 2, the drug-resistant populations from flasks 2 and 3 were continuously exposed to DSM267 at an increased dose (50 nM) for 7 to 8 weeks. Shown are representative dose-response curves for bulk-selected populations from step 1. (F) Shown are dose-response phenotypes of clones isolated from step 1 and step 2 of selection 2 (S2). Each clonal line is labeled on the x axis with a unique identifier that includes the selection number (S), flask number (F), and clone number (C), for example, S2-F2-C1. Individual replicate EC50 values are shown as a scatter plot, with error bars depicting mean and SD. Statistically significant differences in specific clonal phenotypes relative to the wild-type 3D7 A10 parental line are indicated on the graph. Significance was calculated using a nonparametric one-way ANOVA (Kruskal-Wallis) with post hoc multiple comparisons (Dunn’s test): *P < 0.05, **P < 0.01; ns, not significant.
During and after drug exposure, the parasitemia of the cultures was monitored by thin-smear microscopy, which allowed for the identification and quantification of replicating parasites. For pulse selection, P. falciparum asexual blood-stage parasites were consistently visible in thin smears taken from cultures about 2 weeks after exposure to drug in vitro (figs. S1 and S2 and table S1). In all three pulse selection procedures, at least two independent parasite populations became resistant to drug after either the first or second round of drug exposure (Fig. 1, B and E, figs. S1 and S2, and table S1). Drug-resistant parasites arising during pulse selection in vitro exhibited heterogeneous dose-response phenotypes when exposed to DSM265 or DSM267, with half-maximal effective concentration (EC50) values ranging from 2 to ~400 times that of the wild-type parent strain of P. falciparum (Fig. 1, C and F, fig. S2, Table 1, table S2, and data file S1). Whole-genome sequencing revealed that 20 of 21 resistant parasite clones generated during pulse selection had single point mutations in DHODH (Table 1, table S3, and data file S2). These mutations resulted in the following amino acid changes in DHODH: C276Y, L531F, F227L, G181C, and F227Y. One resistant parasite line had an amplification on chromosome 6, which included the dhodh locus (data file S3). CNVs at the dhodh locus were additionally verified by quantitative polymerase chain reaction (qPCR) (fig. S3). Consistent with previous findings, duplication of the dhodh locus conferred a ~2-twofold increase in drug resistance to DHODH inhibitors in P. falciparum parasites in vitro (Table 1, table S2, and data file S1) (27, 29, 30).
Table 1.
Point mutations in drug-resistant parasite lines arising from in vitro selection experiments with resulting EC50 values (nM). EC50 values (nM) represent average of 3 to 10 biological replicates ± SD (table S2 and data file S1). CNVs were determined by whole-genome sequencing and confirmed by qPCR (fig. S3). N/A, not applicable.
| Selection | Flask | Clone ID | DHODH Mutations | Copy No. | DSM265 EC50 | DSM267 EC50 |
|---|---|---|---|---|---|---|
| Parent Lines | N/A | 3D7 A10 | N/A | 1 | 3.55±1.28 | 2.18±0.646 |
| N/A | 3D70087/N9 | N/A | 1 | 9.08±2.41 | 4.57±0.873 | |
| N/A | Dd2 | N/A | 1 | 3.92±2.00 | 3.46±1.07 | |
| Selection 1: 3D7 A10 with DSM265 (Pulse Selection) | Flask 1 | S1-F1-C1 | C276Y | 1 | 45.1±32.5 | 36.2±23.6 |
| S1-F1-C2 | C276Y | 1 | 48.8±21.6 | 34.2±15.4 | ||
| S1-F1-C3 | L531F | 1 | 69.3±29.0 | 35.0±21.4 | ||
| Flask 3 | S1-F3-C1 | WT | 2 | 8.76±4.33 | 5.99±2.72 | |
| S1-F3-C2 | C276Y | 1 | 51.5±29.0 | 37.0±19.6 | ||
| S1-F3-C3 | C276Y | 1 | 45.8±23.4 | 36.8±16.7 | ||
| S1-F3-C4 | C276Y | 1 | 37.2±12.9 | 24.0±6.05 | ||
| Selection 2.1: 3D7 A10 with DSM267 (Pulse Selection) | Flask 2 | S2(1)-F2-C1 | F227L | 1 | 56.3±11.4 | 38.6±16.6 |
| S2(1)-F2-C2 | F227L | 1 | 71.9±54.3 | 35.0±17.8 | ||
| Flask 3 | S2(1)-F3-C2 | L531F | 1 | 63.9±25.9 | 17.2±6.06 | |
| S2(1)-F3-C3 | L531F* | 1 | 49.1±10.2 | 12.8±3.19 | ||
| Selection 2.2: 3D7 A10 with DSM267 (Continuous Selection) | Flask 2 | S2(2)-F2-C1 | F227L+L531F | 1 | 911±864 | 357±307 |
| S2(2)-F2-C2 | F227L | 2 | 91.3±50.9 | 56.6±43.5 | ||
| Flask 3 | S2(2)-F3-C1 | F227Y | 1 | 562±224 | 952±748 | |
| S2(2)-F3-C2 | F227Y | 1 | 1270±257 | >500** | ||
| Selection 3: 3D70087/N9 with DSM265 (Pulse Selection) | Flask 2 | S3-F2-C1 | G181C | 1 | 166±56.8 | 75.3±5.80 |
| S3-F2-C2 | G181C | 1 | 139±76.2 | 61.3±21.3 | ||
| S3-F2-C3 | G181C | 1 | 134±58.6 | 65.6±25.6 | ||
| S3-F2-C4 | F227Y | 1 | 3260±1721 | 5361±2832 | ||
| Flask 3 | S3-F3-C1 | L531F | 1 | 122±49.4 | 39.7±12.3 | |
| S3-F3-C2 | L531F | 1 | 159±25.0 | 42.8±11.7 | ||
| S3-F3-C3 | L531F | 1 | 163±75.3 | 39.2±7.50 | ||
| S3-F3-C4 | L531F | 1 | 186±71.6 | 50.4±11.6 | ||
| S3-F3-C5 | L531F | 1 | 139±53.5 | 34.0±7.78 | ||
| S3-F3-C6 | L531F | 1 | 146±61.6 | 45.3±14.5 | ||
| Selection 4: 3D70087/N9 with DSM265 (Continuous Selection) | Flask 1 | S4-F1-C1 | V532G | 1 | 1010±413 | 495±221 |
| S4-F1-C2 | V532G | 1 | 627±163 | 317±116 | ||
| S4-F1-C3 | V532G | 1 | 690±178 | 363±73.4 | ||
| S4-F1-C4 | V532G | 1 | 846±207 | 463±129 | ||
| Previously-selected PfDHODH mutant lines*** (Parental line indicated) | N/A | N/A | 3D7-E182D | 1 | 174.4±53.7 | 97.1±20.9 |
| N/A | N/A | Dd2-I263F | 1 | > 200** | >200** | |
| N/A | N/A | Dd2-F188I | 1 | 8.96±4.52 | 1.30±0.441 | |
| N/A | N/A | Dd2-F227I | 1 | 51.6±30.3 | 31.5±10.9 | |
| N/A | N/A | Dd2- F227I+L527I | 1 | 99.6±51.7 | 71.0±41.7 |
We also wanted to explore how P. falciparum parasites responded to continuous selective pressure in vitro. We treated resistant parasite populations recovered from the first step of selection 2 with 25 nM DSM267 (which was the EC99 of the parental line) for 7 days, followed by an increase to 50 nM DSM267 for 7 to 8 weeks (fig. S1). Clonal parasite lines from this second selection step had increased EC50 values compared to clones isolated after the first round of drug treatment (Table 1, Fig. 1F, table S2, and data file S1). Whole-genome sequencing revealed single and double point mutations in DHODH, including F227Y and F227L/L531F, as well as one clone showing both dhodh amplification and an F227L mutation (Table 1, table S3, data files S2 and S3, and fig. S3). In a separate selection process, we continuously exposed Pf3D70087/N9 parasites to increasing concentrations of DSM265 from 30 to 200 nM over a 2-month period (selection 4) (fig. S2). Parasites isolated from this selection process showed a stronger drug resistance phenotype compared to those recovered from pulse selection. All four clones characterized from the selection 4 population carried a V532G mutation in DHODH and showed a ~100-fold increase in EC50 (Table 1, fig. S2, table S2, and data files S1 and S2). In addition to these in vitro selections, we assessed the activity of DSM265 and DSM267 against mutant parasite lines previously isolated from in vitro selections using other structural classes of DHODH inhibitors (table S4) (27, 28). We found that four of these lines (3D7-E182D, Dd2-I263F, Dd2-F227I, and Dd2-F227I/L527I) showed cross-resistance with DSM265 (Table 1, table S2, and data file S1).
Establishing a DSM265 therapeutic dose for in vivo resistance selection experiments
For comparisons between in vitro selection and selection of drug-resistant mutant parasites in vivo, we developed an approach based on cycles of drug treatment in mice with severe combined immunodeficiency (SCID) and a null mutation in the interleukin-2 receptor chain [nonobese diabetic (NOD)–scid IL2Rγnul1, hereafter referred to as SCID mice] infected with P. falciparum. The SCID mice were transfused with human erythrocytes, and when the animal’s peripheral blood reached 50% human erythrocytes, they were infected with P. falciparum intravenously (36, 37). We used a therapeutic dose of the DHODH inhibitor DSM265 that decreased parasitemia below the 0.01% limit of detection (LoD) as determined by flow cytometry of infected SCID mouse blood samples. Recrudescent parasites were transferred to naïve SCID mice, and these animals were treated with the same dose of compound. Iterative rounds of drug dosing resulted in parasite populations that were no longer killed by the original drug dose. Samples of these resistant parasite populations were then cloned, sequenced, and phenotyped in drug dose-response assays in vitro (fig. S4).
To establish this system, we first identified the desired therapeutic dose of DSM265 for use in the selections in vivo. We dosed P. falciparum-infected SCID mice with DSM265 once daily for 4 days at a range of doses and then collected blood samples to measure DSM265 concentrations and to establish the PK/PD profile. DSM265 displayed potent in vivo antimalarial activity with an ED90 of 3.6 mg/kg [Area under the curve from 0-23 hours (AUC0–23h), 6.06 μg-hours/ml; Fig. 2, A and B]. Recrudescence of parasites was observed across the full dose range tested, and time to recrudescence was established as weeks after treatment (Fig. 2C). On the basis of these results, we chose the 50 mg/kg dose (~10-fold ED90) for drug resistance selection in vivo.
Fig. 2. DSM265 efficacy and P. falciparum recrudescence in SCID mice.
Figure shows the efficacy of DSM265 against P. falciparum in infected SCID mice (n = 2 mice per group) at indicated doses. Mice were infected with P. falciparum on day 0, and treatment with DSM265 was started on day 3 after infection. Mice were dosed orally once per day for four consecutive days. (A) Shown are the results of efficacy studies used to determine the minimal dose of DSM265 required for parasite clearance. DSM265 was dosed at 1 mg/kg (brown), 10 mg/kg (orange), 25 mg/kg (purple), 50 mg/kg (blue), 75 mg/kg (red), or 100 mg/kg (cyan). (B) Shown are concentrations of DSM265 in blood (ng/ml) taken from mice at 0.5 to 24 hours after the first DSM265 dose. (C) In recrudescence studies, 50 mg/kg was the optimal dose of DSM265 that allowed parasites to recrudesce about 2 weeks after DSM265 treatment.
In vivo drug resistance occurs through the same DHODH mutations identified by in vitro selection
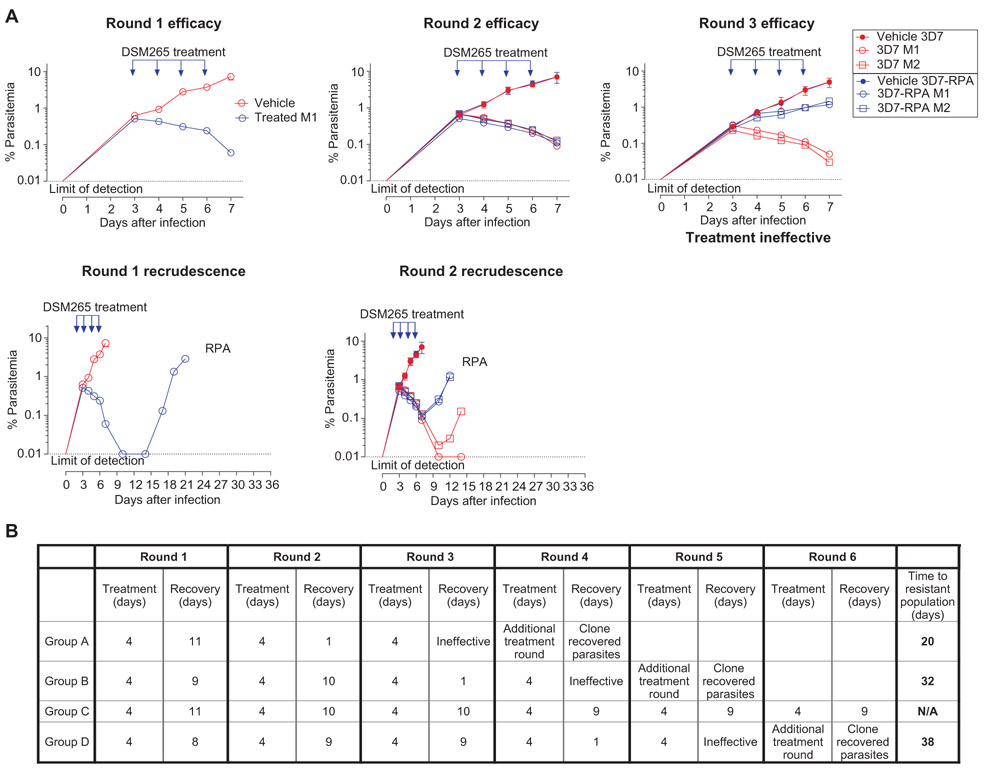
We initially infected four SCID mice with 2 × 107 P. falciparum parasites of the Pf3D70087/N9 strain (groups A, B, C, and D). Each mouse was orally treated with DSM265 (50 mg/kg) once daily for 4 days. In all mice, the parasite population initially decreased below the LoD but was then detected by flow cytometry about 2 weeks after drug treatment (Fig. 3 and fig. S5). Infected red blood cells (iRBCs) from each mouse were then used to infect four naïve mice, two of which were treated with the same dose of DSM265 and two of which received vehicle. The parasite populations from each of the original four mice (A, B, C, and D) were maintained as separate lineages in each of the rounds of selection and were labeled groups A, B, C, and D. This process was repeated until a resistant parasite population emerged in treated animals. Resistant parasites emerged in group A (Fig. 3A) and also in groups B and D but not group C (Fig. 3B and fig. S5).
Fig. 3. Development of DSM265 resistance in SCID mice infected with P. falciparum.
Four P. falciparum–infected SCID mice (experimental animal groups A, B, C, and D) were treated with DSM265 (50 mg/kg), and parasitemia was monitored until recrudescent parasites were observed (round 1). Recrudescent resistant parasites were used to infect naive mice in a second cycle (round 2). Rounds of infection, treatment, and recovery were continued until drug resistance was observed. The parasite populations from groups A to D were maintained as independent lineages starting from the four initially infected mice through each subsequent passage and treatment cycle. (A) Shown are representative results for group A mice for efficacy (top) and parasite recrudescence (bottom). Two of the mice in round 2 were treated orally with the same dose of drug (individual mice M1 and M2, open blue symbols), and the other two mice were treated with vehicle alone as a control (represented as an average, closed blue symbols). Infections with the 3D7 A10 parental line parasites as a control are plotted in red (drug treated, open symbols; vehicle control, closed symbols). In group A, the infected mice were never able to completely clear parasites from round 1 [Resistant population A (RPA)], and the parasites recrudesced faster than did 3D7 A10 wild-type control parasites, indicating that the parasite population had evolved resistance. By round 3, the original dose of DSM265 was completely ineffective. Once resistance to DSM265 was observed, parasites were subjected to an additional round of drug treatment. Genomic DNA then was extracted from the bulk population and subjected to whole-genome sequencing. (B) Shown is a summary of results from all groups of mice (A, B, C, and D) undergoing in vivo resistance selection, with the iterative infection, treatment, and recrudescence periods for each group indicated. The round when resistant parasites were observed (treatment ineffective) and the time to resistance development are also indicated.
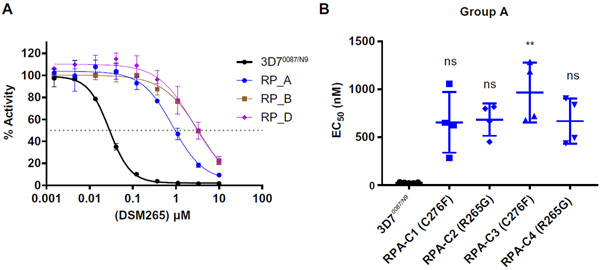
Similar to the in vitro results, a stable resistance phenotype was generated after a short period of time in three of the four groups of mice. For selection group A, reduced efficacy of the drug was observed after only one round of treatment, and treatment was completely ineffective by round 3 (Fig. 3A). For selection groups B and D, treatment was ineffective by rounds four and five, respectively, of drug treatment (Fig. 3B and fig. S5). Drug resistance was never observed for selection group C, and the assay was terminated after six rounds of treatment (Fig. 3B and fig. S5). The drug resistance phenotypes of the parasite populations were confirmed by in vitro dose-response assays (Fig. 4A), and individual clones were isolated by limiting dilution. As observed in our in vitro selections, clonally derived parasites showed heterogeneous dose-response phenotypes, with the EC50 ranging from 16 to 116 times that of the wild-type P. falciparum parental strain (Fig. 4B, fig. S5C, Table 2, table S2 and data file S1).
Fig. 4. Drug resistance phenotypes of parasites in P. falciparum–infected SCID mice.
(A) Resistant P. falciparum parasites (RP) from groups A, B, and D were adapted to in vitro growth and phenotyped using an in vitro dose-response assay. Shown are representative dose-response curves relative to wild-type 3D70087/N9 parasites (wild-type, black symbols; group A parasites, blue symbols; group B parasites, brown symbols; group D parasites, purple symbols). (B) Individual replicate EC50 values are shown for clonal parasite lines isolated from the group A parasite population, with error bars depicting mean and SD. Statistically significant differences in specific drug-resistant phenotypes relative to the wild-type 3D70087/N9 parental line are indicated on the graph. Significance was calculated using a nonparametric one-way ANOVA (Kruskal-Wallis) with post hoc multiple comparisons (Dunn’s test): **P < 0.01.
Table 2.
Point mutations in drug-resistant parasite lines arising from in vivo selection experiments with resulting EC50 values (nM). Point mutation and dose-response phenotype listed for each in vitro–selected clone. EC50 values (nM) represent an average of two to five biological replicates ± SD. Where two biological replicates were performed, only mean is reported (table S2 and data file S1). CNVs were determined by whole-genome sequencing and confirmed by qPCR (fig. S6).
| In vivo selection of 3D70087/N9 treated with DSM265 | ||||
|---|---|---|---|---|
| Clone ID | DHODH mutations |
Copy no. | DSM265 EC50 |
|
| 3D70087/N9 | N/A | 1 | 25.0 ± 4.73 | |
| Group A | RPA-C1 | C276F | 1 | 655 ± 317 |
| RPA-C2 | C276F | 1 | 684 ± 169 | |
| RPA-C3 | R265G | 1 | 966 ± 314 | |
| RPA-C4 | R265G | 1 | 669 ± 238 | |
| Group B | RPB-C1 | G181D | 1 | 3419 |
| RPB-C2 | G181D | 1 | 3173 | |
| RPB-C3 | G181D | 1 | 3346 | |
| RPB-C4 | G181D | 1 | 3098 | |
| Group D | RPD-C1 | G181D | 1 | 3060 |
| RPD-C2 | G181D | 1 | 3360 | |
| RPD-C3 | G181D | 1 | 3240 | |
| RPD-C4 | G181D | 1 | 1800 | |
When comparing the in vivo and in vitro selections, we found overlap in the amino acid residues of DHODH that were mutated. Whole-genome sequencing revealed that two of four clones from group A carried a C276F mutation, which was similar to the C276Y mutation observed in vitro (Table 2, table S3, and data file S2). Both mutations resulted in replacement of the cysteine at position 27 with an amino acid with an aromatic side chain. The other two parasite clones from group A carried an R265G mutation, which was not identified in any of our in vitro selection experiments. However, an R265A mutation was identified after in vitro selection experiments by another group (32). All four clones isolated from in vivo selection groups B and D carried a G181D mutation similar to the G181C mutation identified during our in vitro selection experiments, with the nonpolar glycine being replaced by a charged (G181D) or polar (G181C) amino acid residue (Table 2, table S3, and data file S2).
In addition to whole-genome sequencing of parasite clones from the stably resistant parasite populations, we also collected genomic DNA (gDNA) before each round of drug treatment and performed whole-genome sequencing on the bulk parasite populations (data file S2 and table S3). We calculated allele frequencies based on read counts (table S5). In populations B and D, the G181D mutation predominated before rounds 3 and 4 of selection, respectively, corresponding to when reduced drug efficacy was observed; the G181D mutation reached 100% frequency as drug resistance became stable (table S5). In contrast, for population A, the predominant mutation was R265G, which appeared at 97% frequency after round 1 of selection (table S5). However, the frequency of this allele decreased over the next two rounds of drug treatment to a frequency of 89%. In contrast, the frequency of the C276F mutation increased from 5 to 12% in the same parasite samples (table S5), suggesting that the two parasite lines were engaging in clonal competition in infected SCID mice in vivo (38).
Resistance to DSM265 occurs without a fitness cost to the parasite
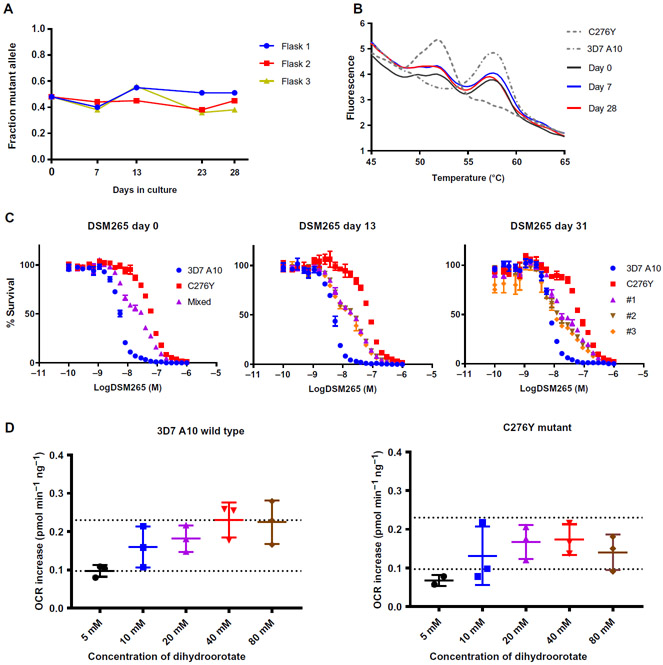
Both our study and a recently published phase 2 clinical trial identified drug-resistant P. falciparum parasites carrying C276 mutations after treatment with DSM265 (33). To assess the relative fitness of P. falciparum parasites carrying the C276Y mutation, we used in vitro competitive growth assays (39). Mutant clone S1-F1-C1 and wild-type parental lines were grown together in mixed culture over a 1-month period in three independent replicate flasks without i exposure to drug. Genomic DNA was collected every three to five P. falciparum replication cycles. Whole-genome sequencing of gDNA i samples allowed calculation of the relative abundance of mutant i versus wild-type alleles based on read counts. Across the three replicate flasks, the allele ratio remained relatively constant over i time (Fig. 5A). The allelic composition was additionally monitored . by modified high-resolution melt assay (40-42), which confirmed I that the allelic composition of the cultures did not change much I over the course of the experimental period (Fig. 5B). In addition to monitoring the genetic composition, we also assessed the dose-response phenotype of the mixed cultures. On day 0, the cultures had an intermediate resistance phenotype relative to the initial . wild-type and mutant populations. After 1 month, all replicate i flasks retained this intermediate phenotype (Fig. 5C). To confirm that there was no loss of fitness, we performed a second independent set of competition assays, which included an additional control flask treated with DSM265. As expected, the mutant allele predominated in this drug-treated flask, whereas all untreated flasks maintained a mixed allele population (fig. S7). Together, these results suggested that the C276Y mutant parasite line was as competitively fit as the 3D7 parental line.
Fig. 5. The C276Y mutation does not decrease fitness in P. falciparum parasites.
The 3D7 A10 parental line of P. falciparum and the C276Y mutant parasite line S1-F1-C1 were cocultured in an in vitro competition assay. (A) The frequency of the C276Y allele (y axis) was calculated on the basis of read counts from whole-genome sequencing of genomic DNA extracted at the indicated culture times (x axis). (B) Shown are representative high-resolution melt profiles of competition assay samples from flask 2. Samples of genomic DNA taken at days 0, 7, and 28 of culture all showed similar melt profiles, indicating that the allelic composition was relatively stable over time. For simplicity, only one representative technical replicate is shown. (C) Shown are dose-response curves revealing an intermediate phenotype of the cocultured parasite populations over time in culture. Parasite cultures were exposed to a 24-point serial dilution of DSM265 doses in a 72-hour assay. (D) Shown is a bioenergetic assay measuring oxygen consumption rate (OCR) for 3D7 A10 wild-type and mutant C276Y (clone S1-F1-C1) P. falciparum strains at varying concentrations of dihydroorotate substrate. Dihydroorotate titration revealed the effect of mutations on DHODH enzyme activity. parasites at the schizont stage were exposed to the indicated concentrations of dihydroorotate substrate, and OCR increases were plotted. Data are shown as a scatter plot of individual biological replicates, with error bars depicting mean and SD (5 mM dose, black; 10 mM dose, blue; 20 mM dose, purple; 40 mM dose, red; 80 mM dose, brown). Dotted lines were added for ease of comparison between plots. Kruskal-Wallis test comparing wild-type and C276Y mutant parasites indicated no statistically significant differences between the two lines at any dose (P = 0.6147, P > 0.999, P > 0.999, P = 0.3032, P = 0.4081 for 5, 10, 20, 40, and 80 mM doses of dihydroorotate, respectively).
The C276Y mutation did not affect DHODH function
As an independent test of the biological impact of DHODH mutations, we next assessed the effect of DHODH mutations on mitochondrial r function in P. falciparum. We recently developed a mitochondrial bioenergetics assay to characterize electron transport chain proteins in saponin-treated whole parasites using the Seahorse Bioscience XFe24 Extracellular Flux Analyzer (43). Saponin causes lysis of the host RBCs but leaves the P. falciparum parasites intact (44). Because the reactions catalyzed by DHODH are directly coupled to mitochondrial respiration, increased DHODH activity can be observed as an elevation 1 of the oxygen consumption rate (OCR). Using OCR as a read-out, we assessed the enzymatic activity of DHODH in whole parasites.
We compared an increase in OCR in response to varying concentrations of dihydroorotate in the 3D7 A10 parental P. falciparum line and the C276Y mutant P. falciparum line. The two lines showed comparable responses to dihydroorotate across all concentrations (no statistically significant differences; P = 0.6147, P > 0.999, P > 0.999, P = 0.3032, P = 0.4081 for 5, 10, 20, 40, and 80 mM doses, respectively) (Fig. 5D and table S6). This was consistent with our observation that the C276Y mutant parasite was as fit as the wild-type parental strain (Fig. 5D and table S6). In addition, we tested a CRISPR-edited 3D7 A10DHODH-E182D mutant parasite line (fig. S8). In previously published work, we showed that a drug-selected E182D P. falciparum mutant exhibited a competitive growth defect and reduced enzymatic efficiency of DHODH (27). Consistent with those previous observations, the CRISPR-edited E182D mutant parasite showed a decreased OCR, particularly at low concentrations of dihydroorotate, although the difference was not statistically significant (P = 0.0675, P = 0.1107, P = 0.1107, P = 0.8902, P = 0.6991 for 5, 10, 20, 40, and 80 mM doses of dihydroorotate, respectively) (fig. S9 and table S6).
Mouse adaptation experiments suggest that C276Y mutant parasites are also fit in vivo
To further assess the fitness of P. falciparum mutant parasites that were resistant to DSM265, we attempted to adapt the E182D and C276Y mutant lines to grow in SCID mice. For these assays, we used the original drug-selected E182D line (table S4) (28) and clone S1-F1-C1 used in other experiments in this study. For each mutant line, we inoculated three SCID mice each with 75 × 106 infected human erythrocytes intravenously. Groups of mice were infected with the corresponding wild-type parental P. falciparum line in parallel. Parasitemia in peripheral blood was followed up for up to 2 months after infection or until 1% parasitemia was achieved.
All mice inoculated with the C276Y mutant parasites showed productive infection within 5 weeks after inoculation as was observed with the corresponding wild-type P. falciparum strain 3D7 A10 (data file S4). To rule out wild-type contamination, we analyzed the in vivo susceptibility of the adapted S1-F1-C1 P. falciparum line to DSM265. A stable resistance phenotype was observed in vivo, with an ED90 of 25 mg kg−1 day−1 (AUC0–23h, 134 μg·hours/ml) (fig. S10). Conversely, we were unable to establish infection with the E182D mutant strain, despite three different attempts and monitoring peripheral blood parasitemia for up to 9 weeks after initial inoculation. The corresponding 3D7 A10 parental P. falciparum line appeared in peripheral blood 5 to 6 weeks after inoculation (data file S4). Overall, this suggested that the competitive fitness of the C276Y mutant line translated into in vivo viability compared to the relatively unfit E182D mutant line.
DISCUSSION
The major finding of our study is that in vitro resistance of P. falciparum asexual blood-stage parasites to the DHODH inhibitor DSM265 can predict drug resistance in infected mice in vivo. Our approach provides a framework for assessing the potential for emergence of drug resistance in P. falciparum during the early stages of drug development. Development of drug resistance may be inevitable, but the rate at which resistance emerges and spreads has implications for public health. An understanding of how parasites develop resistance to new classes of antimalarial drugs both in vitro and in vivo is therefore an important criterion when evaluating the drug development pipeline.
Our study compared in vitro and in vivo selection of resistance to the same compound. The P. falciparum infection mouse model enabled capture of the dynamics of drug resistance development that were lacking from the in vitro experiments (36). We found that selection of resistance to the DHODH inhibitors DSM265 and DSM267 in vitro mirrored the findings in the mouse model. In both systems, parasites recrudesced roughly 2 weeks after drug treatment. Stable drug resistance occurred in most of the selected parasite populations (seven of nine in vitro–selected populations and three of four in vivo–selected populations) and emerged after either one or a few cycles of drug treatment. Together, we identified 13 unique mutations that led to drug resistance (fig. S11). Several of these mutations were also identified in other studies, confirming their contributions to the observed resistance phenotypes (27, 28, 30, 32). There was also similarity in the mutations that arose in vitro compared to in vivo. Of the three residues in DHODH that were mutated during in vivo selections, all were also identified after in vitro selections in our study and in studies by other groups (fig. S11) (30, 32).
Our findings also correlate with results from a recently published phase 2a clinical trial. In that study, two P. falciparum–infected individuals treated with a single dose of DSM265 had recrudescent parasites at day 28 after treatment. Genome sequencing of these recrudescent parasites identified mutations in the dhodh locus. One patient had a clonal parasite population carrying the C276Y mutation. Another had a minor population of parasites harboring the C276F mutation, with the G181S mutation also detected in a subset of reads (33). These DHODH amino acid positions were all mutated in our in vitro and in vivo selection experiments, demonstrating that our approach may be a good model for predicting clinical resistance to DSM265. We identified several mutations that conferred resistance to DHODH inhibitors and saw both in vitro and in vivo that within a single parasite population there could be multiple mutational trajectories simultaneously. Given that there can be 1010 to 1013 parasites in an infected human host (45), it is expected that drug-resistant parasites emerged during the phase 2 clinical trial.
Whether drug resistance readily emerges in natural parasite populations also depends on the fitness of mutant parasites. Previous studies demonstrated that mutations in DHODH, particularly the E182D mutation, can negatively affect competitive fitness (27). In contrast, we showed that the C276Y mutant parasite line was relatively fit in competitive growth assays. We also found that the C276Y mutation did not reduce enzyme efficiency in a mitochondrial bioenergetics analysis and that C276Y mutant parasites could establish a productive infection in vivo. Recent biochemical studies also support the fitness of C276Y mutant parasites because DHODH carrying C276Y or C276F mutations had an overall catalytic efficiency (Kcat/Km) similar to the wild-type enzyme (32). The relative fitness of C276Y mutant parasites may explain why this mutation was so predominant in vitro and why parasites harboring this mutation emerged in patients in the phase 2 clinical trial. The fact that resistance to DSM265 occurred without a fitness cost to the malaria parasite suggests that resistance would be able to readily spread and become stable within natural parasite populations.
Although outside the scope of this study, another important aspect of Plasmodium fitness is transmission by the mosquito vector. Given that we have shown that the C276Y mutant parasite line has oxidative respiration comparable to the wild-type parental strain, we hypothesize that these parasites should be able to survive in the mosquito and be transmitted to the next host efficiently. Future studies are warranted to explore this possibility.
We believe that our in vitro resistance selection approach could prove useful for informing drug development; however, we acknowledge that there may be molecules or drug targets for which in vitro drug resistance development is less likely to reflect resistance development in vivo. For example, there may be cases where a mutation in a target gene is evolutionarily fit under asexual growth conditions in vitro but is not viable in vivo. The SCID mouse model of P. falciparum infection used here captures only some aspects of the dynamics of parasite growth in a human host, given that these mice lack an adaptive immune system. Nevertheless, we suggest that in many cases, in vitro drug resistance selection studies can provide valuable insights and should be considered for both prioritizing new drug candidates and designing combination therapies.
Although this study focuses on mutations in DHODH, future work should explore the potential contribution of genetic changes outside of this locus. We identified additional mutations in our drug-resistant parasite lines (data file S2) that could marginally contribute to the observed drug resistance phenotypes and parasite fitness. Although it is difficult to determine the impact of these additional genetic changes, such mutations would not change the key observation that parasites resistant to DSM265 did not show reduced fitness. Even if the C276Y mutation conferred a fitness cost, if this cost could be compensated for by additional genetic changes arising over the short period of time it took to isolate drug-resistant parasites in vitro, then it is likely that similar compensatory mutations could also evolve in larger, more genetically diverse natural parasite populations.
Our results, in conjunction with a recent phase 2 clinical trial, highlight that drug resistance is a potentially major problem for the clinical use of DSM265. However, extensive characterization of the DHODH resistome, which is now underway by our group and others, may help in the development of therapeutic strategies to suppress the emergence of resistant parasites. We have previously shown that resistance to some DHODH inhibitors induces conformational changes that increase the enzyme’s sensitivity to other structural classes of inhibitors (26, 27). This phenomenon, wherein resistance to one inhibitor causes increased sensitivity to another inhibitor, is known broadly as collateral sensitivity. If the dominant pathways to DSM265 resistance exhibit collateral sensitivity to one or more DHODH inhibitors, then perhaps a combination therapy approach exploiting this collateral sensitivity could provide a way to slow the emergence of drug resistance and increase the useful lifetime of DSM265. Future work could include high-throughput screening to identify inhibitors specific for the mutant forms of the enzyme, focusing on the fittest pathways to resistance such as the C276Y mutation.
MATERIALS AND METHODS
Study design
This study aimed to develop a methodology to inform the likelihood of the development of resistance to next-generation anti-malarial drugs. We performed a proof-of-concept study with DSM265 and DSM267 inhibitors of the Plasmodium DHODH. We conducted four in vitro resistance selections in P. falciparum cultures in vitro totaling nine independent populations. We compared the results of these selections to in vivo selections in NOD-SCID mice with a null mutation in IL2Rγnull, which were transfused with human erythrocytes and infected with P. falciparum intravenously. The mice were dosed using a spray-dried dispersion of DSM265 that was similar to that used in recent phase 2 clinical trials.
For animal studies, female age-matched mice were used. Four mice were initially infected with P. falciparum and treated with DSM265. Blood from each infected mouse after subsequent rounds of DSM265 treatment was inoculated into four more mice infused with human erythrocytes, which then were randomly divided with two mice receiving DSM265 treatment and two mice receiving a vehicle control. In each new round of selection, the wild-type 3D70087/N9 parasite strain was used as a control. Iterative rounds of infection and treatment following this design were continued until the endpoint of the selection. The endpoint was the observance of a stable resistance phenotype, indicated by P. falciparum infection that was no longer responsive to DSM265 treatment. The assay was terminated if a resistance phenotype was not achieved after six rounds of treatment. In addition to the in vitro and in vivo resistance selection studies, we further assessed the likelihood that identified resistance pathways would emerge by characterizing the relative fitness of resistant asexual blood-stage parasites. Fitness was assessed using both in vitro competitive growth assays and a bioenergetics assay to characterize DHODH function.
All experiments were approved by the GlaxoSmithKline (GSK) Diseases of the Developing World (DDW) Ethical Committee on Animal Research, performed at the DDW Laboratory Animal Science facilities accredited by Association for Assessment and Accreditation of Laboratory Animal Care, and conducted in accordance with European Directive 86/609/EEC and the GSK Policy on the Care, Welfare and Treatment of Animals. After infection, mice were randomly assigned to vehicle control or drug treatment groups; no animals were excluded from the analysis. The human erythrocytes were sourced ethically from blood biobanks, and their research use was in accordance with the terms of the informed consent. Erythrocyte concentrates from malaria-negative donors were provided by Biobancos de Castilla y Leon, Barcelona and Centro de Transfusiones de Madrid and the Red Cross Transfusion Blood Bank in Madrid, Spain. Research was conducted according to the GSK’s code of conduct policy POL-GSKF-410 and was in accordance with the terms of the informed consent of each donor.
Reagents
DSM267 and DSM265 were gifts from M. Phillips from University of Texas (UT) Southwestern (35). l-dihydroorotic acid (DHO) was purchased from Sigma-Aldrich (St. Louis, MO). Amodiaquine, artemether, atovaquone, chloroquine, dihydroartemisinin, and mefloquine were purchased from Sigma-Aldrich (St. Louis, MO).
Parasite strains and culturing conditions
The 3D7 A10 clone has been used in previous in vitro drug selection and sequencing efforts (19). The Pf3D70087/N9 line was generated by adapting 3D7 to grow in peripheral blood of engrafted NOD-scid/β2 m−/− mice (34). The Dd2 2D4 clone was derived from MR4 line MRA-156 (MR4, BEI Resources). In addition to the selected lines derived in this study, parasites with point mutations in pfdhodh were generated as described previously (table S4) (27, 28). Parasite strains were cultured in 5% human O+ hematocrit in RPMI 1640 (RPMI) (Life Technologies) supplemented with 28 mM NaHCO3, 25 mM Hepes, and hypoxanthine (50 mg/ml). Depending on the parasite line, the media either contained gentamycin (25 μg/ml), or no antibiotic and was additionally supplemented with 0.5% AlbuMAX II (Life Technologies) and/or O+ human serum (heat inactivated and pooled). Blood and serum products were obtained from Interstate Blood Bank. Cultures were maintained at 37°C in 1.1% O2, 4% CO2, and 95% N2. Parasite populations were synchronized by 5% sorbitol treatment (46).
Selection of drug resistance in vitro
Resistance selections were performed using 100-ml cultures at 3 to 4% starting parasitemia (~109 parasites). Intermittent pulse selections were treated with the EC99 of the selecting agent for 6 to 8 days. Compound pressure was then removed, and cultures were fed with compound-free media daily for the first week and on alternate days thereafter. For continuous selections, cultures were maintained in 25-ml flasks and replenished daily with medium containing the selecting agent. Bulk populations were cloned by limiting dilution in 96-well plates. Pulse treatment selections were cloned in compound-free RPMI. Cultures selected with continuous treatment were cloned at the highest concentration of compound to which they were exposed during the selection.
Efficacy studies in P. falciparum–infected SCID mice
The efficacy of DSM265 was tested in the GSK mouse model of P. falciparum infection (GSKPfalcHuMouse) as previously described (36). Drug treatment was performed on day 3 after infection every 24 hours for four consecutive days by oral gavage in a volume of administration of 10 ml/kg bodyweight, at indicated doses. DSM265 was prepared in 0.5% hydroxypropyl methylcellulose/0.02% Tween 80. Parasitemia was measured as previously described (36). ED90 is the dose of the compound that reduces parasitemia at day 7 of the in vivo assay by 90% with respect to parasitemia in vehicle-treated mice.
The blood concentrations of DSM265 in mice were measured in serial samples of peripheral blood (20 μl) taken by tail puncture at 0.5, 2, 4, 6, 8, and 23 hours after the first administration and 23 hours after the fourth administration. The blood samples were immediately lysed by mixing with 20 μl of distilled water, frozen on dry ice, and stored at −80°C until analysis. The compounds were extracted from 10 μl of each lysate by liquid-liquid extraction in the MultiScreen Solvinert 0.45-μm Hydrophobic Polytetrafluoroethylene 96-well plate system (Millipore) and stored at −80°C until analysis by liquid chromatography–tandem mass spectrometry in API4000 (AB Sciex LLC, Framingham, MA). The compound concentration versus time data were analyzed by noncompartmental analysis using Phoenix Version 6.3 (Pharsight Corporation). Additional statistical analysis was performed with GraphPad Prism Version 6.02 (GraphPad Software Inc.).
Selection of drug resistance in vivo
Four mice infected with 2 × 107 Pf3D70087/N9 parasites were established as the parent generation. The day of infection was designated day 0. From days 3 to 6, each mouse was orally treated daily with DSM265 at a dose of 50 mg/kg free base (10-fold ED90), until the parasite population decreased below LoD. After a recrudescence period, infected RBCs from mice with detectable parasitemia were donated to four mice for an additional passage. Infected mice in the second passage were randomly divided into two groups, one of which received the previous dose of DSM265, and other was treated with vehicle as a control. Drug concentrations in blood were monitored at the end of treatment in each selection cycle. The process was repeated until the resistance phenotype was observed. In each cycle of selection, additional mice infected with the parental P. falciparum Pf3D70087/N9 strain were used as controls. Resistant parasite populations were cryopreserved, cloned in compound-free RPMI, and tested for in vitro dose-response phenotype.
Dose-response assay for in vitro and in vivo parasite lines
For in vitro–selected lines, drug susceptibility was measured by SYBR Green I–based assay (47, 48). Ring-stage parasites were cultured for 72 hours at 1% hematocrit and 1% starting parasitemia in 384-well black clear-bottom plates containing test compounds plated in triplicate in 12-point serial dilutions. Lysis buffer with SYBR Green I fluorescent dye (Invitrogen) was added, and fluorescence readings were taken (excitation at 494 nm, emission at 530 nm). EC50 values were calculated using a nonlinear regression curve fit in GraphPad Prism Software version 7 (GraphPad).
For resistant parasite lines isolated from mice, intraerythrocytic P. falciparum growth inhibition was determined using a modified in vitro [3H]-hypoxanthine incorporation method (49). Briefly, asynchronous cultures of iRBCs at 2% hematocrit and 0.5% parasitemia with 5 μM hypoxanthine were exposed to threefold serial dilutions of the compounds. Ninety-six–well plates (Costar #3894) were incubated for 24 hours at 37°C, 5% CO2, 5% O2, and 95% N2. After 24 hours of incubation, [3H]-hypoxanthine was added, and plates were incubated for an additional 24 hours. After that period, plates were harvested on glass fiber filters (Wallac #1450-421) using a cell harvester 96 (TOMTEC, PerkinElmer). Filters were dried, and melt-on scintillator sheets (MeltiLex A, PerkinElmer #1450-441) were used to determine the incorporation of [3H]-hypoxanthine. Radioactivity was measured using a microbeta counter (PerkinElmer). Data were normalized using the incorporation of the positive control (iRBC without drug). EC50 values were determined using Excel and Grafit 5 software.
gDNA analysis
gDNA was obtained from P. falciparum clones by washing iRBCs with 0.1% saponin and extracting gDNA using the DNeasy Blood and Tissue Kit (Qiagen). Sequencing library preparation and analysis were performed as previously described (19). PCR amplification/Sanger sequencing of the Pfdhodh locus was performed as described (table S7) (27) using BIO-X-ACT Short Mix (Bioline Reagents).
Whole-genome sequencing
Sequencing libraries were prepared with the Nextera XT kit (cat. no. FC-131-1024, Illumina) using the standard dual index protocol, and whole-genome sequencing was performed on an Illumina HiSeq 2500 in RapidRun mode with 100–base pair paired-end reads. Reads were aligned to the P. falciparum 3D7 reference genome (PlasmoDB v. 13.0), and variants were called using a previously established analysis pipeline (19).
CNV assay
CNV at the dhodh locus was assessed as previously described (27, 29). Power SYBR Green Master Mix (Applied Biosystems) was used on a ViiA7 (Applied Biosystems), and relative copy number for 0.1 ng of gDNA was determined by the ΔΔCT method.
Competition growth assays
Synchronized C276Y (S1-F1-C1) and 3D7 A10 parasites were suspended to 1% parasitemia and 5% hematocrit and combined at equal volumes. Replicate 10-ml flasks were maintained for 1 month with gDNA samples collected, and dose-response assays were performed at indicated time points.
High-resolution melt assay
PCR amplification was performed using the mutant allele amplification bias technique as previously described (table S8) (40, 41). Each reaction contained a final concentration of 1.0× LightScanner Master Mix (BioFire Defense, Salt Lake City, UT), 0.025 μM for-ward primer, 0.125 μM reverse primer, 0.1 μM blocked probe, and 1.25 ng of template DNA. Preincubation, amplification, high-resolution melting, and cooling steps were all performed on the 384-well LightCycler 480 System (Roche Molecular Systems Inc.) using default conditions, with an annealing temperature of 56°C for the amplification step. Melt curve genotype analysis was performed using the LightCycler 480 software, and curves were graphed in Prism 7 (GraphPad).
CRISPR-Cas9 mediated introduction of the E182D mutation
Guides targeting the E182D locus (fig. S8) were designed using Benching [Biology Software (2017) retrieved from https://benchling.com] and inserted into a BbsI-digested pDC2-Cas9-U6-hDHFR plasmid behind a T7 promoter. The backbone of the plasmid, which was a gift from M. Lee (Sanger Institute), also expresses the Cas9 enzyme and human dihydrofolate reductase (dhfr) selectable marker, as previously described (50). The homology region (fig. S8) was synthesized by Thermo Fisher Scientific in a pML-RQ (kanR) backbone. Ring-stage parasites were transfected by electroporation as previously described (51, 52), at 0.31 kV, 360 μF using 2-mm cuvettes. Cultures were treated with 5 nM WR99210 for 48 hours and fresh RPMI thereafter.
DHODH biochemical assay using XFe24 analyzer
DHO titration in mitochondria assay solution (MAS) was conducted as previously reported (43). Five different concentrations of DHO were prepared as 10× stocks in MAS and added to the parasites. Extracellular flux analyses were performed with the following settings: mix time, 30 s; wait time, 1 min and 30 s; measure time, 3 min. OCR increase was calculated as the difference of OCR values before and after DHO addition for each concentration.
Statistical analysis
For comparisons between parasite strains, nonparametric one-way analysis of variance (ANOVA; Kruskal-Wallis) tests were performed with post hoc multiple comparisons (Dunn’s test) using GraphPad Prism Software v.7 (GraphPad), with statistical significance defined as P < 0.05.
Supplementary Material
Fig. S1. Study design for in vitro selections.
Fig. S2. Study design for additional in vitro selections.
Fig. S3. Detection of copy number duplication of the dhodh locus in in vitro–isolated clones by real-time qPCR.
Fig. S4. General study design for in vivo selection of drug-resistant P. falciparum in SCID mice.
Fig. S5. Development of resistance to DSM265 in SCID mice infected with P. falciparum for groups B and D.
Fig. S6. Detection of copy number duplication of the dhodh locus in in vivo–isolated clones by real-time qPCR.
Fig. S7. Second competition assay confirming that the C276Y mutation has no fitness cost over time.
Fig. S8. CRISPR-Cas9–mediated introduction of the E182D mutation into the 3D7 A10 parasite strain.
Fig. S9. Bioenergetic analysis of wild-type and CRISPR-edited 3D7 A10DHODH E182D parasite lines.
Fig. S10. Efficacy of DSM265 against in vivo P. falciparum adapted lines in P. falciparum–infected SCID mice.
Fig. S11. Crystal structure of PfDHODH bound to DSM265 (PDB 4RX0) with mutations that confer resistance to DSM265 highlighted.
Table S1. Parasitemia in in vitro pulse selection experiments.
Table S2. Compound concentration (nM) that results in EC50 for all parasite lines described, with an SE and sample size (n) reported.
Table S3. Statistics for whole-genome sequencing.
Table S4. Characteristics of P. falciparum DHODH mutant lines reported in (26, 27).
Table S5. Population-level sequencing of the PfDHODH gene after in vivo resistance selections.
Table S6. OCR from Seahorse bioenergetics assay.
Table S7. Additional primers used for Sanger sequencing of the dhodh locus.
Table S8. Primers and probes for high resolution melt assay.
Data file S4. Detection of parasitemia during infection of SCID mice with DSM265-resistant parasites.
Data file S3. CNV across the Plasmodium genome as detected by whole-genome sequencing reads.
Data file S1. Individual biological replicate values for dose response (EC50) of parasite lines.
Data file S2. Summary of all homozygous variants identified across all selections.
Acknowledgments:
We thank M. Phillips (UT Southwestern) and J. Burrows (Medicines for Malaria Venture) for providing DSM265 and DSM267 and for valuable discussions. We also thank O. Vandal (Bill and Melinda Gates Foundation) and D. Fidock (Columbia University) for helpful discussions and S. Bopp and R. Daniels for help with experimental design. We are also grateful to M. Reynolds and P. Hinkson for technical support.
Funding: This work was funded, in part, by the NIH (grant no. R01 AI093716 (to D.F.W.), a Bill and Melinda Gates Foundation Grand Challenges Exploration grant no. OPP1132451 (to D.F.W., A.K.L., and FJ.G.), and the Harvard Malaria Initiative with support from ExxonMobil Foundation (to D.F.W.). R.E.K.M. was additionally supported by the Harvard Herchel Smith Fellowship, an NIH T32 grant, and funds from the ExxonMobil Foundation.
Footnotes
Competing interests: M.J.L.-M., D.S., A.P.-T., S.V., N.M., and F.J.G. are GlaxoSmithKline employees. D.F.W. is chairwoman, Malaria Policy Advisory Committee, World Health Organization and sits on the advisory boards of Medicines for Malaria Venture and Warren Alpert Foundation. E.A.W. sits on the advisory board of the Tres Cantos Open Lab Foundation. The other authors declare that they no competing interests.
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials. Whole-genome sequencing data for this project can be accessed from the Sequence Read Archive (SRA), accession number SRP158310. Drug-resistant parasite lines can be made available upon request to D.F.W. under a material transfer agreement. The DHODH inhibitors DSM265 and DSM267 can be obtained from Medicines for Malaria Venture (MMV) under a material transfer agreement.
REFERENCES AND NOTES
- 1.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier L Ma, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM,Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Menard D, A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505, 50–55 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaratunga C, Sreng S, Suon S, Phelps ES, Stepniewska K, Lim P, Zhou C, Mao S, Anderson JM, Lindegardh N, Jiang H, Song J, Su XZ, White NJ, Dondorp AM, Anderson J, Fay MP, Mu J, Duong S, Fairhurst RM, Artemisinin-resistant Plasmodium falciparum in Pursat province, western Cambodia: A parasite clearance rate study. Lancet Infect. Dis 12, 851–858 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM; Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium, Evidence of artemisinin-resistant malaria in Western Cambodia. N. Engl. J. Med 359, 2619–2620 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Imwong M, Suwannasin K, Kunasol C, Sutawong K, Mayxay M, Rekol H, Smithuis FM, Hlaing TM, Tun KM, van der Pluijm RW, Tripura R, Miotto O, Menard D, Dhorda M, Day NPJ, White NJ, Dondorp AM, The spread of artemisinin-resistant Plasmodium falciparum in the Greater Mekong subregion: A molecular epidemiology observational study. Lancet Infect. Dis 17, 491–497 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clyde DF, Shute GT, Resistance of East African varieties of Plasmodium falciparum to pyrimethamine. Trans. R. Soc. Trop. Med. Hyg 48, 495–500 (1954). [DOI] [PubMed] [Google Scholar]
- 6.Jones SA, Resistance of P. falciparum and P. malariae to pyrimethamine (daraprim) following mass treatment with this drug; a preliminary note. East Afr. Med. J 31, 47–49 (1954). [PubMed] [Google Scholar]
- 7.Basco LK, Eldin de Pecoulas P, Wilson CM, Le Bras J, Mazabraud A, Point mutations in the dihydrofolate reductase-thymidylate synthase gene and pyrimethamine and cycloguanil resistance in Plasmodium falciparum. Mol. Biochem. Parasitol 69, 135–138 (1995). [DOI] [PubMed] [Google Scholar]
- 8.Wongsrichanalai C, Pickard AL, Wernsdorfer WH, Meshnick SR, Epidemiology of drug-resistant malaria. Lancet Infect. Dis 2, 209–218 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Abdul-Ghani R, Farag HF, Allam AF, Sulfadoxine-pyrimethamine resistance in Plasmodium falciparum: A zoomed image at the molecular level within a geographic context. Acta Trop. 125, 163–190 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Blasco B, Leroy D, Fidock DA, Antimalarial drug resistance: Linking Plasmodium falciparum parasite biology to the clinic. Nat. Med 23, 917–928 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Looareesuwan S, Viravan C, Webster HK, Kyle DE, Hutchinson DB, Canfield CJ, Clinical studies of atovaquone, alone or in combination with other antimalarial drugs, for treatment of acute uncomplicated malaria in Thailand. Am. J. Trop. Med. Hyg 54, 62–66 (1996). [DOI] [PubMed] [Google Scholar]
- 12.Roper C, Pearce R, Bredenkamp B, Gumede J, Drakeley C, Mosha F, Chandramohan D, Sharp, Antifolate antimalarial resistance in southeast Africa: A population-based analysis. The Lancet 361,1174–1181 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Luth MR, Gupta P, Ottilie S, Winzeler EA, Using in vitro evolution and whole genome analysis to discover next generation targets for antimalarial drug discovery. ACS Infect. Dis. 4, 301–314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nzila L Mwai, In vitro selection of Plasmodium falciparum drug-resistant parasite lines. J. Antimicrob. Chemother 65, 390–398 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korsinczky M, Chen N, Kotecka B, Saul A, Rieckmann K, Cheng Q, Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob. Agents Chemother 44, 2100–2108 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Musset L, Bouchaud O, Matheron S, Massias L, Le Bras J, Clinical atovaquone-proguanil resistance of Plasmodium falciparum associated with cytochrome b codon 268 mutations. Microbes Infect. 8, 2599–2604 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Schwöbel B, Alifrangis M, Salanti A, Jelinek T, Different mutation patterns of atovaquone resistance to Plasmodium falciparum in vitro and in vivo: Rapid detection of codon 268 polymorphisms in the cytochrome b as potential in vivo resistance marker. Malar. J 2, 5 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Witkowski B, Lelievre J, Barragan MJL, Laurent V, Su X-Z, Berry A, Benoit-Vical F, Increased tolerance to artemisinin in Plasmodium falciparum is mediated by a quiescence mechanism. Antimicrob. Agents Chemother 54, 1872–1877 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cowell N, Istvan ES, Lukens AK, Gomez-Lorenzo MG, Vanaerschot M, Sakata-Kato T, Flannery EL, Magistrado P, Owen E, Abraham M, LaMonte G, Painter HJ, Williams RM, Franco V, Linares M, Arriaga I, Bopp S, Corey VC, Gnädig NF, Coburn-Flynn O, Reimer C, Gupta P, Murithi JM, Moura PA, Fuchs O, Sasaki E, Kim SW, Teng CH, Wang LT, Akidil A, Adjalley S, Willis PA, Siegel D, Tanaseichuk O, Zhong Y, Zhou Y, Llinas M, Ottilie S, Gamo FJ, Lee MCS, Goldberg DE, Fidock DA, Wirth F, Winzeler EA, Mapping the malaria parasite druggable genome by using in vitro evolution and chemogenomics. Science 359, 191–199 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Corey VC, Lukens AK, Istvan ES, Lee MCS, Franco V, Magistrado P, Coburn-Flynn O, Sakata-Kato T, Fuchs O, Gnadig NF, Goldgof G, Linares M, Gomez-Lorenzo MG, De Cózar C, Lafuente-Monasterio MJ, Prats S, Meister S, Tanaseichuk O, Wree M, Zhou Y, Willis PA, Gamo F-J, Goldberg DE, Fidock DA, Wirth DF, Winzeler EA, A broad analysis of resistance development in the malaria parasite. Nat. Commun 7, 11901 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Phillips MA, Rathod PK, Plasmodium dihydroorotate dehydrogenase: A promising target for novel anti-malarial chemotherapy. Infect. Disord. Drug Targets 10, 226–239 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baldwin J, Michnoff CH, Malmquist NA, White J, Roth MG, Rathod PK, Phillips MA, High-throughput screening for potent and selective inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J. Biol. Chem 280, 21847–21853 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Pavadai E, El Mazouni F, Wittlin S, de Kock C, Phillips MA, Chibale K, Identification of new human malaria parasite Plasmodium falciparum dihydroorotate dehydrogenase inhibitors by pharmacophore and structure-based virtual screening. J. Chem. Inf. Model 56, 548–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Phillips MA, Gujjar R, Malmquist NA, White J, El Mazouni F, Baldwin J, Rathod PK, Triazolopyrimidine-based dihydroorotate dehydrogenase inhibitors with potent and selective activity against the malaria parasite Plasmodium falciparum. J. Med. Chem 51 , 3649–3653 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel V, Booker M, Kramer M, Ross L, Celatka CA, Kennedy LM, Dvorin JD, Duraisingh MT, Sliz P, Wirth DF, Clardy J, Identification and characterization of small molecule inhibitors of Plasmodium falciparum dihydroorotate dehydrogenase. J. Biol. Chem 283, 35078–35085 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ross LS, Lafuente-Monasterio MJ, Sakata-Kato T, Mandt REK, Gamo FJ, Wirth DF, Lukens AK, Identification of collateral sensitivity to dihydroorotate dehydrogenase inhibitors in Plasmodium falciparum. ACS Infect. Dis 4, 508–515 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ross LS, Gamo FJ, Lafuente-Monasterio MJ, Singh OM, Rowland P, Wiegand RC, Wirth DF, In vitro resistance selections for Plasmodium falciparum dihydroorotate dehydrogenase inhibitors give mutants with multiple point mutations in the drug-binding site and altered growth. J. Biol. Chem 289, 17980–17995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukens K, Ross LS, Heidebrecht R, Javier Gamo F, Lafuente-Monasterio MJ, Booker ML, Hartl DL, Wiegand RC, Wirth DF, Harnessing evolutionary fitness in Plasmodium falciparum for drug discovery and suppressing resistance. Proc. Natl. Acad. Sci. U.S.A 111 , 799–804 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guler JL, Freeman DL, Ahyong V, Patrapuvich R, White J, Gujjar R, Phillips MA, DeRisi J, Rathod PK, Asexual populations of the human malaria parasite, Plasmodium falciparum, use a two-step genomic strategy to acquire accurate, beneficial DNA amplifications. PLOS Pathog. 9, e1003375 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Phillips MA, Lotharius J, Marsh K, White J, Dayan A, White KL, Njoroge JW, El Mazouni F, Lao Y, Kokkonda S, Tomchick DR, Deng X, Laird T, Bhatia SN, March S, Ng L, Fidock DA, Wittlin S, Lafuente-Monasterio M, Benito FJG, Alonso LMS, Martinez MS, Jimenez-Diaz MB, Bazaga SF, Angulo-Barturen I, Haselden JN, Louttit J, Cui Y, Sridhar A, Zeeman AM, Kocken C, Sauerwein R, Dechering K, Avery M, Duffy S, Delves M, Sinden R, Ruecker A, Wickham KS, Rochford R, Gahagen J, Iyer L, Riccio E, Mirsalis J, Bathhurst I, Rueckle T, Ding X, Campo B, Leroy, Rogers MJ, Rathod PK, Burrows JN, Charman SA, A long-duration dihydroorotate dehydrogenase inhibitor (DSM265) for prevention and treatment of malaria. Sci. Transl. Med 7, 296ra111 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bedingfield PTP, Cowen D, Acklam P, Cunningham F, Parsons MR, McConkey GA, Fishwick WG, Johnson AP, Factors influencing the specificity of inhibitor binding to the human and malaria parasite dihydroorotate dehydrogenases. J. Med. Chem 55, 5841–5850 (2012). [DOI] [PubMed] [Google Scholar]
- 32.White J, Dhingra SK, Deng X, El Mazouni F, Lee MCS, Afanador GA, Lawong A, Tomchick R, Ng CL, Bath J, Rathod PK, Fidock DA, Phillips MA, Identification and mechanistic understanding of dihydroorotate dehydrogenase point mutations in Plasmodium falciparum that confer in vitro resistance to the clinical candidate DSM265 ACS Infect. Dis 5, 90–101 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Llanos-Cuentas, Casapia M, Chuquiyauri R, Hinojosa JC, Kerr N, Rosario M, Toovey S, Arch RH, Phillips MA, Rozenberg FD, Bath J, Ng CL, Cowell AN, Winzeler, Fidock DA, Baker M, Mohrle JJ, Hooft van Huijsduijnen R, Gobeau N, Araeipour N, Andenmatten N, RQckle T, Duparc S, Antimalarial activity of single-dose DSM265, a novel plasmodium dihydroorotate dehydrogenase inhibitor, in patients with uncomplicated Plasmodium falciparum or Plasmodium vivax malaria infection: A proof-of-concept, open-label, phase 2a study. Lancet Infect. Dis 18, 874–883 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Angulo-Barturen, Jiménez-Díaz MB, Mulet T, Rullas J, Herreros E, Ferrer S, Jiménez E Mendoza A, Regadera J, Rosenthal PJ, Bathurst I, Pompliano DL, de las Heras Gómez, Gargallo-Viola D, A murine model of falciparum-malaria by in vivo selection of competent strains in non-myelodepleted mice engrafted with human erythrocytes. PLOS ONE 3, e2252 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coteron JM, Marco M, Esquivias J, Deng X, White KL, White J, Koltun M, El Mazouni F, Kokkonda S, Katneni K, Bhamidipati R, Shackleford DM, Angulo-Barturen SB Ferrer MB Jimenez-Diaz, Gamo FJ, Goldsmith EJ, Charman WN, Bathurst I, Floyd D, Matthews D, Burrows JN, Rathod PK, Charman SA, Phillips MA, Structure-guided lead optimization of triazolopyrimidine-ring substituents identifies potent Plasmodium falciparum dihydroorotate dehydrogenase inhibitors with clinical candidate potential. J. Med. Chem 54, 5540–5561 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jiménez-Díaz MB, Mulet T, Viera S, Gómez V, Garuti H, Ibáñez J, Alvarez-Doval A, Shultz D, Martinez A, Gargallo-Viola D, Angulo-Barturen I, Improved murine model of malaria using Plasmodium falciparum competent strains and non-myelodepleted NOD-scid IL2Rγnull mice engrafted with human erythrocytes. Antimicrob. Agents Chemother 53, 4533–4536 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peters W, Drug resistance in Plasmodium berghei Vincke and Lips, 1948. I. Chloroquine resistance. Exp. Parasitol 17, 80–89 (1965). [DOI] [PubMed] [Google Scholar]
- 38.Barrick JE, Lenski RE, Genome dynamics during experimental evolution. Nat. Rev. Genet 14, 827–839 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenthal PJ, The interplay between drug resistance and fitness in malaria parasites. Mol. Microbiol 89, 1025–1038 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baniecki ML, Faust AL, Schaffner SF, Park DJ, Galinsky K, Daniels RF, Hamilton E, Ferreira MU, Karunaweera ND, Serre D, Zimmerman PA, Sa JM, Wellems TE, Musset L, Legrand E, Melnikov A, Neafsey DE, Volkman SK, Wirth DF, Sabeti PC, Development of a single nucleotide polymorphism barcode to genotype Plasmodium vivax infections. PLOS Negl. Trop. Dis 9, e0003539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Daniels R, Hamilton EJ, Durfee K, Ndiaye D, Wirth DF, Hartl DL, Volkman SK, Methods to increase the sensitivity of high resolution melting single nucleotide polymorphism genotyping in malaria. J. Vis. Exp, 52839 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Daniels R, Ndiaye D, Wall M, McKinney J, Sene PD, Sabeti PC, Volkman SK,Mboup S, Wirth DF, Rapid, field-deployable method for genotyping and discovery of single-nucleotide polymorphisms associated with drug resistance in Plasmodium falciparum. Antimicrob. Agents Chemother 56, 2976–2986 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sakata-Kato T, Wirth DF, A novel methodology for bioenergetic analysis of Plasmodium falciparum reveals a glucose-regulated metabolic shift and enables mode of action analyses of mitochondrial inhibitors. ACS Infect. Dis. 2, 903–916 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Christophers SR, Fulton JD, Experiments with isolated malaria parasites (Plasmodium Knowlesi) free from red cells. Ann. Trop. Med. Parasitol 33, 161–170 (1939). [Google Scholar]
- 45.Goldberg DE, Siliciano RF, Jacobs WR Jr., Outwitting evolution: Fighting drug-resistant TB, malaria, and HIV. Cell 148, 1271–1283 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lambros C, Vanderberg JP, Synchronization of Plasmodium falciparum erythrocytic stages in culture. J. Parasitol 65, 418–420 (1979). [PubMed] [Google Scholar]
- 47.Johnson JD, Dennull RA, Gerena L, Lopez-Sanchez M, Roncal NE, Waters NC, Assessment and continued validation of the malaria SYBR green I-based fluorescence assay for use in malaria drug screening. Antimicrob. Agents Chemother 51, 1926–1933 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Smilkstein M, Sriwilaijaroen N, Kelly JX, Wilairat P, Riscoe M, Simple and inexpensive fluorescence-based technique for high-throughput antimalarial drug screening. Antimicrob. Agents Chemother 48, 1803–1806 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Desjardins RE, Canfield CJ, Haynes JD, Chulay JD, Quantitative assessment of antimalarial activity in vitro by a semiautomated microdilution technique. Antimicrob. Agents Chemother 16, 710–718 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ng CL, Siciliano G, Lee MCS, de Almeida MJ, Corey VC, Bopp SE, Bertuccini L, Wittlin S, Kasdin RG, Le Bihan A, Clozel M, Winzeler EA, Alano P, Fidock DA, CRISPR-Cas9-modified pfmdr1 protects Plasmodium falciparum asexual blood stages and gametocytes against a class of piperazine-containing compounds but potentiates artemisinin-based combination therapy partner drugs. Mol. Microbiol 101, 381–393 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wu Y, Sifri CD, Lei HH, Su XZ, Wellems TE, Transfection of Plasmodium falciparum within human red blood cells. Proc. Natl. Acad. Sci. U.S.A 92, 973–977 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fidock DA, Wellems TE, Transformation with human dihydrofolate reductase renders malaria parasites insensitive to WR99210 but does not affect the intrinsic activity of proguanil. Proc. Natl. Acad. Sci. U.S.A 94, 10931–10936 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Study design for in vitro selections.
Fig. S2. Study design for additional in vitro selections.
Fig. S3. Detection of copy number duplication of the dhodh locus in in vitro–isolated clones by real-time qPCR.
Fig. S4. General study design for in vivo selection of drug-resistant P. falciparum in SCID mice.
Fig. S5. Development of resistance to DSM265 in SCID mice infected with P. falciparum for groups B and D.
Fig. S6. Detection of copy number duplication of the dhodh locus in in vivo–isolated clones by real-time qPCR.
Fig. S7. Second competition assay confirming that the C276Y mutation has no fitness cost over time.
Fig. S8. CRISPR-Cas9–mediated introduction of the E182D mutation into the 3D7 A10 parasite strain.
Fig. S9. Bioenergetic analysis of wild-type and CRISPR-edited 3D7 A10DHODH E182D parasite lines.
Fig. S10. Efficacy of DSM265 against in vivo P. falciparum adapted lines in P. falciparum–infected SCID mice.
Fig. S11. Crystal structure of PfDHODH bound to DSM265 (PDB 4RX0) with mutations that confer resistance to DSM265 highlighted.
Table S1. Parasitemia in in vitro pulse selection experiments.
Table S2. Compound concentration (nM) that results in EC50 for all parasite lines described, with an SE and sample size (n) reported.
Table S3. Statistics for whole-genome sequencing.
Table S4. Characteristics of P. falciparum DHODH mutant lines reported in (26, 27).
Table S5. Population-level sequencing of the PfDHODH gene after in vivo resistance selections.
Table S6. OCR from Seahorse bioenergetics assay.
Table S7. Additional primers used for Sanger sequencing of the dhodh locus.
Table S8. Primers and probes for high resolution melt assay.
Data file S4. Detection of parasitemia during infection of SCID mice with DSM265-resistant parasites.
Data file S3. CNV across the Plasmodium genome as detected by whole-genome sequencing reads.
Data file S1. Individual biological replicate values for dose response (EC50) of parasite lines.
Data file S2. Summary of all homozygous variants identified across all selections.