Abstract
This paper proposes a representation framework for encoding spatial language in radiology based on frame semantics. The framework is adopted from the existing SpatialNet representation in the general domain with the aim to generate more accurate representations of spatial language used by radiologists. We describe Rad-SpatialNet in detail along with illustrating the importance of incorporating domain knowledge in understanding the varied linguistic expressions involved in different radiological spatial relations. This work also constructs a corpus of 400 radiology reports of three examination types (chest X-rays, brain MRIs, and babygrams) annotated with fine-grained contextual information according to this schema. Spatial trigger expressions and elements corresponding to a spatial frame are annotated. We apply BERT-based models (BERTBASE and BERTLARGE) to first extract the trigger terms (lexical units for a spatial frame) and then to identify the related frame elements. The results of BERTLARGE are decent, with F1 of 77.89 for spatial trigger extraction and an overall F1 of 81.61 and 66.25 across all frame elements using gold and predicted spatial triggers respectively. This frame-based resource can be used to develop and evaluate more advanced natural language processing (NLP) methods for extracting fine-grained spatial information from radiology text in the future.
Keywords: spatial relations, radiology, frame semantics
1. Introduction
Radiology reports are one of the most important sources of medical information about a patient and are thus one of the most-targeted data sources for natural language processing (NLP) in medicine (Pons et al., 2016). These reports describe a radiologist’s interpretation of one or more two- or three-dimensional images (e.g., X-ray, computed tomography, magnetic resonance imaging, ultrasound, positron emission tomography). As a consequence, these reports are filled with spatial relationships between medical findings (e.g., tumor, pneumonia, inflammation) or devices (e.g., tube, stent, pacemaker) and some anatomical location (e.g., right ventricle, chest cavity, T4, femur). These relations describe complex three-dimensional interactions, requiring a combination of rich linguistic representation and medical knowledge in order to process.
Table 1 shows an example radiology report.1 The example demonstrates the large number of medical findings and the relationships with various anatomical entities often found within radiology reports.
Table 1:
Example radiology report containing spatial language. Findings are in green, anatomical locations are in blue, while the spatial expressions are in cyan. Note: XXXX corresponds to phrases stripped by the automatic de-identifier.
| Comparison: XXXX PA and lateral chest radiographs |
| Indication: XXXX-year-old male, XXXX. |
|
Findings: There is XXXX increased opacity
within the right upper lobe with possible mass and associated area of atelectasis or focal consolidation. The cardiac silhouette is within normal limits. XXXX opacity in the left midlung overlying the posterior left 5th rib may represent focal airspace disease. No pleural effusion or pneumothorax. No acute bone abnormality. |
|
Impression: 1. Increased opacity
in the right upper lobe with XXXX associated atelectasis may represent focal consolidation or mass lesion with atelectasis. Recommend chest CT for further evaluation. 2. XXXX opacity overlying the left 5th rib may represent focal airspace disease. |
A flexible way of incorporating fine-grained linguistic representations with knowledge is through the use of frames for spatial relations (Petruck and Ellsworth, 2018). Notably, SpatialNet (Ulinski et al., 2019) extends the use of FrameNet-style frames with enabling the connection of these frames to background knowledge about how entities may interact in spatial relationships.
In this paper, we propose an extension of SpatialNet for radiology, which we call Rad-SpatialNet. Rad-SpatialNet is composed of 8 broad spatial frame types (as instantiated by the relation types), 9 spatial frame elements, and 14 entity types. We also describe an initial study in the annotation of the Rad-SpatialNet schema on a corpus of 400 radiology reports comprising of 3 different imaging modalities from the MIMIC-III clinical note corpus (Johnson et al., 2016). The annotated dataset will soon be made publicly available. There are 1101 sentences that contain at least one spatial relation in the corpus. 1372 spatial trigger expressions (lexical units for spatial frames) are identified in total with the most frequently occurring spatial relation types being ‘Containment’, ‘Description’, and ‘Directional’. Finally, we describe a baseline system for automatically extracting this information using BERT (Devlin et al., 2019) based on deep bi-directional transformer architecture.
2. Related Work
A range of existing work has focused on extracting isolated radiological entities (e.g., findings/locations) utilizing NLP in radiology reports (Hassanpour and Langlotz, 2016; Hassanpour et al., 2017; Cornegruta et al., 2016; Bustos et al., 2019; Annarumma et al., 2019). There exists recently published radiology image datasets labeled with important clinical entities extracted from corresponding report text (Wang et al., 2017; Irvin et al., 2019). Most of these studies have been targeted toward attribute extraction from text without focusing on recognizing relations among these entities. Only a few studies have focused on relation extraction from radiology text (Sevenster et al., 2012; Yim et al., 2016; Steinkamp et al., 2019). However, the datasets were limited to specific report types (e.g., hepatocellular carcinoma) and the relations extracted do not capture spatial information.
Extracting spatial relations has been studied in the general-domain for various purposes such as geographic information retrieval (Yuan, 2011), text-to-scene generation (Coyne et al., 2010; Coyne and Sproat, 2001), and human-robot interaction (Guadarrama et al., 2013). One of the schemas developed for representing spatial language in text is spatial role labeling (Kordjamshidi et al., 2010; Kordjamshidi et al., 2017). In the medical domain, some studies have extracted spatial relations from text including biomedical and consumer health text (Kordjamshidi et al., 2015; Roberts et al., 2015). In radiology, two prior works have identified spatial relations between radiological findings and anatomical locations, however, the reports were specific to appendicitis and did not extract other common clinically significant contextual information linked to the spatial relations (Roberts et al., 2012; Rink et al., 2013).
Frame semantics provide a useful way to represent information in text and has been utilized in constructing semantic frames to encode spatial relations (Petruck and Ellsworth, 2018). The Berkeley FrameNet project (Baker, 2014) contains 29 spatial frames with a total of 409 spatial relation lexical units. A recent work (Ulinski et al., 2019) proposed a framework known as SpatialNet to map spatial language expressions to actual spatial orientations by utilizing resources such as FrameNet (Baker, 2014) and VigNet (Coyne et al., 2011). In this paper, we aim to extend the SpatialNet framework to encode spatial language in radiology and utilize the framework to develop a dataset of reports annotated with detailed spatial information.
3. Rad-SpatialNet Description
Rad-SpatialNet provides a framework description to represent fine-grained spatial information in radiology reports by converting the linguistic expressions denoting any spatial relations to radiology-specific spatial meanings. For this, we extend the core design proposed in the general domain SpatialNet (Ulinski et al., 2019), which is based on FrameNet and VigNet, and tailor the framework specifically to encode spatial language in the domain of radiology. We have presented an overview of radiological spatial relations and the main participating entities in Figure 1. SpatialNet describes spatial frames by linking surface language to lexical semantics and further mapping these frames to represent the real spatial configurations. We update SpatialNet in this work with the aim to disambiguate the various spatial expressions used by radiologists in documenting their interpretations from radiographic images.
Figure 1:
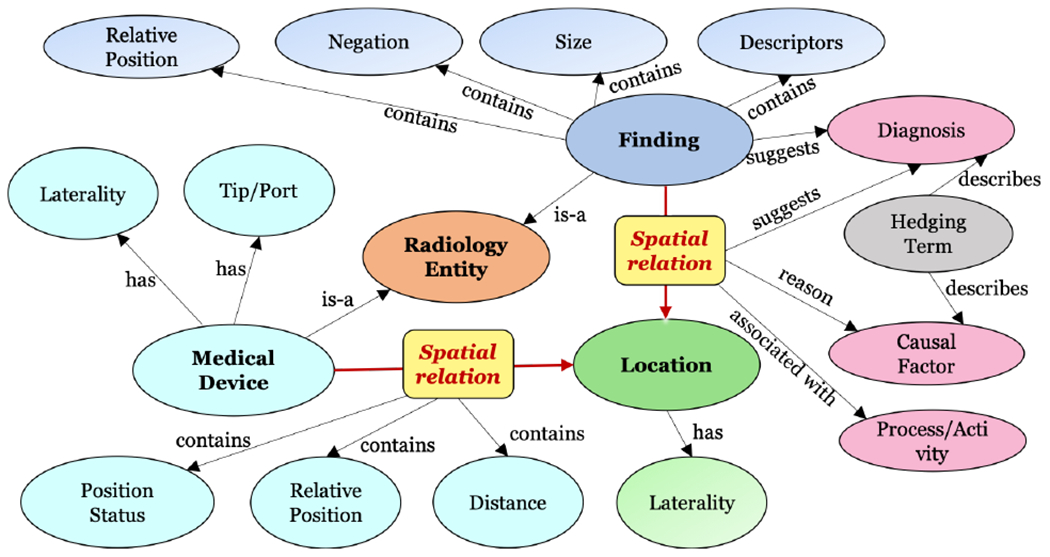
Relationship between entities in Rad-SpatialNet
To build Rad-SpatialNet, we first utilize the language in radiology reports to construct a set of spatial frames leveraging linguistic rules or valence patterns. The fundamental principle in forming the radiology spatial frames is the same used for constructing frames in FrameNet/SpatialNet. However, the main difference is that the target words (lexical units) of the frames are the spatial trigger words which are more common in radiology and are usually prepositions, verbs, and prepositional verbs. We then construct a list of spatial vignettes to transform these high-level spatial frames into more fine-grained frame versions by incorporating semantic, contextual or relation type constraints as well as radiology domain knowledge. The fine-grained frames reveal the true meaning of the spatial expressions from a radiology perspective. We describe these final frames containing the actual spatial configurations as the spatio-graphic primitives. Unlike SpatialNet that uses the VigNet ontology to map the different lexical items into semantic categories, we utilize the publicly available radiology lexicon, RadLex, to map different radiological entities mentioned in the reports to standard terminologies recommended in radiology practice.
The main components involved in the proposed Rad-SpatialNet framework are described in the following subsections.
3.1. Ontology of Radiology Terms
We leverage RadLex (Langlotz, 2006) to map the various radiological entities along with other clinically important information in the report text to standard unified vocabularies to facilitate standardized reporting and decision support in radiology practice as well as research. RadLex consists of a set of standardized radiology terms with their corresponding codes in a hierarchical structure. Rad-SpatialNet utilizes the RadLex ontology to map all possible contextual information with reference to any spatial relation in the reports to the broader RadLex classes which capture the semantic types of the various information. For example, ‘Endotracheal tube’ which is a TUBE and a type of Implantable Device is mapped to the broad RadLex class Medical Device. Similarly, ‘Ground-glass opacity’ belongs to the RadLex class Opacity which falls under the broad class Imaging Observation. Thus, terms such as ‘opacity’, ‘opacification’, and ‘Ground-glass opacity’ are mapped to Imaging Observation. We also link the various modifier or descriptor entities to standard RadLex descriptors. For instance, in ‘roundedparenchymal opacity’, ‘rounded’ is mapped to the Morphologic Descriptor class of RadLex, which is one of the categories of RadLex descriptors. Unlike VigNet, RadLex does not contain any graphical relations representing spatial configurations. So, we utilize Radlex mainly to map the terms in reports to radiology-specific semantic categories and not for creating spatio-graphic primitives. The mapped entities are utilized in the following steps to construct the spatial frames and subsequently the radiology-specific spatio-graphic primitives.
3.2. Spatial Frames
The spatial frames organize information in a radiology report sentence containing any spatial relation between common radiological entities (e.g., imaging observations and anatomical structures) according to the frame semantic principles, similar to FrameNet and SpatialNet. All the spatial frames created are inherited from the Spatial-Contact frame in FrameNet. We adopt similar valence patterns as defined in SpatialNet by specifying various lexical and syntactic constraints to automatically identify the frame elements from a sentence. However, there are differences in the set of frame elements in Rad-SpatialNet compared to SpatialNet. For example, besides Figure and Ground, some of the other common elements in the Rad-SpatialNet frames are Hedge, Diagnosis, Distance, and Relative Position.
To do this, we identify the most frequent words or phrases expressing spatial relations in radiology. Such a word/phrase also forms the lexical unit for a spatial frame. The type or the sense of the spatial trigger is also recognized to include the spatial relation type information in the frame. For example, in the sentence describing the exact position of a medical device - ‘The umbilical venous catheter tip is now 1 cm above the right hemidiaphragm’, ‘above’ is the spatial trigger having a directional sense. This instantiates a spatial frame with Directional as the relation type and above.prep as the lexical unit. Some other common spatial relation types in Rad-SpatialNet are Containment, triggered by lexical units such as in.prep, within.prep, and at.prep; Descriptive, triggered by lexical units such as shows.v, are.v and with.prep; and Spread triggered by lexical units such as extend (into).prep, through-out.prep, and involving.v. The spatial relation types are shown in Table 2 and the elements identified for the spatial frames are described in Table 3.
Table 2:
Broader categories of spatial relations in radiology
| Relation Type | Description |
|---|---|
| Containment | Denotes that a finding/observation/device is contained within an anatomical location (“There is again seen high T2 signal within the mastoid air cells bilaterally”) |
| Directional | Denotes a directional sense in which a radiological entity is described wrt location (“An NGT has its tip below the diaphragm”) |
| Contact | Denotes an entity is in contact with an anatomical structure (“NGT reaches the stomach”) |
| Encirclement | Denotes a finding is surrounding an anatomical location or another finding (“Left temporal hemorrhage with surrounding edema is redemonstrated”) |
| Spread | Denotes traversal of an entity toward an anatomical location (“An NG tube extends to the level of the diaphragms.”) |
| Description | Denotes an anatomical location being described with any abnormality or observation (“There is also some opacification of the mastoid air cells.”) |
| Distance | Denotes a qualitative distance between a radiographic finding and an anatomical location (“There are areas of T2 hyperintensity near the lateral ventricles.”) |
| Adjacency | Denotes a radiographic finding is located adjacent to a location (“There is a small amount of hypodensity adjacent to the body of the right lateral ventricle.”) |
Table 3:
Frame elements in Rad-SpatialNet
| Element | Description |
|---|---|
| Elements with respect to a spatial trigger | |
|
| |
| Figure | The object whose location is described through the spatial trigger (usually refers to finding/location/disorder/device/anatomy/tip/port) |
| Ground | The anatomical location of the trajector described (usually an anatomical structure) |
| Hedge | Uncertainty expressions used by radiologists (e.g., ‘ could be related to’, ‘may concern for’ etc.) |
| Diagnosis | Clinical condition/disease associated with finding/observation suggested as differential diagnoses, usually appears after the hedge related terms |
| Reason | Clinical condition/disease that acts as the source of the finding/observation/disorder |
| Relative Position | Terms used for describing the orientation of a radiological entity wrt to an anatomical location (e.g., ‘posteriorly’ in “Blunting of the cosmiddlehrenic sulci posteriorly is still present”, ‘high’ in “The UV line tip is high in the right atrium.”) |
| Distance | The actual distance of the finding or device from the anatomical location (e.g., ‘1 cm’ in “ETT tube is 1 cm above the carina.”) |
| Position Status | Any position-related information, usually in context to a device (e.g., ‘terminates’ in “A right PIC catheter terminates in the mid SVC.”) |
| Associated Process | Any process/activity associated with a spatial relation (e.g., ‘intubation’ in “may be related to recent intubation”) |
|
| |
| Elements with respect to a radiological entity | |
|
| |
| Status | Indicating status of entities (e.g., ‘stable’, ‘normal’, ‘mild’) |
| Morphologic | Indicating shape (e.g., ‘rounded’) |
| Density | Terms referring to densities of findings/observations (e.g., ‘hypodense’, ‘lucent’) |
| Modality | Indicating modality characteristics (e.g., ‘attenuation’) |
| Distribution | Indicating distribution patterns (e.g., ‘scattered’, ‘diffuse’) |
| Temporal | Indicating any temporality (e.g., ‘new’, ‘chronic’) |
| Composition | Indicating composition of any radiological observation (e.g., ‘calcified’) |
| Negation | The negated phrase related to a finding/observation (e.g., ‘without evidence of’) |
| Size | The actual size of any finding/observation (e.g., ‘14-mm’ describing the size of a lytic lesion) |
| Laterality | Indicating side (e.g., ‘left’, ‘bilateral’) |
| Quantity | Indicating the quantity of any radiological entity (e.g., ‘multiple’, ‘few’) |
The semantic type of the Figure and Ground elements are identified for each spatial frame constructed using the RadLex ontology. For the example above related to the positioning of the umbilical venous catheter tip, the semantic type of Figure is Medical Device and the type of Ground is Anatomical Location. All this information-combining the semantic types and the spatial relation type-are used to further refine the spatial frame Figure-Directional-Ground-SF as Medical Device-Directional-Anatomical Location-SF.
3.3. Spatial Vignettes
The main idea of spatial vignettes are also adopted from SpatialNet. In this paper, we develop vignettes primarily to resolve the ambiguities involved in using the same spatial trigger words or phrases to describe different radiological contexts. In other words, the same spatial expressions might have different spatial configurations based on the context or the radiological entities associated.
The vignettes connect the spatial frames to spatio-graphic primitives utilizing the RadLex ontology, semantic/relation type constraints as well as domain knowledge to generate more accurate spatial representations of radiology language. Consider the following two sentences having the same spatial trigger ‘extends into’:
There is interval increase in the right pleural effusion which extends into the fissure.
There is an NG tube which extends into the stomach.
The first sentence contains a radiologist’s description of a fluid disorder ‘pleural effusion’ with respect to an anatomical reference–fissure in the pleural cavity (a closed space around the lungs), whereas the second sentence describes the positioning of a feeding tube. it is difficult to interpret the actual spatial meaning of the same prepositional verb ‘extends into’ in these two different contexts solely from the lexical information. The spatial vignettes map the spatial frames corresponding to these sentences to different spatio-graphic primitives representing the actual spatial orientation of ‘extends into’ utilizing radiology domain knowledge.
Semantic constraints are applied to the Figure and Ground frame elements, whereas a spatial relation type constraint is also added to the relation sense. If the semantic category of the Figure is Medical Device and the relation type is Directional, with the lexical unit extends (into).prep, then a spatial vignette will generate the spatio-graphic-primitive Medical Device-Terminates Into-Anatomical Location-SGP from the spatial frame Medical Device-Directional-Anatomical Location-SF (corresponding to example 2 above). Another vignette will produce the spatio-graphic primitive Disorder-Extends Into-Anatomical Location-SGP if the semantic category of the Figure is Disorder instead of Medical Device and particularly refers to fluid-related disorders like ‘pleural effusion’(corresponding to example 1 above). The vignettes here determine the spatial meanings of radiology sentences based on both the semantic types of Figure element and specific properties of the Disorder type (for example, fluidity in this case). Similar ambiguities are observed for spatial expressions containing lexcial units such as projects (over).prep and overlying.v as they often occur in both device and observation or disorder-related contexts. Thus, the vignettes differentiate the real orientation of the spatial expression when it is used in context to a medical device versus any other radiographic finding. Consider another set of examples below where both sentences are related to the tip position of two medical devices:
The umbilical arterial catheter tip projects over the T8–9 interspace.
The tip of the right IJ central venous line projects over the upper right atrium.
Here, the spatial vignettes produce a more specific spatial representation of the prepositional verb ‘projects over’ based on the details of anatomical location. A spatial vignette will produce the spatio-graphic primitive Medical Device Tip-In Front Of-Anatomical Location-SGP for the first sentence. IN Front Of is derived as the anatomical structure is corresponding to the ‘T8-9 interspace’ in the spine which is often used as the level of reference to indicate the position of catheters and tubes and these tubes and catheters are in front of the spine. Another vignette will produce Medical Device Tip-Terminates At Level Of-Anatomical Location-SGP for the second sentence as the IJ venous line lies within the internal jugular vein and might go upto the ‘right atrium’.
Consider the following sentences containing ‘overlying.v’ as the lexical unit:
There is a less than 1 cm diameter rounded nodular opacity overlying the 7th posterior rib level.
The left IJ pulmonary artery catheter’s tip is currently overlying the proximal SVC.
If the Figure is Imaging Observation and the Ground is particularly associated with anatomical locations such as ribs, a spatial vignette will produce the primitive Imaging Observation-Projects Over-Anatomical Location-SGP for the first sentence. Ribs are also used the same way as spine to describe the level of objects or pathology. However, for the second sentence, the spatio-graphic primitive will be Medical Device Tip-Terminates At Level Of-Anatomical Location-SGP as the Figure is Medical Device and the Ground (anatomical location) is ‘SVC’.
The following examples contain the spatial trigger ‘of’ connecting two anatomical structures or parts of anatomical structures:
There is increased signal identified within the pons extending to the right side of the midline.
The UA catheter tip overlies the left pedicle of the T9 vertebral body.
In the above cases with respect to the spatial preposition ‘of’, the semantic types of both Figure and Ground are referring to Anatomical Locations. However, there is a difference in the interpretation of the same trigger word ‘of’. The vignette adds a constraint that if the spatial relation type is Descriptive and both Figure and Ground have semantic type as Anatomical Location, then the meaning of the preposition is determined based on the words of the Figure element. If the words are either ‘side’ or ‘aspect’, then ‘of’ refers to a subarea/side of the Ground anatomical location. Whereas for other words, ‘of’ refers to a specific identifiable anatomical structure contained within the Ground element, similar to ‘pedicle’ present at each vertebra in Example 2 above. The spatial vignettes corresponding to the three spatial expressions described above - extends (into).prep, projects (over).prep, and of.prep are illustrated in Figure 2.
Figure 2:
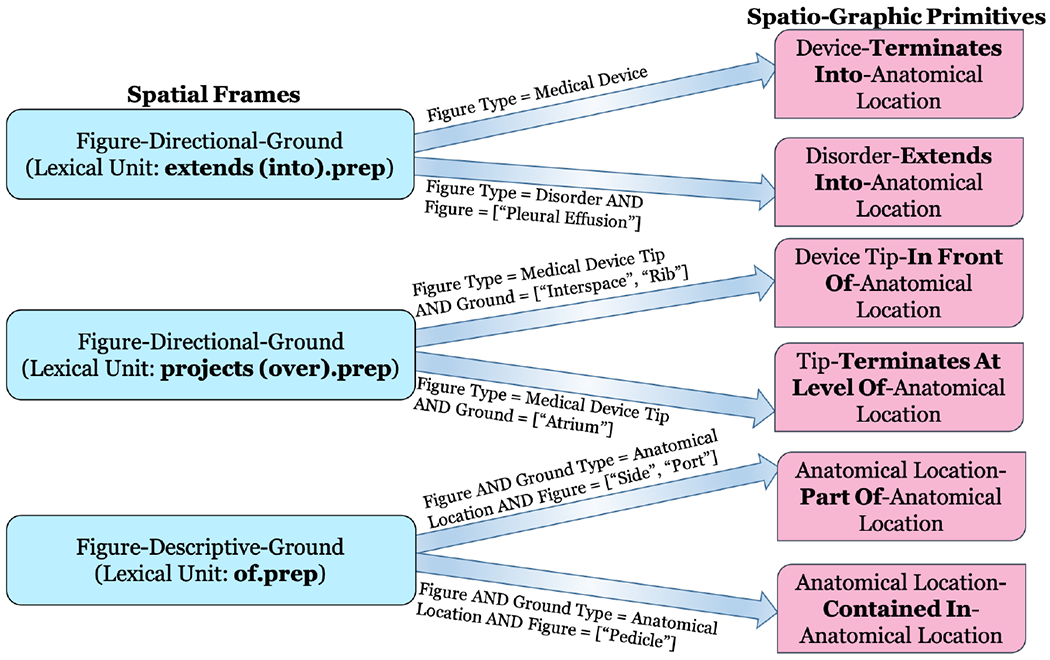
Examples of spatial vignettes for differentiating the spatial meanings of three commonly found spatial expressions in radiology reports.
4. Annotation Process
We annotated a total of 400 radiology reports–Chest X-ray reports (136), Brain MRI reports (127), and Babygram reports (137)–from the MIMIC III clinical database (Johnson et al., 2016). The language used in MIMIC reports is more complex and the report lengths as well as sentence lengths are long compared to other available datasets such as openi chest X-ray reports (Demner-Fushman et al., 2016). We filtered the babygram-related reports following the ‘babygram’ definition, that is, an X-ray of the whole body of an infant (usually newborn and premature infants). Since babygram reports have frequent mentions of medical device positions and involve multiple body organs, we incorporate this modality mainly with the intention to build a corpus with balance between two major spatially-grounded radiological entities–imaging observations/clinical findings and medical devices. We pre-processed the reports to de-identify some identifiable attributes including dates and names and also removed clinically less important contents. All the sentences in the reports containing potential spatial relations were annotated by two human annotators. The annotation was conducted using Brat. The annotations were reconciled three times–following the completion of 50, 200, and 400 reports. Since identifying the spatial frame elements involves interpreting the spatial language from the contextual information in a sentence, the annotations of the first 50 reports (i.e. our calibration phase) differed highly between the annotators. Some of the major disagreements related to spatial relation sense were discussed and the annotation guidelines were updated after first two rounds of annotation. Examples of sample annotations are provided in Figure 3.
Figure 3:
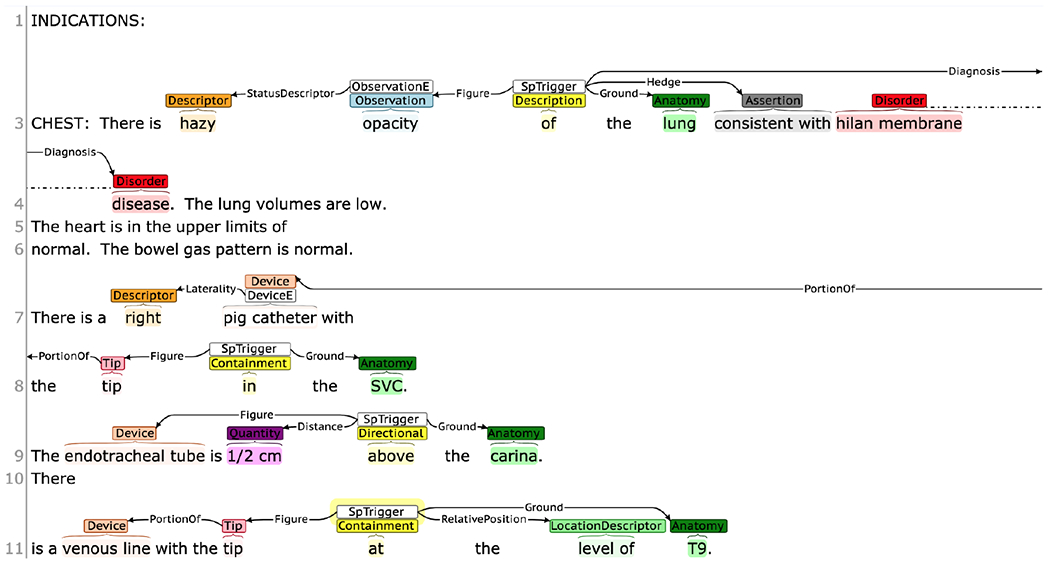
Examples of annotations.
To provide more insights into some of the complexities encountered in the annotation process, we highlight a few cases here. First, oftentimes two anatomical locations are present in a sentence and in such scenarios, an intermediate anatomical location (first location occurrence) is chained as a FIGURE element in context to the broader anatomical location (second location occurrence and the Ground element). However, this chaining is not valid if the two locations are not connected. Consider the sentences below:
There are few small air fluid levels in [mastoid air cells]FirstLocation within the left [mastoid process]SecondLocation.
Area of increased signal adjacent to the left lateral [ventricle]FirstLocation at the level of [corona radiata]SecondLocation.
For the first sentence, ‘mastoid air cells’ is contained within ‘mastoid process’ and these anatomical locations are connected. Therefore, ‘mastoid air cells’ is annotated as Figure element and ‘mastoid process’ as the GROUND in context to the spatial frame formed by the lexical unit ‘within.prep’. Note that ‘mastoid air cells’ is the Ground element associated with the Figure ‘air fluid levels’ through the spatial trigger ‘in’. However, for the second sentence, ‘ventricle’ and ‘corona radiata’ are two separate anatomical references and are not connected. Hence, two separate spatial relations are formed with the radiographic observation ‘signal’, one between ‘signal’ and ‘ventricle’ described through ‘adjacent to’ and the other between ‘signal’ and ‘corona radiata’ described through ‘at’ and the Relative Position ‘level of’. Second, some instances require correct interpretation of whether prepositions such as ‘of’ are Spatial Triggers or are part of location descriptors. Note the examples below:
PICC line with its tip located at the junction of superior vena cava and left brachiocephalic vein.
Abnormal signal in posterior portion of spinal cord.
In the first sentence, ‘junction of’ is annotated as the Relative Position describing the connection between ‘superior vena cava’ and ‘brachiocephalic vein’, whereas in the second sentence, ‘of’ is a Spatial Trigger connecting the Figure–‘portion’ and the Ground–‘spinal cord’. Third, although location descriptor words such as anterior, lateral, and superior are usually annotated as Relative Position, in a few cases they are annotated as Spatial Trigger. For example, note the following two sentences:
There is an area of high signal intensity extending into the [anterior]Relative Position mediastinum.
There is a 6 mm lymph node [anterior to]Spatial Trigger the carina.
In the first sentence, ‘anterior’ is used to describe ‘mediastinum’ and is annotated as a Relative Position, whereas in the second sentence, ‘anterior’ contributes in perceiving the actual spatial sense and hence ‘anterior to’ is annotated as a Spatial Trigger.
5. Results
5.1. Annotation statistics
The inter-annotator agreement results are shown in Table 4. We calculate the overall F1 agreement for annotating the spatial relation types, the main entities, and the spatial frame elements. The agreement measures are particularly low (around 0.4) for Finding/Observations as often there are higher chances of boundary mismatch in the process of separating the descriptor-related words from the main finding or observation term. There are very few instances of Process entities and cases where the disorder terms act as Reasons in the corpus which have resulted in the agreement measures being zero. Ultimately, Rad-SpatialNet requires an extremely knowledge-intensive annotation process, so low agreement at this stage is not unreasonable. Future work will include additiional quality checks to ensure the semantic correctness of the annotations.
Table 4:
Annotator agreement.
| Item | First 50 | Next 150 | Last 200 |
|---|---|---|---|
|
| |||
| Relation Types | |||
|
| |||
| Containment | 0.73 | 0.81 | 0.81 |
| Directional | 0.29 | 0.73 | 0.46 |
| Contact | 0.44 | 0.1 | 0.03 |
| Encirclement | 0.67 | 0.57 | 0 |
| Spread | 0.24 | 0.55 | 0.45 |
| Description | 0.42 | 0.54 | 0.58 |
| Distance | 0 | 0.5 | 0.4 |
| Adjacency | 0 | 0.25 | 0.44 |
|
| |||
| Main entities | |||
|
| |||
| Spatial Trigger | 0.58 | 0.81 | 0.78 |
| Anatomy | 0.48 | 0.68 | 0.76 |
| Device | 0.35 | 0.82 | 0.83 |
| Tip | 0.38 | 0.98 | 0.97 |
| Finding/Observation | 0.28 | 0.43 | 0.38 |
| Descriptors | 0.29 | 0.64 | 0.71 |
|
| |||
| Frame Elements | |||
|
| |||
| Figure | 0.33 | 0.58 | 0.62 |
| Ground | 0.42 | 0.67 | 0.70 |
| Diagnosis | 0 | 0.51 | 0.54 |
| Hedge | 0.19 | 0.48 | 0.45 |
| Reason | 0 | 0.38 | 0 |
| Relative Position | 0.07 | 0.48 | 0.58 |
| Distance | 0.4 | 0.86 | 0.71 |
| Position Status | 0 | 0.62 | 0.42 |
| Associated Process | 0.57 | 0 | 0 |
5.2. Corpus statistics
358 (89.5%) of the reports contain spatial relations. The reconciled annotations contain a total of 1101 sentences with mentions of spatial triggers. There are 1372 spatial trigger terms in total (average of 3.8 triggers per report). The frequencies of the entity types, the types of spatial relations, and the spatial frames are presented in Table 5. The predominant spatial trigger types to instantiate spatial frames are ‘Containment’, ‘Description’, and ‘Directional’. 81 of the 330 DEVICE entities are described using various descriptor terms, 327 of the 436 Observation entities, 240 of the 367 Clinical Findings, 190 of the 390 Disorders, and 564 of the 1492 Anatomical Locations contain descriptors. The distribution of various types of descriptors (consistent with RadLex) across the main radiological entities are shown in Table 6. Note that the figures in Table 5 and Table 6 are considering only those entities and their descriptors which are involved in a spatial relation. There are 1328 frame instances which correspond to the main radiological entities as demonstrated in Table 5. Among the remaining 44 spatial triggers, 11 are related to Port/Lead of devices and 33 describes how a specific part of an anatomical structure is linked to the main part.
Table 5:
General corpus statistics.
| Item | Freq |
|---|---|
| General | |
| Average sentence length | 17.6 |
| Spatial triggers | 1372 |
| Sentences with 1 spatial trigger | 874 |
| Sentences with more than 1 spatial trigger | 227 |
| Entity Types | |
| Medical Device | 330 |
| Tip of Device | 142 |
| Device Port/Lead | 11 |
| Imaging Observation | 436 |
| Clinical Finding | 367 |
| Disorder | 390 |
| Anatomical Location | 1492 |
| Descriptor | 1548 |
| Assertion | 326 |
| Quantity | 67 |
| Location Descriptor | 398 |
| Position Info | 167 |
| Process | 19 |
| Spatial Frames (SFs) | |
| Containment | 642 |
| Description | 387 |
| Directional | 168 |
| Spread | 69 |
| Contact | 54 |
| Adjacency | 32 |
| Encirclement | 14 |
| Distance | 6 |
| Most frequent Lexical Units - Containment SF | |
| ‘in.prep’ | 410 |
| ‘within.prep’ | 134 |
| ‘at.prep’ | 86 |
| Most frequent Lexical Units - Description SF | |
| ‘of.prep’ | 277 |
| ‘are.prep’ | 37 |
| ‘with.prep’ | 17 |
| Most frequent Lexical Units - Directional SF | |
| ‘above.prep’ | 43 |
| ‘projecting (over).prep’ | 24 |
| ‘below.prep’ | 18 |
| Spatial Frames based on semantic types | |
| Medical Device-related | 194 |
| Medical Device Tip-related | 142 |
| Imaging Observation-related | 436 |
| Clinical Finding-related | 344 |
| Disorder-related | 212 |
| Spatial frame elements | |
| Figure | 1491 |
| Ground | 1537 |
| Hedge | 249 |
| Diagnosis | 190 |
| Reason | 33 |
| Relative Position | 398 |
| Distance | 45 |
| Position Status | 167 |
| Associated Process | 21 |
Table 6:
Descriptive statistics of the radiological entities in the annotated corpus. (Obs - Observation, Fndg - Finding, Dis - Disorder, DEVC - Device, Aty - Anatomy)
| Element | Obs | Fndg | Dis | Devc | Aty |
|---|---|---|---|---|---|
|
| |||||
| Status | 231 | 121 | 77 | 1 | 22 |
| Quantity | 50 | 31 | 14 | 5 | 30 |
| Distribution | 43 | 14 | 6 | 0 | 2 |
| Morphologic | 37 | 20 | 6 | 0 | 6 |
| Size Desc. | 33 | 19 | 31 | 1 | 9 |
| Negation | 32 | 50 | 20 | 0 | 1 |
| Temporal | 23 | 26 | 50 | 14 | 0 |
| Laterality | 20 | 3 | 22 | 68 | 499 |
| Size | 19 | 1 | 3 | 0 | 0 |
| Composition | 5 | 8 | 2 | 0 | 2 |
| Density | 5 | 0 | 0 | 0 | 0 |
| Modality | 2 | 0 | 0 | 0 | 0 |
5.3. Baseline system performance
As an initial step of extracting the spatial trigger terms (lexical units of spatial frames) and the associated spatial information (spatial frame elements) in report sentences, we utilize both BERTBASE and BERTLARGE pre-trained language models as our baseline systems. We formulate both the tasks of lexical unit identification and frame elements extraction as sequence labeling task. We initialize the model parameters obtained by pre-training BERTBASE and BERTLARGE on MIMIC-III clinical notes (Si et al., 2019) for 300K steps and fine-tune the models on our constructed corpus. For fine-tuning, we set the maximum sequence length at 128, learning rate at 2e-5, number of training epochs at 4 and use cased version of the models.
10-fold cross validation is performed to evaluate the model’s performance with 80–10-10% training, validation, and test splits of the reports. First, the spatial triggers are extracted in a sentence. Second, we extract the common frame elements with respect to the trigger. For element extraction, we evaluate the system performance using both the gold spatial triggers and the predicted triggers on the test set. The results are shown in Table 7, Table 8, and Table 9.
Table 7:
10 fold CV results for spatial trigger extraction. P - Precision, R - Recall.
| Model | P (%) | R (%) | F1 |
|---|---|---|---|
| BERTBASE (MIMIC) | 92.20 | 43.04 | 57.52 |
| BERTLARGE (MIMIC) | 93.72 | 67.13 | 77.89 |
Table 8:
10 fold CV results for extracting spatial frame elements using BERTBASE (MIMIC). P - Precision, R - Recall.
| Main Frame Elements | Gold Spatial Triggers | Predicted Spatial Triggers | ||||
|---|---|---|---|---|---|---|
| P (%) | R(%) | F1 | P(%) | R(%) | F1 | |
| Figure | 75.14 | 84.10 | 79.35 | 63.26 | 40.48 | 48.00 |
| Ground | 84.96 | 88.90 | 86.87 | 68.83 | 42.95 | 51.64 |
| Hedge | 64.31 | 77.46 | 69.78 | 56.91 | 31.84 | 38.69 |
| Diagnosis | 53.99 | 79.54 | 63.93 | 38.89 | 26.48 | 29.18 |
| Relative Position | 81.27 | 75.56 | 78.12 | 67.01 | 39.91 | 48.81 |
| Distance | 86.64 | 87.50 | 84.87 | 77.17 | 73.67 | 74.29 |
| Position Status | 62.29 | 64.07 | 62.74 | 56.86 | 52.05 | 52.39 |
| Associated Process, Reason | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Overall | 76.53 | 82.69 | 79.48 | 63.98 | 40.73 | 48.55 |
Table 9:
10 fold CV results for extracting spatial frame elements using BERTLARGE (MIMIC). P - Precision, R - Recall.
| Main Frame Elements | Gold Spatial Triggers | Predicted Spatial Triggers | ||||
|---|---|---|---|---|---|---|
| P(%) | R(%) | F1 | P(%) | R(%) | F1 | |
| Figure | 77.42 | 85.75 | 81.35 | 65.51 | 65.44 | 65.12 |
| Ground | 88.92 | 91.57 | 90.22 | 73.31 | 70.21 | 71.51 |
| Hedge | 67.20 | 77.94 | 71.59 | 60.43 | 57.26 | 57.82 |
| Diagnosis | 51.96 | 79.81 | 62.31 | 47.06 | 57.64 | 50.76 |
| Relative Position | 81.31 | 78.42 | 79.57 | 66.02 | 67.76 | 66.33 |
| Distance | 86.83 | 87.50 | 87.00 | 86.50 | 90.24 | 88.05 |
| Position Status | 65.73 | 65.83 | 65.38 | 58.59 | 63.63 | 60.37 |
| Associated Process, Reason | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| Overall | 78.83 | 84.64 | 81.61 | 66.63 | 66.28 | 66.25 |
The results demonstrate that BERTLARGE performs better than BERTBASE for both spatial trigger prediction and frame elements prediction. However, in case of spatial trigger, the recall is low for both the models (Table 7). For element extraction, BERTBASE’s overall F1 combining all the frame elements is 79.48 (using gold spatial triggers) and 48.55 (using predicted triggers), whereas, for BERTLARGE, the difference in the overall F1 between using gold (81.61) versus predicted triggers (66.25) is much lower (15.4 vs 30.9). The F1 values are zeroes for Reason and Associated Process because of few occurrences in the dataset.
6. Discussion
The Rad-SpatialNet framework attempts to capture all possible contextual information in radiology reports from a spatial perspective. The aim is to utilize linguistic information as well as domain knowledge to accurately represent spatial language in radiology. The proposed examples of spatial vignettes described above have been validated by a practicing radiologist (SK). We are currently in the process of developing the valence patterns and vignettes by taking input from radiologists.
The results of the baseline system illustrates that it is difficult to identify the spatial trigger expression. This might be because of their wide variation in the reports and many of these appear as multi-word expressions. We only use the developed corpus to extract the core frame elements in a spatial frame. The results indicate that there is enough scope for improving the predictions by developing more advanced methods in the future.
In this work, we only consider intra-sentence spatial relations. Oftentimes, we encounter inter-sentence relations and scenarios where the differential diagnoses are documented in the sentence following the spatial relation or even far apart in the ‘Impression’ section (around 12.75% of the reports in our corpus). Other complex information in context to a spatial trigger could be considered for later work. Consider the sentence - ‘The tip of the catheter has a mild rightward curve, suggesting that it may be directed into a portal vein.’ Here, information about ‘intermediate position change’ might also be annotated as a frame element. Further, some phrases such as ‘needs repositioning’ can be differentiated from Position Status as Position Recommendation. Thus, further refinements to Rad-SpatialNet are still necessary.
7. Conclusion
This work aims to develop a resource for encoding fine-grained spatial information in radiology reports. For this, we extend an existing spatial representation framework in the open-domain, spatialNet, to accurately represent radiology spatial language. We describe the components of Rad-SpatialNet. Construction of linguistic rules and the spatial vignettes are currently under development. We annotate 400 radiology reports with important spatial information and apply a baseline model based on BERT to extract the spatial triggers and the related elements. The results demonstrate that the task of extracting fine-grained spatial information is challenging and the corpus constructed in this work can serve as an initial resource to develop methods with improved performance in the future.
Acknowledgments
This work was supported in part by the National institute of Biomedical imaging and Bioengineering (NIBIB: R21EB029575), the U.S. National Library of Medicine (NLM: R00LM012104), the Patient-Centered Outcomes Research Institute (PCORI: ME-2018C1-10963), and the Cancer Prevention Research Institute of Texas (CPRIT: RP160015).
Footnotes
Unlike the data annotated in this paper, the example in Table 1 is publicly available without restriction from openi.nlm.nih.gov (image ID: CXR1000_IM-0003-1001).
8. Bibliographical References
- Annarumma M, Withey SJ, Bakewell RJ, Pesce E, Goh V, and Montana G (2019). Automated Triaging of Adult Chest Radiographs with Deep Artificial Neural Networks. Radiology, 291(1):196–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker CF (2014). FrameNet : A Knowledge Base for Natural Language Processing. In Proceedings ofFrame Semantics in NLP: A Workshop in Honor ofChuck Fill-more, number 1968, pages 1–5. [Google Scholar]
- Bustos A, Pertusa A, Salinas J-M, and de la Iglesia-Vayá M. (2019). PadChest: A large chest x-ray image dataset with multi-label annotated reports. arXiv preprint arXiv:1901.07441. [DOI] [PubMed] [Google Scholar]
- Cornegruta S, Bakewell R, Withey S, and Montana G (2016). Modelling Radiological Language with Bidirectional Long Short-Term Memory Networks. In Proceedings of the Seventh International Workshop on Health Text Mining and Information Analysis, pages 17–27. [Google Scholar]
- Coyne B and Sproat R. (2001). WordsEye: An automatic text-to-scene conversion system. In Proceedings of the 28th Annual Conference on Computer Graphics and Interactive Techniques, pages 487–496. [Google Scholar]
- Coyne B, Sproat R, and Hirschberg J (2010). Spatial relations in text-to-scene conversion. In Computational Models of Spatial Language Interpretation, Workshop at Spatial Cognition, volume 620, pages 9–16. [Google Scholar]
- Coyne B, Bauer D, and Rambow O (2011). VigNet: Grounding Language in Graphics using Frame Semantics. In Proceedings of the ACL 2011 Workshop on Relational Models of Semantics, pages 28–36. [Google Scholar]
- Demner-Fushman D, Kohli MD, Rosenman MB, Shooshan SE, Rodriguez L, Antani S, Thoma GR, and McDonald CJ (2016). Preparing a collection of radiology examinations for distribution and retrieval. Journal of the American Medical Informatics Association, 23(2):304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin J, Chang M-W, Lee K, and Toutanova K (2019). BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. In Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, pages 4171–4186, Minneapolis, Minnesota, jun. Association for Computational Linguistics. [Google Scholar]
- Guadarrama S, Riano L, Golland D, Gohring D, Jia Y, Klein D, Abbeel P, and Darrell T (2013). Grounding spatial relations for human-robot interaction. In IEEE International Conference on Intelligent Robots and Systems, pages 1640–1647. [Google Scholar]
- Hassanpour S and Langlotz CP (2016). Information extraction from multi-institutional radiology reports. Artificial Intelligence in Medicine, 66:29–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassanpour S, Bay G, and Langlotz CP (2017). Characterization of Change and Significance for Clinical Findings in Radiology Reports Through Natural Language Processing. Journal of Digital Imaging, 30(3):314–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irvin J, Rajpurkar P, Ko M, Yu Y, Ciurea-Ilcus S, Chute C, Marklund H, Haghgoo B, Ball R, Shpan-skaya K, Seekins J, Mong DA, Halabi SS, Sandberg JK, Jones R, Larson DB, Langlotz CP, Patel BN, Lungren MP, and Ng AY (2019). CheXpert: A Large Chest Radiograph Dataset with Uncertainty Labels and Expert Comparison. In AAAI Conference on Artificial Intelligence. [Google Scholar]
- Johnson AE, Pollard TJ, Shen L, wei H Lehman L, Feng M, Ghassemi M, Moody B., Szolovits P, Celi LA, , and Mark RG. (2016). MIMIC-III, a freely accessible critical care database. Scientific Data, 3:160035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordjamshidi P, Otterlo MV, and Moens M-F (2010). Spatial Role Labeling : Task Definition and Annotation Scheme. In Proceedings of the Language Resources & Evaluation Conference, pages 413–420. [Google Scholar]
- Kordjamshidi P, Roth D, and Moens M-F (2015). Structured learning for spatial information extraction from biomedical text: Bacteria biotopes. BMC Bioinformatics, 16(1):1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordjamshidi P, Rahgooy T, and Manzoor U (2017). Spatial Language Understanding with Multimodal Graphs using Declarative Learning based Programming. In Proceedings of the 2nd Workshop on Structured Prediction for Natural Language Processing, pages 33–43. [Google Scholar]
- Langlotz CP (2006). RadLex: a new method for indexing online educational materials. Radiographics, 26(6):1595–1597. [DOI] [PubMed] [Google Scholar]
- Petruck MR and Ellsworth M (2018). Representing Spatial Relations in FrameNet. In Proceedings of the First International Workshop on Spatial Language Understanding, pages 41–45. [Google Scholar]
- Pons E, Braun LM, Hunink MM, and Kors JA (2016). Natural Language Processing in Radiology: A Systematic Review. Radiology, 279(2). [DOI] [PubMed] [Google Scholar]
- Rink B, Roberts K, Harabagiu S, Scheuermann RH, Toomay S, Browning T, Bosler T, and Peshock R (2013). Extracting actionable findings of appendicitis from radiology reports using natural language processing. In AMIA Joint Summits on Translational Science Proceedings, volume 2013, page 221. [PMC free article] [PubMed] [Google Scholar]
- Roberts K, Rink B, Harabagiu SM, Scheuermann RH, Toomay S, Browning T, Bosler T, and Peshock R (2012). A machine learning approach for identifying anatomical locations of actionable findings in radiology reports. In AMIA Annual Symposium Proceedings, volume 2012, pages 779–788. [PMC free article] [PubMed] [Google Scholar]
- Roberts K, Rodriguez L, Shooshan S, and Demner-Fushman D (2015). Automatic Extraction and PoSt-coordination of Spatial Relations in Consumer Language. In AMIA Annual Symposium Proceedings, volume 2015, pages 1083–1092. [PMC free article] [PubMed] [Google Scholar]
- Sevenster M, Van Ommering R, and Qian Y (2012). Automatically correlating clinical findings and body locations in radiology reports using MedLEE. Journal of Digital Imaging, 25(2):240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si Y, Wang J, Xu H, and Roberts K (2019). Enhancing clinical concept extraction with contextual embeddings. Journal of the American Medical Informatics Association, pages 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkamp JM, Chambers C, Lalevic D, Zafar HM, and Cook TS (2019). Toward Complete Structured Information Extraction from Radiology Reports Using Machine Learning. Journal of Digital Imaging, 32(4):554–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulinski M, Coyne B, and Hirschberg J (2019). SpatialNet: A Declarative Resource for Spatial Relations. In Proceedings of the Combined Workshop on Spatial Language Understanding (SpLU) and Grounded Communication for Robotics (RoboNLP), pages 61–70. [Google Scholar]
- Wang X, Peng Y, Lu L, Lu Z, Bagheri M, and Summers RM (2017). ChestX-ray8: Hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 3462–3471. [Google Scholar]
- Yim W-W, Denman T, Kwan SW, and Yetisgen M (2016). Tumor information extraction in radiology reports for hepatocellular carcinoma patients. In AMIA Joint Summits on Translational Science Proceedings, volume 2016, pages 455–64. [PMC free article] [PubMed] [Google Scholar]
- Yuan Y (2011). Extracting spatial relations from document for geographic information retrieval. In Proceedings - 2011 19th International Conference on Geoinformatics. IEEE. [Google Scholar]


