Abstract
While DNA inside the cells is predominantly canonical right-handed double helix, guanine-rich DNAs have potential to fold into four-stranded structures that contain stacks of G-quartets (G4 DNA quadruplex). Genome sequencing has revealed G4 sequences tend to localize at the gene control regions, especially in the promoters of oncogenes. A growing body of evidence indicates that G4 DNA quadruplexes might have important regulatory roles in genome function, highlighting the need for techniques to detect genome-wide folding of DNA into this structure. Potassium permanganate in vivo treatment of cells results in oxidizing of nucleotides in single-stranded DNA regions that accompany G4 DNA quadruplexes formation, providing an excellent probe for the conformational state of DNA inside the living cells. Here, we describe a permanganate-based methodology to detect G4 DNA quadruplex, genome-wide. This methodology combined with high-throughput sequencing provides a snapshot of the DNA conformation over the whole genome in vivo.
Keywords: Non-B DNA, DNA quadruplex, G4 DNA, Potassium permanganate, Chromatin, High-throughput genomics
1. Introduction
Rather than being a static helix, DNA possesses structural variability. Hydrogen bonding between nucleobases of the complementary DNA strands keeps DNA in the double-stranded right-handed helix: classical B-DNA form. However, DNA elements with special patterns of nucleotides sequence have potential for structural transitions into other DNA forms, non-B-DNA structures. These structures were extensively characterized by biophysical studies, using DNA oligonucleotides or plasmid DNA in various solution conditions. While non-B-DNA structures provide enormous potential for autoregulation of genome function [1], the extent and even existence of such unusual structures inside of living cells is still the matter of some debate. The study of the interplay between DNA conformation and genome biology has been hindered by experimental difficulties associated with detecting non-B-DNA structures and assessing their regulation in vivo, especially in eukaryotic cells [2, 3]. In this chapter, we describe our method to map non-B DNA genome-wide in the living cells, with a focus on G4 DNA quadruplexes. Other groups are currently adopting and adapting this approach allowing researchers to uncover the role of DNA conformational dynamics in genome function.
G4 DNA quadruplexes are four-stranded DNA that stacks planar sets of four mutually Hoogsteen H-bonded guanine bases. Biophysical studies on synthetic oligonucleotide sequences delivered from the eukaryotic genomes indicate the broad variety of G4 structures depending on strand orientation, the size of the single-stranded DNA loops, and solution conditions, such as the selection and concentration of the prevailing cation [4]. These studies have enabled the search for sequences with quadruplex forming potential in genomes [5]. It was shown that DNA elements with predisposition for quadruplex formation (G4 DNA) often reside within regulatory regions, including a significant enrichment of G4 DNA in the promoters of oncogenes [6]. Chemical biological studies have provided crucial insight into G-quadruplex-binding ligands that exhibit pronounced anticancer activities in vivo, ability for transcriptional reprogramming, as well as locus-specific changes in epigenetic information [7, 8]. The potential importance of G4 DNA quadruplex formation within the genomes of living cells and the extensive literature correlating pharmacologic disturbance of quadruplex stability with perturbation of cellular programs stimulated experiments to search for the presence of these structures in vivo [5].
High-affinity G4 DNA quadruplex-recognizing antibodies were used to visualize these structures by immunostaining inside a range of cells [9, 10]. The high stability of G4 DNA quadruplex enabled immunoprecipitation of immunoreactive structures from isolated and fragmented genomic DNA [11]. Deep sequencing of the selected DNA fragments yielded signal that correlated with predicted G4 DNA sequences. Recently, a G4 ChIP–seq protocol was developed that employed an antibody specific to G4 DNA quadruplex to map the genome-wide location of the structures in the chromatin of the formaldehyde fixed cells [12]. However, it should be noted that antibody-based approaches might impact and bias the formation of G-quadruplex-structures: fixing cells, binding of antibodies, enzymatic, chemical, and mechanical factors during genomic DNA isolation might all influence the apparent versus real pattern of G4 DNA quadruplex in the genome [1, 2]. Also, considering the polymorphous nature of G4 and the absence of a structure of anti-G4 complexed with nucleic acid, it remains to be established that the determinants of antibody recognition are universally present and accessible on all formed G4 structures and absent on other structures [4]. Therefore, it was important to develop orthogonal and less chromatin disruptive approaches to map G4 DNA structures in vivo, to further consolidate previous findings. A powerful approach to detect non-B DNA conformation genome-wide at nearly nucleotide resolution relies on the properties of the molecule potassium permanganate (KMnO4) to oxidize unpaired nucleotides of single-stranded DNA (ssDNA) segments that are a pervasive characteristic of non-B DNA structures, G4 DNA included. Though for several decades permanganate-based assays have been used to probe DNA conformation in selective regions of the genome [13–15], we have combined this method with enzymatic footprinting and next generation sequencing to provide a global view of DNA structure in vivo [16, 17].
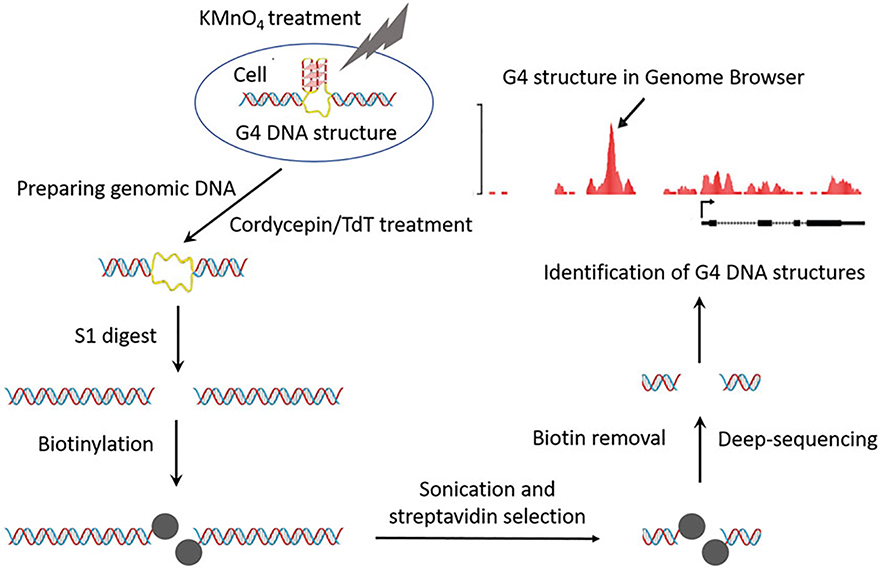
Potassium permanganate is a small molecular which easily penetrates cellular membranes and modifies ssDNA in living cells by oxidizing unpaired pyrimidine bases. Cells treated with permanganate for a very short period remain fully viable after treatment [16]. Thus, marking of single-stranded DNA by this chemical modification reflects the unbiased in vivo pattern of single stranded DNA. In purified genomic DNA, regions with oxidized bases are susceptible to cleavage by single strand specific nuclease—S1 nuclease. After nuclease digestion, double-stranded breaks are produced at the sites of the DNA chemical modification. These breaks are labeled with biotinylated nucleotides during DNA tailing reaction with terminal deoxynucleotidyl transferase (TdT). After DNA sonication, biotinylated DNA fragments are selected with streptavidin beads prior to Illumina library preparation then sequenced using the high-throughput Illumina platform; this is the outline of the method that we call ssDNA-seq [16, 17]. Overlapping the sequencing signal with computationally predicted G4 DNA motifs in the genome delivers a high-resolution map of G4 DNA structures formed in vivo (Fig. 1).
Fig. 1.
G4 DNA structures mapping workflow: from top, counterclockwise direction. Single-stranded DNA in G4 DNA structures is stabilized in live cells by treatment with KMnO4 (described in Subheadings 3.2 and 3.3). Cordycepin and Terminal Transferase treatment of purified genomic DNA is used to block preexisting DNA double stranded breaks (described in Subheading 3.4). Chemically modified DNA is digested with single-strand specific S1 nuclease (described in Subheading 3.5). DNA ends exposed by nuclease treatment are biotinylated (described in Subheading 3.6) and, following sonication, streptavidin selected. Biotin is removed from the selected DNA fragments (described in Subheading 3.7). DNA fragments surrounding G4 DNA structures in vivo are deep sequenced (described in Subheading 3.8). Computational analysis allows genome-wide identification of G4 DNA structures (described in Subheading 3.9)
2. Materials
2.1. Reagents
Proteinase K (Solution, 20 mg/mL).
Phenol:Chloroform:Isoamyl Alcohol 25:24:1, Tris (pH 8.0) saturated.
Ethanol 100%.
Ammonium acetate 7.5 M.
RNase, DNase-free (Solution, 500 μg/mL).
SDS, 10%.
EDTA, 0.5 M.
Glycogen, 2 mg/mL stock solution.
UltraPure Agarose (Invitrogene).
Terminal Transferase (NEB).
Nuclease S1 (Thermo Scientific).
Taq DNA Polymerase with ThermoPol Buffer (NEB).
HiFi HotStart ready Mix (KAPA Biosystems).
Potassium Permanganate, KMnO4.
Cordycepin 5′-triphosphate sodium salt (Sigma Aldrich).
Biotin-16-dUTP, 1 mM (Roche).
100 mM dNTPs, PCR grade (NEB)
10 mM ATP
SYBR Green Nucleic Acid Gel Stain.
MassRuler DNA Ladder Mix (Thermo Fisher Scientific).
Illumina adapter (Adaptor oligo mix).
Illumina genomic SE primers (Fw: 5′-aat gat acg gcg acc acc gag atc tac act ctt tcc cta cac gac gct ctt ccg atc t-3′/Rv: 5′-caa gca gaa gac ggc ata cga gct ctt ccg atc t-3′).
T4 DNA ligase (NEB).
2.2. Buffers
TE buffer: 10 mM Tris–HCl, 1 mM EDTA (pH 8.0).
Low salt buffer: 15 mM Tris–HCl (pH 7.5), 60 mM KCl, 15 mM NaCl, 5 mM MgCl2, 0.5 mM EGTA, 300 mM Sucrose.
Stop Solution: 1% SDS, 50 mM EDTA, 700 mM β-mercaptoethanol.
Elution buffer: 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 1 M NaCl, 2 M β-mercaptoethanol.
TAE buffer: 40 mM Tris (pH 7.6), 20 mM acetic acid, 1 mM EDTA.
10× Terminal Transferase (TdT) reaction buffer: 500 mM Potassium acetate, 200 mM Tris-acetate, 100 mM magnesium acetate, pH 7.9
5× Nuclease S1 buffer: 200 mM sodium acetate (pH 4.5), 1.5 M NaCl, 10 mM ZnSO4
10× (2.5 mM) solution of CoCl2
PBS: 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, without calcium and magnesium, pH 7.4.
2.3. Equipment
Gel electrophoresis apparatus.
Thermomixer.
Ultrasonic sonicator (Bioruptor, Diagenode).
Spectrophotometer, NanoDrop (ND-1000).
Rocker, tube wheel rotator, water bath, aspirator, and thermomixer.
Access to Genome Analyzer IIX (Illumina).
2.4. Kits
QIAquick PCR Purification Kit (Qiagen).
Dynabeads kilobaseBINDER Kit (Thermo Fisher Scientific).
Invitrogen Quant-iT PicoGreen dsDNA Reagent (Thermo Fisher Scientific).
DNA END-Repair kit (Epicentre Biotechnologies).
MinElute Gel Extraction Kit (Qiagen).
MinElute Reaction Cleanup Kit (Qiagen).
3. Methods
3.1. Preparing Stock Solutions
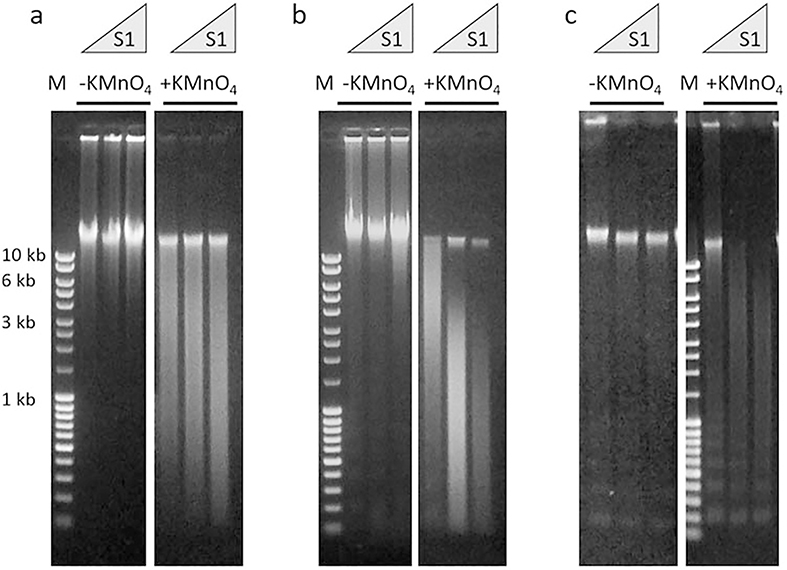
Dissolve KMnO4 in water with constant shaking for at least 1 h (see Note 1). The optimal concentration of potassium permanganate is slightly different for different cell types. In pilot experiments, we routinely check 80 mM, 90 mM and 100 mM stock solutions of permanganate, to achieve the optimal level of DNA chemical modifications (Fig. 2).
Prepare Stop Solution and Elution buffer in the day of experiment.
Prewarm in the water bath PBS buffer, Low Salt buffer, KMnO4 stock solution and Molecular Biology Grade Water to 37 °C.
Fig. 2.
Representative examples of gel electrophoresis analysis of genomic DNA after the S1 nuclease digest described in Subheading 3.5. Cells were treated (+KMnO4) or untreated (−KMnO4) with potassium permanganate. Purified genomic DNA were digested with increasing concentration of S1 nuclease and visualized with SYBR Green after gel electrophoresis. M: DNA mass ladder (MassRuler DNA Ladder Mix). The numbers on the left refer to the molecular weight of the DNA marker in lane M. (a) Optimal S1 nuclease reactivity of the genomic DNA from the cells treated with potassium permanganate. (b) Overexposing the cells to the potassium permanganate results in the excessive chemical modification of the genomic DNA as evident by high S1 nuclease reactivity. (c) Purified genomic DNA shows signs of degradation as apparent by the appearance of a nucleosome-like DNA ladder. Please note that the genomic DNA from the cells not exposed to potassium permanganate is highly resistant to S1 nuclease
3.2. Preparing Genomic DNA from the Cells Treated with Permanganate (Suspension Cells)
Start with 60 × 106 cells (see Note 2). Count cells and transfer the suspension to a centrifuge tube. Recover the cell pellet by centrifugation at 500 × g for 5 min at room temperature (RT). Remove the supernatant by aspiration and gently resuspend the cells in 40 mL PBS buffer (without CaCl2 and MgCl2). Pellet cells by centrifugation at 500 × g for 5 min at RT. Remove the supernatant by aspiration and gently resuspend the cells in 3 mL of Low Salt buffer (see Note 3). Split the cells in the two 50 mL tubes; 1.5 mL of cells suspension in each tube.
A typical experiment requires condition #I, cells not exposed to permanganate; #II, cells exposed to permanganate. Write these numbers on the tubes and keep the cell suspension at 37 °C in a water bath. Add 0.5 mL of H2O to the tube #1 and 0.5 mL of 100 mM KMnO4 solution to the tube #2. Incubate 80 s. Add 2 mL of Stop Solution to each tube. Mix carefully until the cellular lysate is clear (see Note 4). Add DNase free RNase (40 μg/mL), mix, and incubate for 1 h at 37 °C (see Note 5). Then, add Proteinase K (300 μg/mL) and incubate at 42 °C, overnight.
Add an equal volume of Phenol–Chloroform to the cellular lysates. Mix the two phases by slowly inverting the tubes for a few minutes (see Note 4), and separate the two phases by centrifugation at 3500 × g for 10 min at RT. Transfer the aqueous phase to a new 50 mL Falcon tube.
Repeat Subheading 3.2, step 3. Add to the aqueous phases 0.5 volume of 7.5 M ammonium acetate and 2 volumes of ethanol stored at RT. Swirl the tubes to thoroughly mix solutions and precipitate the DNA by centrifugation at 3500 × g for 10 min. Remove ethanol solution using an aspirator. Wash the DNA precipitates with 70% ethanol. Remove as much ethanol as possible and air-dry the pellet for 15 min. Add 1 mL of TE (pH 8.0) to each tube. Place the tubes on a tube wheel rotator and incubate the solution overnight at RT with gentle agitation until the DNA is completely dissolved (see Note 4).
3.3. Preparing Genomic DNA from the Cells Treated with Permanganate (Adherent Cells)
Start with appropriate amount of cell culture flask to have required 60 × 106 cells. Steps below are done for the standard 175 cm2 flask. Label half of the flasks with #I (cells not exposed to permanganate) and another half with #II (cells exposed to permanganate). Remove media and wash the cell layer with 50 mL of prewarmed PBS buffer (without CaCl2 and MgCl2). Add 6 mL of prewarmed Low Salt buffer.
Immediately add 2 mL of H2O to the flasks labeled with #1 and 2 mL of KMnO4 stock solution to the flasks labeled with #2. Incubate 80 s. Add 8 mL of Stop Solution. Mix carefully by flask rocking until cellular lysates is clear. Add RNase-DNase free (40 μg/mL). Remove DNA solution from the flask and incubate for 1 h at 37 °C. Then add Proteinase K (300 μg/mL) and incubate at 42 °C, overnight (see Note 6).
Add an equal volume of Phenol–Chloroform to the cellular lysates. Mix the two phases by slowly inverting the tubes for a few minutes (see Note 4), and separate the two phases by centrifugation at 3500 × g for 10 min at RT. Transfer the aqueous phase to a new 50 mL Falcon tube.
Repeat Subheading 3.3, step 3. Add to the aqueous phases 0.5 volume of 7.5 M ammonium acetate and 2 volumes of ethanol stored at RT. Swirl the tubes to thoroughly mix solutions and precipitate the DNA by centrifugation at 3500 × g for 10 min. Remove ethanol solution using an aspirator. Wash the DNA precipitates with 70% ethanol. Remove as much ethanol as possible and air-dry the pellet for 15 min. Add 1 mL of TE (pH 8.0) to each tube. Place the tubes on a tube wheel rotator and incubate the solution overnight at RT with gentle agitation until the DNA is completely dissolved (see Note 4).
3.4. Blocking Unspecific Breaks in the Genomic DNA
Set up DNA double-stranded breaks blocking reaction by mixing 1 mL of DNA solution with 300 μL of 10× TdT buffer, 300 μL of CoCl2 solution, 80 μL of 4 mM cordycepin 5′-triphosphate, 1000 U of Terminal Transferase and H2O to a final volume of 3 mL. Incubate for 1 h at 37 °C.
Phenol–Chloroform extract and Ethanol precipitate with2.5 M Ammonium Acetate (see Subheading 3.3, step 4 of the current protocol). Repeat Ethanol precipitation with 2.5 M Ammonium Acetate (see Note 7). Dissolve DNA in 0.5 mL of TE buffer.
3.5. Converting the Sites of DNA Chemical Modification into DNA Breaks
Aliquot the DNA solution into three tubes (50 μL in each). Set up S1 nuclease digest of genomic DNA by mixing 50 μL of DNA solution with 60 μL of 5× S1 nuclease buffer and H2O to a final volume of 300 μL. Mark the tubes accordingly and add to them 50 U, 100 U, and 200 U of S1 nuclease. Incubate the reactions at 37 °C for 20 min (see Note 8).
Stop the reaction by extracting the mixtures with 300 μL of Phenol–Chloroform. Add 20 μg of glycogen from a stock solution to the aqueous phase and 30 μL of 3 M Sodium acetate. Mix and add 2 volumes of ethanol stored at +4 °C. Swirl the tubes to thoroughly mix solutions and precipitate the DNA by centrifugation at 15,000 × g for 10 min. Remove ethanol solution using an aspirator. Wash the DNA precipitates with 70% ethanol. Remove as much ethanol as possible and air-dry the pellet for 15 min. Add 30 μL of TE (pH 8.0) to each tube. Place the tubes on a shaker for 1 h to dissolve the DNA.
Run 2 μL of DNA solution from each reaction on a 0.6% agarose gel. Stain the gel with SYBR Green stain according to the manufacturer’s recommendation and by using UV transilluminator check the size of DNA in each reaction (see Fig. 2 for illustrative examples).
For the following biotin-labeling of the DNA breaks, choose S1 nuclease concentrations that produced DNA fragments with average size 2–10 kilobases (from the cells treated with KMnO4). Please compare Fig. 2a, b. Genomic DNA should not show any trace of degradation (Fig. 2c). By scaling up this condition for the remaining 350 μL of DNA solution (see Subheading 3.5, step 1), perform S1 digestion of the genomic DNA (see Subheading 3.5, steps 2 and 3).
3.6. DNA End-Labeling with TdT Tailing Reaction
Set up DNA tailing reaction by mixing 200 μL of DNA solution (samples #I and #II) with 30 μL of 10× TdT buffer, 30 μL of CoCl2 solution, 1 μL of 100 mM dCTP, 1 μL of 100 mM dATP, 4000 U of Terminal Transferase, and H2O to a final volume of 300 μL. Incubate reactions for 5 min at 37 °C.
Add 20 μL of 1 mM Biotin-16-dUTP and incubate at 37 °C for 30 min. Stop the reaction by adding EDTA to a final concentration of 20 μM following extraction with Phenol–Chloroform and ethanol precipitation with ammonium acetate (see Subheading 3.5, step 3). Dissolve DNA pellets in the 300 μL of TE buffer and repeat ethanol precipitation with ammonium acetate (see Note 9). Dissolve DNA pellets in the 300 μL of TE buffer.
Sonicate the biotinylated DNA to generate 200–400 bp DNA fragments. Check the DNA fragment sizes by running 2 μL of DNA solution from each tube on a 1% agarose gel. In our procedure, sonication was performed with an ultrasonic sonicator (Bioruptor, Diagenode) at medium power, by pulsing 20 times for 30 s and cooling in an ice-bath for 30 s between pulses.
3.7. Capturing of Biotinylated DNA by Streptavidin-Coated Beads
Transfer 200 μL of thoroughly resuspended beads (2 mg) to an Eppendorf tubes. Use the magnet to separate beads from supernatant. Add to the beads 300 μL of binding buffer (provided with the beads). Mix for 5 min. Place the tubes on the magnet and remove the supernatant. Resuspend the beads in 300 μL of binding buffer.
Add 300 μL of a solution containing the biotinylated DNA fragments (see Subheading 3.6, step 3) to the beads. Incubate the tubes for 3 h at room temperature on a tube wheel rotator. Aliquot 10 μL of unbound DNA solution and purify it with a QIAquick PCR Purification Kit, eluting DNA samples into 30 μL of TE buffer (see Note 10). Save 2 μL of DNA to use in Subheading 3.7, step 6.
Wash the beads/DNA complex four times in 600 μL Washing solution (provided with the beads) and once in 600 μL TE buffer. To perform each wash, add Washing solution to the beads and incubate the suspension at 50 °C for 5 min with agitation on the thermomixer. Use the magnet to separate the beads from the washing buffer. Add new Washing solution or TE buffer and repeat.
To disrupt biotin-streptavidin complexes, add to the washed beads 100 μL of elution buffer and incubate at 75 °C for 1.5 h with agitation on the thermomixer. Use the magnet to separate the supernatant from the beads. Keep the supernatant. Add to the beads 100 μL of new elution buffer, incubate at 75 °C for 1.5 h, and use the magnet to separate the supernatant from the beads. Pool the supernatants together. Purify DNA fragments with a QIAquick PCR Purification Kit, eluting DNA samples into 30 μL of TE buffer. Save 2 μL of DNA to use in Subheading 3.7, step 6.
To remove the biotinylated tails from DNA, set up S1 nuclease digesting reaction by mixing 28 μL of DNA solution (samples #I, #II, and unbound DNA from the Subheading 3.7, step 2) with 20 μL of 5× Nuclease S1 buffer, 50 U of S1 nuclease, and H2O to a final volume of 100 μL. Incubate reactions for 15 min at 37 °C. Purify DNA with a QIAquick PCR Purification Kit, eluting DNA samples into 30 μL of TE buffer. Aliquot 5 μL of DNA to use in Subheading 3.7, step 6.
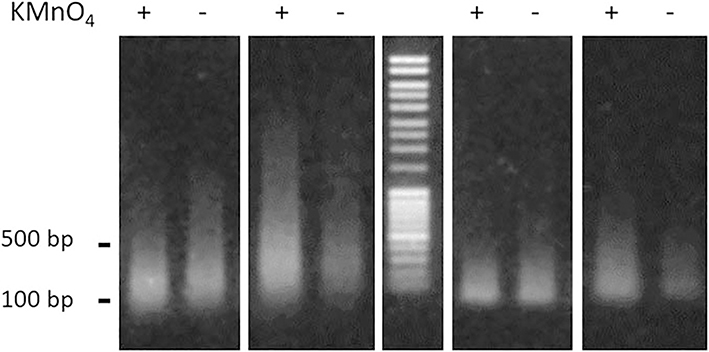
Quantify the recovered DNA. Run DNA samples aliquoted at Subheading 3.7, steps 2, 4, and 5 on a 1% agarose gel. Stain the DNA in the gel with SYBR Green in TAE buffer (see Note 11 and Fig. 3).
Repeat the experiment generating required number of biological replicates.
The DNA samples recovered from the biotin-streptavidin selection are ready for high-throughput sequencing.
Fig. 3.
Representative example of gel electrophoresis analysis of DNA fragments described in Subheading 3.7. From the left to the right: first panel—DNA unbound to the beads; second panel—DNA bound to the beads; third panel—DNA mass ladder (MassRuler DNA Ladder Mix); fourth panel—DNA unbound to the beads treated with S1 nuclease; fifth panel—DNA bound to the beads treated with S1 nuclease. DNA fragments were delivered from the genomic DNA of the cells treated with potassium permanganate (+), or from the cells untreated with potassium permanganate (−). The numbers on the left refer to the molecular weight of the DNA marker
3.8. Template Preparation for Sequencing Analysis
Quantify DNA with PicoGreen dsDNA Reagent according to the manufacturer’s protocol to determine what amount of adaptor to use in the later reaction.
To generate blunt-ended DNA, incubate the DNA for 45 min at room temperature in 25 μL reaction with a mixture of End repair buffer, 0.25 mM of each dNTPs, 1 mM ATP, and 1 μL DNA End-Repair Enzyme mix. Purify the DNA with a MinElute Reaction Cleanup Kit.
In 50 μL reaction, treat the blunt-ended DNA with 15 units of Taq DNA polymerase for 40 min at 70 °C in the presence of 0.2 mM dATP to generate a protruding 3’A base used for adaptor ligation (see Note 12). Purify DNA with a MinElute Reaction Cleanup Kit.
In 20 μL reaction, ligate Illumina genomic adapter to the end of DNA fragments by incubating DNA with Adaptor oligo mix and 1500 units of T4 DNA ligase in the T4 DNA ligase buffer at room temperature for 30 min. Use 1 μL of 1/20 diluted Adaptor oligo mix per 10 ng of DNA quantified at Subheading 3.8, step 1. Purify DNA with a MinElute Reaction Cleanup Kit.
Amplify the DNA for 18 cycles using Illumina genomic SE primers according to the following protocol: 98 °C for 3 min precycle incubation followed by 98 °C for 30 s; 65 °C for 30 s; 72 °C for 30 s and postcycle incubation at 72 °C for 3 min with HiFi HotStart ready Mix.
Run the PCR product through 2% agarose gel and excise the gel slice around 200–300 bps region. Try not to include adaptor dimers located around 140 bps.
Purify the DNA from the gel using MinElute gel extraction kit. Elute the amplicon with 10 μL of 10 mM Tris, pH 8.5.
The purified DNA is used directly for cluster generation and sequencing analysis using the Illumina Genome Analyzer following manufacturer’s protocols.
3.9. Identification of G4 DNA Quadruplex Structures
Find all occurrences of G4 sequence motifs in both strands of a reference genome using QuadParser [18] with at least three guanine bases required in each of four runs of guanine monomer repeat and gap size between repeats of 1–7 bases. Merge overlapped quadruplex motifs on the same strand into a single motif. Precomputed genomic locations of G4 motifs in mouse (mm9) and human (hg19) genomes can be found at https://www.ncbi.nlm.nih.gov/CBBresearch/Przytycka/index.cgi#nonbdna.
Process the raw sequencing data using the built-in Illumina Real-Time Analysis (RTA) software that provides base calls and associated quality scores.
Perform quality control checks (e.g., FastQC) to understand whether the data might have any problems before doing any further analysis.
Align sequencing reads to the reference genome using sequence aligner, for example Bowtie2 [19] or BWA [20].
For each genomic occurrence of G4 sequence motif, count the number of nonredundant reads overlapping two windows of length 500 bp and 1000 bp centered at a given motif using htseq-count script [21]. The 500 bp and 1000 bp windows are called as signal and local window, respectively.
For each G4 motif, compute a p value for observed number of reads in a signal window within a local window using binomial distribution. To find a reasonable p value cutoff for G4 structures use a permutation test, i.e., randomly shuffle read location within local windows and compute p values for number of randomized reads found in signal windows within local windows.
G4 motifs with p value above a cutoff (corresponding to false discovery rate of 5% computed based on randomized data) can be considered as regions forming G4 quadruplex structure (see Note 13).
Footnotes
Keep potassium permanganate solution protected from light. Do not use DEPC-treated water. Do not keep stock solution for extended period—it should be made in the day of experiment.
This number of cells (60 × 106) is required to control all intermediate steps of the procedure and visualize DNA on the agarose gel. We recommend following all the checkpoints for the first experiments. With experience, one can scale-down this amount up to 10 × 106 cells.
This buffer makes the cells swell and take up the KMnO4.
To minimize DNA breakage which might increase background signal after sequencing, cellular lysates and genomic DNA should be handled gently. After the addition of phenol–chloroform, mix the phases by slowly inverting the tubes for a few minutes. The aqueous phase should be removed from the organic phase with a 25-mL pipette, sucking the liquid very slowly. Do not vortex the samples and do not resuspend the DNA pellet by pipetting.
RNase is inhibited by 0.5% SDS, but at this high concentration it will be able to digest most of the RNA in the cellular lysates.
A large amount of RNase is required for experiments with adherent cells. To economize this enzyme, first genomic DNA can be purified without RNase treatment as described in Subheading 3.3, step 2. The DNA solution is then treated with 5 μg/mL of RNase at 37 °C for 1 h. DNA is purified as described in Subheading 3.3, steps 3 and 4.
The double precipitation with ammonium acetate is necessary to remove free cordycepin 5′-triphosphate which might impede with following steps.
Pilot S1 nuclease titration is recommended to estimate the efficiency of DNA chemical modification and to choose optimal nuclease concentration in the preparative DNA digest (see Subheading 3.5, step 4). See also Fig. 2.
The double precipitation with ammonium acetate is necessary to remove free Biotin-16-dUTP which might inhibit interaction between biotinylated DNA and streptavidin-coated beads.
These samples to be used in the Subheading 3.7, step 6 of the current protocol.
This step is performed to check (1) the quality of the final DNA samples, (2) the efficiency of the removal of biotinylated tails from the DNA fragments, (3) low yield of DNA recovery from the cells not treated with potassium permanganate as verification of efficiency DNA chemical modification in vivo.
To avoid freeze and thaw cycles, we aliquot 10 mM dATP to small volumes and store them at −80 °C for single use.
It should be emphasized that transcription bubble formed on DNA by RNA polymerase II may influence identification of G4 structures by increasing false positive calls. Thus, it is recommended to remove from analysis G4 motifs that overlap with binding sites of RNA polymerase II when available [16].
References
- 1.Zaytseva O, Quinn LM (2018) DNA conformation regulates gene expression: the MYC promoter and beyond. Bioessays 40: e1700235 10.1002/bies.201700235 [DOI] [PubMed] [Google Scholar]
- 2.Kouzine F, Levens D (2007) Supercoil-driven DNA structures regulate genetic transactions. Front Biosci 12:4409–4423 [DOI] [PubMed] [Google Scholar]
- 3.van Holde K, Zlatanova J (1994) Unusual DNA structures, chromatin and transcription. Bioessays 16(1):59–68. 10.1002/bies.950160110 [DOI] [PubMed] [Google Scholar]
- 4.Qin Y, Hurley LH (2008) Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie 90(8):1149–1171. 10.1016/j.biochi.2008.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murat P, Balasubramanian S (2014) Existence and consequences of G-quadruplex structures in DNA. Curr Opin Genet Dev 25:22–29. 10.1016/j.gde.2013.10.012 [DOI] [PubMed] [Google Scholar]
- 6.Brooks TA, Kendrick S, Hurley L (2010) Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J 277 (17):3459–3469. 10.1111/j.1742-4658.2010.07759.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guilbaud G, Murat P, Recolin B, Campbell BC, Maiter A, Sale JE, Balasubramanian S (2017) Local epigenetic reprogramming induced by G-quadruplex ligands. Nat Chem 9(11):1110–1117. 10.1038/nchem.2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kendrick S, Muranyi A, Gokhale V, Hurley LH, Rimsza LM (2017) Simultaneous drug targeting of the promoter MYC G-quadruplex and BCL2 i-motif in diffuse large B-cell lymphoma delays tumor growth. J Med Chem 60 (15):6587–6597. 10.1021/acs.jmedchem.7b00298 [DOI] [PubMed] [Google Scholar]
- 9.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A (2001) In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc Natl Acad Sci U S A 98 (15):8572–8577. 10.1073/pnas.141229498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biffi G, Tannahill D, McCafferty J, Balasubramanian S (2013) Quantitative visualization of DNA G-quadruplex structures in human cells. Nat Chem 5(3):182–186. 10.1038/nchem.1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lam EY, Beraldi D, Tannahill D, Balasubramanian S (2013) G-quadruplex structures are stable and detectable in human genomic DNA. Nat Commun 4:1796 10.1038/ncomms2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hansel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, Di Antonio M, Pike J, Kimura H, Narita M, Tannahill D, Balasubramanian S (2016) G-quadruplex structures mark human regulatory chromatin. Nat Genet 48(10):1267–1272. 10.1038/ng.3662 [DOI] [PubMed] [Google Scholar]
- 13.Bui CT, Rees K, Cotton RG (2003) Permanganate oxidation reactions of DNA: perspective in biological studies. Nucleosides Nucleotides Nucleic Acids 22(9):1835–1855. 10.1081/NCN-120023276 [DOI] [PubMed] [Google Scholar]
- 14.Giardina C, Perez-Riba M, Lis JT (1992) Promoter melting and TFIID complexes on drosophila genes in vivo. Genes Dev 6 (11):2190–2200 [DOI] [PubMed] [Google Scholar]
- 15.Johnston BH, Rich A (1985) Chemical probes of DNA conformation: detection of Z-DNA at nucleotide resolution. Cell 42(3):713–724 [DOI] [PubMed] [Google Scholar]
- 16.Kouzine F, Wojtowicz D, Baranello L, Yamane A, Nelson S, Resch W, Kieffer-Kwon KR, Benham CJ, Casellas R, Przytycka TM, Levens D (2017) Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst 4(3):344–356 e347. 10.1016/j.cels.2017.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kouzine F, Wojtowicz D, Yamane A, Resch W, Kieffer-Kwon KR, Bandle R, Nelson S, Nakahashi H, Awasthi P, Feigenbaum L, Menoni H, Hoeijmakers J, Vermeulen W, Ge H, Przytycka TM, Levens D, Casellas R (2013) Global regulation of promoter melting in naive lymphocytes. Cell 153(5):988–999. 10.1016/j.cell.2013.04.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huppert JL, Balasubramanian S (2005) Prevalence of quadruplexes in the human genome. Nucleic Acids Res 33(9):2908–2916. 10.1093/nar/gki609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langmead B, Salzberg SL (2012) Fast gapped-read alignment with bowtie 2. Nat Methods 9 (4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Durbin R (2009) Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25(14):1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anders S, Pyl PT, Huber W (2015) HTSeq—a python framework to work with highthroughput sequencing data. Bioinformatics 31(2):166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]