Abstract
Purpose:
To determine the tumor tissue/cell distribution, functional associations and clinical significance of PD-1, LAG-3 and TIM-3 protein expression in human non-small cell lung cancer (NSCLC).
Experimental Design:
Using multiplexed quantitative immunofluorescence (QIF), we performed localized measurements of CD3, PD-1, LAG-3 and TIM-3 protein in >800 clinically annotated NSCLCs from three independent cohorts represented in tissue microarrays. Associations between the marker’s expression and major genomic alterations were studied in The Cancer Genome Atlas-NSCLC dataset. Using mass cytometry (CyTOF) analysis of leukocytes collected from 20 resected NSCLCs, we determined the levels, co-expression and functional profile of PD-1, LAG-3 and TIM-3 expressing immune cells. Finally, we measured the markers in baseline samples from 90 advanced NSCLC patients treated with PD-1 axis blockers and known response to treatment.
Results:
PD-1, LAG-3 and TIM-3 were detected in tumor infiltrating lymphocytes (TILs) from 55%, 41.5% and 25.3% of NSCLC cases, respectively. These markers showed a prominent association with each other and limited association with major clinicopathologic variables and survival in cases not receiving immunotherapy. Expression of the markers was lower in EGFR-mutated adenocarcinomas and displayed limited association with tumor mutational burden. In single-cell CyTOF analysis, PD-1 and LAG-3 were predominantly localized on T-cell subsets/NKT cells; while TIM-3 expression was higher in NK cells and macrophages. Co-expression of PD-1, LAG-3 and TIM-3 was associated with prominent T-cell activation (CD69/CD137), effector function (Granzyme-B) and proliferation (Ki-67), but also with elevated levels of pro-apoptotic markers (FAS/BIM). LAG-3 and TIM-3 were present in TIL subsets lacking PD-1 expression and showed a distinct functional profile. In baseline samples from 90 advanced NSCLC patients treated with PD-1 axis blockers, elevated LAG-3 was significantly associated with shorter progression-free survival.
Conclusion:
PD-1, LAG-3 and TIM-3 have distinct tissue/cell distribution, functional implications and genomic correlates in human NSCLC. Expression of these immune inhibitory receptors in TILs is associated with prominent activation, but also with a pro-apoptotic T-cell phenotype. Elevated LAG-3 expression is associated with insensitivity to PD-1 axis blockade suggesting independence of these immune evasion pathways.
Introduction
Immunostimulatory therapies blocking the PD-1 axis pathway have become major anti-tumor treatment options in diverse malignancies including non-small cell lung cancer (NSCLC) (1–5). To date, single-agent treatment using monoclonal antibodies targeting PD-1 receptor or its ligand PD-L1 induce lasting clinical responses in ~18% of patients with advanced NSCLC. However, primary resistance occurs in the majority of patients and acquired adaptation of tumors to immune pressure in patients initially responding to therapy has also become a clinical challenge (6–9). Therefore, identification of biomarkers for patient selection and characterization of additional non-redundant actionable immunostimulatory targets is needed.
Various immune and tumor genomic metrics are associated with sensitivity to PD-1 axis blockers including tumor PD-L1 expression, measurement of tumor infiltrating lymphocytes (TILs) or inflammation-related mRNA expression profiles, tumor mutational burden and microsatellite instability (3,4,10–13). To date, however, only detection of PD-L1 protein using immunohistochemistry (IHC) and mismatch repair deficiency are approved by the U.S Food and Drug Administration (FDA) as companion biomarkers for PD-1 blocking antibodies.
Additional immune co-inhibitory receptors beyond PD-1 such as LAG-3 and TIM-3 are induced after T-cell receptor (TCR) stimulation and mediate T-cell suppression/dysfunction (14–17). The potential role of these receptors in mediating tumor immune evasion in human malignancies and their interaction/independence from PD-1 pathway remain poorly understood.
Lymphocyte-activation Gene-3 (LAG-3 or CD223) is a 498-amino acid type I transmembrane protein with high structural homology with CD4 protein and capacity to bind MHC class II molecules (18,19). Alternative ligands have been proposed to explain some of the suppressive effects of LAG-3 in CD8+ cytotoxic cells in the absence of MHC class-II such as Galectin-3, LSECtin and alpha-synuclein fibers (20–22). Recent work from our group identified FGL-1 as a novel high affinity and cell-free ligand for LAG-3 involved in tumor immune evasion (23). Despite being studied in diverse anti-cancer clinical trials (19), the tissue distribution and functional role of LAG-3 in human malignancies has not been clearly defined.
T-cell immunoglobulin and mucin domain-3 (TIM-3) is a 281 amino acid long type I transmembrane protein containing a variable N-terminal Ig domain and a mucin stalk domain (24). An immune suppressive effect of TIM-3 signaling on T-cells was reported to be related with the binding of Galectin-9 and CEACAM1 (25,26). Additional candidate ligands have also shown to be able to modulate TIM-3 functions including phosphatidyl serine (PtdSer) and high mobility group protein B1 (HMGB1) (14).Clinical studies assessing the safety and anti-tumor effect of TIM-3 blockers alone or in combination with other therapies are currently ongoing: NCT02817633, NCT03099109, NCT03066648. The expression, tissue distribution and association of TIM-3 with other immune inhibitory receptors in human lung cancer are not well understood.
Here, we used multiplexed tissue imaging of large tumor collections and single-cell phenotypic analysis of primary cancers to evaluate the distribution and significance of PD-1, LAG-3 and TIM-3 expression in NSCLC. Our results reveal complex functional associations and support independent functions of these receptors to suppress T-cell function.
Materials and Methods
Patients, Cohorts, and Tissue Microarrays
Formalin-fixed paraffin-embedded (FFPE) samples from previously reported retrospective collections of NSCLC not treated with immune checkpoint blockers and represented in tissue microarrays (TMAs) were analyzed (27,28). The first collection includes samples from 426 NSCLC patients seen at Yale Pathology between 1988 and 2012 (Cohort #1). The second cohort includes samples from 304 NSCLC patients collected at Sotiria General Hospital and Patras University General Hospital (Greece) between 1991 and 2001 (Cohort #2). All cases in the cohorts were reviewed by a local pathologist using hematoxylin & eosin stained preparations and tumor histology variant was confirmed by morphology analysis. Tumor cores for TMA construction were obtained from case areas selected by a pathologist to represent the disease. Tumor core selection was not based on specific tumor segments or location. Clinicopathologic information from patients in both cohorts was collected from clinical records and pathology reports. Analysis of mRNA expression and nonsynonymous mutations was performed using the lung cancer dataset from TCGA (Cohort #3, n=370). Another TMA-based cohort from Yale (cohort #4) including retrospective samples from 108 lung adenocarcinomas clinically tested for EGFR and KRAS mutations was also studied. Fresh immune cell/leukocyte extracts from 20 primary resected NSCLCs were also included for mass cytometry analysis (cohort #5). To assess the value of the markers in patients treated with PD-1 axis blockers, we analyzed a combined retrospective cohort from Yale, Cleveland Clinic and Navarra University including 90 baseline/pre-treatment cases treated with PD-1 blocking antibodies (nivolumab or pembrolizumab) or a PD-L1 blocking antibody (atezolizumab) (cohort #6). A summary description of all 6 cohorts/datasets included in the study is provided in Table S1. All the studies were conducted in accordance with recognized ethical guidelines (e.g., Declaration of Helsinki, CIOMS, Belmont Report, U.S. Common Rule) and tissue and clinical information were used after approval from the Yale Human Investigation Committee protocols #9505008219, #1412015109, #1608018220 and #1603017333 or local institutional protocols, which approved the patient consent forms or waiver of consent.
Multiplexed quantitative immunofluorescence (QIF)
A 5-color QIF protocol for FFPE tissue specimens was developed for simultaneous detection of DAPI, CD3, PD-1, LAG-3 and TIM-3 using isotype specific antibodies and different fluorescence conjugates as previously described by our group (28). To reliably measure the markers, we first validated individual assays using control preparations from cell-line transfectants and human tissues. As shown in supplementary Figure S1, PD-1, LAG-3 and TIM-3 were detected exclusively in FFPE cell preparations from HEK293 cells transfected with each respective target, but not in parental cells lacking endogenous expression. The markers were then integrated into a multiplexed panel together with 4’, 6-Diamidino-2-Phenylindole (DAPI) to highlight every cell in the sample and CD3 to map T-cells (Figure 1). For the multiplexed staining, sections were deparaffinized and subjected to antigen retrieval using EDTA buffer (Sigma-Aldrich, St Louis, MO) pH=8.0 and boiled for 20 min at 97°C in a pressure-boiling container (PT module, Lab Vision). Slides were then incubated with dual endogenous peroxidase block (DAKO #S2003, Carpinteria, CA) for 10 min at room temperature and subsequently with a blocking solution containing 0.3% bovine serum albumin in 0.05% Tween solution for 30 minutes. Primary antibodies included CD3 (rabbit polyclonal, Dako), PD-1 (clone EH33), LAG-3 (clone 17B4) and TIM-3 (clone D5D5R). Secondary antibodies and fluorescent reagents used were anti-rabbit Envision (K4003, DAKO) with fluorescein-tyramide (PerkinElmer), anti-mouse IgG2a antibody (Abcam) with Cy3 plus (PerkinElmer), goat anti-rabbit (Abcam) with biotinylated tyramide/Streptavidine-Alexa750 conjugate (PerkinElmer), anti-mouse Envision (K40001) with Cy5-tyramide (PerkinElmer). Residual horseradish peroxidase activity between incubations with secondary antibodies was eliminated by exposing the slides twice for 10 minutes to a solution containing benzoic hydrazide (0.136 mg) and hydrogen peroxide (50 μl). To determine the reproducibility of the QIF assay, we measured serial sections from an index TMA containing positive and negative controls (YTMA345) at different time points. The linear regression coefficients of scores obtained between independent runs were high (R >0.9, P<0.001) (Figure S2), supporting the consistency of the measurements.
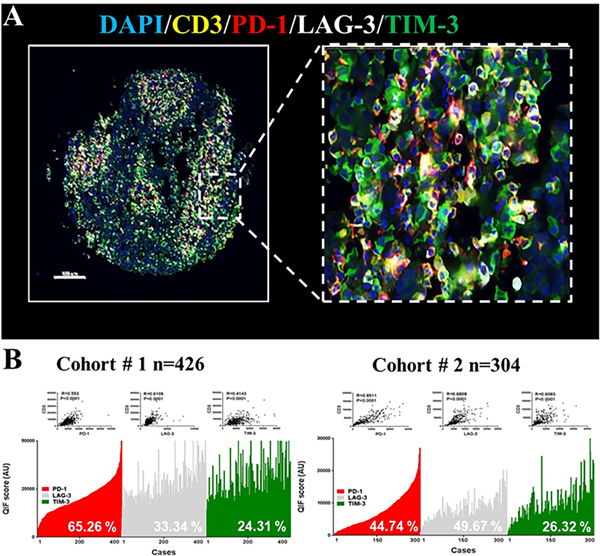
Figure 1: Distribution and frequency of T-cell PD-1, TIM-3 and LAG-3 expression in NSCLC.
A) Representative fluorescence microphotographs showing the simultaneous detection of PD-1 (red), LAG-3 (white), TIM-3 (green) and CD3 (yellow) positive cells in NSCLC. B) Level of the markers measured using QIF in 2 retrospective NSCLC cohorts- Cohort #1 [n=426] (A) and #2 [n=304] (B). The markers were measured in CD3+ T-cells and showed a continuous distribution and strong association with CD3 (insets). The frequency of expression of each marker is indicated with white-colored text within the charts. R= Spearman’s correlation coefficient.
Tissue fluorescence measurement and scoring
Quantitative measurement of the fluorescent signal was performed using the AQUA® method that enables objective and sensitive measurement of targets within user-defined tissue compartments (28). Briefly, the QIF score of each target in CD3+ T-cell compartment from the whole TMA spot was calculated by dividing the target pixel intensities by the area of CD3 positivity. Scores were normalized to the exposure time and bit depth at which the images were captured, allowing scores collected at different exposure times to be comparable. Markers were also measured in the total tissue compartment by collecting the signal score in the area defined by DAPI staining (e.g. all cells in the sample). For graphical representation of the retrospective TMA collections, the scores of LAG-3 and TIM-3 were mean-normalized relative to PD-1 scores to display them in a comparable scale.
Cell preparation and cytometry by time-of-flight (CyTOF) analysis
As previously described (29), primary resected NSCLC tissues were finely minced and mechanically dissociated with the GentleMACS Dissociator (Miltenyi Biotec) in the presence of RPMI 1640 with 0.5% BSA and 5mM EDTA. The resulting cell suspension was filtered using a 70-μm cell strainer (BD Falcon). Cells were centrifuged at 600 g for 7 min at 4 C and re-suspended in PBS with 0.5% BSA and 0.02% NaN3. 2–4*106 cells from each tumor were incubated with antibodies against CD16/32 at 50ug/ml in a total volume of 50μl for 10 min at RT to block Fc receptors. Surface marker metal-conjugated antibodies cocktail were then added, yielding 100 μL final reaction volume and stained for 30min at 4 C. Following staining, cells were washed 2 times with PBS with 0.5% BSA and 0.02% NaN3. Then, cells were re-suspended with RPMI 1640 and 10 μM Cisplatin (Fluidigm) in a total volume of 400ul for 60 seconds before quenching 1:1 with pure FBS to determine viability. Cells were centrifuged at 600 g for 7 min at 4 C and washed once with PBS with 0.5% BSA and 0.02% NaN3. Cells were then fixed using Fixation/Permeabilization Buffer (ebioscience) for 30 min at 4 C. After two washes with permeabilization buffer (ebioscience) cells were incubated with intracellular metal-conjugated antibodies cocktail in 100μl for 30 min at 4 C. A summary of the antibodies/clones used in the mass cytometry analyses is presented in the supplemental Table S2. Antibodies were either purchased pre-conjugated from Fluidigm or purchased purified and conjugated in-house using mass cytometry antibody conjugation kits according to the manufacturer’s instructions. Cells were washed twice in PBS with 0.5% BSA and 0.02% NaN3 and then stained with 1 mL of 1:4000 191/193Ir DNA intercalator (Fluidigm) diluted in PBS with 1.6% PFA overnight. Cells were then washed once with PBS with 0.5% BSA and 0.02% NaN3 and then two times with double-deionized ddH20. Mass cytometry samples were diluted in ddH2O containing bead standards (Fluidigm) to approximately 106 cells per mL and then analyzed on a CyTOF mass cytometer equilibrated with ddH2O. All mass cytometry files were normalized together using the mass cytometry data normalization algorithm (30). For analysis, FCS files were manually pre-gated on Ir193 DNA+CD45+ events, excluding cisplatin+ dead cells, doublets and DNA- negative debris by Cytobank (Santa Clara, CA). The gated CD45+ population was then clustered based on all labeled phenotypic markers using spanning-tree progression analysis of density- normalized events (SPADE) (31). Putative cell populations on the resulting SPADE trees were manually annotated based on the expression of key markers as shown in Supplementary Figure 3.
Cell culture and transfections
For assay validation experiments HEK293 parental cells were transiently transfected with 1ug of full-length cDNA coding each target using Lipofectamine 3000 (Thermo Fisher) for 24 hours. Cells were used fresh for protein extraction and immunoblotting; or fixed in 10% neutral buffered formalin for 8–12 h and embedded in paraffin for quantification by QIF. Cell lines used in this study were purchased from the American Type Culture Collection (ATCC) and authentication was performed every 3–6 months using the GenePrint® 10 System in the Yale University DNA Analysis Facility.
TCGA data analysis for mRNA expression and genomic alterations
We analyzed the NSCLC samples from the Cancer Genome Atlas (TCGA, http://cancergenome.nih.gov/). Briefly, we downloaded the RNA-seq and DNA whole exome sequencing data from 370 NSCLC cases including 250 adenocarcinomas and 120 squamous cell carcinomas. Using data processed through the cBioPortal interface (www.cbioportal.com), DNA segments and RNA transcripts were aligned and DNA variants calling was performed using default TCGA pipelines. We conducted single scatterplot analysis between the mRNA scores of PDCD1 (PD-1), LAG-3 and HAVCR2 (TIM-3) genes. The total number of nonsynonymous mutations detected in the whole exome sequencing data relative to germline DNA was used as the tumor mutational burden.
Statistical Analysis
QIF signals between compartments were analyzed using linear regression, correlation functions and expressed as regression/correlation coefficients. For experiments including numerous fields of view (FOVs) per case slide, we analyzed the top 10% marker scores in each preparation. Patient characteristics were compared using the Student’s t test for continuous variables and chi-square test for categorical variables. Survival functions were compared using Kaplan-Meier estimates and statistical significance was determined using the log-rank test. Correlation studies were performed calculating linear regression coefficients and/or Spearman rho-rank functions. Associations between the markers and statistical significance were determined using JMP Pro. v11 and GraphPad Prism v7.0a software.
Results
Tumor tissue distribution of PD-1, LAG-3 and TIM-3 in human NSCLCs.
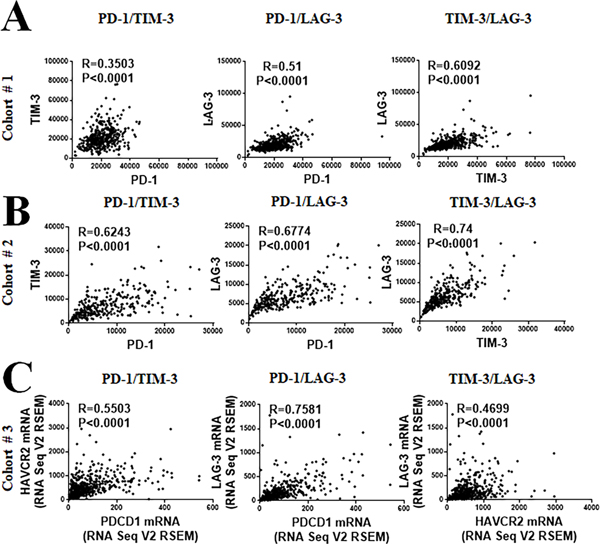
We standardized a QIF panel for simultaneous measurement of the markers DAPI, CD3, PD-1, LAG-3 and TIM-3 and studied 730 retrospectively collected NSCLC samples from 2 independent populations represented in TMA format (Cohort #1 from Yale [N=426] and Cohort #2 from Greece [N=304]). All the markers showed a predominant membranous staining pattern and were detected in CD3+ TILs (Figure 1A). However, PD-1 and LAG-3 were predominantly localized in CD3+ T-cells while TIM-3 was recognized frequently in CD3− populations. Using the visual detection threshold, we detected T-cell PD-1, LAG-3 and TIM-3 expression in 65%, 33% and 24% of NSCLCs in the first cohort; and in 45%, 49% and 26% of cases in the second collection (Figure 1B–C). In both cohorts the levels of T-cell PD-1, LAG-3 and TIM-3 protein were significantly correlated with each other (Spearman’s R=0.3–0.745, P<0.001, Figure 2A–B). The lowest correlation coefficients were between PD-1 and TIM-3. Comparable results were obtained when measuring the expression in the total tissue compartment (e.g. outside T-cells) and between the levels of PD-1 (PDCD1), LAG-3 and TIM-3 (HAVCR2) mRNA transcripts in 406 cases from the NSCLC datasets of The Cancer Genome Atlas (TCGA cohort #3, Figure 2C).
Figure 2: Association between PD-1, LAG-3 and TIM-3 in NSCLC by multiplex QIF and RNA sequencing.
A-B) Histograms showing the association between the protein expression levels of PD-1, LAG-3 and TIM-3 in the NCSLS cohort #1 (A) and #2 (B). C) Association between the transcript levels of HAVCR2 gene (TIM-3), PDCD1 (PD-1) and LAG-3 in NSCLC cases from the TCGA database (Cohort #3, n=406). R= Spearman’s correlation coefficient.
Target expression in individual immune cell populations and functional impact
To determine the expression of the targets and functional associations in specific immune cell subpopulations, we used mass cytometry by time-of-flight (CyTOF) to analyze freshly isolated leukocytes obtained from a collection of 20 primary resected NSCLCs (cohort #4). A panel of 35 phenotypical and functional markers were co-stained (supplementary Table S2) and spanning-tree progression analysis of density-normalized events (SPADE) was used to identify distinct immune cell populations (supplementary Figure S3). PD-1 was predominantly expressed on cytotoxic CD8+ T-cells, CD4+/CD25−/Foxp3− helper cells, regulatory CD4+/CD25+/Foxp3+ T-cells (Tregs) and CD3+/CD56+ NKT cells (Figure 3A, left panel). Virtually no PD-1 signal was detected in B-lymphocytes, NK cells and myeloid cell subsets. LAG-3 expression was seen in all studied T-cell groups with the highest levels in CD8+ T-cells. LAG-3 was also present on NK, NKT cells and granulocytes, but it was low or absent in antigen presenting cells (APCs) and B-lymphocytes (Figure 3A, center panel). TIM-3 was broadly expressed in adaptive and innate immune cells, with detectable levels in all T-lymphocyte subsets, NK, NKT and dendritic cells. Notably, the highest TIM-3 levels were seen in macrophages/NK/NKT cells whilst low/absent expression was found in granulocytes (Figure 3A, right panel).
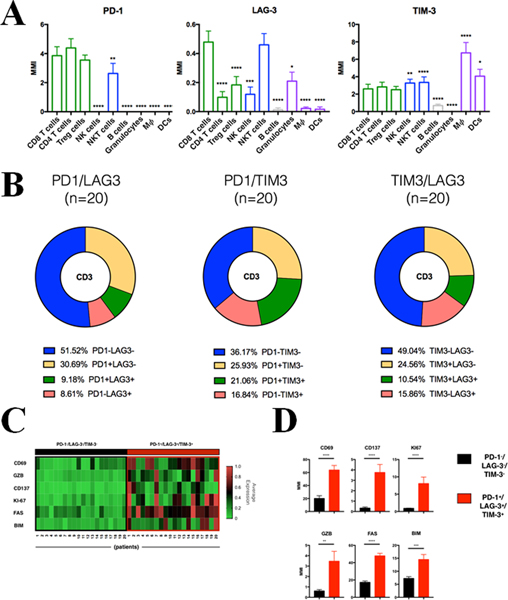
Figure 3. Single-cell multiparametric analysis of PD-1, TIM-3 and LAG-3 expression in immune cell subsets from NSCLC.
CyTOF analysis of tumor infiltrated leukocytes from primary NSCLC surgical specimens (cohort #5, n=20). A) Bar plots depict median PD-1 (left panel), LAG-3 (middle panel) and TIM-3 (right panel) expression levels for each immune cell population defined as indicated in Fig S3. MMI, Median Mass Intensity. B) Frequency of the specific CD3+ subpopulations defined by the combination of PD1 and LAG3 (left), PD1 and TIM3 (middle) and TIM3 and LAG3 (right) expression as indicated. C) Heatmap showing relative normalized expression of activated markers in TILs PD-1−/LAG-3−/TIM-3− (black square) or TILs PD-1+/LAG-3+/TIM-3+ (red square) across 20 lung cancer primary tumors. D) Bar plots of indicated activation markers expression levels for TILs PD-1−/LAG-3−/TIM-3− (black) or TILs PD-1+/LAG-3+/TIM-3+ (red). MMI, median mass intensity; *, P<0.05; **, P<0.01; ***, P<0.005; ****, P<0.001.
Overall, simultaneous co-expression of PD-1, LAG-3 and TIM-3 was seen in 5.4% of CD3+ TILs. PD-1 and LAG-3 were co-expressed in 9.1% of cells, PD-1 and TIM-3 in 21% of cells and LAG-3 and TIM-3 in 10.5% of TILs (Figure 3B, green chart area). The proportion of TILs lacking all 3 markers was 29.4%. Notably, 8.6% of LAG-3+ and 16.8% of TIM-3+ TILs showed absence of PD-1 expression; and 15.8% showed LAG-3 positivity in the absence of TIM-3 (Figure 3B, red chart area).
To evaluate the impact of the markers, we analyzed the expression of functional indicators in individual T-cell populations. Co-expression of PD-1, LAG-3 and TIM-3 was prominently associated with high levels of markers of T-cell activation (CD69, 4–1BB), cytotoxic/effector function (Granzyme-B [GZB]) and proliferation (Ki-67), but also with higher levels of the apoptotic signal receptors (FAS or CD95) and pro-apoptotic proteins (BIM) (Figure 3C–D). TILs with high LAG-3, but low PD-1 and TIM-3 expression (e.g. PD-1−LAG-3+TIM3−) showed higher cytotoxic potential (GZB, CD69 and CD137) than T-cells with elevated TIM-3 alone (e.g. PD1−LAG3−TIM-3+) or those expressing only PD-1 (e.g. PD-1+LAG3−TIM-3−) (supplementary Figure S4 A–B). Notably, co-expression of 2 or more of the immune inhibitory receptors was associated with higher levels of all functional markers (Supplementary Figure S4C).
Clinical significance and molecular associations of PD-1, LAG-3 and TIM-3 expression in NSCLC.
To explore the clinical role of the markers, we studied their association with major clinico-pathological variables and survival in the retrospective cohorts #1 and #2. There was a significant association between the levels of the markers and TIL abundance; and all the markers showed a positive association with each other (supplementary Tables S3–S4). There were no consistent associations between expression of PD-1, LAG-3 or TIM-3 and major clinicopathologic features including age, gender, smoking status, clinical stage and tumor histology variant.
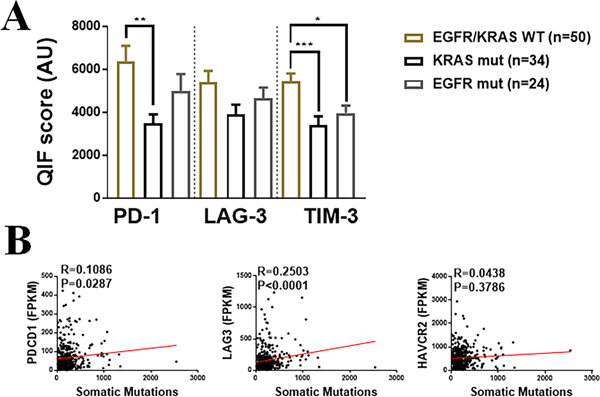
As shown in Figure 4A, the level of PD-1 and TIM-3 were significantly lower in tumor harboring KRAS mutations than in cases with wild type EGFR and KRAS. EGFR mutated lung adenocarcinomas showed significantly lower TIM-3 than tumors lacking mutations in both oncogenes. In the TCGA lung cancer datasets, there was a positive correlation between tumor mutational burden and LAG-3 but this association was not evident for the other markers (Figure 4B).
Figure 4. Association of LAG-3, PD-1, TIM-3 and CD3 with major driver mutations and tumor mutational burden in NSCLC.
A) Levels of PD-1, LAG-3 and TIM-3 in lung adenocarcinoma cohort #3 comprising cases wildtype for EGFR and KRAS driver mutations and with oncogenic KRAS or EGFR variants. B) Association between the markers mRNA levels (FPKMs) and tumor mutational burden (e.g. number of nonsynonymous mutations) in the TCGA NSCLC cohort (Cohort 3, N=406) *=P<0.01; **=P<0.01, ***=P<0.001. R=Spearman’s correlation coefficient.
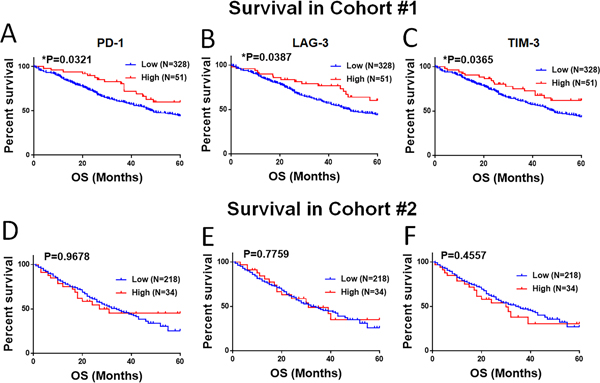
To explore the survival effect of dominant expression of each marker, we assessed the 5-year overall survival in cases with scores above or below the top 15th percentile of the cohort. As shown in Figure 5A–F, prominent T-cell PD-1, LAG-3 and TIM-3 were significantly associated with longer 5-year overall survival in the first cohort, but this was not evident in the second population. Similar associations with survival were seen when stratifying the markers by quartiles (supplementary Figure S5).
Figure 5. Association of PD-1, LAG-3 and TIM-3 with survival in retrospective cohorts.
(A-F) Association of markers with 5-year overall survival in 379 NSCLC cases from cohort # 1 (A-C) and 252 cases from the cohort # 2 (D-F). Elevated expression of the markers was defined as signal above the 85th percentile of the cohort.
PD-1, LAG-3, TIM-3 and sensitivity/resistance to PD-1 axis blockers in NSCLC
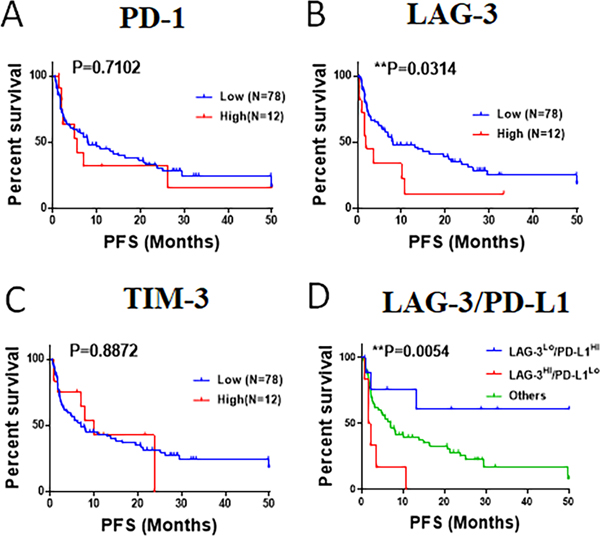
We then studied the association between the baseline level of the markers and survival after treatment with PD-1 axis blockers in 90 patients. Cases with prominent T-cell LAG-3 expression (top 15th percentile of the cohort), showed a significantly shorter progression free survival (log-rank P=0.03 Figures 6A–C). In contrast, elevated levels of T-cell PD-1 or TIM-3 were not significantly associated with survival in the cohort. Notably, cases with high LAG-3 and low tumor PD-L1 expression (<50% tumor proportion score [TPS]) showed a markedly lower progression free survival than cases with low LAG-3 and high (≧50% TPS) tumor PD-L1 expression (Figure 6D).
Figure 6. Association of PD-1, LAG-3 and TIM-3 expression with survival in NSCLC patients treated with PD-1 axis blockers.
(A-C): Charts showing the Kaplan-Meier survival estimates of patients treated with PD-1 axis blockers (cohort #6, n=90). The scores of T-cell PD-1 (A), LAG-3 (B) and TIM-3 (C) were measured using multiplex QIF in pre-treatment samples and stratified using top 15th percentile as cut-point. The differences between groups was compared using the log-rank test. D) Kaplan-Meier survival estimates of patients treated with PD-1 axis blockers stratified by LAG-3 expression (high, above top 15th; low, below or equal top 15th) and PD-L1 expression (high, above 50% tumor proportion score; low, below or equal 50% tumor proportion score).
Discussion
Using QIF and mass cytometry, we determined the expression, functional associations and clinical significance of PD-1, LAG-3 and TIM-3 in human NSCLC. Specifically, we found that all three receptors display variable expression in NSCLC, distinct immune-cell distribution and association with T-cell activation and pro-apoptotic markers. Notably, TILs with elevated LAG-3 showed the most prominent activated and pro-apoptotic phenotype and elevated T-cell LAG-3 (but not PD-1 or TIM-3) was significantly associated with shorter survival after PD-1 axis blockade. Taken together, our findings support a distinct and independent role of these immune inhibitory receptors in lung cancer and show that tumors with dominant LAG-3 expression are less sensitive to PD-1 axis blockers.
Detectable expression of PD-1, LAG-3 and TIM-3 was seen in 55%, 41.5% and 25.3 % of NSCLCs, respectively; and was associated with T-cell inflamed tumors, but not consistently associated with other clinicopathologic variables. A recent study detecting PD-1 and LAG-3 using single-marker IHC in 139 surgically resected lung carcinomas reported PD-1 expression in 43.4% and LAG-3 in 26.9% of cases (32). Here, LAG-3 detection was associated with higher PD-1, non-squamous tumor histology and worse survival. Whereas our study showed no consistent association between elevated LAG-3 and specific tumor histology or with survival in cases not receiving immunotherapy, we found similar frequencies and patterns of tumor LAG-3 and PD-1 protein expression. Although the primary source explaining the partial differences between this study and our findings is uncertain, diverse methodological considerations may account for this including the cases analyzed, modality of marker testing, cut-points used for stratification and assay-specific variables. In our study, we evaluated four different LAG-3 antibodies using control FFPE samples including clones 17B4, D2G40, EPR4392 and 11E3; and selected clone 17B4 for its high specificity and broader dynamic range.
The single-cell studies revealed distinct expression and functional impact of PD-1, LAG-3 and TIM-3 in immune cell subpopulations. While PD-1 and LAG-3 were predominantly localized in NKT and CD8+ T-cells, TIM-3 was commonly seen in macrophages and NK cells. The elevated expression of PD-1 and LAG-3 in cytotoxic T CD8+ and NKT cells is consistent with their expected immune regulatory function in effector cells. Notably, both targets were also highly expressed in Tregs, suggesting that PD-1 and LAG-3 pathways can have additional and cell-specific suppressive functions constituting complex internal systems mediating immune tolerance. Prominent expression of TIM-3 in innate immune cells such as monocytes, dendritic cells and NK cells has been previously reported in non-tumor tissues such as peripheral blood (33–35), but this is the first time to our knowledge it is reported in lung cancer.
KRAS and EGFR are the most commonly mutated driver oncogenes in lung adenocarcinoma. EGFR-mutant tumors display commonly lower TILs, PD-L1 expression and tumor mutational burden than EGFR-wild type tumors (36–38). Consistent with this, patients with EGFR-mutated carcinomas derive less clinical benefit from PD-1 axis blockade (39,40). In our study, we found lower levels of PD-1, LAG-3 and TIM-3 in KRAS and EGFR mutant tumors than in cases lacking mutations in both genes. This supports lower overall immune activation and regulation mediated by PD-1, LAG-3 and TIM-3 in these malignancies and suggests limited therapeutic potential of targeting these receptors. However, a fraction of NSCLCs with KRAS driver mutations may also harbor other genomic variants which could alter the tumor-immune microenvironment (41).
Despite being traditionally considered as exhaustion T-cell markers (42,43), PD-1, LAG-3 and TIM-3 are expressed preferentially in activated TILs. This is consistent with a model where co-inhibitory receptors are up-regulated upon T-cell stimulation in order to limit exaggerated responses and potential tissue damage. In this regard, it has been reported that the expression of these 3 markers is associated and could be used to identify antigen-experienced T-cells in cancer patients (44). However, previous studies have also shown that prominent T-cell activation is associated with a dysfunctional phenotype characterized by engagement of apoptotic programs (45). Consistent with this notion, expression of PD-1, LAG-3 and TIM-3 was associated with elevated expression of key pro-apoptotic targets in lung carcinomas. Additional studies are ongoing to refine the phenotype of cells expressing each immune inhibitory receptor combination and determine how to use this information therapeutically. Studies simultaneously measuring key ligand(s) for PD-1, LAG-3 and TIM-3 are also warranted since functional consequences probably require co-expression of the ligands and receptors in close proximity within the tumor microenvironment.
An intriguing finding of our study is the negative association between LAG-3 overexpression and survival benefit in patients with NSCLC treated with PD-1 axis blockers. This suggests that tumors in which immune evasion is mediated predominantly by LAG-3 are less sensitive to PD-1 axis blockade and opens the possibility of eventually using LAG-3 for selection of patients for immunotherapy. Early results from LAG-3 inhibitors in the CA224-020 clinical trial (NCT01968109) show promising results in advanced melanoma patients with resistance to PD-1 blockers. Here, LAG-3 positivity by IHC in pre-treatment tumor samples is associated with higher response rate supporting a predictive role of LAG-3 expression. In recent work we also found LAG-3 upregulation by protein and mRNA measurements in TILs from 5 of 8 NSCLC patients with acquired resistance to immune checkpoint blockers suggesting a possible role of LAG-3 in this setting (38).
Our study has limitations. The evaluation of three retrospective cohorts (Cohorts # 1–2 and #4) was performed using TMAs that analyze relatively small sample fragments and may over or under-represent the marker measurements. Although not the standard method to measure proteins in tumor tissues clinically, diverse reports from our group and others using TMAs have shown consistent results and significant association with clinical, pathological variables and outcome (46). Additionally, i) each case was represented twice or thrice in the TMA in order to account for possible marker variations across different tumor areas; ii) the data obtained in the TMA cohorts were consistent with results in the TCGA collection (cohort #3) that was conducted using whole tissue sections and mRNA analysis; and iii) the marker scores and distribution obtained using TMAs were similar to those in whole tissue sections used for the cohort #6 analysis. Finally, the collection of patients treated with PD-1 axis blockers (cohort #6) included cases treated with different PD-1 axis blockers and samples collected at different time points before treatment initiation and with multiple previous lines of treatment. These factors are common limitations of retrospective cohort studies and their possible impact in the results are uncertain.
Overall, we have characterized the expression and significance of PD-1, LAG-3 and TIM-3 in a sizable number of NSCLC cases from 6 independent tumor collections. Our results demonstrate dissimilar and non-redundant expression of these targets in TILs from primary lung tumors and provide valuable insights about T-cell activation and dysfunction in this setting.
Supplementary Material
Translational relevance statement:
Our results reveal that PD-1, LAG-3 and TIM-3 show differential tissue/cell distribution, functional impact and clinical significance in NSCLCs. In addition, single-cell studies show that simultaneous co-expression of these immune inhibitory receptors is associated with prominent TIL activation, but also with acquisition of a pro-apoptotic phenotype. Finally, we determine the prognostic value of the markers and identify a negative association between elevated baseline LAG-3 and reduced sensitivity to PD-1 axis blockade. Taken together, our results support independence of these immune inhibitory pathways and expand the current understanding of their interplay in cancer. This information could be used to guide and interpret results from ongoing and future clinical trials.
Acknowledgments:
We would like to Dr. Paula Kavathas (Yale University) for providing access to key resources and Lori Charette from Yale Pathology Tissue Services for excellent support in histology and TMA samples preparation.
Funding: Lung Cancer Research Foundation (LCRF), Yale SPORE in Lung Cancer (P50CA196530), Department of Defense-Lung Cancer Research Program Career Development Award (W81XWH-16-1-0160), sponsored research grant by Tesaro Inc., Yale Cancer Center Support Grant (P30CA016359) a gift by the Grunley Family Fund and a Stand Up To Cancer – American Cancer Society Lung Cancer Dream Team Translational Research Grant (SU2C-AACR-DT1715 and SU2C-AACR-DT22-17). Stand Up To Cancer is a program of the Entertainment Industry Foundation. Research grants are administered by the American Association for Cancer Research, the scientific partner of SU2C. M.F.S. is supported by a Miguel Servet contract from Instituto de Salud Carlos III, Fondo de Investigación Sanitaria (Spain).
Footnotes
Competing interests:
Dr. Kurt Schalper has been speaker or consultant for Merck, Takeda Pharmaceuticals, Shattuck Labs, Pierre-Fabre and Celgene. His laboratory has received research funding from Vasculox/Tioma, Navigate Biopharma, Tesaro Inc, Onkaido Therapeutics/Moderna, Takeda Pharmaceuticals, Surface Oncology, Pierre-Fabre, Merck and Bristol-Myers Squibb. For additional conflict of interest information, detailed forms will be provided by each author specifying their individual disclosures by the time of publication.
References and Notes:
- 1.Gettinger SN, Horn L, Gandhi L, Spigel DR, Antonia SJ, Rizvi NA, et al. Overall Survival and Long-Term Safety of Nivolumab (Anti-Programmed Death 1 Antibody, BMS-936558, ONO-4538) in Patients With Previously Treated Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. 2015;33:2004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rizvi NA, Hellmann MD, Brahmer JR, Juergens RA, Borghaei H, Gettinger S, et al. Nivolumab in Combination With Platinum-Based Doublet Chemotherapy for First-Line Treatment of Advanced Non-Small-Cell Lung Cancer. J Clin Oncol. American Society of Clinical Oncology; 2016;34:2969–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, et al. Pembrolizumab for the Treatment of Non–Small-Cell Lung Cancer. N Engl J Med. Massachusetts Medical Society; 2015;372:2018–28. [DOI] [PubMed] [Google Scholar]
- 4.Herbst RS, Baas P, Kim D-W, Felip E, Pérez-Gracia JL, Han J-Y, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet (London, England). 2016;387:1540–50. [DOI] [PubMed] [Google Scholar]
- 5.Reck M, Brahmer JR. Pembrolizumab in Non–Small-Cell Lung Cancer. N Engl J Med. 2017;376:996–7. [DOI] [PubMed] [Google Scholar]
- 6.Hellmann MD, Rizvi NA, Goldman JW, Gettinger SN, Borghaei H, Brahmer JR, et al. Nivolumab plus ipilimumab as first-line treatment for advanced non-small-cell lung cancer (CheckMate 012): results of an open-label, phase 1, multicohort study. Lancet Oncol. 2017;18:31–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ribas A Adaptive Immune Resistance: How Cancer Protects from Immune Attack. Cancer Discov. 2015;5:915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, et al. Primary Resistance to PD-1 Blockade Mediated by JAK1/2 Mutations. Cancer Discov. 2017;7:188–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao J, Shi LZ, Zhao H, Chen J, Xiong L, He Q, et al. Loss of IFN-γ Pathway Genes in Tumor Cells as a Mechanism of Resistance to Anti-CTLA-4 Therapy. Cell. 2016;167:397–404.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Datar I, Schalper KA. Epithelial-Mesenchymal Transition and Immune Evasion during Lung Cancer Progression: The Chicken or the Egg? Clin Cancer Res. 2016;22:3422–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127:2930–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prat A, Navarro A, Paré L, Reguart N, Galván P, Pascual T, et al. Immune-Related Gene Expression Profiling After PD-1 Blockade in Non-Small Cell Lung Carcinoma, Head and Neck Squamous Cell Carcinoma, and Melanoma. Cancer Res. 2017;77:3540–50. [DOI] [PubMed] [Google Scholar]
- 13.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson AC, Joller N, Kuchroo VK. Lag-3, Tim-3, and TIGIT: Co-inhibitory Receptors with Specialized Functions in Immune Regulation. Immunity. 2016;44:989–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Attanasio J, Wherry EJ. Costimulatory and Coinhibitory Receptor Pathways in Infectious Disease. Immunity. 2016;44:1052–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sakuishi K, Apetoh L, Sullivan JM, Blazar BR, Kuchroo VK, Anderson AC. Targeting Tim-3 and PD-1 pathways to reverse T cell exhaustion and restore anti-tumor immunity. J Exp Med. 2010;207:2187–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldberg M V, Drake CG. LAG-3 in Cancer Immunotherapy. Curr Top Microbiol Immunol. 2010. page 269–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Woo S-R, Turnis ME, Goldberg M V., Bankoti J, Selby M, Nirschl CJ, et al. Immune Inhibitory Molecules LAG-3 and PD-1 Synergistically Regulate T-cell Function to Promote Tumoral Immune Escape. Cancer Res. 2012;72:917–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrews LP, Marciscano AE, Drake CG, Vignali DAA. LAG3 (CD223) as a cancer immunotherapy target. Immunol Rev. 2017;276:80–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu F, Liu J, Liu D, Liu B, Wang M, Hu Z, et al. LSECtin expressed on melanoma cells promotes tumor progression by inhibiting antitumor T-cell responses. Cancer Res. 2014;74:3418–28. [DOI] [PubMed] [Google Scholar]
- 21.Kouo T, Huang L, Pucsek AB, Cao M, Solt S, Armstrong T, et al. Galectin-3 Shapes Antitumor Immune Responses by Suppressing CD8+ T Cells via LAG-3 and Inhibiting Expansion of Plasmacytoid Dendritic Cells. Cancer Immunol Res. 2015;3:412–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mao X, Ou MT, Karuppagounder SS, Kam T-I, Yin X, Xiong Y, et al. Pathological α-synuclein transmission initiated by binding lymphocyte-activation gene 3. Science. 2016;353:aah3374-aah3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang J, Sanmamed MF, Datar Ila, Tianjiao S, Ji L, Sun J, Chen L, Chen Y, Zhu G, Zheng L ZT and BT. Fibrinogen-like protein 1 is a major ligand of LAG3 for T-cell suppression and the evasion of tumor immunity. Cell. in press. [Google Scholar]
- 24.Ocaña-Guzman R, Torre-Bouscoulet L, Sada-Ovalle I. TIM-3 Regulates Distinct Functions in Macrophages. Front Immunol. 2016;7:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y-H, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, et al. CEACAM1 regulates TIM-3-mediated tolerance and exhaustion. Nature. 2015;517:386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashio Y, Nakamura K, Abedin MJ, Seki M, Nishi N, Yoshida N, et al. Galectin-9 induces apoptosis through the calcium-calpain-caspase-1 pathway. J Immunol. 2003;170:3631–6. [DOI] [PubMed] [Google Scholar]
- 27.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, et al. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Investig. 2013;0:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schalper KA, Brown J, Carvajal-Hausdorf D, McLaughlin J, Velcheti V, Syrigos KN, et al. Objective measurement and clinical significance of TILs in non-small cell lung cancer. J Natl Cancer Inst. 2015;107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gettinger SN, Choi J, Mani N, Sanmamed MF5, Datar I, Sowell R, et al. A dormant TIL phenotype defines non-small cell lung carcinomas sensitive to immune checkpoint blockers. Nat Commun. 2018;9:3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Finck R, Simonds EF, Jager A, Krishnaswamy S, Sachs K, Fantl W, et al. Normalization of mass cytometry data with bead standards. Cytometry A. 2013;83:483–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiu P, Simonds EF, Bendall SC, Gibbs KD, Bruggner R V, Linderman MD, et al. Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol [Internet]. 2011 [cited 2018 Oct 10];29:886–91. Available from: http://www.nature.com/articles/nbt.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Yu H, Rozeboom L, Rivard CJ, Ellison K, Dziadziuszko R, et al. LAG-3 Protein Expression in Non-Small Cell Lung Cancer and Its Relationship with PD-1/PD-L1 and Tumor-Infiltrating Lymphocytes. J Thorac Oncol. 2017;12:814–23. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y, Ma CJ, Wang JM, Ji XJ, Wu XY, Moorman JP, et al. Tim-3 regulates pro- and anti-inflammatory cytokine expression in human CD14+ monocytes. J Leukoc Biol. 2012;91:189–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ndhlovu LC, Lopez-Vergès S, Barbour JD, Jones RB, Jha AR, Long BR, et al. Tim-3 marks human natural killer cell maturation and suppresses cell-mediated cytotoxicity. Blood. 2012;119:3734–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gleason MK, Lenvik TR, McCullar V, Felices M, O’Brien MS, Cooley SA, et al. Tim-3 is an inducible human natural killer cell receptor that enhances interferon gamma production in response to galectin-9. Blood. 2012;119:3064–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosell R, Palmero R. PD-L1 expression associated with better response to EGFR tyrosine kinase inhibitors. Cancer Biol Med. 2015;12:71–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akbay EA, Koyama S, Carretero J, Altabef A, Tchaicha JH, Christensen CL, et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 2013;3:1355–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Borghaei H, Paz-Ares L, Horn L, Spigel DR, Steins M, Ready NE, et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N Engl J Med. 2015;373:1627–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gainor JF, Shaw AT, Sequist L V., Fu X, Azzoli CG, Piotrowska Z, et al. EGFR mutations and ALK rearrangements are associated with low response rates to PD-1 pathway blockade in non-small cell lung cancer: A retrospective analysis. Clin Cancer Res. 2016;22:4585–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Skoulidis F, Goldberg ME, Greenawalt DM, Hellmann MD, Awad MM, Gainor JF, et al. STK11/LKB1 Mutations and PD-1 Inhibitor Resistance in KRAS -Mutant Lung Adenocarcinoma. Cancer Discov. 2018;8:822–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giordano M, Henin C, Maurizio J, Imbratta C, Bourdely P, Buferne M, et al. Molecular profiling of CD8 T cells in autochthonous melanoma identifies Maf as driver of exhaustion. EMBO J. 2015;34:2042–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wherry EJ, Ha S-J, Kaech SM, Haining WN, Sarkar S, Kalia V, et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 2007;27:670–84. [DOI] [PubMed] [Google Scholar]
- 44.Gros A, Robbins PF, Yao X, Li YF, Turcotte S, Tran E, et al. PD-1 identifies the patient-specific CD8+ tumor-reactive repertoire infiltrating human tumors. J Clin Invest. 2014;124:2246–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Horton BL, Williams JB, Cabanov A, Spranger S, Gajewski TF. Intratumoral CD8+ T-cell Apoptosis Is a Major Component of T-cell Dysfunction and Impedes Antitumor Immunity. Cancer Immunol Res. 2018;6:14–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsuzaki J, Gnjatic S, Mhawech-Fauceglia P, Beck A, Miller A, Tsuji T, et al. Tumor-infiltrating NY-ESO-1-specific CD8+ T cells are negatively regulated by LAG-3 and PD-1 in human ovarian cancer. Proc Natl Acad Sci U S A. 2010;107:7875–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.