Abstract
Introduction
Dysbetalipoproteinaemia (HLP3) is a disorder characterized by excess cholesterol-enriched, triglyceride-rich lipoprotein remnants in genetically predisposed individuals that powerfully promote premature cardiovascular disease if untreated. The current prevalence of HLP3 is largely unknown.
Material and methods
We performed cross-sectional analysis of 128,485 U.S. adults from the Very Large Database of Lipids (VLDbL), using four algorithms to diagnose HLP3 employing three Vertical Auto Profile ultracentrifugation (UC) criteria and a previously described apolipoprotein B (apoB) method. We evaluated 4,926 participants from the 2011–2014 National Health and Nutrition Examination Survey (NHANES) with the apoB method. We examined demographic and lipid characteristics stratified by presence of HLP3 and evaluated lipid characteristics in those with HLP3 phenotype discordance and concordance as determined by apoB and originally defined UC criteria 1.
Results
In U.S. adults in VLDbL and NHANES, a 1.7–2.0% prevalence is observed for HLP3 with the novel apoB method as compared to 0.2–0.8% prevalence in VLDbL via UC criteria 1–3. Participants who were both apoB and UC criteria HLP3 positive had higher remnant particles as well as more elevated triglyceride/apoB and total cholesterol/apoB ratios (all p < 0.001) than those who were apoB method positive and UC criteria 1 negative.
Conclusions
HLP3 may be more prevalent than historically and clinically appreciated. The apoB method increases HLP3 identification via inclusion of milder phenotypes. Further work should evaluate the clinical implications of HLP3 diagnosis at various lipid algorithm cut-points to evaluate the ideal standard in the modern era.
Keywords: Very Large Database of Lipids (VLDL), National Health and Nutrition Examination Survey, type III hyperlipoproteinemia, dysbetalipoproteinemia, apolipoprotein B
Introduction
Dysbetalipoproteinemia (hyperlipoproteinemia type 3 (HLP3)) is a heritable disorder characterized by excess cholesterol-enriched, triglyceride (TG)-rich remnant lipoproteins. It is one of 5 Fredrickson, Levy, and Lees hyperlipoproteinemia phenotypes described in 1967 using electrophoresis and ultracentrifugation (UC) techniques [1]. HLP3 classically occurs in individuals who are homozygous for a common recessive allele apoE2 via the substitution of cysteine for arginine at position 158 [2]. These mutaions lead to defective removal of chylomicron remnants and very low-density lipoprotein cholesterol (VLDL-C), promoting premature atherosclerotic cardiovascular disease (ASCVD) [2]. Fewer than 5% of apoE2 homozygotes develop hyperlipidemia [3], and it is thought that additional genetic, hormonal, and environmental factors such as obesity and diabetes are required to precipitate a pathologic hyperlipidemic state [4].
Clinicians generally consider familial hyperlipoproteinemias to be rare, and consequently a specific diagnosis is seldom made in clinical practice [5–7]. In the 1960s when HLP3 was first described, lipid fractionation techniques were limited in their ability to assess the prevalence of these phenotypes in large studies; initial research estimated their frequency at 0.1–0.4%. [8–10]. HLP3 cannot be diagnosed from the standard lipid profile comprising cholesterol, high density lipoprotein cholesterol (HDL-C), LDL-C, and TG [11]. Several clinical criteria have been proposed based on UC and apoB measurement [12, 13], however these are not widely used in modern practice. Population-based prevalence studies using classical UC are lacking, and prior studies with selected patients referred for evaluation may improperly estimate true prevalence of HLP3 [8].
In this study, we aimed to describe the prevalence of HLP3 in the modern U.S. adult population with modified UC-based criteria and a recently validated quantitative approach to diagnose familial hyperlipoproteinemia phenotypes that integrates information from the standard lipid profile and apolipoprotein B (apoB), otherwise known as the apoB method [14, 15]. We analyzed participants from the Very Large Database of Lipids (VLDbL) and National Health and Nutritional Examination Survey (NHANES) to evaluate for consistency in apoB method-derived HLP3 prevalence estimates as well as the overall overlap in distribution of apoB, LDL-C, total cholesterol, and TG.
Material and methods
VLDbL study data
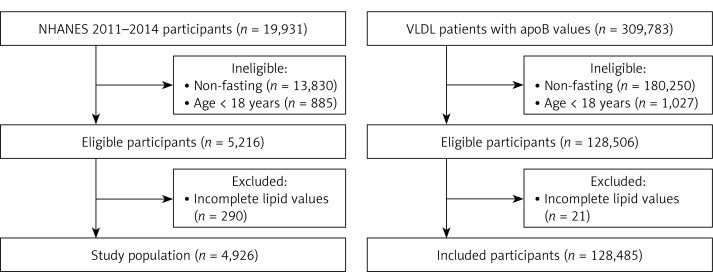
We examined 309,783 participants from VLDbL (clinicaltrials.gov NCT01698489), a database collected from U.S. participants referred for Vertical Auto Profile (VAP) UC lipid profile analysis (Atherotech, Inc., Birmingham, AL) in 2009–2011. We performed a cross-sectional analysis of fasting participants, including those who were 18 years and older with complete demographics (age, sex, diabetes, and hypertension) and lipid values: apoB, HDL-C, VLDL-C, VLDL-C subfraction 3 (VLDL3-C), remnant lipoprotein cholesterol (RLP-C), intermediate density lipoprotein cholesterol (IDL-C), LDL-C, real LDL-C (LDL-Cr), and TG (Figure 1) [16]. LDL-Cr was defined as cholesterol carried in the biochemically defined LDL fraction, distinct from the broader clinical LDL-C definition inclusive of IDL-C and Lipoprotein (a). Laboratory samples were obtained from participants fasting for at least 8 h prior to collection.
Figure 1.
Study population selection criteria
ApoB – apolipoprotein B, NHANES – National Health and Nutrition Examination Survey.
Within VLDbL, cholesterol content of lipoprotein fractions was determined by the VAP method. VAP uses single vertical spin density gradient UC to separate lipoprotein fractions in less than 1 h. Cholesterol content of eluted lipoprotein fractions is measured using a validated colorimetric assay employing a cholesterol oxidase reaction [17]. TG concentrations were directly measured with the Abbott ARCHITECT C-8000 system (Abbott Laboratories, Abbott Park, IL). Yearly quality assessments with VAP and Abbott ARCHITECT-derived values were described previously [16].
ApoB was directly measured through a WHO standardized immunoassay based on immunoturbidimetry and the Abbott ARCHITECT analyzer and reagent. Estimated LDL-C was calculated as TC – HDL-C – TG/adjustable factor, whereby TG and non-HDL-C are used to assign one of 174 participant-specific adjustable factors to estimate VLDL-C in patients with TG < 400 mg/dl [18]. In VLDbL, hypertension was defined as the presence of a diagnosis of hypertension on the laboratory order. Diabetes was defined as those with hemoglobin A1c ≥ 6.5%, fasting glucose ≥ 126 mg/dl, or those with a diagnosis of diabetes on the laboratory order.
NHANES study data
We examined 19,931 participants from NHANES, with a primary focus on individuals from 2011–2012 and 2013–2014 cycles [19]. NHANES is a complex survey conducted by the National Center for Health Statistics to provide a representative sample of the civilian, non-institutionalized U.S. population. The process of sample selection and participation in NHANES was fully described previously [20]. Our analysis included 5,216 fasting participants who were 18 years and older. We further excluded 290 participants with missing apoB, total cholesterol, TG, and LDL-C values (Figure 1).
Participants fasted for at least 8 h prior to sample collection. NHANES study protocols were approved by the institutional review board of the National Center of Health Statistics, and written informed consent was obtained from all participants. Our group had previously demonstrated that lipid distributions in the 2011–2012 NHANES survey were almost identical to those of VLDbL [16].
Within NHANES, TC, HDL-C, and TG were measured spectrophotometrically using oxidation reagents. LDL-C and VLDL-C were calculated using the Martin/Hopkins equation and apoB was measured through immunochemical reactions and light spectroscopy [18, 21, 22]. The full process of lipid measurement, data checking, and quality inspection within NHANES is documented elsewhere [18, 21, 22].
In NHANES, in-home interviews and questionnaires were used to gather demographic information and prescription medication use in the prior 30 days [19, 20]. NHANES race categories included non-Hispanic White, non-Hispanic Black, Mexican American, and other. Lipid-lowering therapies were defined by NHANES standards and included statins, bile acid sequestrants, and cholesterol absorption inhibitors. Height and weight were used to calculate body mass index (BMI), categorized as underweight (BMI < 18.5 kg/m2), normal weight (≥ 18.5 and < 25 kg/m2), overweight (≥ 25 and < 30 kg/m2), and obese (≥ 30 kg/m2). Laboratory blood draws assessed the presence of diabetes through fasting blood glucose values and hemoglobin A1c. Diabetes was defined as fasting a glucose ≥ 126 mg/dl, hemoglobin A1c ≥ 6.5%, or self-report of previous physician diagnosis.
Classification of dyslipidemia
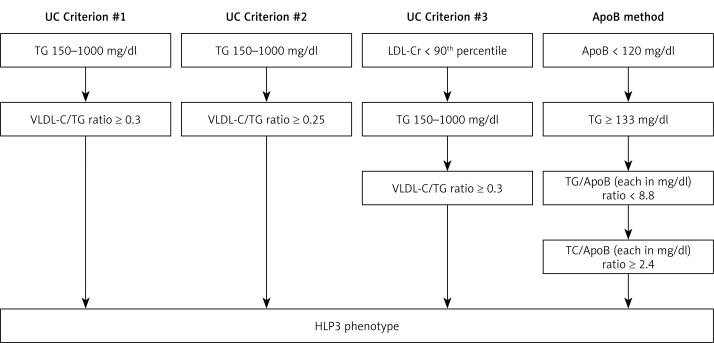
We used UC-based criteria in the VLDbL database to calculate prevalence of HLP3, utilizing TG, TC, VLDL-C, and LDL-C. HLP3 was characterized by meeting one of three criteria: criterion 1: TG 150–1000 mg/dl and VLDL-C/TG ratio ≥ 0.3; criterion 2: TG 150–1000 mg/dl and VLDL-C/TG ratio ≥ 0.25; criterion 3: real low-density lipoprotein-cholesterol (LDL-Cr) < 90th percentile, TG 150–1000 mg/dl, and VLDL-C/TG ratio ≥ 0.3 (Figure 2) [18].
Figure 2.
Dyslipidemia classification schemat
ApoB – apolipoprotein B, HLP3 – type III familial hyperlipoproteinemias phenotype, LDL-Cr – real (biologic) low-density lipoprotein-cholesterol, TG – triglycerides, VLDL-C – very low-density lipoprotein cholesterol.
We then utilized the Sniderman algorithm, an apoB based method derived from a cohort of 1,771 fasting individuals with genetically verified dyslipidemia [14, 15]. Via the apoB algorithm, HLP3 was defined by the following criteria: apoB < 120 mg/dl, TG ≥ 133 mg/dl, TG/apoB < 8.8 (in mg/dl units), and TC/apoB ≥ 2.4 (in mg/dl units) (Figure 2) [23]. We applied this algorithm to VLDbL and NHANES databases to estimate type III prevalence.
Statistical analysis
For primary analysis involving participants from VLDbL, prevalence of HLP3 was calculated and compared between UC and apoB methods. We compared age, sex, and prevalence of co-morbid hypertension and diabetes mellitus between HLP3 and other participants. Median values and interquartile ranges (IQR) of the predominant lipoprotein for each phenotype were analyzed with each method. Comparisons of characteristics by HLP3 status were performed by sample characteristics via χ2 tests and Wilcoxon rank-sum tests.
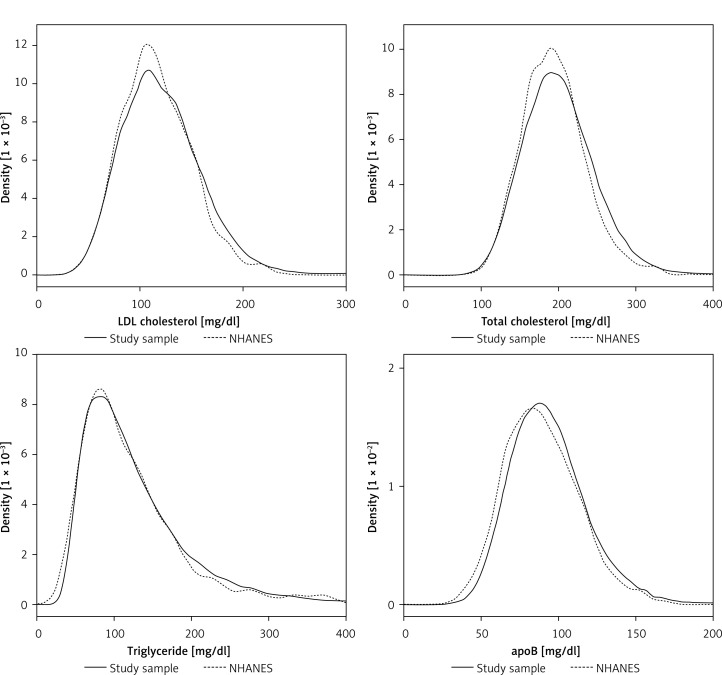
Prior work showed that population lipoprotein cholesterol subtype distributions are nearly identical in VLDbL and NHANES 2007–2008 databases [16]. For the analysis involving participants from NHANES 2011–2014, we first compared lipid distributions in the VLDbL-apoB subset and participants from 2011–2012 NHANES via nearly superimposed kernel density plots as we had done in our previous studies (Figure 3). We accounted for the complex sampling design in NHANES by using sampling weights to generate nationally representative estimates in all analyses. We used the Taylor linearization series to calculate standard errors [19, 20].
Figure 3.
Comparison of lipid and lipoprotein cholesterol distributions between the Very Large Database of Lipids study and 2011–2012 National Health and Nutrition Examination Survey
Lipid distribution between apoB data (n = 128,485) and NHANES 2011–2012 data (n = 2,415). NHANES participants are depicted by dotted lines; VLDbL patients are depicted by solid lines. ApoB – apolipoprotein B, NHANES – National Health and Nutrition Examination Survey.
We examined HLP3 prevalence in NHANES via the apoB method and compared age, sex, race, obesity, diabetes, and treatment of lipid-lowering therapies between HLP3 and other participants. We calculated medians and IQR for each lipid parameter overall and by phenotype. Comparisons of characteristics of the study population by HLP3 status, as determined by the apoB method were performed via χ2 tests and Wilcoxon rank-sum tests.
Lastly, we evaluated participants from VLDbL who were classified as HLP3 via the apoB method and had concordant or discordant HLP3 diagnosis via the original UC criteria [1]. We calculated medians and IQR of lipids characteristics for concordant and discordant participants and compared them via Wilcoxon rank-sum tests.
All statistical analyses were conducted using Stata statistical software version 14.2 (StataCorp, College Station, TX). All statistical tests were two-sided with significance at p < 0.05.
Results
Study populations
A total of 128,485 VLDbL samples were analyzed. Median age of participants was 57 years (IQR: 46–68 years) with 45% men. Diabetes was present in 20.1% and hypertension in 25.5% of participants. Median TC, estimated LDL-C, LDL-Cr, IDL-C, VLDL-C, VLDL3-C, RLP-C, apoB, and TG among all VLDbL participants were 197 (IQR: 168–228), 117 (93–144), 95 (73–119), 13 (9–19), 22 (17–29), 13 (10–16), 26 (20–35), 92 (76–108), and 110 (78–159) mg/dl, respectively.
A total of 4,926 NHANES participants from the 2011–2014 cycles were analyzed. Median age was 46 years (IQR: 32–60 years) and 48% were men. By BMI category, 1.7% were underweight, 29.5% normal weight, 32.2% overweight, and 36.6% obese. Roughly 1 in 3 (33.4%) were on lipid-lowering therapy, with 31.4% on a statin, 1.7% on a cholesterol absorption inhibitor, and 0.5% on a bile acid sequestrant. Median TC, estimated LDL-C, VLDL-C, apoB, and TG among all NHANES participants were 187 (IQR: 162–214), 111 (89–136), 20 (16–26), 87 (72–105), and 99 (68–147) mg/dl, respectively.
HLP3 phenotype prevalence by clinical characteristic
Utilizing the UC criteria in VLDbL, HLP3 phenotype prevalence was 0.20% (n = 255) via criterion 1, 0.80% (n = 1,032) via criterion 2, and 0.18% via criterion 3 (n = 232) (Table I). Utilizing the apoB method, HLP3 was found in 1.74% of VLDbL (n = 2,239) and 1.97% of NHANES (n = 102) (Tables II, III).
Table I.
Prevalence and characteristics of the Very Large Database of Lipids study population by HLP3 status (UC criteria)
| Parameter | UC Criterion #1 | UC Criterion #2 | UC Criterion #3 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| HLP3 n = 255 (0.20%) | No HLP3 n = 128,230 (99.8%) | P-value | HLP3 n = 1,032 (0.80%) | No HLP3 n = 127,453 (99.2%) | P-value | HLP3 n = 232 (0.18%) | No HLP3 n = 128,253 (99.8%) | P-value | |
| Age, % (n): | < 0.001 | < 0.001 | < 0.001 | ||||||
| 18–39 | 6.7 (17) |
15.3 (19,592) |
8.7 (90) |
15.3 (19,519) |
6.5 (15) |
15.3 (19,594) |
|||
| 40–75 | 87.4 (223) |
73.6 (94,166) |
81.2 (837) |
73.6 (93,552) |
88.4 (205) |
73.6 (94,184) |
|||
| > 75 | 5.9 (15) |
11.1 (14,178) |
10.1 (104) |
11.1 (14,089) |
5.2 (12) |
11.1 (14,181) |
|||
| Sex, % (n): | 0.397 | 0.004 | 0.652 | ||||||
| Male | 42.1 (107) |
44.8 (57,105) |
49.2 (503) |
44.7 (56,709) |
43.3 (100) |
44.8 (57,112) |
|||
| Female | 57.9 (147) |
55.2 (70,445) |
50.8 (519) |
55.3 (70,073) |
56.7 (131) |
55.2 (70,461) |
|||
| Presence of diabetes, % (n) | 23.1 (59) |
20.1 (25,830) |
0.234 | 21.5 (222) |
20.1 (25,667) |
0.273 | 22.8 (53) |
20.1 (25,836) |
0.306 |
| Presence of hypertension, % (n) | 25.9 (66) |
25.6 (32,757) |
0.902 | 24.5 (253) |
25.6 (32,570) |
0.446 | 26.3 (61) |
25.5 (32,762) |
0.794 |
| Total cholesterol, median (IQR) | 248 (202–290) |
197 (168–228) |
< 0.001 | 249 (209–286) |
197 (168–228) |
< 0.001 | 240 (199–277) |
197 (168–228) |
< 0.001 |
| ApoB, median (IQR) | 84 (69–118) |
92 (76–108) |
0.078 | 115 (87–138) |
91 (76–108) |
< 0.001 | 82 (68–104) |
92 (76–108) |
< 0.001 |
| HDL-C, median (IQR) | 48 (41–57) |
52 (43–64) |
< 0.001 | 46 (40–53) |
52 (43–64) |
< 0.001 | 48 (41–57) |
52 (43–64) |
< 0.001 |
| VLDL-C, median (IQR) | 71 (58–99) |
22 (17–29) |
< 0.001 | 56 (46–72) |
22 (17–29) |
< 0.001 | 73 (59–103) |
22 (17–29) |
< 0.001 |
| VLDL3-C, median (IQR) | 41 (34–53) |
13 (10–16) |
< 0.001 | 31 (26–39) |
13 (10–16) |
< 0.001 | 41 (35–54) |
13 (10–16) |
< 0.001 |
| RLP-C, median (IQR) | 88 (73–107) |
26 (20–35) |
< 0.001 | 67 (57–82) |
26 (20–34) |
< 0.001 | 88 (72–108) |
26 (20–35) |
< 0.001 |
| IDL-C, median (IQR) | 45 (37–55) |
13 (9–19) |
< 0.001 | 35 (29–44) |
13 (9–19) |
< 0.001 | 45 (36–54) |
13 (9–19) |
< 0.001 |
| LDL-C, median (IQR) | 105 (80–145) |
117 (92–144) |
0.007 | 138 (99–174) |
116 (92–143) |
< 0.001 | 101 (77–137) |
117 (92–144) |
< 0.001 |
| LDL-Cr, median (IQR) | 48 (34–92) |
95 (73–119) |
< 0.001 | 97 (54–132) |
95 (73–119) |
0.300 | 45 (32–72) |
95 (73–119) |
< 0.001 |
| TG, median (IQR) | 202 (171–266) |
110 (78–158) |
< 0.001 | 194 (167–245) |
109 (78–157) |
< 0.001 | 202 (171–271) |
110 (78–158) |
< 0.001 |
ApoB – apolipoprotein B, HDL-C – high density lipoprotein cholesterol, IDL-C – intermediate density lipoprotein cholesterol, IQR – interquartile range, LDL-C – low-density lipoprotein cholesterol, LDL-Cr – real low-density cholesterol, TG – triglycerides, RLP-C – remnant lipoprotein cholesterol, VLDL-C – very low-density lipoprotein cholesterol, VLDL3-C – very-low density cholesterol subfraction 3.
Table II.
Prevalence and characteristics of the Very Large Database of Lipids study population by HLP3 status (apoB method)
| Parameter | HLP3 n = 2,239 (1.74%) | No HLP3 n = 126,246 (98.26%) | P-value |
|---|---|---|---|
| Age, % (n): | < 0.001 | ||
| 18–39 | 11.7 (262) | 15.4 (19,347) | |
| 40–75 | 76.2 (1,700) | 73.6 (92,689) | |
| > 75 | 12.1 (270) | 11.0 (13,923) | |
| Sex, % (n): | < 0.001 | ||
| Male | 27.9 (622) | 45.1 (56,590) | |
| Female | 72.1 (1,606) | 54.9 (68,986) | |
| Presence of diabetes, % (n) | 17.8 (399) | 20.2 (25,490) | 0.006 |
| Presence of hypertension, % (n) | 25.3 (567) | 25.6 (32,256) | 0.808 |
| Total cholesterol, median (IQR) | 206 (177–239) | 197 (168–228) | < 0.001 |
| ApoB, median (IQR) | 79 (66–93) | 92 (77–109) | < 0.001 |
| HDL-C, median (IQR) | 67 (56–78) | 52 (42–64) | < 0.001 |
| VLDL-C, median (IQR) | 30 (26–40) | 22 (17–29) | < 0.001 |
| VLDL3-C, median (IQR) | 16 (14–22) | 13 (10–16) | < 0.001 |
| RLP-C, median (IQR) | 34 (27–46) | 26 (20–34) | < 0.001 |
| IDL-C, median (IQR) | 17 (13–25) | 13 (9–19) | < 0.001 |
| Estimated LDL-C, median (IQR) | 107 (85–134) | 117 (94–144) | < 0.001 |
| LDL-Cr, median (IQR) | 73 (48–99) | 95 (73–119) | < 0.001 |
| TG, median (IQR) | 161 (144–196) | 109 (78–157) | < 0.001 |
ApoB – apolipoprotein B, estimated LDL-C – low-density lipoprotein cholesterol via Martin/Hopkins equation estimation, HDL-C – high-density lipoprotein cholesterol, HLP3 – type III familial hyperlipoproteinemias, IQR – interquartile range, LDL-Cr – real low-density cholesterol, RLP-C – remnant lipoprotein cholesterol, TG – triglyceride, VLDL-C – very-low density lipoprotein cholesterol, VLDL3-C – very-low density cholesterol subfraction 3.
Table III.
Prevalence and characteristics of the NHANES 2011–2014 study population by HLP3 status (apoB method)
| Parameter | HLP3 n = 102 (1.97%) | No HLP3 n = 4,824 (98.03%) | P-value |
|---|---|---|---|
| Age, % (n): | 0.539 | ||
| 18–39 | 31.3 (35) | 38.6 (1,806) | |
| 40–75 | 60.2 (59) | 55.1 (2,626) | |
| > 75 | 8.5 (8) | 6.3 (392) | |
| Sex, % (n): | 0.745 | ||
| Male | 46.4 (49) | 48.4 (2,353) | |
| Female | 53.6 (53) | 51.6 (2,471) | |
| Race, % (n): | 0.121 | ||
| Non-Hispanic white | 62.4 (44) | 66.2 (1,975) | |
| Non-Hispanic black | 12.9 (22) | 11.5 (1,017) | |
| Mexican American | 4.3 (4) | 8.6 (601) | |
| Others | 20.4 (32) | 13.6 (1,231) | |
| BMI category, % (n): | 0.920 | ||
| Underweight | 1.9 (3) | 1.7 (95) | |
| Normal | 29.9 (38) | 29.5 (1,452) | |
| Overweight | 29.8 (28) | 32.3 (1,508) | |
| Obese | 38.4 (33) | 36.5 (1,769) | |
| Presence of diabetes, % (n) | 17.1 (17) | 12.7 (798) | 0.284 |
| Lipid-lowering therapy, % (n) | 31.0 (16) | 33.4 (969) | 0.759 |
| Statin use, % (n): | 31.0 (16) | 31.4 (902) | 0.965 |
| Cholesterol absorption inhibitors | 0.6 (1) | 1.7 (42) | 0.312 |
| Bile acid sequestrants | 0.0 (0) | 0.5 (11) | 0.647 |
| Total cholesterol, median (IQR) | 189 (162–218) | 187 (162–214) | 0.185 |
| ApoB, median (IQR) | 69 (63–81) | 88 (72–105) | < 0.001 |
| HDL-C, median (IQR) | 56 (44–63) | 51 (43–62) | 0.039 |
| VLDL-C, median (IQR) | 28 (25–36) | 20 (16–26) | < 0.001 |
| Estimated LDL-C, median (IQR) | 103 (80–121) | 112 (89–136) | 0.015 |
| TG, median (IQR) | 171 (150–239) | 98 (68–144) | < 0.001 |
ApoB – apolipoprotein B, estimated LDL-C – low-density lipoprotein cholesterol via Martin/Hopkins equation estimation, HDL-C – high-density lipoprotein cholesterol, HLP3 – type III familial hyperlipoproteinemias, IQR – interquartile range, LDL-Cr – real low-density cholesterol, NHANES – National Health and Nutrition Examination Survey, RLP-C – remnant lipoprotein cholesterol, TG – triglyceride, VLDL-C – very-low density lipoprotein cholesterol.
Across both cohorts, the majority of type III phenotypes occurred in the 40–75 year age range. Age appeared equally distributed between HLP3 and other participants in NHANES, with an increased proportion of participants 40–75 years old noted among HLP3 as compared to other participants in VLDbL across both diagnostic methods (all p < 0.001). Female sex was more common in HLP3 as compared to other participants in VLDbL with the apoB method (p < 0.001) and UC criterion 2 (p = 0.004); the female to male ratio was larger with the apoB method as compared to each of the UC criteria. Prevalence of diabetes mellitus was lower among VLDbL participants with apoB method diagnosis of HLP3 as compared to other participants (p = 0.006), with no clear difference across groups defined by the UC criteria. There was no significant difference across VLDbL diagnostic methods in terms of prevalence of hypertension by HLP3 status, regardless of the definition. Using NHANES data, there were no notable differences by HLP3 status in terms of race, BMI category, and frequency of lipid-lowering therapy.
Lipid characteristics by HLP3 status
In VLDbL, participants classified as HLP3 via UC had higher TC, IDL-C, VLDL-C, and TG (all p < 0.001) in addition to lower HDL-C (p < 0.001) as compared to other participants across criteria (Table I). HLP3 diagnosed via UC criterion 1 had lower LDL-C (p = 0.007), criterion 2 had higher LDL-C and apoB (p < 0.001), and criterion 3 had lower LDL-C and apoB (p < 0.001) as compared to other participants. HLP3 diagnosed via the apoB method had higher TC, HDL-C, VLDL-C, IDL-C, and TG (all p < 0.001) and lower apoB and LDL-C (all p < 0.001) as compared to other participants (Table II). As compared with those without HLP3, those with HLP3 had median TG levels 83.6%, 78.0%, and 83.6% vs. 47.7% higher for the UC criteria 1–3 vs. apoB method.
In NHANES, participants classified as HLP3 via the apoB method had higher HDL-C, VLDL-C, and TG as compared to other participants, but lower apoB and LDL-C (p ≤ 0.039) (Table III). Median TG in HLP3 participants was notably 74.5% higher than other participants in NHANES.
Of individuals diagnosed by the apoB method with HLP3 in NHANES, 77.7% were diagnosed via a TG cut-point of ≥ 150 mg/dl, whereas 22.3% were diagnosed via an intermediate TG cut-point of 133–149 mg/dl. This was compared to 64.9% and 35.1%, respectively, within VLDbL. Among individuals within VLDbL who were classified as HLP3 via the apoB method, we examined those who were concordant and discordant by UC criteria 1 (Table IV). Participants who were both apoB and UC criteria HLP3 positive had higher remnant particles as well as more elevated TG/apoB and total cholesterol/apoB ratios (all p < 0.001) than those who were apoB method positive and UC criteria 1 negative.
Table IV.
Lipid characteristics among the very large database of lipids study patients with apoB method diagnosed HLP3 and concordant/discordant UC Criteria (ApoB method type III phenotype, n = 2,239)
| Lipid characteristics [mg/dl] Median (IQR) | UC Concordant (n = 181) | UC Discordant (n = 2,058) | P-value, UC Concordant vs. Discordant |
|---|---|---|---|
| Total cholesterol | 223 (192–259) | 205 (176–238) | < 0.001 |
| ApoB | 76 (64–88) | 79 (66–93) | 0.026 |
| HDL-C | 48 (41–58) | 68 (58–79) | < 0.001 |
| VLDL-C | 76 (62–104) | 29 (25–36) | < 0.001 |
| IDL-C | 45 (35–54) | 17 (12–23) | < 0.001 |
| LDL-C | 91 (73–110) | 102 (77–129) | < 0.001 |
| Estimated LDL-C | 133 (109–160) | 105 (83–132) | < 0.001 |
| TG | 202 (170–263) | 158 (143–189) | < 0.001 |
| VLDL3-C | 43 (35–55) | 16 (14–19) | < 0.001 |
| RLP-C | 88 (73–108) | 33 (26–42) | < 0.001 |
| TG/apoB | 2.9 (2.4–3.5) | 2.1 (1.7–2.7) | < 0.001 |
| TC/apoB | 2.9 (2.7–3.3) | 2.5 (2.5–2.7) | < 0.001 |
ApoB – apolipoprotein B, HDL-C – high-density lipoprotein cholesterol, HLP3 – type III familial hyperlipoproteinemias, IDL-C – intermediate density lipoprotein cholesterol, LDL-C – low-density lipoprotein cholesterol, RLP-C – remnant lipoprotein cholesterol, TG – triglyceride, VLDL-C – very-low density lipoprotein cholesterol, VLDL3-C – very-low density cholesterol subfraction 3.
Discussion
HLP3 prevalence
In these large, representative U.S. adult cohorts, we found that the HLP3 phenotype is much more prevalent than historically credited when using the apoB method. With the apoB method we found a prevalence of approximately 2% in NHANES and 1.8% in VLDbL as compared to prior smaller studies suggesting 0.1–0.4% using electrophoresis and UC [8–10]. Utilizing Vertical Auto Profile in VLDbL, UC-defined HLP3 prevalences of 0.2–0.8% appear more consistent with prior prevalence estimates.
There did not appear to be a significant association with HLP3 prevalence in either cohort in terms of race or BMI, however female sex was represented in greater proportion in HLP3 samples in VLDbL via the apoB method. Age group 40–75 years old was represented in greater proportion in HLP3 samples in VLDbL, possibly due to a combination of environmental risk factors such as obesity and diabetes being more prevalent later in life as well as reduced lifespan due to atherogenic remnants. Greater than 2/3 of the NHANES population was obese or overweight and 1/8 had diabetes mellitus; furthermore, HLP3 participants had higher TG as compared to the general study population across all diagnostic methods. It is likely that genetic, hormonal, and environmental factors such as high fat diet and current obesity and diabetes epidemics are contributing to its increased phenotypic expression in the United States [3, 4, 24].
Comparison of UC and ApoB methods
We compared three variations of the UC criteria, the first and second of which were credited by Fredrickson et al. in 1975 as being diagnostic of HLP3 and possible HLP3 [1]. The third is a variation on criteria 1 proposed by our group that additionally excludes LDL-Cr greater than the 90th percentile. Of the UC criteria, criterion 2 appears to encompass the highest proportion of participants in our study, with a relatively higher apoB and LDL-C in addition to lower VLDL-C and TG. We observed a 3–10 fold increased prevalence with the apoB method as compared to all UC criteria; as such, it seems that there is increased sensitivity with the apoB algorithm as compared to even the most liberal of the UC criteria.
Prior work from Sniderman et al. identified that the apoB method increases sensitivity compared to UC at the expense of broadening identification to potentially less severe clinical phenotypes or detecting modulation of lipoprotein production or metabolism by additional genes; they argued that marked variance in plasma cholesterol and TGs among patients with HLP3 would be better captured via the apoB method [11]. The apoB algorithm achieves increased sensitivity via more lenient percentile equivalents than UC, as an apoB of 120 mg/dl roughly corresponds to the 80th percentile and TG of 133 mg/dl to the 50th percentile in men 50–60 years old [24, 25]. A considerable additional number of participants were detected in VLDbL and NHANES above a TG cutoff of ≥ 150 mg/dl as compared to 133–149 mg/dl.
HLP3 prevalence estimates varied substantially across UC criteria and between UC and apoB methods. Among various UC critreria, relatively minor changes in VLDL-C/TG ratio cut-points appear to have a noticable effect on prevalence estimates, such as in this case where decreasing the threshold from 0.3 to0.25 in UC criteria 2 increases prevalence by four-fold; UC criteria 2 accordingly generated the mildest clinical UC phenotype given its relatively higher average apoB, LDL, LDL-Cr and lower VLDL, VLDL3, RLP, and IDL when compared to UC criteria 1 and 2.
Along these lines, lipid analysis demonstrated that apoB method diagnosed HLP3 patients tend to have milder phenotypes than those diagnosed via UC criteria, as evidenced by larger differences in remnant particle cholesterol concentrations between HLP3 and non-HLP3 as compared to those identified with the apoB method. Participants with apoB and UC criteria concordance had higher IDL-C, VLDL-C, VLDL3-C, and RLP-C as well as more elevated TG/apoB and TC/apoB ratios as compared to those only identified via the apoB method, further supporting that the UC method captures more severe HLP3 phenotypes. The presence of elevated HDL-C in HLP3 patients compared to non-HLP3 patients in the apoB method raises concern for a higher number of false positives, as HDL-C should not be proportionally elevated in HLP3 compared to other dyslipidemia conditions associated with elevated TG.
Implications of ApoB method on HLP3 detection and treatment
Statin prescription appears to be similar in NHANES HLP3 participants, NHANES non-HLP3 participants, as well as the general U.S. adult population [26], suggesting that HLP3 patients are being treated similarly to normal dyslipidemic phenotypes in the general population. High TG levels in HLP3 are known to respond to a combination of dietary intervention and aggressive medical therapy with statins, fibrates, omega-3 derivatives, and niacin [14, 27, 28]. More potent, novel agents are in development that directly target apolipoprotein CIII and angiopoietin-like 3 and 4 to preferentially decrease TG-rich lipoproteins [29–31]. Dyslipidemia phenotyping may consequently aid in patient selection for more rapid initiation of conventional lipid therapies, quicker addition of combination therapy, and selective use of emerging dyslipidemia therapies to reduce lifetime cumulative exposure to atherogenic lipoproteins.
As medicine moves toward a precision approach, tailoring individual therapies toward specific dyslipidemia phenotypes could allow for personalized therapy. UC, the current gold standard to diagnose dysbetalipoproteinemia, is not readily available nor practical in clinical practice. The apoB algorithm is quicker, less costly, and utilizes components which are standardized by the Centers for Disease Control and Prevention and World Health Organization. Implementing apoB testing on a population level may lead to increased identification of previously “subclinical” dysbetalipoproteinemia phenotypes which may confer high atherogenic risk. These methods may additionally be used as screening tools to evaluate for clinical HLP3 in affected individuals and family members, prompting referral to a lipidologist for genetic analysis and treatement.
Study limitations
Our study was subject to several limitations. In the NHANES lipid protocol, chylomicrons were not directly isolated and were presumed to be reflected in the VLDL-C fraction. We were unable to account for secondary etiologies of dyslipidemia such as medication adverse effects or systemic conditions, nor test genetics or family histories, limiting our assessment of primary versus secondary causes of dyslipidemia. VLDbL utilized VAP UC instead of traditional beta quantification UC, the original method used to characterize HLP3 [1]. Given that VAP is validated against traditional UC [32, 33], it is unlikely that its use significantly changed the results; it would be reasonable, however, to repeat analyses with both VAP and beta quantification UC to assess if results differ significantly. Finally, we restricted our study to analyze several variations on previously defined UC criteria and the original Sniderman apoB algorithm. Attempts have been made to improve upon the apoB method, such as the Boot et al. non-HDL/apoB substitution for TC/apoB criterion [34]. Further work should be done to define the optimal diagnostic algorithm for HLP3 identification.
In conclusion, HLP3 may be more prevalent than historically and clinically appreciated. Although the Sniderman apoB algorithm appears to increase rate of diagnosis via its higher sensitivity, it does so at the expense of capturing cases with less severe lipid phenotypes. Since the apoB method can be automated and scaled, its use could open the door towards a precision medicine approach to dyslipidemia management that takes HLP3 into account. Further work should evaluate the clinical implications of HLP3 classification at various lipid algorithm cut-points to evaluate the ideal diagnostic standard in the modern era.
Acknowledgments
The Very Large Database of Lipids has received support from the David and June Trone Family Foundation. Dr. Martin has received research support from the PJ Schafer Cardiovascular Research Fund, American Heart Association, Aetna Foundation, CASCADE FH, Maryland Innovation Initiative, iHealth, Stanford MedX, Nokia, Google, and Apple.
Conflict of interest
VAP, VS, JP, RMV, MLE, EB, RQ, EG, MB, RSB, DM and ADS have no relevant disclosures. PPT: Speakers Bureau; Amarin, Akcea, Amgen, Kowa, Merck, Nova Nordisk, Regeneron, Sanofi (Consultant/Advisory Board). SRJ: Co-inventor for a method to estimate LDL cholesterol levels, patent application pending; funding from the David and June Trone Family Foundation. DS: personal fees from Amgen Inc, Akcea Therapeutics, Medicure, Sanofi, Regeneron; personal fees from the National Lipid Association outside the submitted work. SSM: Co-inventor for a method to estimate LDL cholesterol levels, patent application pending; Consultant/Advisory Board for Sanofi/Regeneron, Amgen, Quest Diagnostics, Akcea, Novo Nordisk, Esperion.
References
- 1.Fredrickson DS, Levy RI, Lees RS. Fat transport in lipoproteins: an integrated approach to mechanisms and disorders. N Engl J Med. 1967;276:215–25. doi: 10.1056/NEJM196701262760406. [DOI] [PubMed] [Google Scholar]
- 2.Mahley RW, Huang Y, Rall SC. Pathogenesis of type III hyperlipoproteinemia (dysbetalipoproteinemia). Questions, quandaries, and paradoxes. J Lipid Res. 1999;40:1933–49. [PubMed] [Google Scholar]
- 3.Schaefer JR. Unraveling hyperlipidemia type III (dysbetalipoproteinemia), slowly. Eur J Hum Genet. 2009;17:541–2. doi: 10.1038/ejhg.2008.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Beer F, Stalenhoef AFH, Hoogerbrugge N, et al. Expression of type III hyperlipoproteinemia in apolipoprotein E2 (Arg158 --> Cys) homozygotes is associated with hyperinsulinemia. Arterioscler Thromb Vasc Biol. 2002;22:294–9. doi: 10.1161/hq0202.102919. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins PN, Brinton EA, Nanjee MN. Hyperlipoproteinemia type 3: the forgotten phenotype. Curr Atheroscler Rep. 2014;16:440. doi: 10.1007/s11883-014-0440-2. [DOI] [PubMed] [Google Scholar]
- 6.Sniderman AD. Type III hyperlipoproteinemia: the forgotten, disregarded, neglected, overlooked, ignored but highly atherogenic, and highly treatable dyslipoproteinemia. Clin Chem. 2019;65:225–7. doi: 10.1373/clinchem.2018.298026. [DOI] [PubMed] [Google Scholar]
- 7.Soran H, Adam S, Mohammad JB, et al. Hypercholesterolaemia – practical information for non-specialists. Arch Med Sci. 2018;14:1–21. doi: 10.5114/aoms.2018.72238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wood PD, Stern MP, Silvers A, Reaven GM, von der Groeben J. Prevalence of plasma lipoprotein abnormalities in a free-living population of the Central Valley, California. Circulation. 1972;45:114–26. doi: 10.1161/01.cir.45.1.114. [DOI] [PubMed] [Google Scholar]
- 9.Jones GJL, Hewitt D, Godin GJ, et al. Plasma lipoprotein levels and the prevalence of hyperlipoproteinemia in a Canadian working population. Can Med Assoc J. 1980;122:37–46. [PMC free article] [PubMed] [Google Scholar]
- 10.Gibson TC, Whorton EB. The prevalence of hyperlipidemia in a natural community. J Chronic Dis. 1973;26:227–36. doi: 10.1016/0021-9681(73)90103-3. [DOI] [PubMed] [Google Scholar]
- 11.Sniderman AD, Graaf J de, Thanassoulis G, Tremblay AJ, Martin SS, Couture P. The spectrum of type III hyperlipoproteinemia. J Clin Lipidol. 2018;12:1383–9. doi: 10.1016/j.jacl.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Blom DJ, O’Neill FH, Marais AD. Screening for dysbetalipoproteinemia by plasma cholesterol and apolipoprotein B concentrations. Clin Chem. 2005;51:904–7. doi: 10.1373/clinchem.2004.047001. [DOI] [PubMed] [Google Scholar]
- 13.Sathiyakumar V, Park J, Quispe R, et al. Impact of novel low-density lipoprotein-cholesterol assessment on the utility of secondary non-high-density lipoprotein-C and apolipoprotein B targets in selected worldwide dyslipidemia guidelines. Circulation. 2018;138:244–54. doi: 10.1161/CIRCULATIONAHA.117.032463. [DOI] [PubMed] [Google Scholar]
- 14.Sniderman A, Couture P, de Graaf J. Diagnosis and treatment of apolipoprotein B dyslipoproteinemias. Nat Rev Endocrinol. 2010;6:335–46. doi: 10.1038/nrendo.2010.50. [DOI] [PubMed] [Google Scholar]
- 15.Holewijn S, Sniderman AD, den Heijer M, Swinkels DW, Stalenhoef AFH, de Graaf J. Application and validation of a diagnostic algorithm for the atherogenic apoB dyslipoproteinemias: ApoB dyslipoproteinemias in a Dutch population-based study. Eur J Clin Invest. 2011;41:423–33. doi: 10.1111/j.1365-2362.2010.02426.x. [DOI] [PubMed] [Google Scholar]
- 16.Martin SS, Blaha MJ, Toth PP, et al. Very large database of lipids: rationale and design. Clin Cardiol. 2013;36:641–8. doi: 10.1002/clc.22214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rautela GS, Liedtke RJ. Automated enzymic measurement of total cholesterol in serum. Clin Chem. 1978;24:108–14. [PubMed] [Google Scholar]
- 18.Martin SS, Blaha MJ, Elshazly MB, et al. Comparison of a novel method vs the friedewald equation for estimating low-density lipoprotein cholesterol levels from the standard lipid profile. JAMA. 2013;310:2061–8. doi: 10.1001/jama.2013.280532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Centers for Disease Control and Prevention National Health and Nutrition Examination Survey: Sample Design, 2011-2014. https://wwwn.cdc.gov/nchs/data/series/sr02_162.pdf.
- 20.Centers for Disease Control and Prevention National Health and Nutrition Examination Survey: Analytic Guidelines, 2011-2012. https://wwwn.cdc.gov/nchs/data/nhanes/2011-2012/analytic_guidelines_11_12.pdf. Accessed October 24, 2018.
- 21.Centers for Disease Control and Prevention Laboratory procedure manual. Apolipoprotein B. https://wwwn.cdc.gov/nchs/data/nhanes/2011-2012/labmethods/apob_g_met.pdf. Accessed October 24, 2018.
- 22.Centers for Disease Control and Prevention National Health and Nutrition Examination Survey: LDL & Triglycerides. https://wwwn.cdc.gov/Nchs/Nhanes/2011-2012/TRIGLY_G.htm. Accessed October 24, 2018.
- 23.Sniderman A, Tremblay A, Bergeron J, Gagné C, Couture P. Diagnosis of type III hyperlipoproteinemia from plasma total cholesterol, triglyceride, and apolipoprotein B. J Clin Lipidol. 2007;1:256–63. doi: 10.1016/j.jacl.2007.07.006. [DOI] [PubMed] [Google Scholar]
- 24.Miller M, Stone NJ, Ballantyne C, et al. Triglycerides and cardiovascular disease: a scientific statement from the American Heart Association. Circulation. 2011;123:2292–333. doi: 10.1161/CIR.0b013e3182160726. [DOI] [PubMed] [Google Scholar]
- 25.Cromwell WC, Otvos JD, Keyes MJ, et al. LDL Particle number and risk of future cardiovascular disease in the Framingham Offspring Study – implications for LDL management. J Clin Lipidol. 2007;1:583–92. doi: 10.1016/j.jacl.2007.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salami JA, Warraich H, Valero-Elizondo J, et al. National trends in statin use and expenditures in the US adult population from 2002 to 2013: insights from the Medical Expenditure Panel Survey. JAMA Cardiol. 2017;2:56–65. doi: 10.1001/jamacardio.2016.4700. [DOI] [PubMed] [Google Scholar]
- 27.Marais AD, Solomon GA, Blom DJ. Dysbetalipoproteinaemia: a mixed hyperlipidaemia of remnant lipoproteins due to mutations in apolipoprotein E. Crit Rev Clin Lab Sci. 2014;51:46–62. doi: 10.3109/10408363.2013.870526. [DOI] [PubMed] [Google Scholar]
- 28.Hoogwerf BJ, Bantle JP, Kuba K, Frantz ID, Hunninghake DB. Treatment of type III hyperlipoproteinemia with four different treatment regimens. Atherosclerosis. 1984;51:251–9. doi: 10.1016/0021-9150(84)90172-2. [DOI] [PubMed] [Google Scholar]
- 29.Gaudet D, Brisson D, Tremblay K, et al. Targeting APOC3 in the familial chylomicronemia syndrome. N Engl J Med. 2014;371:2200–6. doi: 10.1056/NEJMoa1400284. [DOI] [PubMed] [Google Scholar]
- 30.Graham MJ, Lee RG, Brandt TA, et al. Cardiovascular and metabolic effects of ANGPTL3 antisense oligonucleotides. N Engl J Med. 2017;377:222–32. doi: 10.1056/NEJMoa1701329. [DOI] [PubMed] [Google Scholar]
- 31.Farnier M. Future lipid-altering therapeutic options targeting residual cardiovascular risk. Curr Cardiol Rep. 2016;18:65. doi: 10.1007/s11886-016-0743-8. [DOI] [PubMed] [Google Scholar]
- 32.Chung BH, Wilkinson T, Geer JC, Segrest JP. Preparative and quantitative isolation of plasma lipoproteins: rapid, single discontinuous density gradient ultracentrifugation in a vertical rotor. J Lipid Res. 1980;21:284–91. [PubMed] [Google Scholar]
- 33.Kulkarni KR. Cholesterol profile measurement by vertical auto profile method. Clin Lab Med. 2006;26:787–802. doi: 10.1016/j.cll.2006.07.004. [DOI] [PubMed] [Google Scholar]
- 34.Boot CS, Middling E, Allen J, Neely RDG. Evaluation of the non-HDL cholesterol to apolipoprotein B ratio as a screening test for dysbetalipoproteinemia. Clin Chem. 2019;65:313–20. doi: 10.1373/clinchem.2018.292425. [DOI] [PubMed] [Google Scholar]