Abstract
Introduction
Liver cirrhosis (LC) is a heterogeneous liver disease, the last stage of liver fibrosis, and the major risk factor for hepatocellular carcinoma (HCC). Our study aimed to evaluate the expression of microRNAs and the endothelial vascular growth factor (VEGFA) gene in LC and HCC.
Material and methods
The sample group consisted of 46 tissue samples: 21 of LC, 15 of HCC, and 10 of non-tumoural and non-cirrhotic liver tissue (control group). MiRNAs were chosen based on a mirDIP prediction database as regulators of the VEGFA gene. Gene expression of VEGF and miRNAs was quantified by real-time quantitative polymerase chain reaction. VEGFA protein expression was evaluated by ELISA.
Results
VEGFA gene expression was significantly overexpressed in LC compared to the control group (p < 0.0001). Hsa-miR-206 (p = 0.0313) and hsa-miR-637 (p = 0.0156) were down-expressed in LC. In HCC, hsa-miR-15b (p = 0.0010), hsa-miR-125b (p = 0.0010), hsa-miR-423-3p (p = 0.0010), hsa-miR-424 (p = 0.0313), hsa-miR-494 (p < 0.0001), hsa-miR-497 (p < 0.0001), hsa-miR-612 (p = 0.0078), hsa-miR-637 (p < 0.0001), and hsa-miR-1255b (p = 0.0156) presented down-expression.
Conclusions
Overexpression of VEGFA in LC suggests impairment of angiogenesis in this tissue. The differential expression of microRNAs in LC and HCC observed in our study can lead to the evaluation of possible biomarkers for these diseases.
Keywords: liver diseases, VEGFA, microRNAs, biomarkers
Introduction
Liver cirrhosis (LC) is a heterogeneous liver disease, the last stage of liver fibrosis, and the major risk factor for hepatocellular carcinoma (HCC). The major risk factors for LC are hepatitis B and C (HBV and HCV, respectively), alcoholic liver disease, and non-alcoholic steatohepatitis (NASH). LC is the major risk factor for HCC. HCC is the sixth most common type of cancer worldwide and the second leading cause of death, with high malignancy. LC is defined by regeneration nodules involved in fibrosis and the healing process, which lead to substitution of hepatic tissue and function loss due to chronic injury of the liver [1, 2]. Complications associated with this disease are ascites, oesophageal varices, and encephalopathy. Prognosis is related to disease staging as assessed by standard scales such as the model for end-stage liver (MELD) and Child-Pugh. Treatment consists of alcohol abstinence, a specific diet, and liver transplantation [3, 4].
HCC is more frequent in developing countries, where 83% of cases occurred in Asia in 2012 [5]. LC, alcohol abuse, aflatoxins exposure, HBV and HCV infections, autoimmune hepatitis (AIH), and NASH are important risk factors for HCC development [6–8]. Prevention is based on HBV vaccination and the use of agents that reduce carcinogenesis in patients with a higher chance of developing the disease [9].
HCC tissue is highly vascularized; therefore, the angiogenic process represents an important role in the formation, proliferation, and metastasis of the tumour. The VEGFA gene is responsible for translating vascular endothelial growth factor A, which is an important mitogen of endothelial cells and acts on new vessel formation in the tumour [10]. Studies report that VEGFA expression is increased in HCC, and most types of cancer express this growth factor. Furthermore, VEGFA presents oncogenicity once it increases cell proliferation [11, 12].
In addition, post-transcriptional regulatory processes are performed by non-coding RNAs, microRNAs (miRNA, miR), which are single-stranded molecules of approximately 22 nucleotides in length. MiRNA is responsible for inhibiting protein translation by binding to target messenger RNA (mRNA), leading to degradation or only inhibition. Changes in gene expression of regulatory factors may reduce or elevate the expression of target genes and lead to tumour development as well as its growth and proliferation [13–15]. MiRNA regulation is modified by changes in gene expression, and imbalances may increase carcinogenesis in tissues. MiRNAs regulate the VEGFA gene in tissues, and alteration in this pathway is related to pathogenicity and growth of injured tissues [16]. Therefore, the aim of this study was to evaluate the expression of the VEGFA gene and 17 miRNAs predicted to be regulators of VEGFA in LC and HCC samples.
Material and methods
Samples
The sample group consisted of 46 diagnosticated tissue samples: 21 of LC, 15 of HCC, and 10 of non-tumoural and non-cirrhotic liver tissue (control group). These samples were collected in videolaparoscopic hepatectomy, biopsy, and liver transplantation procedures performed at the Surgical Centre of Hospital de Base de São José do Rio Preto (FAMERP/FUNFARME) after sample donors signed the informed consent form (previously approved by the Research Ethics Committee, CEP/FAMERP, no. 6228/2011). After collection, these samples were stored in RNAlater (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s instructions and stored at –20ºC until RNA and protein isolation.
In silico miRNA analysis
MiRNAs were chosen based on a mirDIP prediction database (ophid.utoronto.ca/mirDIP/). Seventeen miRNAs were selected (hsa-miR-1, hsa-miR-15b, hsa-miR-125b, hsa-miR-206, hsa-miR-423-3p, hsa-miR-424, hsa-miR-452, hsa-miR-494, hsa-miR-497, hsa-miR-576-5p, hsa-miR-612, hsa-miR-637, hsa-miR-874, hsa-miR-889, hsa-miR-920, hsa-miR-1255a, hsa-mir-1255b) that were predicted to be regulators of the VEGFA gene. Selected miRNAs presented the maximum score for prediction at choice time.
Total RNA and protein isolation
Total RNA and protein were isolated using the mirVana PARIS kit (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s instructions. Total RNA quantification was performed using Picodrop P200 (Picodrop Limited, Cambridgeshire, UK), and 230/280 and 260/280 ratios were measured to evaluate purity and precipitated salt contamination, respectively. Total protein of samples was quantified by the BCA Protein Assay Method (Pierce, Rockford, USA). During isolation and quantification, samples were kept in ice to avoid degradation and stored at –80ºC.
Reverse transcription polymerase chain reaction
Total RNA was reverse-transcribed for VEGFA gene expression analysis using a high capacity reverse transcription kit (Thermo Fisher Scientific, Carlsbad, CA) according to the manufacturer’s protocol. A TaqMan miRNA Reverse Transcription kit (Thermo Fisher Scientific, Carlsbad, CA) was used to evaluate miRNA expression, according to the manufacturer’s protocol.
RT-qPCR
The analysis of VEGFA gene expression was performed using the TaqMan Gene Expression Assay (Hs00900055_m1) (Thermo Fisher Scientific, Carlsbad, CA). Gene expression data were normalized using RPLPO, GAPDH, and HPRT1 as reference genes. The TaqMan MicroRNA Custom Assay (Thermo Fisher Scientific, Carlsbad, CA) was used to analyse miRNA expression. The assay contained primers and probes for the 17 miRNAs predicted to be VEGFA regulators. U6, RNU44, and RNU48 were used as reference genes for miRNA analysis. Relative quantification (RQ) was calculated in relation to non-tumoural and non-cirrhotic liver tissue (calibrator) using the 2–ΔΔCt method [17]. The reactions were performed on a StepOne Plus Real-Time PCR System (Applied Biosystems, Foster City, CA) and cycled following the manufacturer’s instructions.
VEGFA protein quantification
VEGFA protein quantification was performed by enzyme-linked immunosorbent assay (ELISA), using the ELISA Duo Set (R&D Systems, Minneapolis, USA) according to the manufacturer’s instructions. Optical density was determined using a reader set to 450 nm (Thermo Fisher Scientific, Carlsbad, CA) using SkanIt Software (Thermo Fisher Scientific, Carlsbad, CA). The reactions were duplicated.
Statistical analysis
Statistical tests were performed using GraphPad Prism 6. The d’Agostino and Pearson normality test was used to test the data for normal distribution. Grubbs’ outliers test was performed to check the normally distributed data for outliers. A t-test, Wilcoxon test, and Mann-Whitney test were performed for comparisons between the groups. P-values of < 0.05 were considered significant.
Results
Characterization of LC and HCC groups is presented in Table I. The variables evaluated included age, alcohol and tobacco consumption, HBV and HCV virus infections, and autoimmune hepatitis (AIH).
Table I.
Characteristics of LC and HCC patients
| Variables | Sample groups | |
|---|---|---|
| LC | HCC | |
| Age (median) | 59 | 58 |
| Alcohol consumption | 56.52% | 53.34% |
| Tobacco consumption | 56.52% | 40% |
| HBV | 26.08% | 20% |
| HCV | 43.47% | 33.34% |
| AIH | 4,34% | 13.34% |
| Liver cirrhosis | – | 93.34% |
AIH – autoimmune hepatitis, HBV – hepatitis B virus, HCV – hepatitis C virus.
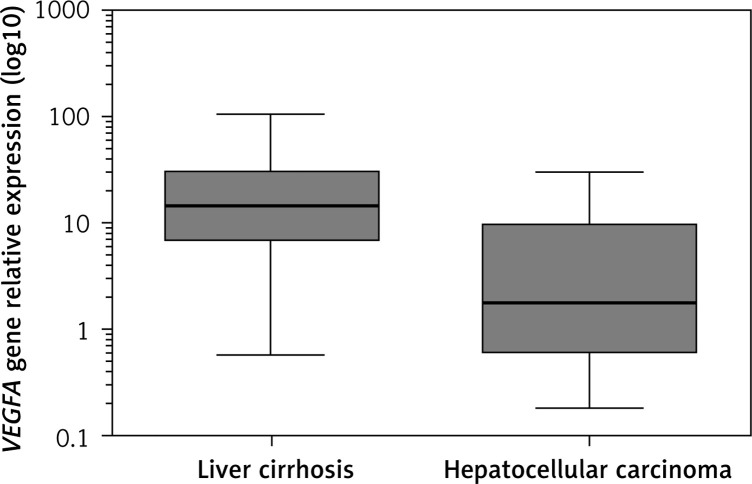
Analysis of VEGFA gene expression showed significant overexpression in LC compared to the control group, with RQ median of 14.55 (p < 0.0001). Nine samples of HCC presented high expression of VEGFA (56.25%) in relation to the control group but without statistical significance (RQ median = 1.874, p = 0.1040). VEGFA gene expression was 12.97-fold higher in LC than in HCC (p = 0.0001) (Figure 1).
Figure 1.
Representative RQ median of VEGFA gene expression in liver cirrhosis and hepatocellular carcinoma tissues
VEGFA gene expression was negatively correlated with tumour size (R = –0.52, p = 0.048). Tumours smaller than 2.1 cm (median) presented high expression of VEGFA, while tumours larger than 2.1 cm presented low expression (Table II).
Table II.
Expression of VEGFA gene according to tumour size
| Sample | Tumour size | RQ |
|---|---|---|
| T1 | 1.2 | 20.79 |
| T2 | 1.3 | 9.71 |
| T3 | 1.4 | 30.80 |
| T4 | 1.4 | 1.34 |
| T5 | 1.4 | 12.88 |
| T6 | 2.0 | 0.57 |
| T7 | 2.0 | 0.71 |
| T8 | 2.1 | 0.60 |
| T9 | 2.5 | 0.79 |
| T10 | 2.8 | 1.81 |
| T11 | 3.1 | 5.06 |
| T12 | 3.8 | 0.18 |
| T13 | 4.3 | 1.89 |
| T14 | 5.0 | 2.02 |
| T15 | 8.5 | 0.31 |
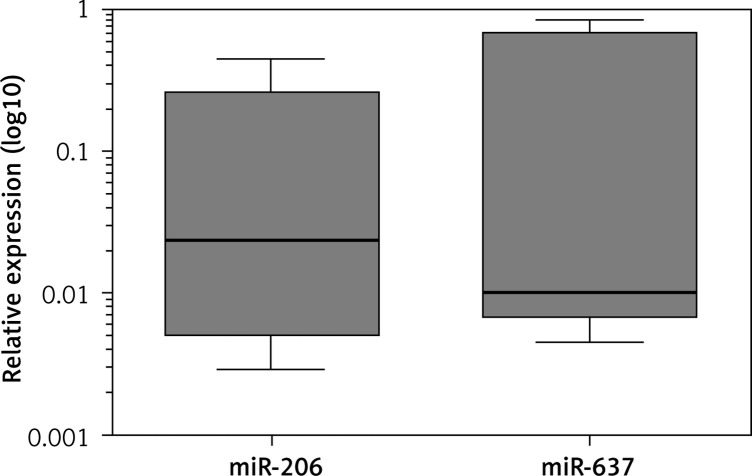
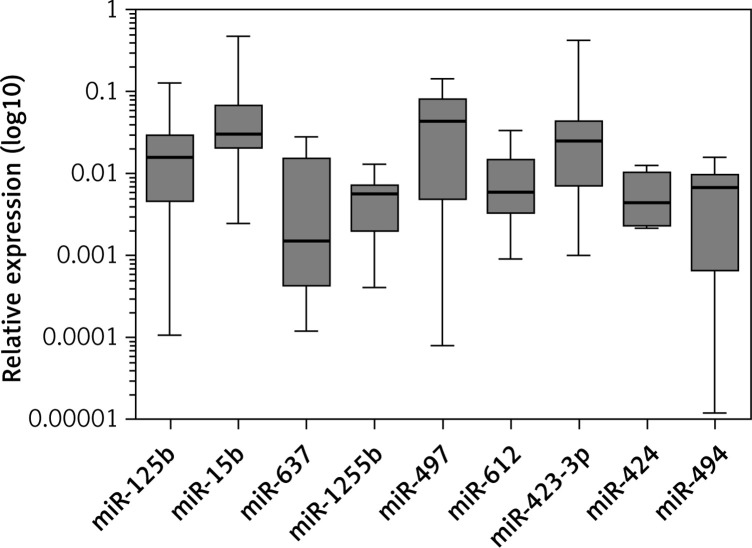
In relation to miRNAs, 15 miRNAs showed expression in both sample tissues. Hsa-miR-206 (p = 0.0313) and hsa-miR-637 (p = 0.0156) were down-expressed in LC (Figure 2). In HCC, hsa-miR-15b (p = 0.0010), hsa-miR-125b (p = 0.0010), hsa-miR-423-3p (p = 0.0010), hsa-miR-424 (p = 0.0313), hsa-miR-494 (p < 0.0001), hsa-miR-497 (p < 0.0001), hsa-miR-612 (p = 0.0078), hsa-miR-637 (p < 0.0001), and hsa-miR-1255b (p = 0.0156) presented down-expression (Figure 3, Table III).
Figure 2.
Differential expression of miRNAs in LC
Figure 3.
Differential expression of miRNAs in HCC
Table III.
miRNAs differentially expressed in LC and HCC
| miRNA | Tissue sample | Median or mean | P-value | Function related | Detected expression | Reference |
|---|---|---|---|---|---|---|
| hsa-miR-206 | LC | 0.02369 | 0.0313 | Cell proliferation suppression, apoptosis | Low | [35, 36] |
| hsa-miR-637 | LC and HCC | 0.01019 and 0.006758 | 0.0156 and < 0.0001 | LIF gene inhibition, induced STAT3 phosphorylation | Low | [33] |
| hsa-miR-15b | HCC | 0.03091 | 0.0010 | Apoptosis, proliferation, survival and angiogenesis | Low | [37, 38] |
| hsa-miR-125b | HCC | 0.01632 | 0.0010 | Cell invasion, tumour metastasis, VEGF regulation | Low | [37, 42] |
| hsa-miR-423-3p | HCC | 0.02558 | 0.0010 | Cell proliferation, cell cycle | Low | [46, 48] |
| hsa-miR-424 | HCC | 0.004519 | 0.0313 | Angiogenesis regulation and cell invasion | Low | [51, 53] |
| hsa-miR-494 | HCC | 0.006319 | < 0.0001 | Cell cycle, sorafenib resistance, PTEN regulation | Low | [59] |
| hsa-miR-497 | HCC | 0.04621 | < 0.0001 | Invasion, proliferation, cell growth and apoptosis | Low | [63–66] |
| hsa-miR-612 | HCC | 0.00605 | 0.0078 | Metastasis suppressor | Low | [69] |
| hsa-miR-1255b | HCC | 0.00568 | 0.0156 | No functional study in the literature | Low | – |
Regarding protein expression, there was no significant difference in VEGFA expression between HCC and the control group (p = 0.9999) or between LC and the control group (p = 0.1309). However, VEGFA expression was higher in LC than in HCC (77.04 μg/ml vs. 54.16 μg/ml, respectively; p = 0.0078). Therefore, LC tissues presented VEGFA protein expression 0.4-fold higher than tumour samples.
Discussion
VEGFA is the most important angiogenic growth factor in physiological and pathological states. Increased expression of VEGFA is observed in a variety of solid tumours [10, 18–20], including HCC [21, 22]. HCC samples presented high expression of VEGFA in relation to the control group in our study but without statistical significance. In LC samples VEGFA was significantly overexpressed. Ninety-three percent of HCC patients have cirrhosis, which is a pre-cancer stage and represents the main risk factor for this tumour type [23].
High expression of VEGFA in LC samples indicates the presence of angiogenic factors that contribute to new vessel formation and substitution of liver tissue for fibrous tissue [24, 25]. Campagnolo et al. [22] observed that VEGFA and EGFL7 gene expression levels were 120-fold and 25-fold higher, respectively, in LC tissue samples compared to normal tissue [22]. Von Marschall et al. [26] found VEGFA expression in 73% of LC tissue samples, and this increase may be related to the formation of fibrous tissue (necrotic foci), which secretes VEGFA. Yang et al. [27] verified that high expression of VEGFA was associated with fast tissue repair and more fibrogenic activity. El-Assal et al. [28] also observed a significantly high expression in LC tissue (RQ mean = 56.4) in relation to control (RQ mean = 42.0). All studies confirm our findings.
We observed that tumours larger than 2.1 cm presented down-expression of the VEGFA gene. Yamaguchi et al. [29] showed that tumours larger than 3 cm down-expressed VEGFA. In HCC, tumours larger than 5 cm presented fewer blood vessels than smaller tumours [28–30]. This inverse relationship may occur because small tumours require the formation of new blood vessels for growth, while large tumours may secrete other growth factors such as TGF-β, bFGF, TNF-α, and IL-8 [25].
The VEGFA gene is regulated at all stages of gene expression. Numerous studies have identified miRNAs targeting VEGFA [31]. We performed extensive research on miRNA databases and selected 17 miRNAs for investigation in LC and HCC samples. These miRNAs were predicted to be regulators of VEGFA gene expression according to the maximum score on prediction.
In both samples, HCC and LC, hsa-miR-637 presented reduced expression. This miRNA was found at reduced levels in HCC cell lines, and its increase resulted in cell death, whereas its suppression reduced the apoptosis rate to 8%. These results suggest that hsa-miR-637 plays a protective role in HCC and cirrhosis. It was verified that hsa-miR-637 is responsible for the inhibition of the LIF (leukaemia inhibitory factor) gene, which translates a cytokine responsible for inducing Stat3 phosphorylation. This transcription factor regulates several growth factors, including the VEGFA gene [32]. Reduced expression of this miRNA in glioma cells was associated with a poor prognosis by promoting growth, migration, and invasion of these cells [33]. Reduced expression of hsa-miR-637 may be related to tumour initiation in LC patients. It is known that LC is the major risk factor for HCC and a pre-cancer stage.
Hsa-miR-206 was down-regulated in LC in this study. In HCC samples, this miRNA did not show differential expression. This miRNA was found differentially expressed in LC and HCC in a previous study but was more highly expressed in HCC than in LC [34]. On the other hand, it was observed at low levels in HCC and was related to low tumour differentiation [35]. The increase of hsa-miR-206 levels in HCC cell lines resulted in suppression of cell proliferation, invasion, and migration and promoted apoptosis [35]. Overexpression of miR-206 was associated with promotion of apoptosis, induction of the cell cycle, and inhibition of migration in HepG2 cells [36]. Therefore, the role of this miRNA is not completely elucidated in cirrhosis and HCC.
In addition to hsa-miR-637, HCC samples also presented down-expression of hsa-miR-15b, hsa-miR-125b, hsa-miR-423-3p, hsa-miR-424, hsa-miR-494, hsa-miR-497, hsa-miR-612, and hsa-miR-1255b. In acute liver failure, the reduction of hsa-miR-15b levels led to a decrease of apoptosis and proliferation and survival of tumour cells [37]. In HCC, this miRNA was down-expressed and was related to tumour proliferation [38]. Furthermore, the high expression of this miRNA reduced HCC recurrence after resection, showing that it could be an important prognostic marker for this tumour type [38]. It was observed that hsa-miR-15b regulated the VEGFA gene in nasopharyngeal carcinoma in hypoxic conditions [39, 40]. In glioma, miR-15b was responsible for reduction of angiogenesis, targeting other genes (NRP-2 and MMP-3) [41].
As in our study, hsa-miR-125b was down-expressed in HCC in other studies [37, 42]. This miRNA was responsible for repression of cell invasion and tumour metastasis by preventing the formation of blood vessels [43]. Therefore, the down-expression observed in our study corroborates the hypothesis that hsa-miR-125b could play a role in the protection of hepatic tissue. The relation of the VEGFA gene and this miRNA was found in recovery of denatured dermis, where this miRNA presented reduced levels after the third day of burning, increasing VEGFA gene expression for new vessel formation for reconstitution of injured tissue [44]. In rats, low levels of this miRNA were observed in biliary ducts and were associated with the HDC gene (L-histamine decarboxylase), which is a down-regulator of VEGFA, influencing its expression in biliary synthesis [45].
High expression of hsa-miR-423-3p increased cell proliferation in breast cancer cell lines [46], laryngeal carcinoma [47], and HCC [48]. In HCC cells, miR-423 promoted cell growth and cell cycle progression at the G1/S transition [48]. This miRNA was considered a new oncogenic miRNA in HCC because of its role in suppression of the tumour suppressor p21Cip1/ Waf1 [48]. In colorectal cancer, this miRNA was found at high expression levels and promoted cell cycle progression [49]. The overexpression of miR-423-3p in several cancer types suggests a role in tumour development and progression. We observed down-expression of miR-423-3p in HCC. Our results suggest that this miRNA did not play an essential role in tumour promotion in our study and should be thoroughly investigated in HCC.
Hsa-miR-424 was related to the regulation of angiogenesis in endothelial cells, targeting the VEGFR2 gene [50]. Corroborating our results, this miRNA was observed in down-expression in HCC [51, 52], and these levels of hsa-miR-424 had no influence on the suppression of cell migration and invasion, contributing to tumour development [51]. Another study demonstrated that the survival rate of HCC patients is lower in those with low serum levels of this miRNA [53]. Hsa-miR-424 was associated with the lysosome pathway and the D-glutamine and D-glutamate metabolism pathway. The lysosome pathway is linked to cell metabolism, cell growth, dead cell clearance, and ubiquitination [52]. Impaired lysosomal maturation can contribute to the carcinogenesis of HBV-related HCC [36]. The down-expression of this miRNA is responsible for acquisition and subsequent maintenance of the tumour stem cell phenotype via the target PBX3 gene, enabling HCC cells to reprogramme into tumour stem cells [54]. Furthermore, overexpression of miR-424 represses the cell cycle/E2F pathway by targeting Akt3 and E2F3, resulting in inhibition of HCC growth and an increase of the survival rate after liver resection [55]. In addition, down-regulation of miR-424 results in increased expression of c-Myb, a pro-invasive molecule that contributes to malignant transformation, cell growth, angiogenesis, and resistance to apoptosis [56–58].
Hsa-miR-494 regulates the cell cycle in HCC, and its suppression reduced cell transformation. It was also observed that in high expression, miR-494 contributed to cell proliferation and increased resistance to sorafenib. A study revealed that the suppression of the PTEN gene by this miRNA increased proliferation, invasion, and migration of HCC cells [59]. Up regulation of miRNA-494 has been identified as a potential non-invasive biomarker for HCC until now [60–62], although we found down-regulation of this miRNA in our HCC samples. Therefore, further studies are needed to clarify the role of miRNA-494 in HCC.
In HCC and breast cancer, hsa-miR-497 showed low expression and was related to apoptosis and the suppression of invasion, proliferation, and cell growth [63]. This miRNA was suggested to be an angiogenesis inhibitor in HCC, directly targeting the VEGFA gene, as well as in ovarian cancer [63–66]. Moreover, miR-497 led to significant up-regulation of CHEK1, a Ser/Thr kinase that mediates cell-cycle arrest after DNA damage. CHEK1 overexpression resulting from the down-regulation of miR-497 in HCC is responsible for tumour development and poor prognosis [67]. Our result regarding this miRNA corroborates this hypothesis.
In our study, hsa-miR-612 was down-expressed in HCC. This result corroborates a previous study that observed inhibitory effects of this miRNA on cell proliferation, migration, invasion, and metastasis of HCC [68]. MiR-612 was able to suppress the stemness of HCC via Wnt/b-catenin signalling, reduce the number and size of tumourspheres, and prevent the capacity of tumourigenesis in NOD/SCID mice [68]. Furthermore, miR-612 was observed to have a role in suppression of HCC stemness via the Sp1/Nanog signalling pathway [69]. MiR-612 inhibited the protein level of Sp1 and reduced the transcription activity of Nanog in a recent study. Nanog is a transcription factor in embryonic stem cells (ESCs) and is thought to play a critical role in maintenance of pluripotency [70].
Regarding hsa-miR-1255b, there are no data in the literature about functional studies with this miRNA. The expression levels of miR-1255b have not been examined so far in cancer cells. The down-expression of this miRNA in HCC samples in our study suggests a possible role in liver tumourigenesis and should be investigated in further studies as a novel biomarker of HCC.
In addition to VEGFA and miRNAs, other biomarkers have been used for the identification of cellular and genetic alterations and neoplasms as well as the prognosis and diagnosis of diseases, such as α-fetoprotein (a serologic marker) [71], cyclooxygenase-2 (COX-2, an important enzyme in carcinogenesis in HCC) [72], B-catenin and E- and N-cadherins (involved in cellular alterations and indicators of epithelial-mesenchymal transition) [73], vitamin D deficiency (mainly in women with cardiovascular risk) [74], and presenilin 2 (PSEN2, a possible tumour suppressor in renal cancer) [75].
In conclusion, our results showed overexpression of the VEGFA gene in LC and down-expression of miRNAs in LC and HCC. These miRNAs are involved in important pathways for cancer development. It is important to emphasize that these miRNAs could be potential biomarkers for LC and HCC, which can direct the treatment of these diseases. The differential expression of mR-1255b was identified for the first time in HCC and suggests a novel biomarker for this tumour type. The discovery of biomarkers is relevant for diagnosis and prognosis of these diseases. Functional studies involving miRNAs and target genes can help in the development of new adjuvant therapies in LC and HCC treatment.
Acknowledgments
We greatly appreciate the support of the following: The São Paulo Research Foundation-FAPESP (grant number 2012/10321-6), National Council for Scientific and Technological Development-CNPq (process number 310582/2014-8), Higher Education Improvement Coordination-CAPES (Financial code 001), and the Faculty of Medicine os São José do Rio Preto (FAMERP) and Regional Faculty of Medicine of São José do Rio Preto Foundation (FUNFARME) for their institutional support, UPGEM-Genetics and Molecular Biology Research Unit and GETF-Study Group of Liver Tumors. Thanks also to Dr. Alexandre Lins Werneck for assistance with the English revision.
Both Eny Maria Goloni-Bertollo PhD and Renato Ferreira da Silva PhD coordinated and carried out the project.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Schuppan D, Afdhal NH. Liver cirrhosis. Lancet. 2008;371:838–51. doi: 10.1016/S0140-6736(08)60383-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Raphe R, Duca WJ, Arroyo-Jr PC, Silva RC, Silva RF. Hepatocellular carcinoma: risk factors, diagnosis, staging and treatment in a Referral Centre. J Cancer Ther. 2013;4:384–93. [Google Scholar]
- 3.Befeler AS, Palmer DE, Hoffman M, Longo W, Solomon H, Di Bisceglie AM. The safety of intra-abdominal surgery in patients with cirrhosis. Model for end-stage liver disease score is superior to child-turcotte-pugh classification in predicting outcome. Arch Surg. 2005;140:650–4. doi: 10.1001/archsurg.140.7.650. [DOI] [PubMed] [Google Scholar]
- 4.Forrest E. Alcohol and the liver. Medicine. 2015;43:581–4. [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136:E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142:1264–73. doi: 10.1053/j.gastro.2011.12.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kar P. Risk Factors for hepatocellular carcinoma in india. J Clin Exp Hepatol. 2014;4:S34–42. doi: 10.1016/j.jceh.2014.02.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bodzin AS, Butsuttil RW. Hepatocellular carcinoma: advances in diagnosis, management, and long-term outcome. World J Hepatol. 2015;7:1157–67. doi: 10.4254/wjh.v7.i9.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buitrago-Molina LE, Vogel A. mTor as a potential target for the prevention and treatment of hepatocellular carcinoma. Curr Cancer Drug Targets. 2012;12:1045–61. doi: 10.2174/156800912803988011. [DOI] [PubMed] [Google Scholar]
- 10.Biselli-Chicote PM, Oliveira AR, Pavarino EC, Goloni-Bertollo EM. VEGF gene alternative splicing: pro- and anti-angiogenic isoforms in cancer. J Cancer Res Clin Oncol. 2012;138:363–70. doi: 10.1007/s00432-011-1073-2. [DOI] [PubMed] [Google Scholar]
- 11.Bupathi M, Kaseb A, Meric-Bernstam F, Naing A. Hepatocellular carcinoma: where there is unmet need. Molecular Oncology. 2015;9:1501–9. doi: 10.1016/j.molonc.2015.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Llovet JM. Focal gains of VEGFA: candidate predictors of sorafenib response in hepatocellular carcinoma. Cancer Cell. 2014;25:560–2. doi: 10.1016/j.ccr.2014.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chekulaeva M, Filipowicz W. Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr Opin Cell Biol. 2009;21:452–60. doi: 10.1016/j.ceb.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 14.Li M, Fu W, Wo L, Shu X, Liu F, Li C. miR-128 targets genes in tumorigenesis and metastasis. Exp Cell Res. 2013;319:3059–64. doi: 10.1016/j.yexcr.2013.07.031. [DOI] [PubMed] [Google Scholar]
- 15.Lu Y, Zhang L, Waye MMY, Waye MM, Fu WM, Zhang JF. miR-218 mediates tumorigenesis and metastasis: perspectives and implications. Exp Cell Res. 2015;224:173–82. doi: 10.1016/j.yexcr.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 16.Wang W, Zhang E, Lin C. MicroRNAs in tumor angiogenesis. Life Sci. 2015;136:28–35. doi: 10.1016/j.lfs.2015.06.025. [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real time PCR quantitative and the 2-ΔΔCt method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 18.Schneider BP, Miller KD. Angiogenesis of breast cancer. J Clin Oncol. 2005;23:1782–90. doi: 10.1200/JCO.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 19.Ferroni P, Spila A, Martini F, et al. Prognostic value of vascular endothelial growth factor tumor tissue content of colorectal cancer. Oncology. 2005;69:145–53. doi: 10.1159/000087838. [DOI] [PubMed] [Google Scholar]
- 20.Jaiswal SG, Gadbail AR, Chaudhary MS, Jaiswal GR, Gawande M. Correlation of serum levels of vascular endothelial growth factor with TNM staging, histopathologic grading, and surgical therapy for oral squamous cell carcinoma. Quintessence Int. 2011;42:771–9. [PubMed] [Google Scholar]
- 21.Moon WS, Rhyu KH, Kang MJ, et al. Overexpression of VEGF and angiopoietin 2: a key to high vascularity of hepatocellular carcinoma? Mod Pathol. 2003;16:552–7. doi: 10.1097/01.MP.0000071841.17900.69. [DOI] [PubMed] [Google Scholar]
- 22.Campagnolo L, Telesca C, Massimiani M, et al. Different expression of VEGF and EGFL7 in human hepatocellular carcinoma. Dig Liver Dis. 2016;48:76–80. doi: 10.1016/j.dld.2015.09.019. [DOI] [PubMed] [Google Scholar]
- 23.El-Serag HB. Hepatocellular carcinoma. N Engl J Med. 2011;365:1118–27. doi: 10.1056/NEJMra1001683. [DOI] [PubMed] [Google Scholar]
- 24.Corpechot C, Barbu V, Wendum D, et al. Hypoxia-induced VEGF and collagen I expressions are associated with angiogenesis and fibrogenesis in experimental cirrhosis. Hepatol. 2002;35:1010–21. doi: 10.1053/jhep.2002.32524. [DOI] [PubMed] [Google Scholar]
- 25.Giatromanolaki A, Kotsiou S, Koukourakis MI, Sivridis E. Angiogenic factor expression in hepatic cirrhosis. Mediator Inflamm. 2007;2007:67187. doi: 10.1155/2007/67187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Von Marschall Z, Cramer T, Höcker M, Finkenzeller G, Wiedenmann B, Rosewicz S. Dual mechanism of vascular endothelial growth factor upregulation by hypoxia in human hepatocellular carcinoma. Gut. 2001;48:87–96. doi: 10.1136/gut.48.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang L, Kwon J, Popov Y, et al. Vascular endothelial growth factor promotes fibrosis resolution and repair in mice. Gastroenterology. 2014;146:1339–50. doi: 10.1053/j.gastro.2014.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.El-Assal ON, Yamanoi A, Soda Y, et al. Clinical significance of microvessel density and vascular endothelial growth factor expression in hepatocellular carcinoma and surrounding liver: possible involvement of vascular endothelial growth factor in the angiogenesis of cirrhotic liver. Hepatology. 1998;27:1554–62. doi: 10.1002/hep.510270613. [DOI] [PubMed] [Google Scholar]
- 29.Yamaguchi R, Yano H, Iemura A, Ogasawara S, Haramaki M, Kojiro M. Expression of vascular endotelial growth fator in human hepatocelular carcinoma. Hepatology. 1998;28:68–77. doi: 10.1002/hep.510280111. [DOI] [PubMed] [Google Scholar]
- 30.Jeng KS, Sheen IS, Wang YC, et al. Prognostic significance of preoperative circulating vascular endothelial growth factor messenger RNA expression in resectable hepatocellular carcinoma: a prospective study. World J Gastroenterol. 2004;10:643–8. doi: 10.3748/wjg.v10.i5.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arcondéguy T, Lacazette E, Millevoi S, Prats H, Touriol C. A mRNA processing, stability and translation: a paradigm for intricate regulation of gene expression at the post-transcriptional level. Nucleic Acids Res. 2013;41:7997–8010. doi: 10.1093/nar/gkt539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santoni M, Conti A, Piva F, et al. Role of STAT3 pathway in genitourinary tumors. Future Sci OA. 2015;1:FSO15. doi: 10.4155/fso.15.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Que T, Song Y, Liu Z, et al. Decreased miRNA-637 is an unfavorable prognosis marker and promotes glioma cell growth, migration and invasion via direct targeting Akt1. Oncogene. 2015;34:4952–63. doi: 10.1038/onc.2014.419. [DOI] [PubMed] [Google Scholar]
- 34.Tan Y, Lin B, Ye Y, Wen D, Chen L, Zhou X. Differential expression of serum microRNAs in cirrhosis that evolve into hepatocellular carcinoma related to hepatitis B virus. Oncol Rep. 2015;33:2863–70. doi: 10.3892/or.2015.3924. [DOI] [PubMed] [Google Scholar]
- 35.Yungiao L, Vanke H, Jun X, Tangmeng G. MicroRNA-206, down-regulated in hepatocellular carcinoma, suppresses cell proliferation and promotes apoptosis. Hepatogastroenterology. 2014;61:1302–7. [PubMed] [Google Scholar]
- 36.Liu W, Xu C, Wan H, et al. MicroRNA-206 overexpression promotes apoptosis, induces cell cycle arrest and inhibits the migration of human hepatocellular carcinoma HepG2 cells. Int J Mol Med. 2014;34:420–8. doi: 10.3892/ijmm.2014.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.An F, Gong B, Wang H, et al. miR-15b and miR-16 regulate TNF mediated hepatocyte apoptosis via BCL2 in acute liver failure. Apoptosis. 2012;17:702–16. doi: 10.1007/s10495-012-0704-7. [DOI] [PubMed] [Google Scholar]
- 38.Chung GE, Yoon J, Myung SJ, et al. High expression of microRNA-15b predicts a low risk of tumor recurrence following curative resection. Oncol Rep. 2010;23:113–9. [PubMed] [Google Scholar]
- 39.Hua Z, Lv Q, Ye W, et al. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Madanecki P, Kapoor N, Bebok Z, Ochocka R, Collawn JF, Bartoszewski R. Regulation of angiogenesis by hypoxia: the role of microRNA. Cell Mol Biol Lett. 2013;18:47–57. doi: 10.2478/s11658-012-0037-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng X, Chopp M, Lu Y, Buller B, Jiang F. MiR-15b and miR-152 reduce glioma cell invasion and angiogenesis via NRP-2 and MMP-3. Cancer Letters. 2013;329:146–54. doi: 10.1016/j.canlet.2012.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C, Yen C, Chou R, et al. Downregulation of microRNA-15b by hepatitis B virus X enhances hepatocellular carcinoma proliferation via fucosyltransferase 2-induced Globo H expression. Int J Cancer. 2014;134:1638–47. doi: 10.1002/ijc.28501. [DOI] [PubMed] [Google Scholar]
- 43.Zhou HC, Fang JH, Shang LR, et al. MicroRNAs miR-125b and miR-100 suppress metastasis of hepatocellular carcinoma by disrupting the formation of vessels that encapsulate tumour clusters. J Pathol. 2016;240:450–60. doi: 10.1002/path.4804. [DOI] [PubMed] [Google Scholar]
- 44.Zhou S, Zhang P, Liang P, Huang X. The expression of miR-125b regulates angiogenesis during the recovery of heat-denatured HUVECs. Burns. 2015;41:803–11. doi: 10.1016/j.burns.2014.10.012. [DOI] [PubMed] [Google Scholar]
- 45.Meng F, Onori P, Hargrove L, et al. Regulation of the histamine/VEGF axis by miR-125b during cholestatic liver injury in mice. Am J Pathol. 2014;184:662–73. doi: 10.1016/j.ajpath.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 46.Zhao H, Gao A, Zhang Z, et al. Genetic analysis and preliminary function study of miR-423 in breast cancer. Tumor Biol. 2015;36:4763–71. doi: 10.1007/s13277-015-3126-7. [DOI] [PubMed] [Google Scholar]
- 47.Guan G, Zhang D, Zheng Y, et al. microRNA-423-3p promotes tumor progression via modulation of AdipoR2 in laryngeal carcinoma. Int J Clin Exp Pathol. 2014;7:5683–91. [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, Huang S, Wu S, et al. MicroRNA-423 promotes cell growth and regulates G1/S transition by targeting p21Cip1/Waf1 in hepatocellular carcinoma. Carcinogenesis. 2011;32:1641–7. doi: 10.1093/carcin/bgr199. [DOI] [PubMed] [Google Scholar]
- 49.Li H, Zhang H, Chen Y, et al. MiR-423-3p enhances cell growth through inhibition of p21cip1/waf1 in colorectal cancer. Cell Physiol Biochem. 2015;37:1044–54. doi: 10.1159/000430230. [DOI] [PubMed] [Google Scholar]
- 50.Chamorro-Jorganes A, Araldi E, Penalva LOF, et al. MicroRNA-16 and microRNA-424 regulate cell-autonomous angiogenic functions in endothelial cells via targeting vascular endothelial growth factor receptor-2 and fibroblast growth factor receptor-1. Arterioscler Thromb Vasc Biol. 2011;31:2595–606. doi: 10.1161/ATVBAHA.111.236521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu L, Ding GF, He C, Sun L, Jiang Y, Zhu L. MicroRNA-424 is down-regulated in hepatocellular carcinoma and suppresses cell migration and invasion through c-Myb. PLoS One. 2014;9:e91661. doi: 10.1371/journal.pone.0091661. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 52.Lu M, Kong X, Wang H, Huang G, Ye C, He Z. A novel microRNAs expression signature for hepatocellular carcinoma diagnosis and prognosis. Oncotarget. 2017;8:8775–84. doi: 10.18632/oncotarget.14452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yao H, Liu X, Chen S, Xia W, Chen X. Decreased expression of serum miR-424 correlates with poor prognosis of patients with hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8:14830–5. [PMC free article] [PubMed] [Google Scholar]
- 54.Han H, Du Y, Zhao W, et al. PBX3 is targeted by multiple miRNAs and is essential for liver tumour-initiating cells. Nat Commun. 2015;6:8271. doi: 10.1038/ncomms9271. [DOI] [PubMed] [Google Scholar]
- 55.Yang H, Zheng W, Shuai X, et al. MicroRNA-424 inhibits Akt3/E2F3 axis and tumor growth in hepatocellular carcinoma. Oncotarget. 2015;6:27736–50. doi: 10.18632/oncotarget.4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ramsay RG, Gonda TJ. MYB function in normal and cancer cells. Nat Rev Cancer. 2008;8:523–34. doi: 10.1038/nrc2439. [DOI] [PubMed] [Google Scholar]
- 57.Gonda TJ, Leo P, Ramsay RG. Estrogen and MYB in breast cancer: potential for new therapies. Expert Opin Biol Ther. 2008;8:713–7. doi: 10.1517/14712598.8.6.713. [DOI] [PubMed] [Google Scholar]
- 58.Pattabiraman DR, Gonda TJ. Role and potential for therapeutic targeting of MYB in leukemia. Leukemia. 2013;27:269–77. doi: 10.1038/leu.2012.225. [DOI] [PubMed] [Google Scholar]
- 59.Liu K, Liu S, Zhang W, et al. miR-494 promotes cell proliferation, migration and invasion, and increased sorafenib resistance in hepatocellular carcinoma by targeting PTEN. Oncol Rep. 2015;34:1003–10. doi: 10.3892/or.2015.4030. [DOI] [PubMed] [Google Scholar]
- 60.Lim L, Balakrishnan A, Huskey N, et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis throuth suppression of mutated in colorectal cancer. Hepatology. 2014;59:202–15. doi: 10.1002/hep.26662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fornari F, Ferracin M, Trerè D, et al. Circulating microRNAs, miR-939, miR-595, miR-519d and miR-494, identify cirrhotic patients with HCC. PLoS One. 2015;10:e0141448. doi: 10.1371/journal.pone.0141448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chuang KH, Whitney-Miller CL, Chu CY, et al. MicroRNA-494 is a master epigenetic regulator of multiple invasion-suppressor microRNAs by targeting ten eleven translocation 1 in invasive human hepatocellular carcinoma tumors. Hepatology. 2015;62:466–80. doi: 10.1002/hep.27816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen L, Li J, Xu L, et al. MiR-497 induces apoptosis of breast cancer cells by targeting Bcl-w. Exp Ther Med. 2012;3:475–80. doi: 10.3892/etm.2011.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li D, Zhao Y, Liu C, et al. Analysis of miR-195 and miR-497 expression, regulation and role in breast cancer. Clin Cancer Res. 2011;17:1722–30. doi: 10.1158/1078-0432.CCR-10-1800. [DOI] [PubMed] [Google Scholar]
- 65.Furuta M, Kozaki K, Tanimoto K, et al. The tumor-suppressive miR-497-195 cluster targets multiple cell-cycle regulators in hepatocellular carcinoma. PLoS One. 2013;8:e60155. doi: 10.1371/journal.pone.0060155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang W, Ren F, Wu Q, Jiang D, Li H, Shi H. Micro-RNA-497 suppresses angiogenesis by targeting vascular endothelial growth factor A through the PI3K/AKT and MAPK/ERK pathways in ovarian cancer. Oncology Reports. 2014;32:2127–33. doi: 10.3892/or.2014.3439. [DOI] [PubMed] [Google Scholar]
- 67.Xie Y, Wei RR, Huang GL, Zhang MY, Yuan YF, Wang HY. Checkpoint kinase 1 is negatively regulated by miR-497 in hepatocellular carcinoma. Med Oncol. 2014;31:844. doi: 10.1007/s12032-014-0844-4. [DOI] [PubMed] [Google Scholar]
- 68.Tao Z, Wan J, Zeng L, et al. MiR-612 suppresses the invasive-metastatic cascade in hepatocellular carcinoma. J Exp Med. 2013;210:789–803. doi: 10.1084/jem.20120153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu Y, Liu DL, Dong LL, et al. miR-612 suppresses stem cell-like property of hepatocellular carcinoma cells by modulating Sp1/Nanog signaling. Cell Death Dis. 2016;7:e2377. doi: 10.1038/cddis.2016.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pan G, Thomson JA. Nanog and transcriptional networks in embryonic stem cell pluripotency. Cell Res. 2007;17:42–9. doi: 10.1038/sj.cr.7310125. [DOI] [PubMed] [Google Scholar]
- 71.Zhao YJ, Ju Q, Li GC. Tumor markers for hepatocellular carcinoma. Mol Clin Oncol. 2013;1:593–8. doi: 10.3892/mco.2013.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chen G, Li X, Yang J, et al. Prognostic significance of cyclooxygenase-2 expression in patients with hepatocellular carcinoma: a meta-analysis. Arch Med Sci. 2016;12:1110–7. doi: 10.5114/aoms.2016.61916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kasprzak A, Rogacki K, Adamek A, et al. Tissue expression of beta-catenin and E- and N-cadherins in chronic hepatitis C and hepatocellular carcinoma. Arch Med Sci. 2017;13:1269–80. doi: 10.5114/aoms.2017.65272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Faridi K, Lupton J, Martin S, et al. Vitamin D deficiency and non-lipid biomarkers of cardiovascular risk. Arch Med Sci. 2017;13:732–7. doi: 10.5114/aoms.2017.68237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jędroszka D, Orzechowska M, Bednarek A. Predictive values of Notch signalling in renal carcinoma. Arch Med Sci. 2017;13:1249–54. doi: 10.5114/aoms.2017.65649. [DOI] [PMC free article] [PubMed] [Google Scholar]