Abstract
Tumor stem cells or cancer initiating cells (CICs) are single tumor cells that can regenerate a tumor or a metastasis. The identification and isolation of CICs remain challenging, and a variety of putative CIC markers have been described. We hypothesized that cell lines of the NCI60 panel contain CICs and express putative CIC markers. We investigated expression of putative CIC surface markers (CD15, CD24, CD44, CD133, CD166, CD326, PgP) and the activity of aldehyde dehydrogenase in the NCI60 panel singly and in combination by six-color fluorescence-activated cell sorting analysis. All investigated markers were expressed in cell lines of the NCI60 panel. Expression levels of individual markers varied widely across the 60 cell lines, and neither single marker expression nor simple combinations nor co-expression patterns correlated with the colony-formation capacity of cell lines. Rather, marker expression patterns correlated with tumor types in multidimensional analysis. Whereas some expression patterns correlated with tumor entities such as basal breast cancer, other expression patterns occurred across different tumor types and largely related to expression of a more mesenchymal phenotype in individual breast, lung, renal, and melanoma cell lines. Our data for the first time demonstrate that tumor cell lines display CIC markers in a complex pattern that relates to the tumor type. The complexity and tumor type specificity of marker display creates challenges for the application of cell sorting and other approaches to isolation of putative tumor stem cell populations and suggests that therapeutic targeting strategies will need to take this into account.
Keywords: Cancer stem cell, Cell surface marker, Fluorescence-activated cell sorting, Cell culture, NCI60 panel
Introduction
Studies of selected human tumor cell lines and fresh surgical specimens suggest that expression of certain cell surface and functional markers define a subset of cancer cells that is responsible for tumor recurrence following chemotherapy and growth of metastases (tumor stem cell hypothesis) [1]. Identification of cancer stem cells or cancer initiating cells (CICs) has been reported for an array of tumor entities including some of the most common cancers, for example, colon, breast, prostate, and melanoma [2–5]. Isolated by surface antigen expression, these CICs derived from patient biopsies are operationally defined by their ability to form tumors at limiting dilutions in immune-compromised mice and to recapitulate the morphologic heterogeneity of the original tumor. It is thought that CICs are relatively drug resistant by virtue of quiescence, expression of drug-resistance mechanisms such as ABC transporters, and possibly their location in tissue niches with restricted drug access. Thus, improved cancer therapy may require novel therapy targeting CICs.
CICs are currently identified by expression of cell surface or functional markers. Many of these markers, which are overlapping between tumor entities, are derived from those already established for studying normal hematopoietic or embryonic stem cells. Surface markers used for the identification of CICs include CD24 (ligand for P-selectin), CD44 (hyaluronan receptor), CD 15 (SSEA-1, Lewis X), CD133 (prominin1), and CD166 (ALCAM). CD24lo CD44+ subpopulations with CIC characteristics have been described for breast tumors [2, 6] and prostate cancer [3], whereas in pancreatic cancer and colon cancer, a subpopulation of CD24+CD44+ cells has been reported to contain CICs [7, 8]. CD133 has been used for identifying CICs in colon cancer [4], prostate cancer [9], hepatocellular carcinoma [10, 11], and other tumor types [12–14], and CD166 has been described for the identification of colorectal CICs [15]. The glycoprotein CD15 is a stage-dependent embryonic marker that is expressed on stem cells of the nervous system [16, 17]; expression of CD15 correlates with metastases and poor prognosis in colon cancer [18, 19] and progression of prostate cancer [20], making it a potential marker for CICs. Other markers that have also been evaluated for the identification of normal tissue stem cells and that may reflect CIC markers associated with solid tumors include CD326 (EpCAM) and CD29 (integrin b1). CD326 is expressed by mammary epithelial progenitor cells [21] and has been used to capture and isolate circulating breast cancer cells (http://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfPCD/classification.cfm?ID=4179). CD29 is highly expressed on mammary, colonic, and neuronal stem cells and is involved in proliferation and migration of these cells [8, 22–25].
Functional assays used for the identification of CICs include ABC transporter and aldehyde dehydrogenase activity. The expression of ABCG2, an ABC transporter with drug efflux function, is evaluated functionally by drug-exclusion studies that identify a side population (Hoechst 33,342 negative population) with CIC characteristics [26]. Similarly, P-glycoprotein (MDR1), another ABC transporter, may be expressed, and its presence may explain the observed drug resistance in certain ovarian cancer, breast cancer, and melanoma patients [27, 28]. Aldehyde dehydrogenase, an intracellular enzyme involved in retinoid metabolism, is expressed in cells with stem cell characteristics [29] and has been implicated as a marker for cancer initiating cells in multiple myeloma [30], breast cancer [31], colon cancer [32], and lung cancer [33]. Finally, the clonogenic capacity of cells can be assessed by colony-formation assays in vitro and tumor formation in vivo.
The NCI60 tumor cell line panel includes cell lines derived from hematopoietic malignancies and solid tumors including small- and non-small-cell lung cancer, central nervous system (CNS), colon, breast, ovarian, and prostate cancer, and melanoma. Established from tumor cell lines available in the late 1980s as a drug-screening tool, the cell lines of the NCI60 panel have been used for large-scale drug screening, creating an unprecedented public database of drug sensitivity information, and have been extensively characterized for molecular characteristics including basal patterns of gene expression [34, 35].
We hypothesized that tumor cell lines of the NCI60 panel harbor tumor stem cell populations that can be isolated and characterized to enable research specifically directed towards discovering new agents for targeting tumor stem cells. Therefore, as a first step, we evaluated expression of putative tumor stem cell markers in the NCI60, first as individual markers and subsequently for patterns of marker co-expression.
Materials and Methods
NCI60 Cell Culture
Adherent 2D Culture
Cells were cultured in RPMI 1,640 supplemented with 2 mM L-glutamine, 5% fetal bovine serum (FBS), and 0.1% gentamicin (Invitrogen, Carlsbad, CA, http://www.invitrogen.com) at 37C, 5% CO2. For experiments, cells (passage 3 to passage 6 from the cryopreserved cell bank) were grown to 90% confluence and brought into single-cell suspension using Cell-Dissociating Buffer (Invitrogen).
Anchorage Independent Colonosphere Culture
Cells were cultured serum-free in RPMI1640 supplemented with 15% knock-out supplement and 2 mM glutamine (Invitrogen). Cells were passaged by dissociating them into single-cell suspension using cell-dissociation buffer and resuspending aliquots in serum-free culture medium.
Colony-Forming Units
One hundred cells were cultured in 100 mm tissue culture dishes. After 2 weeks, cell colonies were visualized with Coomassie Blue solution (0.5% Coomassie Brilliant Blue G250 in methanol/acetic acid/water 3/1/6), and visible colonies were counted. Assays were performed in triplicate.
Soft Agar Cloning Assays
The soft agar colony formation assay was performed as previously described [36], but 5% defined fetal bovine serum (Thermo Scientific, Waltham, MA, www.thermo.com) or 15% knock-out supplement (Invitrogen) were used. Briefly, triplicate 35 mm dishes were coated with a 1 ml base layer containing 0.70% agarose (Seaplaque: Lonza, Rockland, ME, http://www.lonza.com). On day 0, cells were dissociated and subcultured by layering various cell concentrations in 0.5 ml culture medium containing 0.36% agarose onto the base coat. Culture dishes were refrigerated (4C) for 15 minutes, brought to room temperature for 10 minutes, and then overlaid with 0.5 ml culture medium and incubated at 37°C, 5% CO2. Colonies were visualized using 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromid (MTT) (1 mg/ml) at the indicated time points. Cumulative counts of colonies (diameters > 60 μm) and cumulative volumes of colony-forming units (diameters > 20 μl) for 35 contiguous fields (equivalent to 51% of the cell layer culture volume) were determined with an Omnicon FAS-II (Bausch & Lomb, Rochester, NY, http://www.bausch.com) following calibration with an Omnicon test plate 3 and polystyrene microspheres embedded in soft agar matrix. Selective scoring of viable cell groups was achieved by adjustment of the instrument detection threshold to exclude images of nonstained cellular material and debris. Data were analyzed using Excel software.
ALDH and CD133 Double Labeling
ALDH activity and CD133 expression were analyzed using the Aldefluor Assay Kit (Stem Cell Technologies, Vancouver, BC, Canada, http://www.stemcell.com) and CD133/1 antibody (Miltenyi, Auburn, CA, http://www.miltenyibiotec.com) according to the manufacturer. Briefly, cells were suspended in assay buffer and incubated with the ALDH substrate Aldefluor or with Aldefluor and the ALDH inhibitor DEAB (negative control) for 30 minutes at 37C. Cells were subsequently washed and labeled with Allophycocyanin (APC)-conjugated CD133/1 for 30 minutes on ice. Cells were washed with assay buffer twice and analyzed using FACSCalibur (BD Bioscience, San Diego, http://www.bdbiosciences.com) and CellQuest software (BD Bioscience). Data were analyzed using FlowJo (Tree Star Inc., Ashland, OR, http://www.treestar.com). Experiments were performed in triplicate.
CD15, CD24, and CD44 Triple Labeling
Cells (106 cells/ml) were resuspended in FACS buffer (BD Bioscience, San Jose, CA) and incubated with fluorescein isothiocyanate (FITC)-conjugated CD15, R-phycoerythin (PE)-conjugated CD24, and Allophycocyanin (APC)-conjugated CD44 according to the manufacturer (all BD Bioscience, San Jose, CA) for 30 minutes on ice, washed with FACS buffer twice, and fixed with 5% formalin for later analysis. Data were collected using FACSCalibur (BD Bioscience) and CellQuest software (BD Bioscience) and analyzed using FlowJo (Tree Star Inc.). Experiments were performed in triplicate.
Six-Color Analysis
NCI60 Cell Culture for Six-Color Analysis
Cells were cultured in RPMI 1,640 supplemented with 2 mM L-glutamine, 5% FBS. For experiments, cells (less than 20 passages from the cryopreserved cell bank) were grown to 80% confluence and brought into single-cell suspension using TrypLE Express--Trypsin Replacement Enzyme (Invitrogen).
Instrumentation
A BD FACSAria and Diva 5.0 (BD Biosciences, San Jose, CA) were used for six-color FACS analysis. Laser alignment (blue, 488 nm 100 mW; red, 638 nm 30 mW; violet, 405 nm 50 mW) was verified with Rainbow Beads (Spherotech, Lake Forest, IL, http://www.spherotech.com) prior to running tumor cell samples. The six colors used in the analysis were compensated to account for spectral overlap emitted by the fluorochromes within each laser as well as across the lasers using a semi-automated process using compensation beads (BD Biosciences) coated with anti-mouse Ig antibodies. The beads were incubated with each individual antibody used in the six-color panel for 15 minutes at room temperature. Unstained and stained beads were run individually, 5,000 events were recorded, and the data were imported into the Diva software compensation matrices for automatic compensation.
Tumor Cell Labeling
106 cells were incubated with antibody solution or isotype control for 15 minutes on ice, washed, and resuspended in staining buffer (2% [v/v] fetal calf serum in phosphate-buffered saline) and analyzed using the FACSAria. Antibodies and specific antibody concentrations used (CD24PE-Cy5, CD44~Pacific Blue, CD133~PE, CD166~APC, CD326~Alexa700, and PgP~FITC) are listed in supporting information Table 1. Gates were set with isotype controls such that, for each cell, less than 1% of the total cell population was false-positive. Labeled cells were then analyzed (10,000 events), and data were saved as FCS and txt files using WinList 3.0 (Verity House, http://www.vsh.com), and the txt file was imported into JMP-7 for statistical analysis.
Xenograft Model
In order to investigate tumorigenicity of cell populations, cells were brought into single-cell suspension and diluted such that the indicated cell number was suspended in 100 ul of a 1/1 mixture of culture medium and Matrigel (BD Biosciences, San Jose, CA). Cells were injected subcutaneously into the right axillary region of female NOD.SCID mice (Animal Production Program, NCI-Frederick, http://web.ncifcrf.gov) using 27G needles. Tumor growth was assessed weekly with tumor mass calculated from bidimensional caliper measurements using the formula [tumor length x (tumor width)]]/2 = tumor weight in mg [37]. Masses that were >150 mg and grew progressively during the observation period (120 days) were defined as tumors.
Statistical Analysis
Data from the six-color analysis was subjected to k-means clustering and principal component analysis using JMP-7 (SAS Institute, Research Triangle, NC, http://www.sas.com). For each cell, six fluorescence intensity measurements as well as forward- and side-scatter data were utilized. For each cell line, 2–4 staining/analysis runs were included in the analysis depending on the consistency of the staining results. Linear regressions were performed using GraphPad Prism 5.0b (GraphPad Software, LA Jolla, CA, http://www.graphpad.com).
Results
Surface Marker Expression and ALDH Activity Vary Greatly Within the 60 Cell Line Panel
In order to identify a CIC marker signature in the NCI60 panel, we first evaluated expression of surface markers that had previously been described for the identification of CICs [14]. Expression of CD44, CD24, CD133, and CD15 varied greatly even between cell lines of the same tumor type (Table 1, supporting information Fig. S1). For example, the size of the CD133 positive population varied from 0.1% (SF-295, SF-539) to 1.4% (SF-268) in cell lines derived from CNS tumors, and from 0.2% (HCT 15) to 74.5% (HT29) in cells derived from colon tumors. Likewise, ALDH activity varied greatly, and sizes of cell populations with high ALDH activity ranged from 0.04% (COLO205, HCC-2998) to 38.8% (HT−−29) (Table 1).
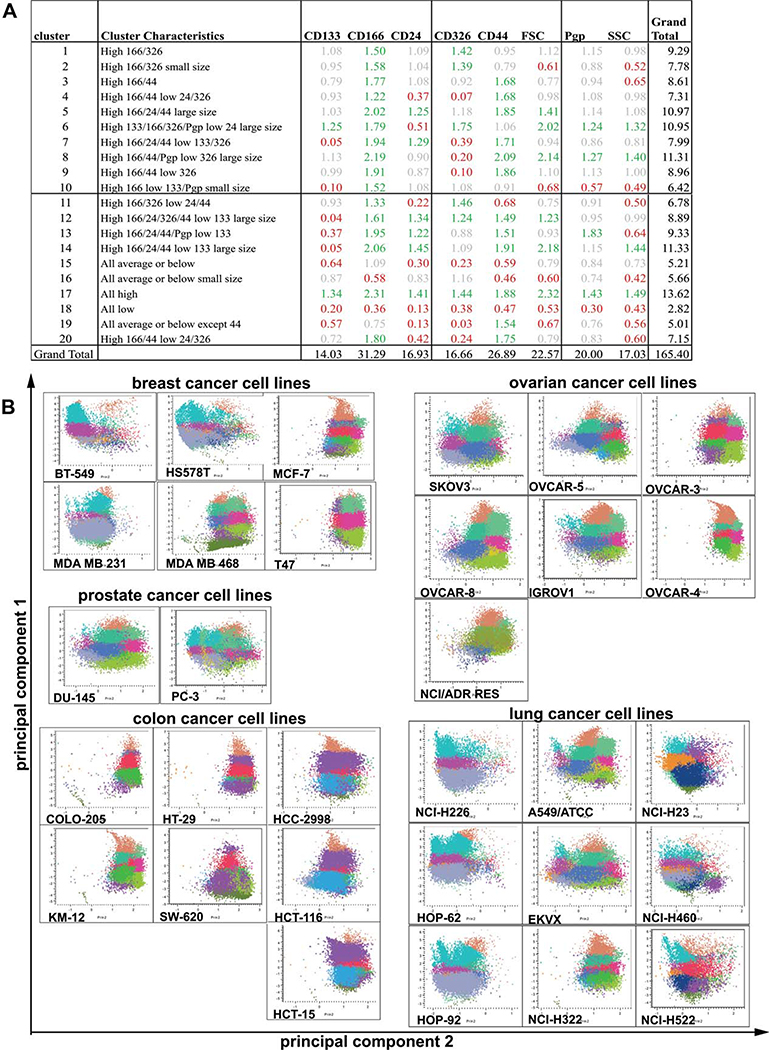
Table 1.
Expression of putative tumor stem cell markers in the NCI60 tumor cell line panel. Expression levels of CD15/CD24/CD44 or expression levels of CD133 and ALDH activity were analyzed by FACS analysis (50,000 cells/sample), and the size of marker positive and marker negative populations was determined using FlowJo (Treestar Inc). For each marker, the size of the marker positive subpopulation was determined (percent positive cells), and data were centered to the mean. FACS analysis was performed in triplicate
| Tumor Type | Cell Line Name | CD44+ | CD24+ | CD133+ | CD15+ | ALDH+ | CD44+ CD24[minus] | CFU (2-D) | soft agar assay |
|---|---|---|---|---|---|---|---|---|---|
| hematopoeitic | CCRF-CEM | 8.44 | 0.53 | 0.04 | 7.59 | 0.03 | 8.29 | 6 | |
| hematopoeitic | HL-60 | 43.01 | 0.03 | 0.07 | 56.77 | 0.04 | 42.98 | 32 | |
| hematopoeitic | K-562 | 0.13 | 0.31 | 0.09 | 44.91 | 12.86 | 0.11 | 64 | |
| hematopoeitic | MOLT-4 | 0.57 | 0.49 | 0.05 | 0.39 | 1.32 | 0.54 | 28 | |
| hematopoeitic | RPMI-8226 | 0.09 | 0.19 | 0.05 | 1.68 | 0.00 | 0.06 | 18 | |
| hematopoeitic | SR | 0.08 | 0.22 | 0.07 | 0.21 | 0.00 | 0.06 | 12 | |
| NSC Lung | A549/ATCC | 84.41 | 71.73 | 3.64 | 2.05 | 2.55 | 17.84 | 94 | 63 |
| NSC Lung | EKVX | 77.00 | 81.47 | 1.95 | 0.48 | 9.12 | 7.86 | 35 | 0 |
| NSC Lung | HOP-62 | 81.41 | 4.35 | 1.91 | 0.87 | 2.46 | 77.46 | 45 | 37 |
| NSC Lung | HOP-92 | 95.44 | 1.61 | 0.25 | 1.85 | 0.48 | 93.99 | 15 | |
| NSC Lung | NCI-H226 | 89.32 | 16.41 | 0.15 | 0.90 | 14.76 | 74.05 | 20 | 0 |
| NSC Lung | NCI-H23 | 30.95 | 4.30 | 0.11 | 43.51 | 0.01 | 27.97 | 42 | 27 |
| NSC Lung | NCI-H322M | 54.49 | 52.65 | 3.45 | 88.27 | 0.01 | 23.09 | 20 | 29 |
| NSC Lung | NCI-H460 | 67.44 | 27.81 | 0.38 | 50.55 | 1.61 | 49.94 | 161 | 55 |
| NSC Lung | NCI-H522 | 1.40 | 81.25 | 0.18 | 2.41 | 35.74 | 0.02 | 29 | 1 |
| Colon | COLO 205 | 60.97 | 58.36 | 5.78 | 75.50 | 0.04 | 18.39 | 43 | 77 |
| Colon | HCC-2998 | 3.19 | 5.35 | 0.60 | 54.26 | 0.04 | 1.95 | 8 | 56 |
| Colon | HCT-116 | 58.62 | 0.78 | 63.18 | 0.14 | 0.56 | 57.93 | 126 | 102 |
| Colon | HCT-15 | 14.14 | 22.41 | 0.17 | 49.74 | 0.03 | 7.44 | 120 | 63 |
| Colon | HT-29 | 71.39 | 55.45 | 74.51 | 52.78 | 38.80 | 20.99 | 112 | 71 |
| Colon | KM12 | 48.87 | 9.06 | 10.71 | 10.65 | 3.42 | 40.29 | 29 | 37 |
| Colon | SW-620 | 11.49 | 52.84 | 22.04 | 90.05 | 0.11 | 2.25 | 158 | 60 |
| CNS | SF-268 | 86.02 | 18.77 | 1.39 | 0.25 | 0.03 | 68.12 | 37 | 8 |
| CNS | SF-295 | 98.07 | 24.70 | 0.09 | 4.37 | 0.03 | 73.56 | 65 | 54 |
| CNS | SF-539 | 84.22 | 33.08 | 0.13 | 0.48 | 0.00 | 52.76 | 30 | 18 |
| CNS | SNB-19 | 65.79 | 20.00 | 0.50 | 1.06 | 0.01 | 47.64 | 90 | 75 |
| CNS | SNB-75 | 88.23 | 20.36 | 0.25 | 0.39 | 0.69 | 68.13 | 10 | 8 |
| CNS | U251 | 77.53 | 4.98 | 0.55 | 0.12 | 0.75 | 72.82 | 38 | 99 |
| Melanoma | LOX IMVI | 67.54 | 2.11 | 0.48 | 22.45 | 2.59 | 65.84 | 2 | 50 |
| Melanoma | M14 | 95.03 | 2.80 | 0.45 | 0.11 | 2.27 | 92.27 | 74 | 92 |
| Melanoma | MALME-3M | 97.37 | 0.19 | 0.30 | 0.19 | 6.56 | 97.20 | 29 | 13 |
| Melanoma | SK-MEL-2 | 71.24 | 0.60 | 1.92 | 0.19 | 1.36 | 70.79 | 14 | 8 |
| Melanoma | SK-MEL-28 | 90.39 | 1.29 | 21.25 | 0.30 | 9.12 | 89.18 | 18 | 36 |
| Melanoma | SK-MEL-5 | 83.09 | 3.71 | 1.74 | 0.50 | 0.00 | 79.89 | 29 | 37 |
| Melanoma | UACC-257 | 87.94 | 12.52 | 3.84 | 0.26 | 8.62 | 75.66 | 27 | 22 |
| Melanoma | UACC-62 | 67.28 | 4.24 | 0.21 | 0.58 | 0.15 | 63.35 | 34 | 19 |
| Melanoma | MDA-MB-435 | 76.91 | 0.77 | 0.40 | 0.95 | 6.62 | 76.24 | 117 | |
| Ovarian | IGROV-1 | 66.38 | 78.70 | 6.81 | 19.77 | 9.86 | 2.29 | 10 | 1 |
| Ovarian | OVCAR-3 | 12.70 | 91.33 | 22.46 | 88.38 | 0.97 | 0.05 | 19 | 0 |
| Ovarian | OVCAR-4 | 77.81 | 11.98 | 1.77 | 0.35 | 0.65 | 44.55 | 22 | 0 |
| Ovarian | OVCAR-5 | 98.97 | 54.69 | 19.36 | 0.54 | 0.04 | 44.42 | 41 | 5 |
| Ovarian | OVCAR-8 | 89.94 | 94.43 | 0.09 | 93.32 | 0.00 | 2.61 | 118 | 84 |
| Ovarian | SK-OV-3 | 90.74 | 58.49 | 0.78 | 8.16 | 0.00 | 34 | 19 | |
| Ovarian | NCI/ADR-RES | 73.62 | 64.05 | 0.54 | 84.19 | 0.96 | 19.67 | 19 | |
| Renal | 786-0 | 94.59 | 96.32 | 0.93 | 15.01 | 2.58 | 0.99 | 37 | 2 |
| Renal | A-498 | 77.86 | 43.71 | 0.39 | 7.03 | 5.08 | 37.07 | 20 | 0 |
| Renal | ACHN | 77.55 | 81.56 | 0.45 | 0.44 | 16.48 | 1.16 | 68 | 1 |
| Renal | CAKI-1 | 78.71 | 67.50 | 0.23 | 3.16 | 13.16 | 20.40 | 39 | 0 |
| Renal | RXF-393 | 93.59 | 63.98 | 0.20 | 5.34 | 0.00 | 30.26 | 3 | 1 |
| Renal | SN12C | 14.63 | 93.43 | 57.47 | 41.57 | 0.00 | 0.04 | 17 | 34 |
| Renal | TK-10 | 94.31 | 91.45 | 0.75 | 91.84 | 1.66 | 6.25 | 53 | 4 |
| Renal | UO-31 | 89.70 | 21.47 | 5.93 | 4.98 | 0.02 | 68.74 | 28 | 0 |
| Prostate | PC-3 | 74.93 | 14.00 | 0.78 | 45.00 | 22.32 | 62.61 | 26 | |
| Prostate | DU145 | 66.56 | 38.92 | 0.87 | 68.37 | 0.08 | 34.27 | 53 | |
| Breast | MCF7 | 1.37 | 20.44 | 1.24 | 33.66 | 0.40 | 0.11 | 74 | |
| Breast | MDA MB 231/ATCC | 90.19 | 4.22 | 0.38 | 27.39 | 0.15 | 86.33 | 100 | |
| Breast | Hs-578T | 67.80 | 28.23 | 1.03 | 0.45 | 0.10 | 41.06 | 14 | |
| Breast | BT-549 | 45.27 | 12.91 | 4.54 | 1.01 | 0.04 | 33.40 | 68 | |
| Breast | T-47D | 24.21 | 72.03 | 1.11 | 69.69 | 0.03 | 2.67 | 74 |
Multidimensional Analysis of Surface Markers Defines Patterns of Co-Expression of Stem Cell Markers that are Related to Tumor Type
Next, we asked whether it was possible to identify patterns of marker co-expression defined by some of the most widely reported cell surface markers. We simultaneously determined expression of CD44 (Pacific Blue), CD24 (PE-Cy7), CD133 (PE), CD166 (APC), CD326 (EpCAM, Alexa Fluor 700), and PgP (FITC), and forward- and side-scatter for each cell line and defined naturally occurring co-expression patterns using k-means cluster analysis followed by hierarchical clustering of the percentage population in each of the k-means clusters.
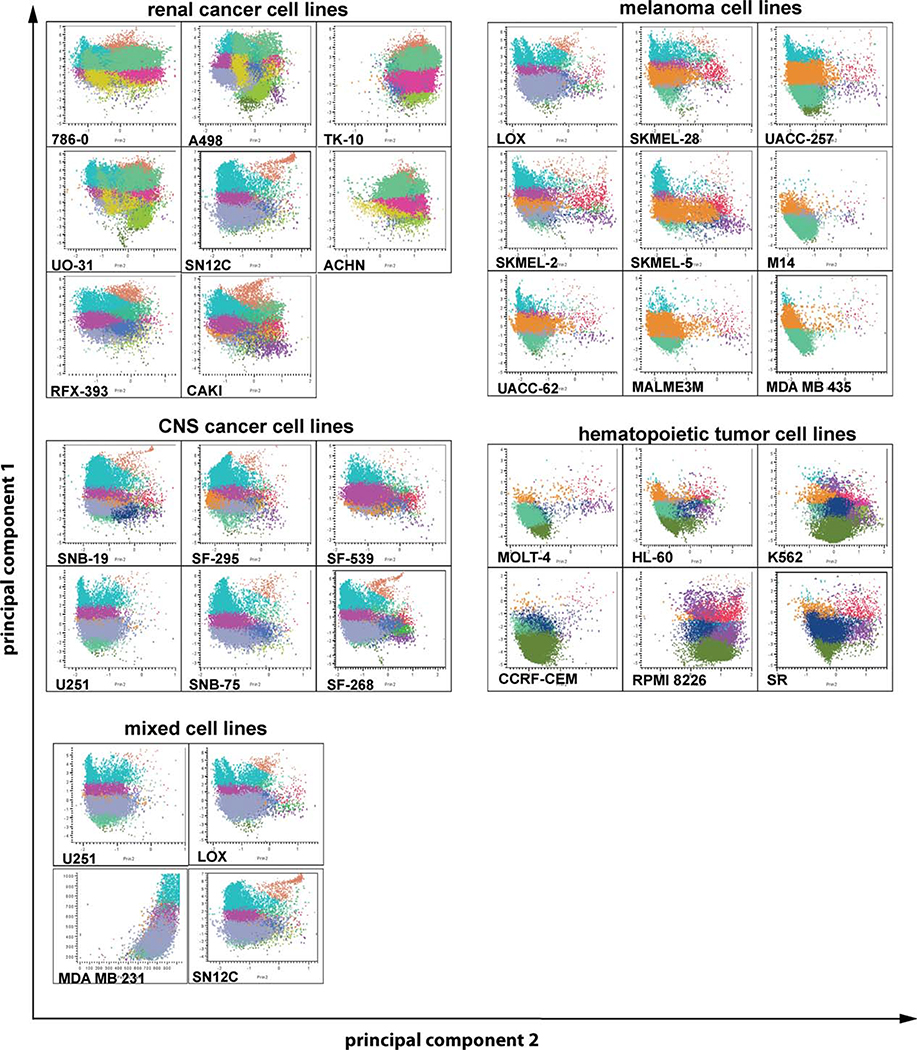
A cluster number of 20 (Fig. 1A) was determined to provide adequate resolution for definition of naturally occurring co-expression patterns. The patterns of co-expression for each cell line, organized by tumor type, are illustrated in Figures 1 and 2; cells were color coded by cluster and plotted on a two-dimensional graph based on the first two principal components of the data. By inspection, it is apparent that similarities in patterns of co-expression exist within and between tumor types. For example, the breast cancer cell lines BT549, MDA MB 231, and Hs578T and the cell lines MCF-7 and T47D show similar cluster patterns that are not related to the cluster pattern of MDA MB 468 cells (Fig. 1B, supporting information Tables S2–S4). Whereas the two prostate cancer cell lines DU145 and PC-3 have different cluster patterns (Fig. 1B, supporting information Tables S2–S4), the cluster pattern of DU145 cells is similar to the ovarian cancer cell lines SKOV3, OVCAR-5, OVCAR-8, and IGROV1 (Fig. 1B, supporting information Tables S2–S4). Likewise, several clusters of similar appearance were identified for colon cancer cell lines (COLO 205, KM12 and HT29, and HCC-2998, HCT-116, and HCT-15, Fig. 1B, supporting information Tables S2–S4), lung cancer cell lines (NCI-H226, HOP-62, HOP-92, Fig. 1B, supporting information Tables S2–S4), renal cancer cell lines (Fig. 2, supporting information Tables S2–S4), melanoma cell lines (Fig. 2, supporting information Tables S2–S4), CNS cancer cell lines (Fig. 2, supporting information Tables S2–S4), and hematopoietic neoplasia (Fig. 2, supporting information Tables S2–S4). Interestingly, although some cluster patterns were related to the tumor type, for example, all melanoma cell lines but LOX are MB 231 cells (breast cancer), and SN12C (renal cancer) closely related (Fig. 2, supporting information Tables S2-- showed closely related cluster patterns (Fig. 2, supporting in S4), U251 cells (CNS cancer), LOX cells (melanoma), MDA formation Tables S2–S4). This indicates that the expression of putative CIC markers is not only related to the tumor type but might also be influenced by properties of the tumor cells such as an epithelial versus mesenchymal phenotype of cell.
Figure 1.
(A): Cluster mean characteristics expressed as a ratio to the population means. Data from the six-color analysis was subjected to k-means clustering and principal component analysis using JMP-7 (SAS Institute, Research Triangle, NC, http://www.sas.com). For each cell, six fluorescence intensity measurements as well as forward- and side-scatter data were utilized. For each cell line, at least two staining/analysis runs were included in the analysis. Cluster characteristics are tabulated as ratio of the cluster mean to the overall average. Thus, ratios > 1 indicate values higher than the overall average and ratios < 1 indicate lower values. Color coding: green >1.5; gray 1.5–0.7; red <0.7. (B): Cluster pattern of breast, prostate, ovarian, colon, and lung cancer cell lines. Data were analyzed using k-means clustering, and the resulting clusters were color-coded. Plots were generated using the first two principal components and grouped by tumor type. Correlation between cluster patterns was analyzed for each tumor type using JMP-7. Breast cancer cell lines are organized to illustrate the partitioning of stem cell marker co-expression with the molecular taxonomy of breast cancer. Cell lines BT549, HS578T, and MDA-MB-231 fall into the “normal-like” category; cell lines MCF-7 and T47 are “luminal type”, and MDA-MB-468 is “basal type”.
Figure 2.
Cluster pattern of renal cancer, melanoma, CNS cancer, and hematopoietic tumor cell lines. Data were analyzed using hierarchical k-means clustering, and the resulting 20 k-means clusters were color-coded. Plots were generated using the first two principal components and grouped by tumor type. The correlation between cluster patterns was analyzed for each tumor type using JMP-7 (SAS Institute, Research Triangle, NC, http://www.sas.com). The bottom panel shows cell lines from different tumor types (U251, CNS cancer; LOX, melanoma; MDAMB231, breast cancer; SN12C, renal cancer) that have related cluster patterns. Abbreviations: CNS, central nervous system.
Surface Marker Expression Does Not Correlate with Colony-Formation Capacity of Cell Lines
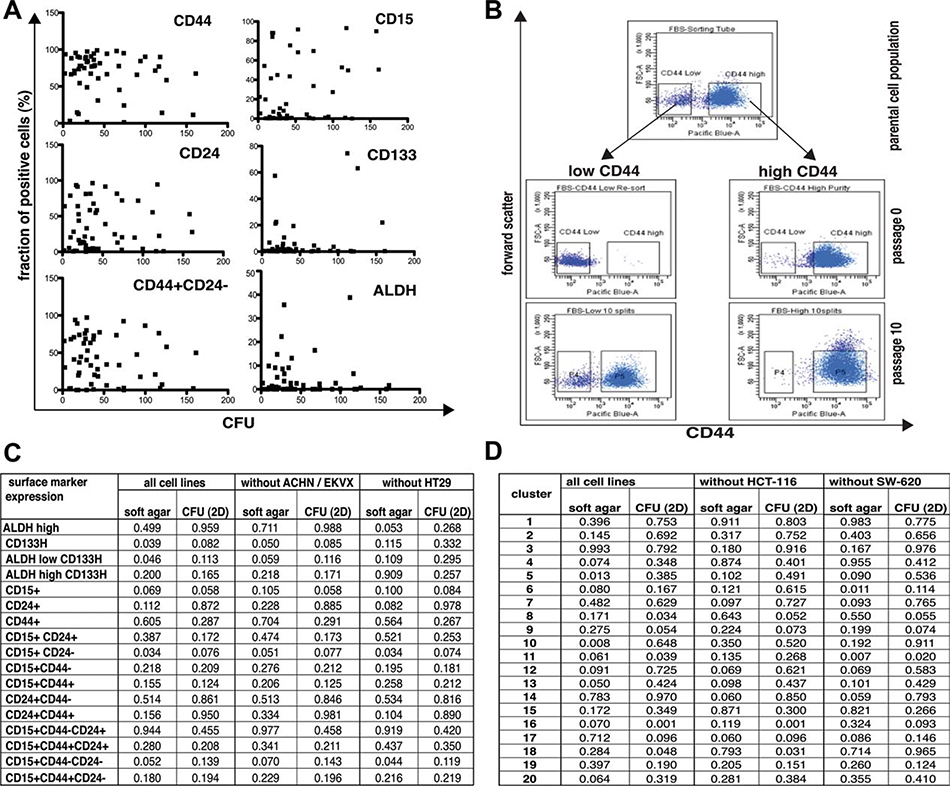
CICs have stem-like properties and can regenerate all cell types of the original tumor from a single cell. In vitro, CICs form colonies from a single cell. Therefore, we determined expression of CIC markers in cell lines in correlation with the capacity to form colonies from single cells in vitro (Fig. 3). The colony-formation capacity of cell lines in 2D culture and soft agar assays varied greatly between and within tumor types (Fig. 3A, Table 1).
Figure 3.
Expression of putative tumor stem cell markers does not correlate with colony-forming capacity of cell lines. (A): Expression of CD44, CD24, CD15, CD133, and ALDH does not correlate with colony-forming capacity in 2D adherent culture. Expression levels of CD15, CD24, CD44, CD133, and ALDH activity were analyzed by fluorescence-activated cell sorting (FACS) analysis (50,000 cells/sample) and the size of marker positive and marker negative populations was determined using FlowJo (Treestar Inc., Ashland, OR, http://www.treestar.com). To determine colony-forming units in cell lines, 100 cells were cultured in 100 mm tissue culture plates for 2 weeks, cell colonies were stained with Coomassie Brilliant Blue, and the number of visible colonies was determined. Data are presented as the average of three independent experiments. Scatter plots were generated using Graphpad Prism 5.0b. (B): CD44- (“low CD44”) OVCAR-5 cells generate CD44+ (“high CD44”) cells, while CD44+ OVCAR-5 cells do not generate CD44- cells. OVCAR-5 cells that were grown under adherent culture conditions were FACS sorted into CD44- and CD44+ subpopulations, controlled for purity of the population by FACS analysis of an aliquot, and cultured in 2D adherent culture for 10 passages before expression of CD44 was re-analyzed by FACS. (C): Co-expression of CD133/ALDH and CD24/CD44/CD15 does not correlate with colony-forming potential of cell lines in 2D adherent culture or anchorage-independent soft agar assays. Data acquisition was performed as described above. Additionally, 5,000 cells were embedded in soft agar and colonies counted after 7 days. Data were analyzed for all cell lines (“all cell lines”) or after ACHN/EKVX or HT29 cells that appeared to be outliers in the graphs were excluded to test robustness of the dataset. Linear regressions were performed using GraphPad Prism 5.0b, and p-values are listed. (D): Cluster patterns of surface markers do not correlate with colony-forming potential of cell lines in 2D adherent culture or anchorage-independent soft agar assays. Data acquisition was performed as described above. Additionally, 5,000 cells were embedded in soft agar and colonies were counted after 7 days. Data were analyzed for all cell lines (“all cell lines”) or after HCT-116 or SW-620 cells that appeared to be outliers in the graphs were excluded to test robustness of the dataset. Linear regressions were performed using GraphPad Prism 5.0b, and p-values are listed.
Although we observed a statistical correlation between clonogenicity of cell lines in soft agar assays and expression of CD15, CD24, CD44, CD133, or ALDH, these correlations depended on only a few cell lines; for example, removal of the HT29 data set resulted in loss of the correlation of clonogenicity with CD133 expression, and removal of ACHN/EKVX cells from the panel resulted in loss of the correlation between clonogenicity of cell lines in soft agar assays and expression of CD15/CD24 in soft agar assays (Fig. 3C), indicating that there is no robust correlation between surface marker expression and clonogenic potential of cell lines under adherent and anchorage independent (soft agar) culture conditions.
In an attempt to further evaluate if CD44 subpopulations have clonogenic properties as described in literature [2, 6, 8], we sorted OVCAR-5 cells, which were the only cell line that had distinct CD44+ and CD44- subpopulations, and carried the respective subpopulations over 10 passages. We found that, contrary to published data, it was the CD44- population that demonstrated stem cell properties as indicated by the ability to regenerate both the CD44+ and CD44- populations, whereas the CD44+ population remained CD44+ over the same ten-passage interval (Fig. 3B). Interestingly, the cloning efficiency as determined by soft agar assays was significantly higher (p < .001) for the CD44+ population (10.2 ± 0.4%) that could not regenerate the parental CD44 expression pattern than for the CD44- population (4.9 ± 0.4%). Thus, the clonogenic potential of cell lines and subpopulations was not closely coupled to regenerative capacity.
We next investigated if cluster size correlates with clonogenicity of cell lines. We again could identify several clusters that correlated with clonogenicity of cell lines in soft agar assays or in 2D culture, namely, cluster 5, cluster 8, cluster 10, cluster 11, cluster 13, cluster 16, and cluster 18 (Fig. 3D). However, the statistical correlation of clonogenicity and cluster size of these cell lines critically depended on the cluster pattern obtained for HCT-116 or SW-620 cells, that is, after removal of data for HCT-116 cells (clusters 5, 8, 10, 11) or SW620 cells (clusters 5, 8, 10, 16) from the dataset, clonogenic capacity in soft agar assays or 2D culture no longer correlated with the fraction of cells in these clusters (Fig. 3D), again indicating that there is no robust correlation between surface marker expression pattern and clonogenic potential of cell line under adherent or anchorage-independent (soft agar) culture conditions.
In summary, our data indicate that the expression of the surface markers CD15, CD24, CD44, CD133, CD166, CD326, PgP, and aldehyde dehydrogenase activity, singly or in combination, does not correlate with the clonogenic potential of tumor cell lines and may, therefore, not be suitable as a universal signature to detect CICs in cell lines derived from different tumor types.
Anchorage-Independent Growth of Tumor Cell Lines Alters Surface Marker Expression and Tumorigenicity of Colon Cancer Cell Lines
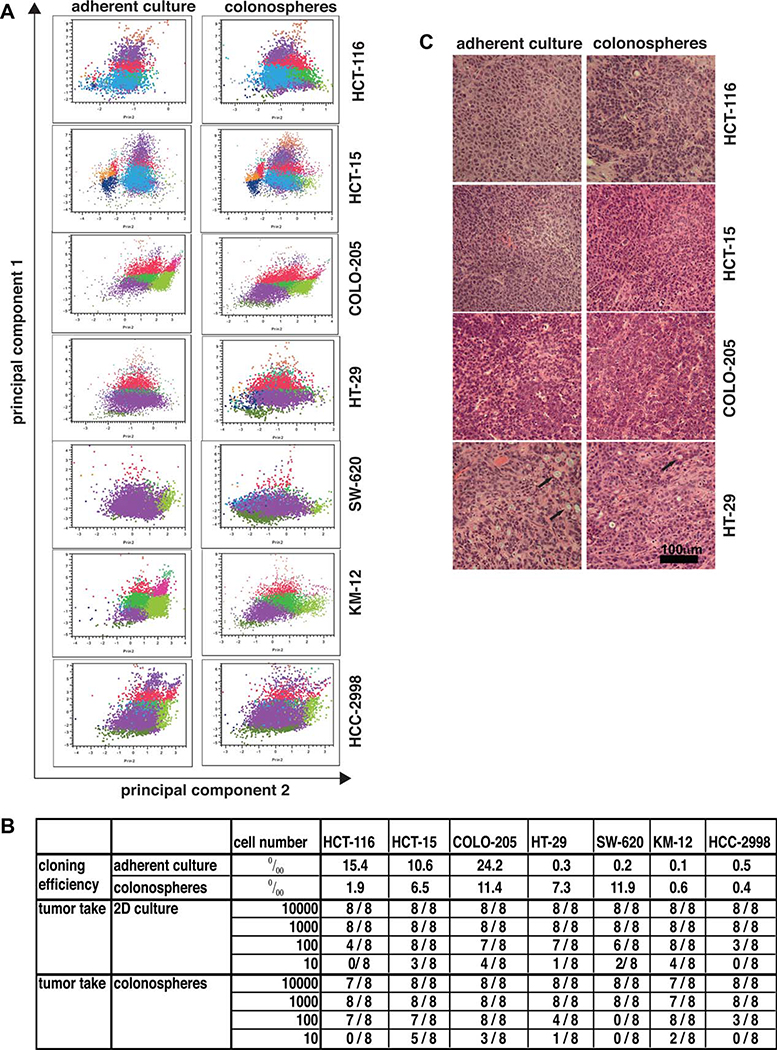
It has been postulated that anchorage-independent culture of tumor cells would enrich cultures for CICs. As a strategy to enrich cultures for tumor stem cells, “colonospheres” were generated for all of the cell lines by plating the routine stock in serum-free RPMI-1640 supplemented with 15% knock-out supplement (Invitrogen) and 2 mM l-glutamine. Under these conditions, the cells attached minimally and grew as floating masses. For each cell line, colonosphere cultures were expanded and cryopreserved, and cells from these frozen stocks were used for assessment of surface marker display and tumorigenicity.
Six-color multidimensional fluorescence-activated cell sorting (FACS) analysis demonstrated that the cluster pattern of cells derived from colonospheres differed from that derived from adherent cultures of colon cancer cell lines (Fig. 4A), but no consistent patterns of change were apparent. Thus, we next investigated if clonogenic potential and tumorigenicity of colon cancer cell lines is changed by anchorage-independent culture. We found that the influence of culture conditions on the clonogenic potential of a cell line was cell-line dependent (Fig. 4B). Whereas HT-29, SW-620, and KM-12 cells had a higher clonogenic capacity in soft agar assays when previously grown as colonospheres as compared to adherent cultures, HCT-116, HCT-15, and COLO-205 cells formed more colonies in soft agar assays when grown previously in adherent culture as compared to colonosphere cultures, indicating that serum-free, anchorage-independent culture conditions do not necessarily increase the fraction of colony-forming units in tumor cell cultures.
Figure 4.
Anchorage-independent growth of tumor cell lines alters surface marker expression and tumorigenicity of colon cancer cell lines. Colon cell lines that were grown in 2D adherent cultures or anchorage-independent as colonospheres were brought into single-cell suspension, and were analyzed by multidimensional FACS analysis, by colony formation assays (soft agar), or were injected subcutaneously injected into NOD/SCID mice. (A): Cluster pattern of colon cell lines grown in adherent culture or anchorage-independent as colonospheres. Cells were brought into single-cell suspension, and six-color fluorescence-activated cell sorting analysis was performed. The correlation between cluster patterns was analyzed for each tumor type using JMP-7 (SAS Institute, Research Triangle, NC, http://www.sas.com). Please note that the color coding of the clusters is not identical with the color coding in Figure 3. (B): Influence of culture conditions on colony-forming capacity and tumor-forming capacity of colon cancer cell lines. Colon cell lines that were grown in 2D adherent cultures or anchorage-independent as colonospheres were brought into single-cell suspension and either used for soft agar assays (data present the average of three independent experiments), or the cell number indicated in the table was injected subcutaneously into NOD/SCID mice and animals were subsequently observed for tumor growth. (C): In vitro culture conditions of colon cancer cell lines do not influence the histology of xenograft tumors derived from colon cancer cell lines. Histology (H&E staining) of colon xenograft tumors derived from HCT-116, HCT-15, COLO-205, and HT29 showed comparable, relatively undifferentiated carcinomas whether generated from adherent cultures or colonospheres. Mucin producing cells were occasionally observed in HT-29 tumors (arrows).
We next determined if serum-free, anchorage-independent growth influences the number of tumor-initiating cells in colon cancer cultures. Injection of single-cell suspensions into animals in a limiting dilution protocol revealed that, similar to our in vitro results, the effect of culture conditions on the number of tumor cells necessary to initiate tumor growth in vivo was cell-line dependent (Fig. 4B). In limiting dilution experiments, tumor take after injection of 100 or 10 tumor cells was higher when HCT-116 or HCT-15 cells were grown in anchorage-independent colonosphere cultures as compared to adherent cultures prior to injection. However, for SW-620 and HT-29 cell lines, the tumor take was higher when cells were grown under adherent culture conditions as compared to colonosphere cultures prior to injection into animals. Histologically, no differences were observed between tumors derived from cells grown under adherent conditions or as colonospheres (Fig. 4C).
Taken together, our results indicate that the effect of adherent and serum-free, anchorage-independent culture conditions on in vitro clonogenic potential and in vivo tumor-forming capacity of tumor cell lines is cell-line dependent. Furthermore, the clonogenic potential in vitro does not necessarily predict tumor-formation capacity in vivo as apparent for SW-620, which, after being grown in anchorage-independent culture, has a higher clonogenic capacity in vitro but lower tumor-forming capacity in vivo (Fig. 4B).
Hierarchical Clustering Demonstrates That Expression Patterns of Putative CIC Markers Relate to Tumor Types
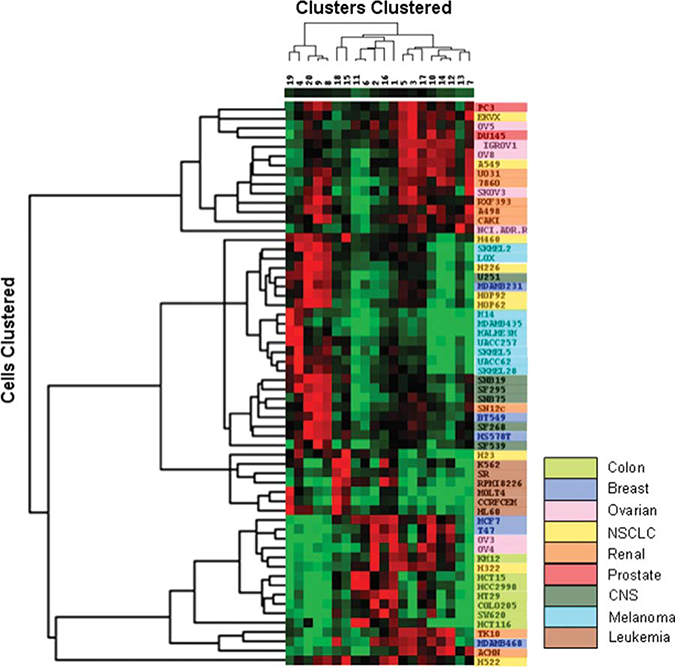
Application of hierarchical clustering methods to the k-means derived clustering patterns by cell line resulted in generation of the heat map shown in Figure 5. It is apparent that certain tumor types presented very homogeneous patterns and close clustering whereas others were more heterogeneous.
Figure 5.
Heat map generated using “Cluster” and “Treeview” (Eisen et al. (1998) PNAS 95:14863). Percentage populations in each of the 20 k-means defined clusters for each of the cell lines were log (natural) transformed (zero values set to 0.001%) and then subjected to hierarchical (average linkage) clustering by cluster and cell line. The dendrogram at the left of the figure defines clusters by cell line and the vertical dimension reflects clustering by k-means cluster. The scale for the heat map ranges from 0 = bright green to maximum = bright red. Cell lines are color coded by tumor panel to facilitate recognition of panel-related associations. Abbreviations: CNS, central nervous system.
Specifically, the first branch at the top of the cell line dendrogram contains prostate, lung, ovarian, and renal lines with substantial similarity of co-expression pattern. Clusters 3, 5, and 7, which have high expression of CD44 and CD166 in common, dominated this pattern (Fig. 1A). Additionally, cluster 5 also shows high expression of CD24. Cluster 7 shows high levels of CD44, CD166, and CD24 with low expression of CD133 and EpCAM. For the majority of the tumor cell lines, a relatively small number of clusters, for example, 3–4, contained the vast majority of cells. However, some patterns were more complex with cells distributed over 6–7 clusters. The lung cancer cell lines A549 and EKVX are highly correlated examples of such a complex pattern (Fig. 1B).
At the bottom of the dendrogram is a very homogeneous clustering of colon tumor cell lines flanked by some breast, ovarian, renal, and a single lung cancer cell line. The colon tumor co-expression patterns were dominated by a high percentage of cells in clusters 1, 2, 6, and 11 (Fig. 1, supporting information Table S2). These clusters have high levels of CD166 and EpCAM in common and differ in other aspects. Cluster 6 is notable in that CD166, CD133, EpCAM, and PgP are all highly expressed in cells of large size with low CD24 levels (Fig. 1).
Tumor cell lines considered to be of mesenchymal origin, that is, hematopoietic, CNS, and melanoma cell lines, are clustered in the center of the dendrogram and associated with subsets of the lung, renal, and breast cancer panels. The cell lines of the renal panel, together with the ovarian, non small cell lung cancer (NSCLC)2, and breast tumor panels were relatively heterogeneous in terms of their co-expression patterns and are thus scattered across the dendrogram (Fig. 5).
Discussion
Expression of Putative Cancer Stem Cell Markers Does Not Correlate with Clonogenic Capacity of Tumor Cell Lines
Existence of tumor stem cells or cancer initiating cells (CICs) that are drug- and radiation-resistant and that are crucial for tumor recurrence has been discussed widely over the past decade [1, 14]. Although the concept of stem cell-like tumor cells is intriguing, identification and isolation of such cells is difficult. In analogy to adult tissue stem cells, it has been postulated that CICs can self-renew by symmetric mitosis and can re-establish all tumor cell types found in the original tumor mass by asymmetric mitosis that generates one daughter cell that retains CIC properties and another that is differentiated [38]. Stem cells are typically prospectively identified by expression of surface markers or functional markers such as ABC transporters or ALDH, and clonogenic potential and the capacity to regenerate tumors is subsequently demonstrated for the identified subset of tumor cells.
When examined individually, putative stem cell markers were heterogeneously expressed across the NCI60 tumor cell lines tested (Figs. 1, 2), and marker expression was influenced by culture conditions (Fig. 4). We found that expression patterns of putative stem cell markers did not significantly correlate with clonogenic capacity of cell lines (Fig. 4). This was particularly surprising for the functional markers ALDH and PgP that have been widely described as stem cell markers [26, 28, 29, 33]. Although used as stem cell markers, ALDH and the ABC transporter PgP are enzymes involved in drug metabolism and transmembrane drug transport and are known to confer drug resistance. Thus, assays used to measure ALDH and ABC transporter activity also assess viability and drug resistance of cells and, therefore, may identify cell populations with high clonogenic capacity because these subpopulaions contain viable cells that show increased resistance to the toxic substances used in these assays.
Evaluation of pairs of markers such as the CD44+/ CD24- phenotype [2, 38], which was previously reported for breast cancer stem cells, and clusters of surface markers as established by six-color multidimensional analysis, showed a similar degree of heterogeneity and again no simple relationship to cloning efficiency could be identified.
Moreover, in our hands, the correlation between cloning efficiency and tumorigenic capacity of colon cell lines was poor, and both parameters were influenced by culture conditions. This is particularly concerning since clonogenic capacity in vitro is often used as an indicator for increased tumorigenicity in vivo, and both parameters are thought to be indicative for the size of the fraction of tumor stem cells in cultures. Interestingly, anchorage-independent culture, which is thought to enrich cultures for tumor stem cells, did not necessarily increase clonogenic or tumorigenic efficiency of colon cancer cultures. Taken together, our results indicate that, whereas the formation of tumors from low numbers of tumor cells supports the hypothesis that cancer cell lines contain CICs, putative CIC marker expression, clonogenic potential, and tumorigenic potential of tumor cell lines are influenced by culture conditions and do not directly correlate with each other. Thus, we could not identify a universal marker signature for tumor stem cells or even one applicable to a particular tumor type.
Expression of Putative Cancer Stem Cell Markers Correlates with Molecular Taxonomy of Breast Cancer Cell Lines
Multidimensional analysis for patterns of expression of putative tumor stem cell markers revealed the existence of patterns of co-expression that were related to tumor or spanned multiple tumor types. The breast cancer panel (Figs. 1, 5) perhaps represents the best example of a heterogeneous pattern of marker co-expression. This heterogeneity tracks precisely with the molecular taxonomy of breast cancer as defined by patterns of gene expression. A recently examined large panel of breast cancer cell lines was classified by gene expression into luminal, HER2, basal, and “normal-like” groupings and was further characterized for expression of EpCAM (CD326) [39]. Among the breast cancer cell lines also used in the NCI60 panel, MDA-MB-468 was classified as basal type lacking EpCAM expression; BT549, Hs578T, and MDA-MB-231 were classified as normal-like and also lacking EpCAM expression; and MCF-7 and T47D were classified as luminal and positive for EpCAM expression. It is noteworthy that the tumor stem cell marker co-expression patterns of the luminal and basal cell lines cluster with neoplasms of epithelial origin at the bottom of the dendrogram (Fig. 5), whereas normal-like breast cancer cell lines are interdigitated among non-epithelial tumors in the middle of the dendrogram. This clustering is leveraged by EpCAM expression, which we found to be consistent with the report by Sieuwerts et al. [39].
The mesenchymal pattern presents a high percentage of cells in clusters 4, 8, 9, and 20. In addition to MDA-MB-231, LOX melanoma, renal cell line SN12C, and all CNS tumor cell lines manifest this pattern. The observation of a pattern common to subsets of breast, ovarian, and lung cancers and malignancies such as melanomas and CNS tumors may reflect a “de-differentiated” state as long recognized by pathologists in certain carcinomas at the level of the light microscope or may simply be a manifestation of expression of common pathways important for maintenance of stem cell character.
Previously, patterns of basal gene expression in the NCI60 panel, as determined by array technology, were found to correlate with tumor type [35]. For example, comprehensive examination of expression of ABC transporters also led to identification of patterns related to tumor type, notably an association of ABCB5 with melanoma [40]. The ABC transporter expression-based cell line clustering, as well as that based on drug uptake transporters [41], show considerable similarity to the pattern based on stem cell marker co-expression reported here and may provide a mechanistic basis for drug-sensitivity phenotypes that allow cells to re-establish tumors after chemotherapy.
Taken together, the expression of CIC markers in the immortal NCI60 cell line panel suggests the presence of CICs or stemlike cells and the multidimensional analysis of putative CIC marker expression indicates that CIC marker expression correlates with tumor type. Application of multidimensional analysis to fresh tumor specimens will be necessary to establish the general significance of the patterns seen in the NCI60 tumor cell lines.
Conclusion
Our results with the NCI60 tumor cell line panel indicate that display of tumor stem cell markers is complex and follows patterns of co-expression related to tumor type. This result extends to molecular subtypes in the case of breast cancer. The complexity, tumor-type specificity, and dependence on culture conditions of marker display indicate that tumor stem cell signatures may vary between tumor types and cultures, explaining the challenge to the tumor stem cell field to identify robust markers for the prospective identification of cancer initiating cells or tumor stem cells.
Supplementary Material
Acknowledgments
We thank Dr. Anne Monks and Curtis Hose for useful discussions and assistance with creation of the heat map shown in Figure 5, Tom Silvers for assistance with data processing, Dr. Larry Rubinstein for advice regarding statistical treatment of the data, and Dr. Bill Matsui for introducing us to the use of Aldefluor. This research was supported in part by the Intramural Research Program of the NIH, NCI, and in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported [in part] by the Developmental Therapeutics Program in the Division of Cancer Treatment and Diagnosis of the National Cancer Institute. NCI-Frederick is accredited by AAALACi and follows the Public Health Service Policy on the Care and Use of Laboratory Animals. All animals used in this research project were cared for and used humanely according to the following policies: The U.S. Public Health Service Policy on Humane Care and Use of Animals (1996); the Guide for the Care and Use of Laboratory Animals (NIH publication No. 86–23, 1985); and the U.S. Government Principles for Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training (1985).
Footnotes
Disclosure of Potential Conflicts of Interest
The authors indicate no potential conflicts of interest.
Disclosure of potential conflicts of interest is found at the end of this article.
References
- 1.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer 2008;8:755–768. [DOI] [PubMed] [Google Scholar]
- 2.Al-Hajj M, Wicha MS, Benito-Hernandez A et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A 2003;100:3983–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hurt EM, Kawasaki BT, Klarmann GJ et al. CD44+ CD24(−) prostate cells are early cancer progenitor/stem cells that provide a model for patients with poor prognosis. Br J Cancer 2008;98:756–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O’Brien CA, Pollett A, Gallinger S et al. A human colon cancer cell capable of initiating tumour growth in immunodefificient mice. Nature 2007;445:106–110. [DOI] [PubMed] [Google Scholar]
- 5.Fang D, Nguyen TK, Leishear K et al. A tumorigenic subpopulation with stem cell properties in melanomas. Cancer Res 2005;65: 9328–9337. [DOI] [PubMed] [Google Scholar]
- 6.Fillmore C, Kuperwasser C. Human breast cancer stem cell markers CD44 and CD24: Enriching for cells with functional properties in mice or in man? Breast Cancer Res 2007;9:303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee CJ, Dosch J, Simeone DM. Pancreatic cancer stem cells. J Clin Oncol 2008;26:2806–2812. [DOI] [PubMed] [Google Scholar]
- 8.Vermeulen L, Todaro M, de Sousa Mello F et al. Single-cell cloning of colon cancer stem cells reveals a multi-lineage differentiation capacity. Proc Natl Acad Sci U S A 2008;105:13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins AT, Berry PA, Hyde C et al. Prospective identification of tumorigenic prostate cancer stem cells. Cancer Res 2005;65: 10946–10951. [DOI] [PubMed] [Google Scholar]
- 10.Ma S, Chan KW, Hu L et al. Identification and characterization of tumorigenic liver cancer stem/progenitor cells. Gastroenterology 2007; 132:2542–2556. [DOI] [PubMed] [Google Scholar]
- 11.Suetsugu A, Nagaki M, Aoki H et al. Characterization of CD133+ hepatocellular carcinoma cells as cancer stem/progenitor cells. Biochem Biophys Res Commun 2006;351:820–824. [DOI] [PubMed] [Google Scholar]
- 12.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med 2008;86:1025–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mizrak D, Brittan M, Alison MR. CD133: Molecule of the moment. J Pathol 2008;214:3–9. [DOI] [PubMed] [Google Scholar]
- 14.Klonisch T, Wiechec E, Hombach-Klonisch S et al. Cancer stem cell markers in common cancers—Therapeutic implications. Trends Mol Med 2008;14:450–460. [DOI] [PubMed] [Google Scholar]
- 15.Dalerba P, Dylla SJ, Park IK et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A 2007; 104:10158–10163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Capela A, Temple S. LeX is expressed by principle progenitor cells in the embryonic nervous system, is secreted into their environment and binds Wnt-1. Dev Biol 2006;291:300–313. [DOI] [PubMed] [Google Scholar]
- 17.Lanctot PM, Gage FH, Varki AP. The glycans of stem cells. Curr Opin Chem Biol 2007;11:373–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Irimura T, Nakamori S, Matsushita Y et al. Colorectal cancer metastasis determined by carbohydrate-mediated cell adhesion: Role of sialyl-LeX antigens. Semin Cancer Biol 1993;4:319–324. [PubMed] [Google Scholar]
- 19.Nakamori S, Kameyama M, Imaoka S et al. Increased expression of sialyl Lewisx antigen correlates with poor survival in patients with colorectal carcinoma: clinicopathological and immunohistochemical study. Cancer Res 1993;53:3632–3637. [PubMed] [Google Scholar]
- 20.Dimitroff CJ, Lechpammer M, Long-Woodward D et al. Rolling of human bone-metastatic prostate tumor cells on human bone marrow endothelium under shear flow is mediated by E-selectin. Cancer Res 2004;64:5261–5269. [DOI] [PubMed] [Google Scholar]
- 21.Stingl J, Raouf A, Emerman JT et al. Epithelial progenitors in the normal human mammary gland. J Mammary Gland Biol Neoplasia 2005; 10:49–59. [DOI] [PubMed] [Google Scholar]
- 22.Shackleton M, Vaillant F, Simpson KJ et al. Generation of a functional mammary gland from a single stem cell. Nature 2006;439:84–88. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto K, Beauchamp RD, Whitehead RH. Identification and isolation of candidate human colonic clonogenic cells based on cell surface integrin expression. Gastroenterology 2002;123:1941–1948. [DOI] [PubMed] [Google Scholar]
- 24.Jacques TS, Relvas JB, Nishimura S et al. Neural precursor cell chain migration and division are regulated through different beta1 integrins. Development 1998;125:3167–3177. [DOI] [PubMed] [Google Scholar]
- 25.Campos LS, Leone DP, Relvas JB et al. Beta1 integrins activate a MAPK signalling pathway in neural stem cells that contributes to their maintenance. Development 2004;131:3433–3444. [DOI] [PubMed] [Google Scholar]
- 26.Wu C, Alman BA. Side population cells in human cancers. Cancer Lett 2008;268:1–9. [DOI] [PubMed] [Google Scholar]
- 27.Bourguignon LY, Peyrollier K, Xia W et al. Hyaluronan-CD44 interaction activates stem cell marker Nanog, Stat-3-mediated MDR1 gene expression, and ankyrin-regulated multidrug efflux in breast and ovarian tumor cells. J Biol Chem 2008;283:17635–17651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keshet GI, Goldstein I, Itzhaki O et al. MDR1 expression identifies human melanoma stem cells. Biochem Biophys Res Commun 2008; 368:930–936. [DOI] [PubMed] [Google Scholar]
- 29.Moreb JS. Aldehyde dehydrogenase as a marker for stem cells. Curr Stem Cell Res Ther 2008;3:237–246. [DOI] [PubMed] [Google Scholar]
- 30.Matsui W, Wang Q, Barber JP et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res 2008;68:190–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginestier C, Hur MH, Charafe-Jauffret E et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007;1:555–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chu P, Clanton DJ, Snipas TS et al. Characterization of a subpopulation of colon cancer cells with stem cell-like properties. Int J Cancer 2009;124:1312–1321. [DOI] [PubMed] [Google Scholar]
- 33.Ucar D, Cogle CR, Zucali JR et al. Aldehyde dehydrogenase activity as a functional marker for lung cancer. Chem Biol Interact 2009;178: 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shoemaker RH. The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–823. [DOI] [PubMed] [Google Scholar]
- 35.Ross DT, Scherf U, Eisen MB et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet 2000;24: 227–235. [DOI] [PubMed] [Google Scholar]
- 36.Alley MC, Pacula-Cox CM, Hursey ML et al. Morphometric and colorimetric analyses of human tumor cell line growth and drug sensitivity in soft agar culture. Cancer Res 1991;51:1247–1256. [PubMed] [Google Scholar]
- 37.Plowman J, Hollingshead M, Simpson-Herren L et al. Human tumor xenograft models in NCI drug development In: Teicher BA, ed. Cancer Therapeutics: Experimental and Clinical Agents. New Jersey: Humana Press; 1997:101–125. [Google Scholar]
- 38.Dontu G, Al-Hajj M, Abdallah WM et al. Stem cells in normal breast development and breast cancer. Cell Prolif 2003;36 Suppl 1: 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sieuwerts AM, Kraan J, Bolt J et al. Anti-epithelial cell adhesion molecule antibodies and the detection of circulating normal-like breast tumor cells. J Natl Cancer Inst 2009;101:61–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Szakács G, Annereau JP, Lababidi S et al. Predicting drug sensitivity and resistance: Profiling ABC transporter genes in cancer cells. Cancer Cell 2004;6:129–137. [DOI] [PubMed] [Google Scholar]
- 41.Okabe M, Szakacs G, Reimers MA et al. Profiling SLCO and SLC22 genes in the NCI-60 cancer cell lines to identify drug uptake transporters. Mol Cancer Ther 2008;7:3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.