Graphical abstract
Keywords: Remdesivir, Lopinavir/ritonavir, Chloroquine/hydroxychloroquine, SARS-CoV-2, Antiviral mechanism
Abstract
Coronavirus disease 2019 (COVID-19) is a kind of viral pneumonia with an unusual outbreak in Wuhan, China, which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). There is currently no licensed antiviral treatment available to prevent human CoV infection. The widespread clinical use and existing knowledge on antiviral mechanisms of remdesivir, lopinavir/ritonavir and chloroquine/hydroxychloroquine in the treatment of previous epidemic diseases, namely, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), may be helpful in the combat with novel SARS-CoV-2 infection. Recent clinical evidence didn’t confirm the beneficial role of lopinavir/ritonavir and chloroquine/hydroxychloroquine for COVID-19 patients and their use was reassessed. We provide an overview of the current evidence into the mechanisms of action of these available drugs which are repurposed for treatment of the new virus. Available data identifies remdesivir as an adenosine analogue that can target the RNA-dependent RNA polymerase and block viral RNA synthesis. It has been a promising antiviral drug against a wide array of RNA viruses. 3CLpro is a major CoV protease that cleaves the large replicase polyproteins during viral replication and can be targeted by the protease inhibitor lopinavir/ritonavir but the clinical effects are controversial. Chloroquine/Hydroxychloroquine could impair the replication of SARSCoV-2 by multiple mechanisms and their immunomodulatory properties could ameliorate clinical manifestations that are mediated by immune reactions of the host although its beneficial effects are under question and need to be proven at the clinical level. Existing in vitro and in vivo evidence delineate the molecular mechanisms of these drugs in CoV-infected cells. Numerous studies demonstrated the ability of remdesivir to inhibit SARS-CoV-2 replication but future research would be needed to understand the exact mode of action of lopinavir/ritonavir and chloroquine/hydroxychloroquine in SARS-CoV-2 infected cells and to use this knowledge in the treatment of the current COVID-19.
1. Introduction
Recently, the current pandemic of coronavirus disease 2019 (COVID-19), characterized by a pulmonary infection in humans, is caused by a novel virus strain from family Coronaviridae known as severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The previous outbreak of severe acute respiratory syndrome (SARS) in 2002–2003 and Middle East respiratory syndrome (MERS) in 2012 has demonstrated the lethality of coronaviruses when they cross the species barrier and infect humans [1,2]. So far, six coronaviruses infecting humans have been identified and the novel coronavirus is the seventh one described to date as being responsible for a respiratory infection. SARS-CoV and MERS-CoV and the new SARS-CoV-2 belong to the betacoronavirus family [[3], [4], [5], [6]]. The coronaviruses have the largest genome (around 30 K) among the RNA viruses. SARS-CoV-2 was closely related (from 82.3%–88% identity) to two bat-derived severe acute respiratory syndrome (SARS)-like coronaviruses, bat-SL-CoVZC45 and bat-SL-CoVZXC21, but it was more distant from SARS-CoV (from 77.2%–79%) and MERS-CoV (about 50 %) [6,7]. Furthermore, the performed bioinformatic analysis showed that the nucleotide sequence of SARS-CoV-2 is similar to those of other betacoronaviruses with nucleotide identities of ≥ 83.6 % [8].
There are currently no effective, licensed therapies for human coronaviruses (HCoV) infections and existing treatment strategies are generally limited to symptomatic treatment and supportive care [9,10]. In the absence of a specific treatment for this novel virus, the effort of researchers is focused on understanding and controlling the disease and on preventing and controlling the replication and spread of the virus.
To devise therapeutic strategies to counteract the SARS-CoV-2 infection, numerous potential treatment options are being evaluated in ongoing clinical trials. Many antiviral and immunological treatments being investigated against coronaviruses are summarized by WHO in Landscape analysis of therapeutics as of March 21, 2020. The real-time dashboard of completed, ongoing and planned clinical trials for COVID-19 [11] includes drugs and promising therapies such as remdesevir, lopinavir/ritonavir, hydroxychloroquine, IL-6 inhibitors (tocilizumab and sarilumab), convalescent plasma therapy, stem-cell transfusion, vaccine candidates, traditional Chinese medicines which are of top 15 interventions of the presented network. Among them, remdesivir, an analogue of adenosine, seems to have a more promising future due to proven in vitro and in vivo antiviral efficacy. Till the beginning of June 2020, promising therapies involving lopinavir/ritonavir and chloroquine or hydroxychloroquine were part of treatment guidelines in many countries, but currently they are excluded from COVID-19 treatment protocols because of uncertainty regarding their risks and benefits and it is recommended that they should be used only in the context of clinical trials [[12], [13], [14]]. In spite of its known in vitro/in vivo efficacy and safety profiles, some trials evaluating these drugs for COVID-19 infection treatment such as SOLIDARITY (WHO), RECOVERY (UK; NTC04381936) and DisCoVeRy (INSERM; NTC04315948) discontinued hydroxychloroquine and lopinavir/ritonavir arms. The interim trial results showed that hydroxychloroquine and lopinavir/ritonavir produced little or no reduction in the mortality of hospitalized COVID-19 patients when compared to standard of care. Nevertheless, some countries worldwide continue to recommend chloroquine/hydroxychloroquine as a treatment option [[15], [16], [17], [18]].
The existing drugs that target viral proteins (associated with enzymatic activities or blocking viral replication machinery) or host proteins (involved in viral life cycle, regulating the function of the immune system or other cellular processes in host cells) have great potential and are available on the market.
Our review aims to highlight the potential molecular mechanisms of the therapeutic options available for the cure of other health conditions and their repurposing for the treatment of this novel coronavirus SARS-CoV-2.
2. Selected treatments of SARS-CoV-2
2.1. Remdesivir (GS-5734) – polymerase inhibitor
Remdesivir (RDV) is an investigational compound with a broad spectrum of antiviral activities against RNA viruses, including SARS-CoV and MERS-CoV. GS-5734 was originally developed for the treatment of the Ebola virus disease. GS-5734, the single Sp isomer of the 2-ethylbutyl l-alaninate phosphoramidate prodrug, effectively bypasses the rate-limiting first phosphorylation step of the Nuc (nucleoside ribose analogue). The mechanism of action of Nuc requires intracellular anabolism to the active triphosphate metabolite (NTP), which is expected to interfere with the activity of viral RNA-dependent RNA-polymerases (RdRp) [19]. GS-5734 selectively inhibits Ebola virus replication by targeting its RdRp and inhibiting viral RNA synthesis following efficient intracellular conversion to NTP in non-human primates. This compound shows a broad spectrum of antiviral activities against several RNA viruses, including respiratory syncytial virus (RSV), Junín virus, Lassa fever virus, and Middle East respiratory syndrome virus, but was inactive against alphaviruses or retroviruses [20]. Furthermore, remdesivir dose-dependently inhibits endemic human CoV-229E and CoV-OC43 replications, which typically cause upper respiratory infection in children but can cause more severe lower respiratory infection in adults with underlying respiratory conditions (i.e. asthma, COPD) and the elderly as well as a member of the deltacoronavirus genus, PDCoV, which have the most divergent RdRp of known CoV as compared to SARS- and MERS-CoV [21]. An in silico test of the COVID-19 RdRp built model suggested the effectiveness of remdesivir as a potent drug [22]. SARS-CoV and SARS-CoV-2 both belong to the betacoronaviruses of the B lineage and the RdRp amino acid sequences of the two viruses are 96 % identical whereas MERS-CoV belongs to the betacoronaviruses of the C lineage and is only 71 % identical with SARS-CoV-2 [23].
Another in vitro and in vivo proof came from Sheahan et al. who examined if GS-5734 could inhibit replication of SARS-CoV and MERS-CoV in primary human airway epithelial (HAE) cell cultures. They found out a dose-dependent reduction in replication with average IC50 values of 0.069 μM (SARS-CoV) and 0.074 μM (MERS-CoV). Moreover, the compound inhibits a broad range of diverse CoV including circulating human, zoonotic bat CoV and prepandemic zoonotic CoV. With both prophylactic and therapeutic (+1dpi) dosing of GS-5734, a reduction in replication below a disease-causing threshold in mouse model of SARS-CoV pathogenesis was demonstrated. Therapeutic GS-5734 substantially reduced the SARS-CoV induced weight loss in infected animals and significantly suppressed virus lung titers (P = 0.0059) thus demonstrating that therapeutic administration of GS-5734 can reduce disease and suppress replication during an ongoing infection [24]. Furthermore, remdesivir has the potential to block SARS-CoV-2 infection in vitro at low-micromolar concentration [25,26] and in treatment of MERS-CoV and SARS-CoV infections in vivo it demonstrated a significant improvement of pulmonary pathology in mice [24,27].
The RNA-dependent RNA polymerase (RdRp)-mediated mechanism of CoV inhibition by GS-5734 is proven, even in the setting of intact exoribonuclease (ExoN)-mediated proofreading. Using the model β-coronavirus murine hepatitis virus (MHV), it was demonstrated that GS-5734 dramatically inhibited viral replication and viral RNA synthesis in wild-type (WT) virus, while an nsp14 ExoN(-) mutant lacking proofreading demonstrated increased susceptibility to GS-5734 (4.5-fold more active). This suggests that GS-5734 is recognized, at least partially, by a functional ExoN, but that the ExoN activity is not sufficient to prevent potent inhibition of CoV replication. The results provide strong evidence that RdRp is the target for remdesivir and support the hypothesis that GS-5734 directly inhibits viral RNA synthesis [28].
The mechanism of inhibition of RdRp of MERS-CoV by remdesivir was studied by Gordon et al. They co-expressed the MERS-CoV nonstructural proteins nsp5, nsp7, nsp8, and nsp12 (RdRp) in insect cells as a part of a polyprotein. Co-expression of the MERS nsp5 protease with nsp7, nsp8 and nsp12 in insect cells yielded a stable complex composed of nsp8 and nsp12. The triphosphate form of the inhibitor (RDV-TP) is utilized as a substrate and competes with its natural counterpart ATP and they observed that incorporation of the nucleotide analogue was significantly more efficient. Once added into the growing RNA chain, the inhibitor does not cause immediate chain termination. The presence of the 3′-hydroxyl group allows the addition of three more nucleotides until RNA synthesis is arrested at position i+3. Therefore, the main possible mechanism of action is delayed RNA chain termination [29]. Recently the same authors obtained almost identical results with SARS-CoV, MERS-CoV, and SARS-CoV-2 RdRps. They provided evidence that all three coronavirus RdRp complexes terminated RNA synthesis at position i+3 [23].
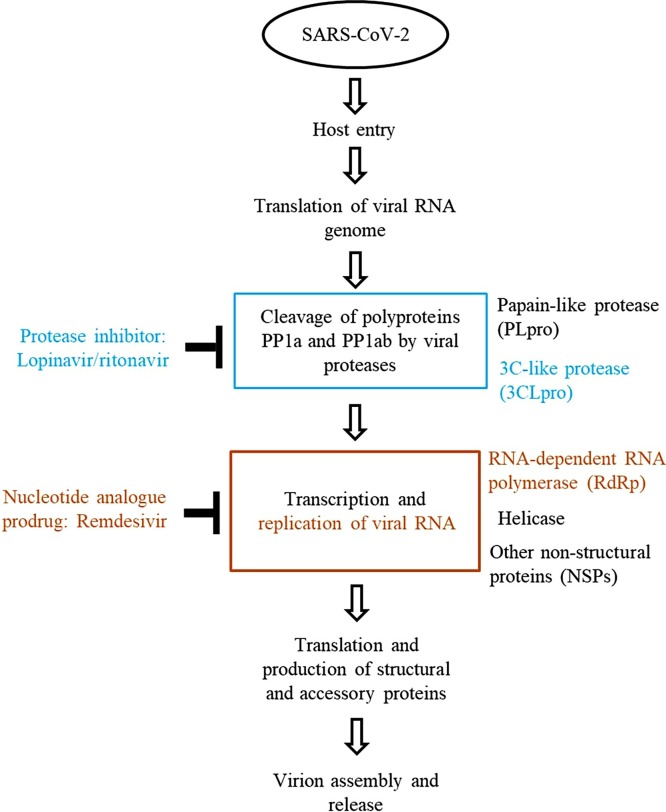
Almost all viruses encode polymerases in the central steps of replication and transcription, therefore, polymerases are becoming the most attractive and suitable targets for antiviral development. There are two major types of polymerase inhibitors: (i) nucleoside and nucleotide substrate analogs and (ii) allosteric inhibitors. Nucleoside analogs are first triphosphated by the host cell to produce the active inhibitor and then act as an inhibitor by competing with the natural nucleoside triphosphates and terminating the growing viral nucleic acids. To date, most of the approved antiviral drugs for anti-HIV therapy utilize this mechanism [9]. Remdesivir is a nucleotide analogue with a proved mechanism of action as an inhibitor of RNA-dependent RNA polymerases. This mechanism is probably involved in an antiviral activity against SARS-CoV-2. Biochemical data provided by Gordon et al. [23,29] suggested a unifying mechanism of inhibition of SARS-CoV, MERS-CoV, and SARS-CoV-2 (Fig. 1 ) and future emerging CoVs may be similarly susceptible to the inhibition by remdesivir [21].
Fig. 1.
Inhibition of viral infection by lopinavir/ritonavir and remdesivir.
2.2. Lopinavir/Ritonavir – protease inhibitor
The proteases encoded by most viruses play a crucial role in the viral life cycle. The protease inhibitors (PIs) bind competitively to the substrate site of the viral protease. This enzyme is responsible for the post translational proteolysis of a polyprotein precursor and the release of functional viral proteins, allowing them to function correctly and individually in replication/transcription and maturation. Inhibition results in the production of immature virus particles [9,30]. Coronavirus proteases, of which there are two in SARS-CoV (a papain-like cysteine proteinase (PLpro, nsp3) and a 3C-like proteinase (3CLpro or Mpro, nsp5)) and three in several other coronaviruses, cleave the ORF-1 polypeptide as it is translated, enabling the formation of the viral replication complex [31]. The substrate-binding pockets are highly conserved among CoV 3CLpro, suggesting the possibility for a wide-spectrum inhibitor design targeting this region in the 3CLpro of all CoVs. It is postulated that the 3CLpro-inhibiting activity of lopinavir/ritonavir contributes at least partially to its anti-CoV effects [32,33]. In silico binding studies of the drugs using the identified crystal structure of Mpro and employing the Hex program to conduct the docking of the ligands to the SARS-CoV main proteinase revealed that lopinavir and ritonavir could basically bind to the active site of SARS main proteinase, but the efficacy of lopinavir/ritonavir was predicted to be poor [34]. According to the latest report of the structure of 3CLpro from SARS-CoV2 (PDB code 6LU7) and the available structure of 3CLpro from SARS-CoV (PDB code 1UK4), the two main proteases differ by only 12 amino acids. Comparing ligand binding free energies for the main proteases has confirmed that good binders for SARS-CoV are, in general, also good binders for SARS-CoV2 3CLpro [35].
Protease inhibitors - a class of drugs best known for success against HIV - block the final step of virion assembly in the treatment of human immunodeficiency virus infection with proven efficacy [36,37]. The combination of lopinavir with ritonavir is widely used as a boosted protease inhibitor in the treatment of HIV infection. Because of low oral bioavailability of lopinavir and its extensive metabolism by the CYP3A4 isoenzyme, lopinavir needs to be coadministered with ritonavir to achieve drug concentrations high enough to inhibit viral replication [27,[38], [39], [40]].
So far, the reported results from studies in different cell lines, animal models, and patients for lopinavir/ritonavir are not so convincing in their inhibition action in human coronaviruses. Screening the library of 348 FDA-approved drugs for anti-MERS-CoV activity in cell culture has identified four compounds (chloroquine, chlorpromazine, loperamide, and lopinavir) which inhibit MERS-CoV replication (50 % effective concentration, EC50 3−8 μmol/L) in vitro. Lopinavir inhibited MERS-CoV and SARS-CoV replication with comparable efficacy (EC50 = 8.0 μM and a maximal protective effect were observed at a dose of 12 μM) [41]. It was previously shown that lopinavir, but not ritonavir, inhibit SARS-CoV chymotrypsin-like (3CL) protease at the concentration of 50 μM [42]. Moreover, it was suggested that lopinavir blocks a postentry step in the MERS-CoV replicative cycle in vitro [41]. The detectable antiviral activities of ribavirin, rimantadine, lopinavir, and baicalin were shown by using the fRhK-4 cell line [43] and in Vero E6 cells infected with SARS-CoV-2 lopinavir inhibit replication with EC50 at 26.1 μM [26].
During the SARS outbreak, treatment with lopinavir in combination with ritonavir, was explored with some success in nonrandomized clinical trials [37,41]. Patients with SARS-CoV treated with lopinavir/ritonavir showed a progressive decrease of viral load and reduction of the composite adverse outcome at day 21 [44]. Recently, the antiviral activity of remdesivir and IFN-β was found to be superior to that of lopinavir/ritonavir-IFN-β against MERS-CoV in vitro and in vivo [27].
The efficacy of lopinavir/ritonavir with or without ribavirin is evaluated in SARS-CoV-2 patients under randomized control trials. Currently, it was demonstrated that this combination has no benefits in adult patients with severe Covid-19 [45]. Although protease inhibitors are a common class of medication used in the treatment of HIV-1 infection their efficacy in human coronavirus infections is not convincing. Moreover, several anti-HIV PIs are also known to influence other intracellular pathways. It was demonstrated that HIV protease inhibitors (indinavir, saquinavir, and lopinavir), independently from any viral infection, can hinder lymphocyte apoptosis by influencing mitochondrial homeostasis. In view of the weak antiviral activity of protease inhibitors, further studies should be done to ascertain whether the clinical benefit could be attributed to their anti-apoptotic rather than their antiviral activity [46,47]. Hence, even if the molecular target of lopinavir/ritonavir is the main protease (3CLpro) in SARS-CoV-2 infected cells (Fig. 1) there are no biochemical and molecular studies confirming the interaction and associating this with clinical efficacy of the protease inhibitor.
2.3. Chloroquine/hydroxychloroquine
Chloroquine (CHQ) was introduced into clinical practice in 1947 as a prophylactic treatment for malaria. Hydroxychloroquine (HCQ) differs from chloroquine by the presence of a hydroxyl group at the end of the side chain: the N-ethyl substituent is β-hydroxylated. Currently, CHQ and its hydroxyl form, HCQ, are used as anti-inflammatory agents for the treatment of rheumatoid arthritis, lupus erythematosus, and amoebic hepatitis [48,49]. In addition, CHQ has been studied as a potent antiviral agent against HIV-1/AIDS [[50], [51], [52]], HCoV-229E [53], SARS-CoV [54,55], influenza A H5N1virus [49], Influenza A and B [56,57] and many other RNA and DNA viruses [58]. Many recent reports and published studies suggested that CHQ and HCQ were associated with reduced progression of the COVID-19 and decreased duration of the symptoms [[59], [60], [61], [62]]. There are, in fact, overall more than 200 trials currently underway around the world on its impact either as a prophylactic or treatment for COVID-19. It is noteworthy that the usefulness of hydroxychloroquine and chloroquine is intensively investigated [63].
Chloroquine was found to exert an antiviral effect during pre- and post-infection conditions suggesting to have both prophylactic and therapeutic advantages. Time-of-addition assay demonstrated that CHQ functioned at both entry and post-entry stages of the SARS-CoV-2 infection in Vero E6 cells [25,64]. However, it did not reduce viral replication in SARS-CoV infected mice [65]. Hydroxychloroquine is significantly more potent than CHQ in vitro (EC50 values: 0.72 and 5.47 μM, respectively) and has a lower potential for drug-drug interactions than chloroquine. Pharmacokinetic models demonstrate that hydroxychloroquine sulfate is significantly superior (5 days in advance) to chloroquine phosphate in inhibiting SARS-CoV-2 in vitro [66,67] and was demonstrated to be much less (∼40 %) toxic than CHQ in animals [68]. On the other hand, data presented by Liu et al. demonstrated that the antiviral effect of HCQ against SARS-CoV-2 infection was comparable with CHQ in vitro (CC50 273.20 μM and 249.50 μM for CHQ and HCQ, respectively). Moreover, they suggested that both CHQ and HCQ blocked the transport of SARS-CoV-2 from early endosomes (EEs) to endolysosomes (ELs) and caused noticeable size/morphological changes in EEs and ELs. They surmised that endosome maturation might be blocked at intermediate stages of endocytosis, resulting in failure of further transport of virions to the ultimate releasing site [64].
Hydroxychloroquine shares the same mechanism of action as chloroquine. Apart from the probable role of CHQ and HCQ as antiviral agents, their mechanisms of action are not fully understood and it was demonstrated that they have multiple effects on mammalian cells [54].
ACE2 is known to be a cell receptor for SARS-CoV [69]. The high similarities of the amino acid sequences and predicted protein structures of the receptor-binding domain of SARS-CoV-2 and SARS-CoV suggest that SARS-CoV-2 may efficiently use human ACE2 as a receptor for cellular entry and employ the cellular serine protease TMPRSS2 for S protein priming [6,7,70]. Zhou et al. confirmed that SARS-CoV-2 used the ACE2 receptor to enter cells and did not use other coronavirus receptors, such as aminopeptidase N (APN) and dipeptidyl peptidase 4 (DPP4) [71]. So the primary mechanism by which cell infection is prevented by these drugs may be at the stage of binding with the surface receptor and endosome-mediated viral entry.
Two independent in vitro studies confirmed that CHQ inhibits the replication of the SARS-CoV. Chloroquine inhibits the early stage of SARS-CoV replication in Vero E6 cells with a 50 % effective concentration of 8.8 ± 1.2 μM/L. The antiviral activity of CHQ was indicative at the time point at virus attachment or penetration [55]. Vincent et al. established that the drug might interfere with terminal glycosylation of the cellular receptor, ACE2. When CHQ was added prior to infection, the impairment of terminal glycosylation of ACE2 may result in reduced binding affinities between ACE2 and SARS-CoV spike protein and negatively influence the initiation of SARS-CoV infection. When CHQ or NH4Cl were added after infection, these agents could rapidly raise the pH and interrupt on-going fusion events between the virus and endosomes, thus inhibiting the infection. On the basis of in vitro experiments they suggested that the primary mechanism by which infection was prevented was the poor affinity of SARS-CoV spike protein to under-glycosylated ACE2 [54].
In vitro studies with HIV infected cells also identified that inhibition of glycosylation might be a possible mechanism of action of CHQ. CHQ inhibits HIV replication at a postintegration stage, resulting in the production of immature virions [72]. It was demonstrated that the sole mechanism explaining the anti-HIV activity of CHQ was a decrease in the infectivity of the newly produced virus associated with defective production of the heavily glycosylated 2G12 epitope of gp120 [73]. According to in vitro results, the antiretroviral effects of CHQ are attributable to the inhibition of viral particle glycosylation. These effects appeared to be specific since the CHQ concentrations effective in vitro neither affected any other step in HIV-1 replication nor were cytotoxic [51,52]. Thus, there is direct evidence that CHQ is an inhibitor of glycosylation of gp120 and these alterations may be responsible for the decreased infectivity of HIV grown in the presence of chloroquine.
When added after the initiation of infection, these drugs might affect the endosome-mediated fusion and subsequent virus replication [54]. SARS-CoV pseudoviruses may enter cells via receptor-dependent, clathrin- and caveolae-independent, pH-sensitive endocytosis, likely through a process involving lipid rafts [74]. A later study, however, suggests that the entry of coronaviruses into the host cells occurs through clathrin-mediated endocytosis. Murine hepatitis virus (MHV), a prototypic member of the CoV family, requires trafficking to lysosomes for proteolytic cleavage at the FP proximal position of its spike (S) protein membrane fusion to occur [75]. Many authors indicated that S protein cleavage is an important step for fusion activity and subsequent internalization of the SARS-CoV virus genome into cells [[76], [77], [78]]. Adding CHQ prior to infection results to inhibition of endosome maturation and strongly decreased MHV infection and fusion, which was not observed when the drug was added at 2 hpi, indicating that the compound mainly affects MHV entry [75].
Chloroquine is a weak base that is known to increase the pH of lysosomal [79] and trans-Golgi network (TGN) vesicles, leading to the dysfunction of enzymes necessary for proteolytic processing and post-translational modifications of newly synthesized viral proteins. Chloroquine is able to prevent the processing of prM protein to M protein in flavivirus-infected mammalian and mosquito cells by raising the pH of the post-Golgi vesicles in which this cleavage occurs. As a result, virions from infected cells, which had been treated with acidotropic amines late in the virus replication cycle contained prM protein rather than M protein and this reduced the infectivity of the virus [80]. The chloroquine-mediated rise in endosomal pH modulates iron metabolism in a variety of cell types. Decreasing in intracellular concentration of iron affects the function of several cellular enzymes involved in pathways leading to the replication of cellular DNA and to the expression of different genes [[81], [82], [83]].
Autophagy is a lysosome-dependent degradative pathway. CHQ and its analogue HCQ are known clinically relevant autophagy inhibitors. CHQ is a weak base that inhibits lysosomal acidification, which prevents the fusion of autophagosomes with lysosomes and subsequent autophagic degradation [79,84,85]. Inhibition of autophagy with CHQ stimulates superoxide generation, ubiquitin-conjugated protein accumulation, and apoptosis in a colon cancer xenograft model [86]. CHQ treatment clearly inhibited autophagy in mouse lung and efficiently ameliorated acute lung injury and dramatically improved the survival rate in mice infected with live avian influenza A H5N1 virus [49]. H5N1 virus-induced autophagic cell death in alveolar epithelial cells through a pathway involving the kinase Akt, the tumor suppressor protein TSC2, and the mammalian target of rapamycin and autophagy-blocking agents might be useful as prophylactics and therapeutics against infection of humans by the H5N1 virus [87]. Furthermore, Prentice et al. suggested that authophagy was induced by the coronavirus mouse hepatitis virus (MHV) and was required for formation of double membrane-bound MHV replication complexes which significantly enhanced the efficiency of replication. Replication of the virus was impaired in ATG5 knockout embryonic stem cells [88]. The same authors also examined the SARS-CoVs and found out similar colocalization of the key viral replication proteins with endogenous LC3, a protein marker for autophagosome [89,90]. It could be assumed that autophagy inhibitors like CHQ could inhibit virus replication. At present, the exact role of autophagy in CoV infection remains debatable, and there is much evidence suggesting that the endocytic pathway plays a key role in mediating viral entry for many CoVs, including SARS-CoVs, MERS-CoVs and possibly SARS-CoV-2 [90].
The anti-inflammatory properties of CHQ/HCQ should also be considered. Several studies have suggested that multiple organ failure observed in fatal cases are most likely associated with not only the direct viral infection and destruction of susceptible cells (e.g., endothelial cells) but also the effects of proinflammatory cytokines, chemokines and other mediators released from infected and activated cells such as monocytes and macrophages [91]. The clinical worsening of individuals with SARS in week 2 is apparently unrelated to uncontrolled SARS coronavirus replication but may be related to immunopathological damage [92]. Another study reveals that the presence of viral elements within endothelial cells and the accumulation of inflammatory cells led to endotheliitis in several organs as a direct consequence of viral involvement and to host inflammatory response [93].
Moreover, CHQ has immunomodulatory effects, suppressing the production/release of tumour necrosis factor-α and interleukin-6, which mediate the inflammatory complications of several viral diseases [83]. Chloroquine/HCQ was reported to inhibit the production of soluble mature TNF in macrophage cell line [94], inhibit TNF-α receptor in human histocytic U-937 cells [95], inhibit TNF-α, IFN-γ and IL-6 in peripheral blood mononuclear cells (PBMC) [96], reduce TNF-α production and lipopolysaccharide (LPS)-induced IL-1β release in human monocytic cells [97]. It is suggested that CHQ exerts anti-inflammatory and immunomodulatory effects predominantly by pretranslational and nonlysosomotropic mechanisms [98]. Chloroquine-induced inhibition of TNF and IL-6 production is not mediated through a lysosomotropic mechanism, and chloroquine probably acts on TNF secretion by disrupting iron homeostasis [99]. Besides its antiviral activity and due to its suppressive effects on the production/release of TNF-α and interleukin 6, CHQ/HCQ may be effectively used in the treatment of viral infections characterized by symptoms associated with inflammatory processes and/or immune-hyperactivation. Anti-inflammatory effects of CHQ remain poorly understood and beside the regulation of pro-inflammatory cytokines, CHQ can also act on the immune system through cell signaling. CHQ inhibits the activation of p38 MAPK in HCoV-229E-infected cells and evokes the activation of ERK independently of infection. These results suggested that CHQ may inhibit CoV replication by suppressing the p38 activation [100]. Additionally, CHQ strongly inhibited phosphorylation of mitogen-activated protein kinase (MAPK) p38, and to a lesser extent c-Jun N-terminal kinase and extracellular signal-regulated kinase ½ [97]. Chloroquine could also inhibit innate immune responses trough downregulation of TLR9 signaling pathways requiring endocytosis and acidification of endosomes within plasmacytoid dendritic cells (pDCs) [101] and act as novel antagonists to chemokine receptor CXCR4 that suppress pancreatic cancer cell proliferation [102]. On the other hand, another hypothesized mechanism of CHQ is via the inhibition of antigen degradation and improving the cross-presentation efficiency of DCs in vitro. in vivo evidence suggested that a short course of treatment with CHQ followed by a booster dose of a soluble antigen immunization can efficiently enhance human CD8 + T cell responses [103] and single vaccination with inactivated influenza virus combined with chloroquine treatment elicits a higher T cell immunity in mice [104].
Regulation of NLRP3 inflammasome activation may offer a promising therapeutic approach by inhibiting or slowing down the process of acute respiratory distress syndrome. HCQ is a known NLRP3 inhibitor, and its potential clinical effectiveness is certainly based on the downregulation of IL‐1β expression [105]. The major pro-inflammatory cytokine Interleukin-1-beta (IL-1β) is elevated in plasma from hospitalized COVID-19 patients and its associated signaling pathway seems to drive SARS-CoV-2 pathogenicity. IL-1β secretion is primarily initiated by inflammasomes. Therefore, inflammasome inhibitors may provide valuable therapeutic tools for COVID-19 patients by preventing hyperinflammatory syndromes [106]. At a severe stage of SARS-CoV-2 infection, CHQ and HCQ treatment may be beneficial to reduce massive cytokine release by their anti-inflammatory and immunomodulatory effects [107].
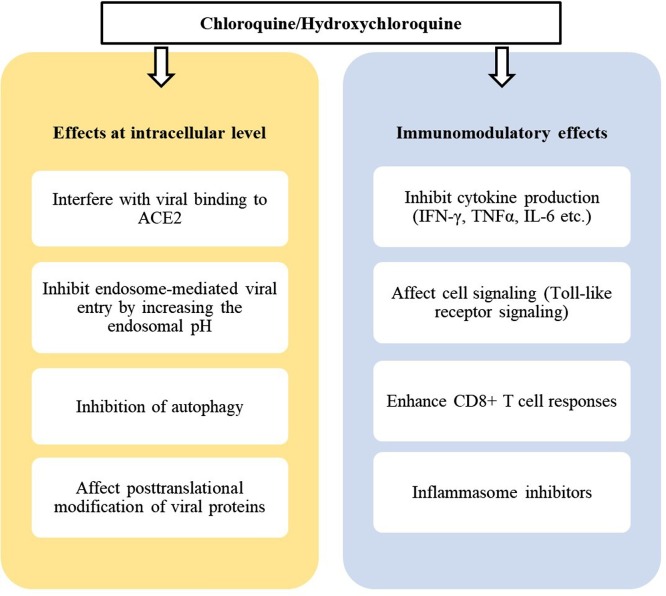
In summary, the multiple molecular mechanisms of CHQ/HCQ action remain under investigation. The exact mode of action of CHQ/HCQ has not yet been identified in SARS-CoV-2 infected patients, and probably multiple pathways could be involved (Fig. 2 ).
Fig. 2.
Multiple effects of chloroquine/hydroxychloroquine.
3. Conclusion
The SARS-CoV-2 is the cause of the coronavirus disease 2019 (COVID-19) that has been declared a global pandemic by the World Health Organization (WHO) in 2020. Despite some clinical characteristics that differentiate COVID-19 from SARS-CoV, MERS-CoV, and seasonal influenza [108] the pathogen SARS-CoV-2 has the same phylogenetic similarity to SARS-CoV and MERS- CoV. Most of the encoded proteins exhibited high sequence identity between SARS-CoV-2 and the related bat-derived coronaviruses (bat-SL-CoVZC45, and bat-SL-CoVZXC21). A notable difference was a longer spike protein encoded by SARS-CoV-2 compared with the bat SARS-like coronaviruses, SARS-CoV, and MERS-CoV. In addition, SARS-CoV-2 was distinct from SARS-CoV in a phylogeny of the complete RNA-dependent RNA polymerase (RdRp) gene [7]. Moreover, the receptor-binding domain of SARS-CoV-2, which directly engages the ACE2 receptor for cell entry, was more closely related to those of SARS-CoVs (73.8–74.9 % amino acid identity) [6,7].
Since the outbreak, researchers have released many agents that could have potential efficacy against COVID-19. There is currently no clinically proven specific antiviral agent available for SARS-CoV-2 infection like SARS-CoV and MERS-CoV. Certain agents like chloroquine, hydroxychloroquine, lopinavir/ritonavir, and remdesivir are being used in ongoing clinical trials all over the world with hopes to further delineate their role in the treatment and prophylaxis of COVID-19. Furthermore, due to their availability and using for decades and proven safety records, it is reasonable to suggest that they may be appropriate for treatment of COVID-19.
Remdesivir, an adenosine analogue with well-studied mechanism of action in CoV infections, can target the RNA-dependent RNA polymerase and block viral RNA synthesis and has been a promising antiviral drug. Antiviral studies in cell culture and animal models, the available human safety data as well as the clear mechanism of action characterize RDV as a direct-acting antiviral.
Since some authors found that lopinavir–ritonavir treatment did not significantly accelerate clinical improvement [45] hence, antiviral effects as an inhibitor of the SARS-CoV main 3CL protease should be further investigated.
Although CHQ and HCQ are well-known drugs for the treatment of patients with rheumatic diseases for many years their exact mechanisms of action are not fully understood. At the molecular level, CHQ and HCQ interfere with lysosomal activity and autophagy, disrupt membrane stability and alter signaling pathways and transcriptional activity. Direct and indirect mechanisms of these drugs inhibit immune activation by reducing Toll-like receptor signaling and cytokine production and enhance CD8 + T cell responses [103,109]. Evidence of in vitro efficacy at a clinically achievable dose and high tissue concentration as well as preliminary clinical evidence of efficacy, suggested that CHQ/HCQ could be used for antiviral treatment. According to two open-label, non-randomized clinical trials conducted by French researchers, hydroxychloroquine in combination with azithromycin holds the potential for reducing viral load and accelerating recovery of COVID-19 patients [61,62]. Recently announced interim results from Solidarity trial suggested that hydroxychloroquine and lopinavir/ritonavir provided no additional benefit compared to standard-of-care. Using the anti-inflammatory capacity of CHQ/HCQ and boosting the specific antiviral immunity at the early stages of the disease through these agents may prevent progression to the severe phase of the disease [107]. Zn supplement can favour COVID-19 treatment using some of these anti-viral drugs because Zn could be stopping SARS-CoV-2 replication in infected cells, if combined with chloroquine as an ionophore and may boosting immune effects of the drugs [110].
Nevertheless, it seems that the mechanism of action of these treatment therapies is the same in human CoVs. Future research would be needed to understand the exact mode of acting of these drugs and their potential to achieve efficient and safe control of the SARS-CoV-2 infection.
Funding
Tchaikapharma High Quality Medicines Inc. funded this review article.
Declaration of Competing Interest
KHU, EPF and VGP are employees of Tchaikapharma High Quality Medicines Inc.
Acknowledgement
We are grateful to Lyudmila Koroteeva for language proofreading of the manuscript.
References
- 1.Schoeman D., Fielding B.C. Coronavirus envelope protein: current knowledge. Virol. J. 2019;16:69. doi: 10.1186/s12985-019-1182-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J. Med. Virol. 2020;92:479–490. doi: 10.1002/jmv.25707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zumla A., Hui D.S., Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. doi: 10.1016/S0140-6736(15)60454-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park S.E. Epidemiology, virology, and clinical features of severe acute respiratory syndrome-coronavirus-2 (SARS-CoV-2; Coronavirus Disease-19) Clin. Exp. Pediatr. 2020;63:119–124. doi: 10.3345/cep.2020.00493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hui D.S. Epidemic and emerging coronaviruses (Severe Acute Respiratory Syndrome and Middle East Respiratory Syndrome) Clin. Chest Med. 2017;38:71–86. doi: 10.1016/j.ccm.2016.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G., Hu Y., Tao Z.W., Tian J.H., Pei Y.Y., Yuan M.L., Zhang Y.L., Dai F.H., Liu Y., Wang Q.M., Zheng J.J., Xu L., Holmes E.C., Zhang Y.Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lu R., Zhao X., Li J., Niu P., Yang B., Wu W., Huang B.H., Zhu N., Bi Y., Ma X., Zhan F., Wang L., Hu T., Zhou H., Hu Z., Zhou W., Zhao L., Chen L., Meng Y., Wang J., Lin Y., Yuan J., Xie Z., Ma J., Liu W.J., Wang D., Xu W., Holmes E.C., Gao G.F., Wu G., Chen W., Shi W., Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan J.F.W., Kok K.H., Zhu Z., Chu H., To K.K.W., Yuan S., Yuen K.Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020;9:221–236. doi: 10.1080/22221751.2020.1719902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lou Z., Sun Y., Rao Z. Current progress in antiviral strategies. Trends Pharmacol. Sci. 2014;35:86–102. doi: 10.1016/j.tips.2013.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.CDC (Centers for Disease Control and Prevention) 2020. About Coronavirus Disease 2019: Prevention and Treatment.https://www.cdc.gov/coronavirus/about/prevention.html Available from: [Google Scholar]
- 11.Thorlund K., Dron L., Park J., Hsu G., Forrest J.I., Mills E. A real-time dashboard of clinical trials for COVID-19. Lancet Digital Health. 2020;2:E286–E287. doi: 10.1016/S2589-7500(20)30086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Van Ierssel S., Dauby N., Bottieau E., et al. 2020. Interim Clinical Guidance for Adults with Suspected or Confirmed COVID-19 in Belgium.https://asprtracie.hhs.gov/technical-resources/resource/8036/interim-clinical-guidance-for-adults-with-suspected-or-confirmed-covid-19-in-bum ver.8. (Added 4/7/2020) [Google Scholar]
- 13.The National Institutes of Health, Coronavirus Disease . 2019. (COVID-19) Treatment Guidelines.https://www.covid19treatmentguidelines.nih.gov Last Updated: May 12, 2020. [PubMed] [Google Scholar]
- 14.Infectious Diseases Society of America . IDSA; 2020. Infectious Diseases Society of America Guidelines on the Treatment and Management of Patients with COVID-19.https://www.idsociety.org/practice-guideline/covid-19-guideline-treatment-and-management/ 4/11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.published by China National Health Commission; 2020. Chinese Clinical Guidance for COVID-19 Pneumonia Diagnosis and Treatment, Version 7.http://kjfy.meetingchina.org/msite/news/show/cn/3337.html on March 4. [Google Scholar]
- 16.Turkish Ministry of Health . 2020. COVID-19 (SARS-CoV-2 Enfeksiyonu) Rehberi, ERİŞKİN HASTATEDAVİSİ, 2 Ağustos.https://covid19bilgi.saglik.gov.tr/tr/covid-19-rehberi.html Ankara. [Google Scholar]
- 17.2020. Lineamientos de manejo hospitalario del paciente con COVID-19 – Sociedad Peruana de Peumología (SPN)https://www.ersnet.org/covid-19-guidelines-and-recommendations-directory Peru. [Google Scholar]
- 18.National Organization of Public Health (EODY) and Hellenic Thoracic Society (HTS); Greece: 2020. Management Algorithm for Patients with Confirmed COVID-19 in a Hospital Setting.https://www.ersnet.org/covid-19-guidelines-and-recommendations-directory [Google Scholar]
- 19.Cho A., Saunders O.L., Butler T., Zhang L., Xu J., Vela L.E., Feng J.Y., Ray A.S., Kim C.U. Synthesis and antiviral activity of a series of 10-substituted4-aza-7,9-dideazaadenosine C-nucleosides. Bioorg. Med. Chem. Lett. 2012;22:2705–2707. doi: 10.1016/j.bmcl.2012.02.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warren T.K., Jordan R., Lo M.K., Ray A.S., Mackman R.L., Soloveva V., et al. Therapeutic efficacy of the small molecule GS-5734 against Ebola virus in rhesus monkeys. Nature. 2016;531:381–385. doi: 10.1038/nature17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown A.J., Won J.J., Graham R.L., Dinnon K.H., III, Sims A.C., Feng J.Y., Cihlar T., Denison M.R., Baric R.S., Sheahan T.P. Broad spectrum antiviral remdesivir inhibits human endemic and zoonotic deltacoronaviruses with a highly divergent RNA dependent RNA polymerase. Antiviral Res. 2019;169 doi: 10.1016/j.antiviral.2019.104541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elfiky A.A. Anti-HCV, nucleotide inhibitors, repurposing against COVID-19. Life Sci. 2020;248 doi: 10.1016/j.lfs.2020.117477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon C.J., Tchesnokov E.P., Woolner E., Perry J.K., Feng J.Y., Porter D.P., Götte M. Remdesivir is a direct-acting antiviral that inhibits RNA-dependent RNA polymerase from severe acute respiratory syndrome coronavirus 2 with high potency. J. Biol. Chem. 2020;295(20):6785–6797. doi: 10.1074/jbc.RA120.013679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sheahan T.P., Sims A.C., Graham R.L., Menachery V.D., Gralinski L.E., Case J.B., Leist S.R., Pyrc K., Feng J.Y., Trantcheva I., Bannister R., Park Y., Babusis D., Clarke M.O., Mackman R.L., Spahn J.E., Palmiotti C.A., Siegel D., Ray A.S., Cihlar T., Jordan R., Denison M.R., Baric R.S. Broad-spectrum antiviral GS-5734 inhibits both epidemic and zoonotic coronaviruses. Sci. Transl. Med. 2017;9:eaal3653. doi: 10.1126/scitranslmed.aal3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir, and chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro. Cell Res. 2020;30:269–271. doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choy K.T., Wong A.Y.L., Kaewpreedee P., Sia S.F., Chen D., Hui K.P.Y., Chu D.K.W., Chan M.C.W., Cheung P.P.H., Huang X., Peiris M., Yen H.L. Remdesivir, lopinavir, emetine, and homoharringtonine inhibit SARS-CoV-2 replication in vitro. Antiviral Res. 2020;178 doi: 10.1016/j.antiviral.2020.104786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheahan T.P., Sims A.C., Leist S., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Clarke M.O., Spahn J.E., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Agostini M.L., Andres E.L., Sims A.C., Graham R.L., Sheahan T.P., Lu X., Smith E.C., Case J.B., Feng J.Y., Jordan R., Ray A.S., Cihlar T., Siegel D., Mackman R.L., Clarke M.O., Baric R.S., Denison M.R. Coronavirus susceptibility to the antiviral remdesivir (GS-5734) is mediated by the viral polymerase and the proofreading exoribonuclease. mBio. 2018;9 doi: 10.1128/mBio.00221-18. e00221-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gordon C.J., Tchesnokov E.P., Feng J.Y., Porter D.P., Götte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J. Biol. Chem. 2020;295:4773–4779. doi: 10.1074/jbc.AC120.013056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weller I.V.D., Williams I.G. ABC of AIDS antiretroviral drugs. BMJ. 2001;322:1410–1412. doi: 10.1136/bmj.322.7299.1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong R.T. Drug targets in severe acute respiratory syndrome (SARS) virus and other coronavirus infections. Infect. Disord. Drug Targets. 2009;9:223–245. doi: 10.2174/187152609787847659. [DOI] [PubMed] [Google Scholar]
- 32.Yang H., Xie W., Xue X., Yang K., Ma J., Liang W., Zhao Q., Zhou Z., Pei D., Ziebuhr J., Hilgenfeld R., Yuen K.Y., Wong L., Gao G., Chen S., Chen Z., Ma D., Bartlam M., Rao Z. Design of wide-spectrum inhibitors targeting coronavirus main proteases. PLoS Biol. 2005;3:e324. doi: 10.1371/journal.pbio.0030324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zumla A., Chan J.F.W., Azhar E.I., Hui D.S.C., Yuen K.Y. Coronaviruses - drug discovery and therapeutic options. Nat. Rev. Drug Discov. 2016;15:327–347. doi: 10.1038/nrd.2015.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang X.W., Yap Y.L. Old drugs as lead compounds for a new disease? Binding analysis of SARS coronavirus main proteinase with HIV, psychotic and parasite drugs. Bioorg. Med. Chem. 2004;12:2517–2521. doi: 10.1016/j.bmc.2004.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.M. Macchiagodena , M. Pagliai , P. Procacci . Inhibition of the Main Protease 3CLPro of the Coronavirus Disease 19 via Structure-Based Ligand Design and Molecular Modeling 18 doi: 10.1016/j.cplett.2020.137489 2020; 137489. [DOI] [PMC free article] [PubMed]
- 36.Vastag B. Old drugs for a new bug: influenza, HIV drugs enlisted to fight SARS. JAMA. 2003;290:1695–1696. doi: 10.1001/jama.290.13.1695. [DOI] [PubMed] [Google Scholar]
- 37.Tai D.Y.H. Pharmacologic treatment of SARS: current knowledge and recommendations. Ann. Acad. Med. Sing. 2007;36:438–443. https://www.ncbi.nlm.nih.gov/pubmed/17597972 [PubMed] [Google Scholar]
- 38.Kaletra . 2020. Highlights of Prescribing Information.https://www.rxabbvie.com/pdf/kaletratabpi.pdf [Google Scholar]
- 39.López Aspiroz E., Santos Buelga D., Cabrera Figueroa S., López Galera R.M., Ribera Pascuet E., Domínguez-Gil Hurlé A., García Sánchez M.J. Population pharmacokinetics of lopinavir/ritonavir (Kaletra) in HIV-infected patients. Ther. Drug Monit. 2011;33:573–582. doi: 10.1097/FTD.0b013e31822d578b. [DOI] [PubMed] [Google Scholar]
- 40.Cooper C.L., van Heeswijk R.P.G., Gallicano K., Cameron D.W. A review of low-dose ritonavir in protease inhibitor combination therapy. Clin. Infect. Dis. 2003;36:1585–1592. doi: 10.1086/375233. [DOI] [PubMed] [Google Scholar]
- 41.de Wilde A.H., Jochmans D., Posthuma C.C., Zevenhoven-Dobbe J.C., van Nieuwkoop S., Bestebroer T.M., van den Hoogen B., Neyts J., Snijder E.J. Screening of an FDA-approved compound library identifies four small-molecule inhibitors of middle east respiratory syndrome coronavirus replication in cell culture. Antimicrob. Agents Chemother. 2014;58:4875–4884. doi: 10.1128/AAC.03011-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu C.Y., Jan J.T., Ma S.H., Kuo C.J., Juan H.F., Cheng Y.S.E., Hsu H.H., Huang H.C., Wu D., Brik A., Liang F.S., Liu R.S., Fang J.M., Chen S.T., Liang P.H., Wong C.H. Small molecules targeting severe acute respiratory syndrome human coronavirus. Proc. Natl. Acad. Sci. U. S. A. 2004;101:10012–10017. doi: 10.1073/pnas.0403596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F., Chan K.H., Jiang Y., Kao R.Y.T., Lu H.T., Fan K.W., Cheng V.C.C., Tsui W.H.W., Hung I.F.N., Lee T.S.W., Guan Y., Peiris J.S.M., Yuen K.Y. In vitro susceptibility of 10 clinical isolates of SARS coronavirus to selected antiviral compounds. J. Clin. Virol. 2004;31:69–75. doi: 10.1016/j.jcv.2004.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chu C.M., Cheng V.C.C., Hung I.F.N., Wong M.M.L., Chan K.S., Kao R.Y.T., Poon L.L.M., Wong C.L.P., Guan Y., Peiris J.S.M., Yuen K.Y., on behalf of the HKU/UCH SARS Study Group Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2004;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G., et al. A Trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matarrese P., Gambardella L., Cassone A., Vella S., Cauda R., Malorni W. Mitochondrial membrane hyperpolarization hijacks activated T lymphocytes toward the apoptotic-prone phenotype: homeostatic mechanisms of HIV protease inhibitors. J. Immunol. 2003;170:6006–6015. doi: 10.4049/jimmunol.170.12.6006. [DOI] [PubMed] [Google Scholar]
- 47.Cheng V.C.C., Chan J.F.W., To K.K.W., Yuen K.Y. Clinical management and infection control of SARS: lessons learned. Antiviral Res. 2013;100:407–419. doi: 10.1016/j.antiviral.2013.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Solomon V.R., Lee H. Chloroquine and its analogs: a new promise of an old drug for effective and safe cancer therapies. Eur. J. Pharmacol. 2009;625:220–233. doi: 10.1016/j.ejphar.2009.06.063. [DOI] [PubMed] [Google Scholar]
- 49.Yan Y., Zou Z., Sun Y., Li X., Xu K.F., Wei Y., Jin N., Jiang C. Anti-malaria drug chloroquine is highly effective in treating avian influenza A H5N1 virus infection in an animal model. Cell Res. 2013;23:300–302. doi: 10.1038/cr.2012.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Boelaert J.R., Piette J., Sperber K. The potential place of chloroquine in the treatment of HIV-1-infected patients. J. Clin. Virol. 2001;20:137–140. doi: 10.1016/s1386-6532(00)00140-2. [DOI] [PubMed] [Google Scholar]
- 51.Savarino A., Lucia M.B., Rastrelli E., Rutella S., Golotta C., Morra E., Tamburrini E., Perno C.F., Boelaert J.R., Sperber K., Cauda R. Anti-HIV effects of chloroquine inhibition of viral particle glycosylation and synergism with protease inhibitors. J. Acquir. Immune Defic. Syndr. 2004;35:223–232. doi: 10.1097/00126334-200403010-00002. [DOI] [PubMed] [Google Scholar]
- 52.Savarino A., Di Trani L., Donatelli I., Cauda R., Cassone A. New insights into the antiviral effects of chloroquine. Lancet Infect. Dis. 2006;6:67–69. doi: 10.1016/S1473-3099(06)70361-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Blau D., Holmes K. Human Coronavirus HCoV-229E enters susceptible cells via the endocytic pathway. Adv. Exp. Med. Biol. 2001;494:193–198. doi: 10.1007/978-1-4615-1325-4_31. [DOI] [PubMed] [Google Scholar]
- 54.Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:69. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keyaerts E., Vijgen L., Maes P., Neyts J., Van Ranst M. In vitro inhibition of severe acute respiratory syndrome coronavirus by chloroquine. Biochem. Biophys. Res. Commun. 2004;323:264–268. doi: 10.1016/j.bbrc.2004.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shibata M., Aoki H., Tsurumi T., Sugiura Y., Nishiyama Y., Suzuki S., Maeno K. Mechanism of uncoating of influenza B virus in MDCK cells: action of chloroquine. J. Gen. Virol. 1983;64:1149–1156. doi: 10.1099/0022-1317-64-5-1149. [DOI] [PubMed] [Google Scholar]
- 57.Ooi E.E., Chew J.S., Loh J.P., Chua R.C. In vitro inhibition of human influenza A virus replication by chloroquine. Virol. J. 2006;3:39. doi: 10.1186/1743-422X-3-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Devaux C.A., Rolain J.M., Colson P., Raoult D. New insights on the antiviral effects of chloroquine against coronavirus: what to expect for COVID-19? Int. J. Antimicrob. Agents. 2020;12 doi: 10.1016/j.ijantimicag.2020.105938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao J., Tian Z., Yang X. Breakthrough: Chloroquine phosphate has shown apparent efficacy in treatment of COVID-19 associated pneumonia in clinical studies. Biosci. Trends. 2020;14:72–73. doi: 10.5582/bst.2020.01047. [DOI] [PubMed] [Google Scholar]
- 60.Chen J., Liu D., Liu L., Liu P., Xu Q., Xia L., Ling Y., Huang D., Song S., Zhang D., Qian Z., Li T., Shen Y., Lu H. A pilot study of hydroxychloroquine in treatment of patients with common coronavirus disease-19 (COVID-19) J. Zhejiang Univ. (Med. Sci.) 2020;49:215–219. doi: 10.3785/j.issn.1008-9292.2020.03.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gautret P., Lagier J.G., Parola P., Hoang V.T., Meddeb L., Mailhe M., Doudier B., Courjon J., Giordanengo V., Vieira V.E., Dupont H.T., Honore S., Colson P., Chabriere E., La Scola B., Rolain J., Brouqui P., Raoult D. Hydroxychloroquine and azithromycin as a treatment of COVID-19: results of an open-label non-randomized clinical trial. Int. J. Antimicrob. Agents. 2020;20 doi: 10.1016/j.ijantimicag.2020.105949. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 62.Gautret P., Lagier J.G., Parola P., Hoang V.T., Meddeb L., Sevestre J., Mailhe M., Doudier B., Aubry C., Amrane S., Seng P., Hocquart M., Eldin C., Finance J., Vieira V.E., Dupont H.T., Honore S., Stein A., Million M., Colson P., Chabriere E., La Scola B., Veit V., Jacquier A., Deharo J.C., Drancourt M., Fournier P.E., Rolain J.M., Brouqui P., Raoult D. Clinical and microbiological effect of a combination of hydroxychloroquine and azithromycinin 80 COVID-19 patients with at least a six-day follow up: a pilot observational study. Travel Med. Infect. Dis. 2020;34 doi: 10.1016/j.tmaid.2020.101663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Eynde J.J.V. COVID-19: An update about the Discovery clinical trial. Pharmaceuticals. 2020;13:98. doi: 10.3390/ph13050098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Liu J., Cao R., Xu M., Wang X., Zhang H., Hu H., Li Y., Hu Z., Zhong W., Wang M. Hydroxychloroquine, a less toxic derivative of chloroquine, is effective in inhibiting SARS-CoV-2 infection in vitro. Cell Discov. 2020;6:16. doi: 10.1038/s41421-020-0156-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barnard D.L., Day C.W., Bailey K., Heiner M., Montgomery R., Lauridsen L., Chan P.K.S., Sidwell R.W. Evaluation of immunomodulators, interferons and known in vitro SARS-CoV inhibitors for inhibition of SARS-CoV replication in BALB/c mice. Antivir. Chem. Chemother. 2006;17:275–284. doi: 10.1177/095632020601700505. [DOI] [PubMed] [Google Scholar]
- 66.Yao X., Ye F., Zhang M., Cui C., Huang B., Niu P., Liu X., Zhao L., Dong E., Song C., Zhan S., Lu R., Li H., Tan W., Liu D. In Vitro antiviral activity and projection of optimized dosing design of hydroxychloroquine for the treatment of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) Clin. Infect. Dis. 2020;9(March):ciaa237. doi: 10.1093/cid/ciaa237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jean S.S., Lee P.I., Hsueh P.R. Treatment options for COVID-19: the reality and challenges. J. Microbiol. Immunol. Infect. 2020;53(3):436–443. doi: 10.1016/j.jmii.2020.03.034. S1684-1182(20)30094-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McChesney E.W. Animal toxicity and pharmacokinetics of hydroxychloroquine sulfate. Am. J. Med. 1983;75:11–18. doi: 10.1016/0002-9343(83)91265-2. [DOI] [PubMed] [Google Scholar]
- 69.Li W., Moor M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Choe H., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Nature. 2003;426:450–454. doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.H., Nitsche A., Müller M.A., Drosten C., Pöhlmann S. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai W.P., Nara P.L., Kung H.F., Oroszlan S. Inhibition of human immunodeficiency virus infectivity by chloroquine. AIDS Res. Hum. Retroviruses. 1990;6:481–489. doi: 10.1089/aid.1990.6.481. [DOI] [PubMed] [Google Scholar]
- 73.Savarino A., Gennero L., Chen H.C., Serrano D., Malavasi F., Boelaert J.R., Sperber K. Anti-HIV effects of chloroquine: mechanisms of inhibition and spectrum of activity. AIDS. 2001;15:2221–2229. doi: 10.1097/00002030-200111230-00002. [DOI] [PubMed] [Google Scholar]
- 74.Wang H., Yang P., Liu K., Guo F., Zhang Y., Zhang G., Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Burkard C., Verheije M.H., Wicht O., van Kasteren S.I., van Kuppeveld F.J., Haagmans B.L., Pelkmans L., Rottier P.J.M., Bosch B.J., de Haan C.A.M. Coronavirus cell entry occurs through the endo-lysosomal pathway in a proteolysis-dependent manner. PLoS Pathog. 2014;10 doi: 10.1371/journal.ppat.1004502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Watanabe R., Matsuyama S., Shirato K., Maejima M., Fukushi S., Morikawa S., Taguchi F. Entry from the cell surface of severe acute respiratory syndrome coronavirus with cleaved S protein as revealed by pseudotype virus bearing cleaved S protein. J. Virol. 2008;82:11985–11991. doi: 10.1128/JVI.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kam Y.W., Okumura Y., Kido H., Ng L.F.P., Bruzzone R., Altmeyer R. Cleavage of the SARS coronavirus spike glycoprotein by airway proteases enhances virus entry into human bronchial epithelial cells in vitro. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Belouzard S., Chu V.C., Whittaker G.R. Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc. Natl. Acad. Sci. U. S. A. 2009;106:5871–5876. doi: 10.1073/pnas.0809524106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Poole B., Ohkuma S. Effect of weak bases on the intralysosomal pH in mouse peritoneal macrophages. J. Cell Biol. 1981;90:665–669. doi: 10.1083/jcb.90.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Randolph V.B., Winkler G., Stollar V. Acidotropic amines inhibit proteolytic processing of flavivirus prM protein. Virology. 1990;174:450–458. doi: 10.1016/0042-6822(90)90099-D. [DOI] [PubMed] [Google Scholar]
- 81.Byrd T.F., Horwitz M.A. Chloroquine inhibits the intracellular multiplication of Legionella pneumophila by limiting the availability of iron a potential new mechanism for the therapeutic effect of chloroquine against intracellular pathogens. J. Clin. Invest. 1991;88:351–357. doi: 10.1172/JCI115301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Legssyer R., Josse C., Piette J., Ward R.J., Crichton R.R. Changes in function of iron-loaded alveolar macrophages after in vivo administration of desferrioxamine and/or chloroquine. J. Inorg. Biochem. 2003;94:36–42. doi: 10.1016/s0162-0134(02)00633-5. [DOI] [PubMed] [Google Scholar]
- 83.Savarino A., Boelaert J.R., Cassone A., Majori G., Cauda R. Effects of chloroquine on viral infections: an old drug against today’s diseases? Lancet Infect. Dis. 2003;3:722–727. doi: 10.1016/S1473-3099(03)00806-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Carew J.S., Espitia C.M., Esquivel J.A., Mahalingam D., Kelly K.R., Reddy G., Giles F., Nawrocki S.T. Lucanthone is a novel inhibitor of autophagy that induces cathepsin D-mediated apoptosis. J. Biol. Chem. 2011;286:6602–6613. doi: 10.1074/jbc.M110.151324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Glaumann H., Ahlberg J. Comparison of different autophagic vacuoles with regard to ultrastructure, enzymatic composition, and degradation capacity-formation of crinosomes. Exp. Mol. Pathol. 1987;41:346–362. doi: 10.1016/0014-4800(87)90018-9. [DOI] [PubMed] [Google Scholar]
- 86.Carew J.S., Medina E.C., Esquivel J.A., Mahalingam D., Swords R., Kelly K., Zhang H., Huang P., Mita A.C., Mita M.M., Giles F.J., Nawrocki S.T. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J. Cell. Mol. Med. 2010;14:2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sun Y., Li C., Shu Y., Ju X., Zou Z., Wang H., et al. Inhibition of autophagy ameliorates acute lung injury caused by avian influenza A H5N1 infection. Sci. Signal. 2012;5:ra16. doi: 10.1126/scisignal.2001931. [DOI] [PubMed] [Google Scholar]
- 88.Prentice E., Jerome W.G., Yoshimori T., Mizushima N., Denson M.R. Coronavirus replication complex formation utilizes components of cellular autophagy. J. Biol. Chem. 2004;279:10136–10141. doi: 10.1074/jbc.M306124200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Prentice E., McAuliffe J., Lu X., Subbarao K., Denison M.R. Identification and characterization of severe acute respiratory syndrome coronavirus replicase proteins. J. Virol. 2004;78:9977–9986. doi: 10.1128/JVI.78.18.9977-9986.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yang N., Shen H.M. Targeting the endocytic pathway and autophagy process as a novel therapeutic strategy in COVID-19. Int. J. Biol. Sci. 2020;16:1724–1731. doi: 10.7150/ijbs.45498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Al-Bari A.A. Targeting endosomal acidification by chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5 doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F.N., Poon L.L.M., Law K.I., Tang B.S.F., Hon T.Y.W., Chan C.S., Chan K.H., Ng J.S.C., Zheng B.J., Ng W.L., Lai R.W.M., Guan Y., Yuen K.Y., members of the HKU/UCH SARS Study Group Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Varga Z., Flammer A.J., Steiger P., Haberecker M., Andermatt R., Zinkernagel A.S., Mehra M.R., Schuepbach R.A., Ruschitzka F., Moch H. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:P1417–P1418. doi: 10.1016/S0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jeong J.Y., Jue D.M. Chloroquine inhibits processing of tumor necrosis factor in lipopolysaccharide-stimulated RAW 264.7 macrophages. J. Immunol. 1997;158:4901–4907. https://www.jimmunol.org/content/158/10/4901 [PubMed] [Google Scholar]
- 95.Jeong J.Y., Choi J.W., Jeon K.I., Jue D.M. Chloroquine decreases cell-surface expression of tumor necrosis factor receptors in human histocytic U-937 cells. Immunology. 2002;105 doi: 10.1046/j.0019-2805.2001.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van den Borne B.E., Dijkmans B.A., de Rooij H.H., le Cessie S., Verweij C.L. Chloroquine and hydroxychloroquine equally affect tumor necrosis factor-alpha, interleukin 6, and interferon-gamma production by peripheral blood mononuclear cells. J. Rheumatol. 1997;24:55–60. https://www.ncbi.nlm.nih.gov/pubmed/9002011 [PubMed] [Google Scholar]
- 97.Seitz M., Valbracht J., Quach J., Lotz M. Gold sodium thiomalate and chloroquine inhibit cytokine production in monocytic THP-1 cells through distinct transcriptional and posttranslational mechanisms. J. Clin. Immunol. 2003;23:477–484. doi: 10.1023/B:JOCI.0000010424.41475.17. [DOI] [PubMed] [Google Scholar]
- 98.Weber S.M., Levitz S.M. Chloroquine interferes with lipopolysaccharide-induced TNF-a gene expression by a nonlysosomotropic mechanism. J. Immunol. 2000;165:1534–1540. doi: 10.4049/jimmunol.165.3.1534. [DOI] [PubMed] [Google Scholar]
- 99.Picot S., Peyron F., Donadille A., Vuillez J.P., Barbe G., Ambroise-Thomas P. Chloroquine-induced inhibition of the production of TNF, but not of IL-6, is affected by disruption of iron metabolism. Immunology. 1990;80:127–133. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1422100/ [PMC free article] [PubMed] [Google Scholar]
- 100.Kono M., Tatsumi K., Imai A.M., Saito K., Kuriyama T., Shirasawa H. Inhibition of human coronavirus 229E infection in human epithelial lung cells (L132) by chloroquine: involvement of p38 MAPK and ERK. Antiviral Res. 2008;77:150–152. doi: 10.1016/j.antiviral.2007.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lund J., Sato A., Akira S., Medzhitov R., Iwasaki A. Toll-like receptor 9–mediated recognition of Herpes simplex virus-2 by plasmacytoid dendritic cells. J. Exp. Med. 2003;198:513–520. doi: 10.1084/jem.20030162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kim J., Yip M.L.R., Shen X., Li H., Hsin L.Y.C., Labarge S., Heinfich E.L., Lee W., Lu J., Vaidehi N. Identification of anti-malarial compounds as novel antagonists to chemokine receptor CXCR4 in pancreatic cancer cells. PLoS One. 2012;7 doi: 10.1371/journal.pone.0031004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Accapezzato D., Visco V., Francavilla V., Molette C., Donato T., Paroli M., Mondelli M.U., Doria C., Torrisi M.R., Barnaba V. Chloroquine enhances human CD8+ T cell responses against soluble antigens in vivo. JEM. 2005;202:817–828. doi: 10.1084/jem.20051106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Garulli B., Di Mario G., Sciaraffia E., Accapezzato D., Barnaba V., Castrucci M.R. Enhancement of T cell-mediated immune responses to whole inactivated influenza virus by chloroquine treatment in vivo. Vaccine. 2013;31:1717–1724. doi: 10.1016/j.vaccine.2013.01.037. [DOI] [PubMed] [Google Scholar]
- 105.Lucchesi A., Silimbani P., Musuraca G., Cerchione C., Martinelli G., di Carlo P., Napolitano M. Clinical and biological data on the use of hydroxychloroquine against SARS‐CoV‐2 could support the role of the NLRP3 inflammasome in the pathogenesis of respiratory disease. J. Med. Virol. 2020;24 doi: 10.1002/jmv.26217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Theobald S.J., Simonis A., Kreer C., Zehner M., Fischer J., Albert M.-C., Malin J.J., Gräb J., Winter S., de Silva U.S., Bӧll B., Kӧhler P., Gruell H., Suàrez I., Hallek M., Fätkenheuer G., Jung N., Cornely O., Lehmann C., Kashkar H., Klein F., Rybniker J. 2020. The SARS-CoV-2 Spike Protein Primes Inflammasome-mediated interleukin-1-beta Secretion in COVID-19 Patient-derived Macrophages. [DOI] [Google Scholar]
- 107.Li X., Wang Y., Agostinis P., Rabson A., Melino G., Carafoli E., Shi Y., Sun E. Is hydroxychloroquine beneficial for COVID-19 patients? Cell Death Dis. 2020;11:512. doi: 10.1038/s41419-020-2721-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., et al. Clinical characteristics of coronavirus disease 2019 in China. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schrenmeier E., Dӧrner T. Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 2020;16:155–166. doi: 10.1038/s41584-020-0372-x. [DOI] [PubMed] [Google Scholar]
- 110.Rahman M.T., Idid S.Z. Can Zn be a critical element in COVID-19 treatment? Biol. Trace Elem. Res. 2020;26(May):1–9. doi: 10.1007/s12011-020-02194-9. [DOI] [PMC free article] [PubMed] [Google Scholar]