Abstract
Transgene insertion patterns are critical for the analysis of transgenic animals because the influence of transgenes may change depending on the insertion pattern (such as copy numbers and orientations of concatenations) and the insertion position in the genome. We previously reported a genomic walking strategy to locate transgenes in the genomes of transgenic mice (Exp. Anim. 53: 103–111, 2004) and to analyze transgene insertion patterns (Exp. Anim. 55: 65–69, 2006). With such strategies, however, we could not determine the copy number of transgenes or global genome modification induced by transgene insertion due to read-length limitation. In this study, we used a long-read sequencer (MinION, Oxford Nanopore Technologies) to overcome this limitation. We obtained 922,210 reads using MinION with genomic DNA from a transgenic mouse strain (4C30, Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 87: 550–562, 2011). Among the reads, we found one 21,457-bp read containing the transgene using a local BLAST search. Nucleotide dot plot analysis revealed that the transgene was inserted in the genome as a tandem concatemer with an almost entire construct (15–3,508 of 3,508 bp) and a partial fragment (4–660, 657 bp). Ensembl’s BLAST search against the C57BL/6N genome revealed a 9,388-bp deletion at the insertion position in the intron of the Sgcd gene, confirming that mutations such as a large genomic deletion could occur at the time of transgene insertion. Thus, long-read sequencers are useful tools for the analysis of transgene insertion patterns.
Keywords: long-read sequencer, mice, nanopore, transgene insertion pattern
Introduction
To analyze transgenic animals as research models in a wide variety of fields, we need to consider how transgenes are integrated into the host genome. This is because the influence of transgenes may change depending on the insertion pattern (such as copy numbers and orientations of each concatenated transgene) and the insertion position in the genome [15, 21, 31, 40]. However, it is challenging to determine transgene insertion patterns because transgene insertion is random and unpredictable, and the inserted transgenes, which often form a multiple tandem concatemer [3], are long (kilobases or longer) in most cases.
To date, transgenes have been mapped in host genomes mainly by in situ hybridization [36] and/or by determining genome sequences flanking the transgene; e.g., genomic walking [35, 38], inverse PCR [25], microarray hybrid capture [10], and targeted locus amplification [4, 15, 21]. We previously reported a genomic walking strategy to locate transgenes in the genomes of transgenic mice [30] and to analyze insertion patterns of transgenes [38]. However, transgene insertion patterns, especially copy numbers, often cannot be determined by the methods described above because these methods have a length limit of sequences that can be analyzed and a low capability to distinguish sequence repeats and their orientations. The copy numbers of transgenes are often determined by Southern blotting [1], quantitative PCR [11, 20, 37], and, more recently, digital PCR [7]. These methods, however, are not useful for determining the detailed structure of transgenes, such as the orientation and precise length of each copy of an integrated transgene construct. To determine such global structures, we need a method that can determine very long sequences, since large-scale rearrangements of both transgenes and host genomes often occur at transgene insertion sites [15].
To overcome this difficulty in the present study, we used a long-read sequencer, which has developed considerably in recent years, for analysis of transgene integration. Among the currently available long-read sequencers, we chose a MinION [8, 18] (Oxford Nanopore Technologies, Oxford Science Park, UK; ONT) because we can start using the sequencer with a comparatively low initial cost (~$1,500) and the sequencer can theoretically read the entire length of a given DNA (e.g., >30 kb or more). Nicholls et al. [28] reported a similar study using the MinION with post-sequence assembly by minimap2 [23]. In the present study, we used a local BLAST search to identify DNA fragments containing transgenes without a sequence assembly, and then confirmed precise sequences of detected transgenes and their flanking genome by additional PCR-based analysis and Sanger sequencing. This is because a local BLAST search is a practical method that does not require much more calculation power than the combination of whole-genome assembly and subsequent homology search. Specifically, we analyzed the transgene insertion pattern in the 4C30 mouse strain, our transgenic model of dilated cardiomyopathy (DCM) harboring pCAGGS-based transgenes [39], with the MinION and compared the results with those obtained by the previous genomic walking method [30]. Our preliminary experiments with genomic walking [17] and quantitative PCR (data not shown) suggested that the 4C30 strain had only one copy of the transgene in its genome. Therefore, we tried to find one insertion site by MinION. We also discuss the transgene insertion pattern as well as changes in the genome at the time of transgene insertion.
Materials and Methods
Transgenic mouse strain
The present study used the 4C30 mouse strain raised in-house and homozygous for transgenes containing ST3GalII cDNA (Fig. 1A) [39]. The strain had been maintained for more than 15 years by confirmation of the transgene transmission by a flanking primer method [17] based on our previous study [30]. All animal experiments including tissue sample collection were conducted in accordance with the guidelines for animal experiments of the National Institutes of Biomedical Innovation, Health, and Nutrition (Osaka, Japan).
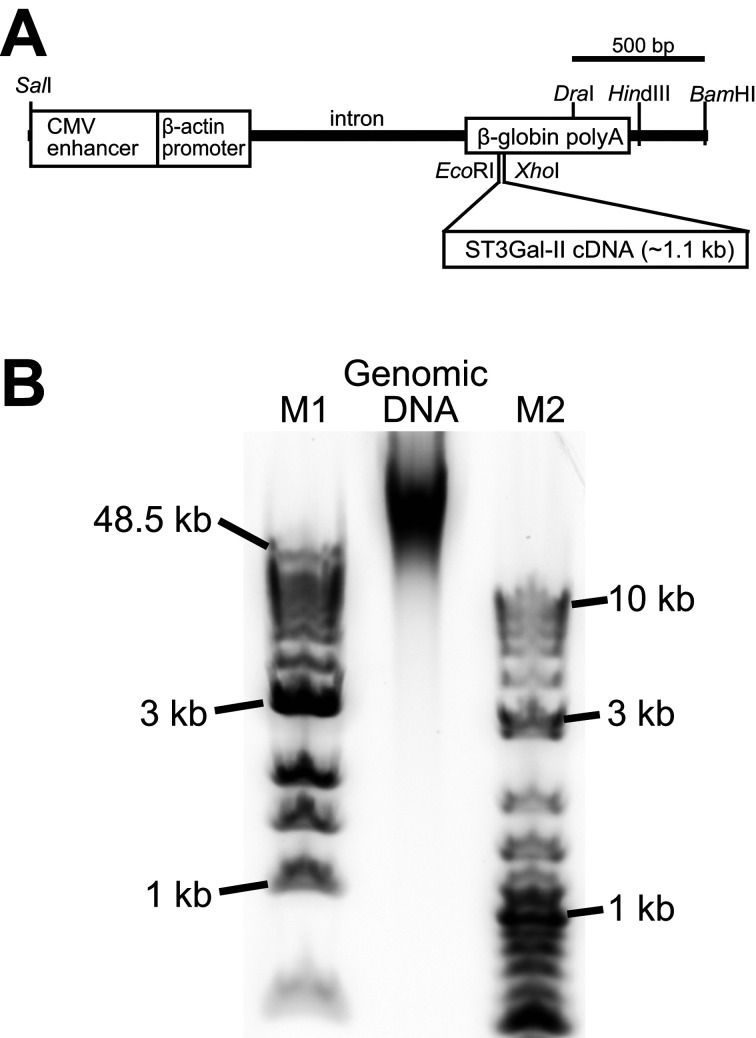
Fig. 1.
Transgene structure and DNA size of a genomic DNA sample. (A) Transgene structure used for production of 4C30 mice (accession number: LC495729) [39]. The transgene (3,508 bp) was a SalI-BamHI fragment derived from the pCAGGS vector containing cytomegalovirus enhancer, chicken β-actin promoter, and rabbit β-globin polyadenylation signal sequences [29]. The cDNA of mouse ST3 β-galactoside α-2,3-sialyltransferase 2 (ST3GalII, a.k.a., St3gal2 and Siat5) [22], a mouse sialyltransferase, was inserted into EcoRI-XhoI sites in the β-globin polyadenylation signal sequence. (B) DNA size distribution of a genomic DNA extracted from a 4C30 mouse kidney. Note that most genomic DNAs in this sample were longer than 50 kbp. The genomic DNA sample (~0.4 µg) and molecular markers (~0.5 µg) were separated with 1% agarose gel electrophoresis (E-gel EX 1%, Thermo Fisher Scientific). The electropherogram was recorded using a laser scanner (FX-Pro, Bio-rad). M1: 1 kb extended DNA ladder (NEB), M2: 2-log DNA ladder (NEB).
Long-read sequencing by a nanopore sequencer (MinION)
Genomic DNA was extracted from the kidney of a 4C30 mouse using smart DNA prep (m) (Analytik Jena AG, Jena, Germany) according to the manufacturer’s instructions. DNA concentration was measured using a Qubit 2 fluorometer and Qubit dsDNA BR Assay kit (Invitrogen, Carlsbad, CA, USA). Length distribution of the genomic DNA was confirmed by agarose gel electrophoresis (Fig. 1B). A library for nanopore sequencing was prepared with 6 µg of genomic DNA using a Rapid sequencing kit (SQK-RAD004, ONT) and then applied to a MinION equipped with a flow-cell (FLO-MIN106D, ONT). NinKNOW software (Version 3.3.16.0, ONT) was used for driving 24-h sequencing runs and following base-calls.
Local BLAST search, dot plot analysis, and mapping on the C57BL/6N genome
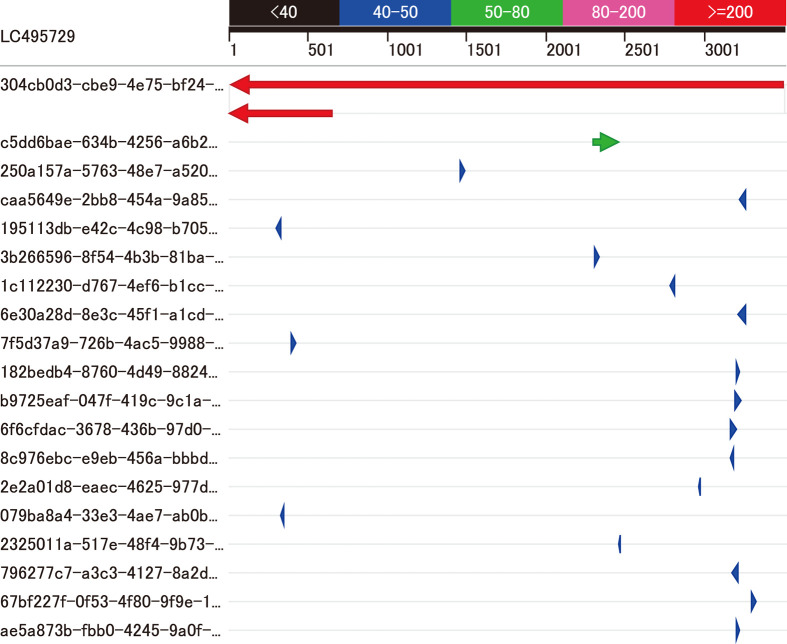
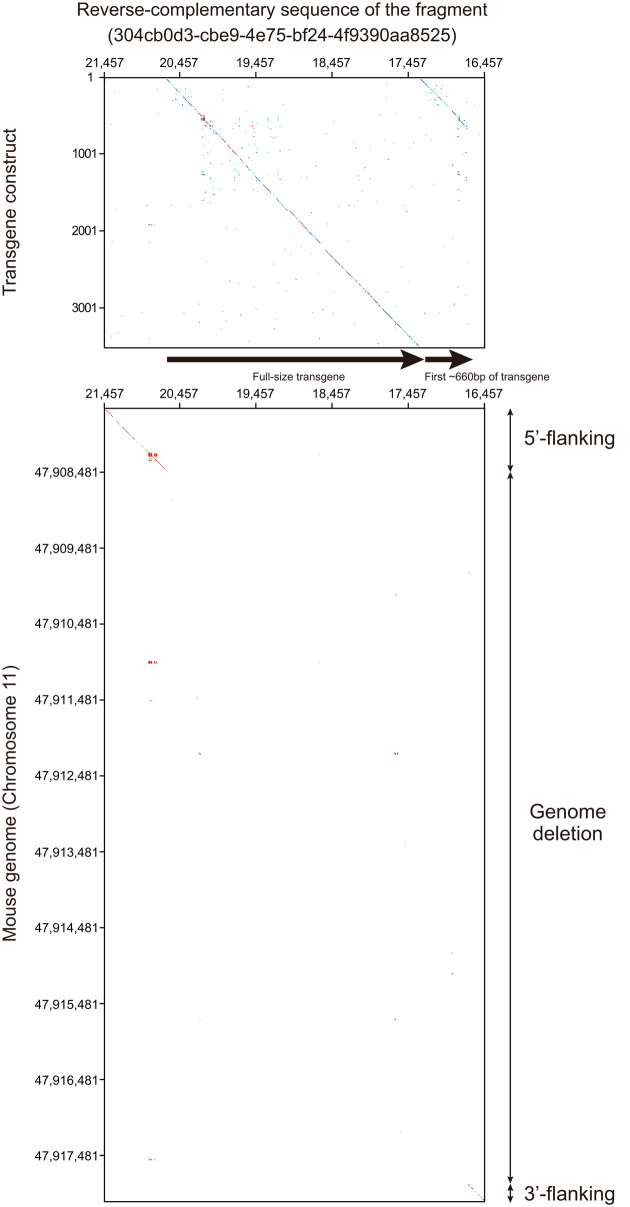
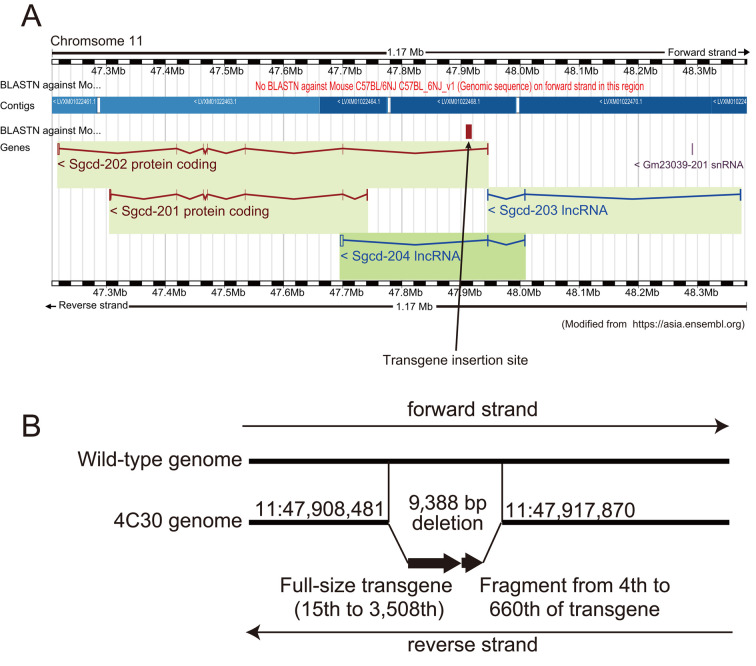
In the reads obtained by the MinION (Table 1), we identified one fragment containing the transgene sequence using a local BLAST search (Blast +) [6] incorporated in a genetic information analysis software (GENETYX Version 14, GENETYX Corp., Tokyo, Japan) with default settings (Table 2 and Fig. 2). Then, we obtained a mouse genomic sequence containing flanking sequences of the transgene using Ensembl’s BLAST search (http://asia.ensembl.org/Mus_musculus/Tools/Blast) against the C57BL/6NJ genome database (assembly: C57BL_6NJ_v1) Then, we determined the broad-area structure by nucleotide dot plot analysis [12] of the transgene and mouse genome sequences against the fragment using GENETYX HarrPlot 3.1.1 program with a Nucleotide Huge Plot method (Fig. 3). In addition, we confirm the genomic position of the transgene insertion by Ensembl’s BLAST search using the sequence flanking the transgene in the mouse genome (Fig. 4A).
Table 1. Read length distribution.
| Read length (kb) | Reads |
|---|---|
| 0 to 20 | 904,588 |
| 20 to 40 | 14,936 |
| 40 to 60 | 2,013 |
| 60 to 80 | 461 |
| 80 to 100 | 133 |
| 100 to 120 | 56 |
| 120 to 140 | 12 |
| 140 to 160 | 5 |
| 160 to 180 | 2 |
| 180 to 200 | 1 |
| 200 to 220 | 2 |
| 220 to 240 | 1 |
| Total reads | 922,210 |
This read length distribution was summarized from the sequence summary information produced by MinKNOW software driving MinION.
Table 2. Results of a local Blast search for the transgene sequence against all reads obtained by MinION.
| ID | Sequence ID | Expect | Score (Bits) |
Identity (%) |
Total Score |
Query Coverage (%) |
Max Identity (%) |
Start | End |
|---|---|---|---|---|---|---|---|---|---|
| #1 | 304cb0d3-cbe9-4e75-bf24-4f9390aa8525 | 0 | 3,458 | 84 | 4,189 | 99 | 86 | 15 | 3,507 |
| 0 | 731 | 86 | 9 | 660 | |||||
| #2 | c5dd6bae-634b-4256-a6b2-25dd05c3c733 | 4.00E-06 | 61 | 69 | 61 | 4 | 69 | 2,301 | 2,470 |
| #3 | 250a157a-5763-48e7-a520-3e7a6f070eb4 | 0.007 | 50 | 89 | 50 | 1 | 89 | 1,460 | 1,496 |
| #4 | caa5649e-2bb8-454a-9a85-83c5f1440288 | 0.025 | 48 | 83 | 48 | 1 | 83 | 3,217 | 3,264 |
| #5 | 195113db-e42c-4c98-b705-a4445236ee84 | 0.088 | 46 | 89 | 46 | 1 | 89 | 301 | 336 |
| #6 | 3b266596-8f54-4b3b-81ba-d988673b44c8 | 0.31 | 45 | 83 | 45 | 1 | 83 | 2,304 | 2,345 |
| #7 | 1c112230-d767-4ef6-b1cc-e4da6f79e35f | 1.1 | 43 | 87 | 43 | 0 | 87 | 2,785 | 2,817 |
| #8 | 6e30a28d-8e3c-45f1-a1cd-1382e9598b2e | 1.1 | 43 | 75 | 43 | 1 | 75 | 3,205 | 3,262 |
| #9 | 7f5d37a9-726b-4ac5-9988-ab1b0b46dda3 | 1.1 | 43 | 86 | 43 | 1 | 86 | 396 | 433 |
| #10 | 182bedb4-8760-4d49-8824-d333dae0488d | 1.1 | 43 | 96 | 43 | 0 | 96 | 3,200 | 3,225 |
| #11 | b9725eaf-047f-419c-9c1a-66eef58ead55 | 3.8 | 41 | 81 | 41 | 1 | 81 | 3,189 | 3,236 |
| #12 | 6f6cfdac-3678-436b-97d0-a88af6d2d49b | 3.8 | 41 | 79 | 41 | 1 | 79 | 3,161 | 3,209 |
| #13 | 8c976ebc-e9eb-456a-bbbd-d05d5f1fd751 | 3.8 | 41 | 87 | 41 | 0 | 87 | 3,161 | 3,193 |
| #14 | 2e2a01d8-eaec-4625-977d-ea3277e9976f | 3.8 | 41 | 92 | 41 | 0 | 92 | 2,958 | 2,984 |
| #15 | 079ba8a4-33e3-4ae7-ab0b-20e141e227f4 | 3.8 | 41 | 93 | 41 | 0 | 93 | 328 | 355 |
| #16 | 2325011a-517e-48f4-9b73-70648b2bf499 | 3.8 | 41 | 92 | 41 | 0 | 92 | 2,454 | 2,480 |
| #17 | 796277c7-a3c3-4127-8a2d-9e5aac5dce08 | 3.8 | 41 | 77 | 41 | 1 | 77 | 3,167 | 3,222 |
| #18 | 67bf227f-0f53-4f80-9f9e-1dfd2dcc7b28 | 3.8 | 41 | 82 | 41 | 1 | 82 | 3,291 | 3,330 |
| #19 | ae5a873b-fbb0-4245-9a0f-6b669b9a9ac6 | 3.8 | 41 | 87 | 41 | 0 | 87 | 3,201 | 3,232 |
Fig. 2.
Local BLAST search. A local BLAST search for the transgene sequence (LC495729) against all reads obtained by MinION (Table 2) revealed that one fragment (sequence ID: 304cb0d3-cbe9-4e75-bf24-4f9390aa8525, #1 in Table 2) had two sequences homologous to the transgene (red arrows in the first and second rows). The homologous sequence in the third row (green arrow, #2 in Table 2) was an exon of the endogenous St3Gal2 gene. Homologies of the remaining sequences (#3 to #19 in Table 2) were coincidences because BLAST search in the Ensembl database confirmed the existence of the entire sequence of each fragment in the mouse reference genome sequence.
Fig. 3.
Nucleotide dot plot analysis. Nucleotide dot plots of DNA sequences of transgene construct (LC495729) and mouse reference genome against the reverse complementary sequence of the fragment (304cb0d3-cbe9-4e75-bf24-4f9390aa8525) hit by the local BLAST search (Fig. 2) are shown in the upper and lower panels, respectively. The upper panel reveals that the tandem concatenation of a whole transgene and the first ~660 bp of the transgene was integrated in the 4C30 genome. The lower panel indicates that the ~10 kilobases of the genome sequence was deleted between 5’- and 3’-flanking regions of the transgene insertion site.
Fig. 4.
Deduced pattern of transgene insertion in the 4C30 genome. (A) A BLAST search of the 5’ flanking sequence of the fragment (304cb0d3-cbe9-4e75-bf24-4f9390aa8525) against the C57BL/6NJ mouse genome on the Ensembl website revealed that the transgenes were inserted in the δ-sarcoglycan (Sgcd) gene; i.e., in the first intron of the Sgcd-202 coding gene and 5’ upstream of the Sgcd-201 coding gene, both of which were encoded in the reverse strand in the 4C30 mouse genome. (B) In more detail, nearly the full length (15–3,508) followed by the first 657 bp (4–660) of the 3,508-bp transgene were inserted between nucleotides 47,908,481 and 47,917,870 on chromosome 11 (positions were based on the C57BL/6NJ genome retrieved from the Ensembl database). The 9,388-bp genome sequence from nucleotides 47,908,482 to 47,917,869 was lost at the insertion site of the 4C30 genome. The genome sequence determined in this study can be obtained from DDBJ (accession number: LC496472).
Sequence confirmation by additional PCR and sequencing
The DNA sequence of the structure determined by dot plot analysis was confirmed by additional multiple overlapping PCRs covering between 5’- and 3’-flanking regions of the transgene and subsequent Sanger sequencing (Fig. 4B). Primers used for this confirmation step were designed by Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi).
Availability of sequence information
All read data generated by MinION have been deposited to the DDBJ Sequence Read Archive (accession number: DRA008802). The DNA sequence of the transgene construct (Fig. 1A), as well as the genome sequence containing inserted transgenes and their flanking sequences determined in the present study (Fig. 4B), have been deposited to DDBJ Annotated/Assembled Sequences (accession numbers: LC495729 and LC496472, respectively).
Results
Long-read sequencing by a nanopore sequencer (MinION)
We analyzed high-molecular-weight genomic DNA (>50 kb) from a kidney of a 4C30 mouse (Fig. 1B) with MinION. After a 24-h run, we obtained 922,210 reads, ranging from 7 to 238,124 bp with an n50 of 7,607 bp, totaling 2,985,697,306 bp (Table 1).
Local BLAST search, dot plot analysis, and mapping on the C57BL/6N genome
We found one fragment containing the transgene sequence with a local BLAST search against all reads obtained with the MinION (Table 2 and Fig. 2). Small homologous sequences contained in other fragments were probably accidental similarities to the mouse genome because BLAST search against the C57BL/6N genome showed such homologous sequences were those that initially existed in the mouse genome, not new sequences produced by the insertion of transgene fragments.
A nucleotide dot plot of the transgene construct and the mouse genome sequences against the fragment revealed that a tandemly concatenated sequence of nearly full size and the first 660 bp of the transgene were inserted into the genome of the 4C30 strain mouse with 10 kb genome deletion (Fig. 3). A BLAST search against the C57BL/6N genome revealed that the transgene was inserted into one site, the intron of the Sgcd gene on chromosome 11 (Fig. 4A).
Sequence confirmation by additional PCR analysis and sequencing
We confirmed the sequence of the transgene and its flanking genome by additional PCR analysis and Sanger sequencing. The conclusive location in the 4C30 mouse genome and the transgene insertion pattern are illustrated in Fig. 4. The position of the transgene insertion was the same as we previously reported [30].
Discussion
Our results clearly indicate that a long-read DNA sequencer in combination with a local BLAST search is a valuable tool for determining the whole range of transgene insertion patterns in a transgenic animal genome. Long-read sequencers like nanopore sequencers have an advantage over PCR-based methods, such as genomic walking, because long-read sequencers can read DNA sequences much longer than those of the average PCR-based sequencing method. Long-read sequencers can also determine the copy number of transgenes because the sequencers can determine the DNA sequences of a fragment or fragments containing a whole transgene insertion sequence, despite being tandemly repeated in the host genome [28]. However, confirmation of the DNA sequence is necessary with further PCR analysis and Sanger sequencing and/or short-read next-generation sequencers because of the lower accuracy of long-read DNA sequencers (approx. 85–95%) [14, 19] compared with short-read DNA sequencers.
A local BLAST search was adequate for determining the transgene insertion site in the mouse genome in the present study. We believe that de novo assembly, which requires high CPU power, will not be necessary for similar purposes because we should focus on fragments containing transgenes only. However, assembly might be required, as reported by Nicholls et al. [28], when many transgene copies are inserted into the genome and the overall insertion is too long to be covered in single fragment reads. We can expect to obtain fragment reads containing an entire range of the transgene insertion in the host genome by nanopore sequencers with carefully prepared high-molecular-weight genome DNA from transgenic animals because nanopore sequencers can read >100 kb of DNA fragments (the maximum read length in the present study was 238 kb), which can, in many cases, contain the entire range of transgenes in the genome. For collection of long genome DNA, methods using magnet beads are recommended instead of the methods with spin columns because less fragmentation of DNA can be achieved with less physical stress in the magnet-bead method [34].
We should consider the low coverage in the genome analysis with MinION. MinION has a total read length of about 3 Gb in our experience, which is insufficient to read the entire mouse genome by a single run. In order to search the entire genome for transgene insertion positions, we need to accumulate the read data by repeating the sequence runs until we obtain reads covering the entire genome while checking the actual coverage by mapping the sequence data to the genome. However, more than 80% of the transgenic mouse strains have only one insertion site [27]. Especially in the case of the 4C30 strain, our preliminary data with genomic walking [17] and quantitative PCR (data not shown) suggested that there was only one insertion site. Therefore, in most cases, it is likely that it is practically sufficient to repeat long-read sequencing until one insertion position is detected. Thus, in the insertion position search, we would be able to reduce the repeat of sequencing by utilizing not only long-read sequencer data but also various related information on copy numbers (such as data with Southern blot and quantitative PCR).
The insertion pattern of the transgene in 4C30 mice (Fig. 4B) was typical of those found in most transgenic animals; i.e., transgenesis might be accomplished in two steps: extrachromosomal concatenation of vector constructs, followed by insertion into the chromosome [2]. Before concatenation of the construct, end-nibbling may occur to various degrees, especially on the 5’ ends of transgenes [32]. The second copy, which lost the latter part of the construct, may have been created by exposure to UV transillumination during gel purification of the transgene construct [16], as discussed in a previous study [38].
It is unknown what the effect would be following an approximately 9-kb deletion at the transgene insertion site, which was found for the first time in the intron of the Sgcd gene in DCM model 4C30 mice. Since the Sgcd gene is one of the causal genes for DCM [33], we compared the expression of the Sgcd gene in the hearts of 4C30 and wild-type mice. We found no differences in expression of Sgcd at the mRNA or protein levels between the two types of mice [39]. Since only transcripts of Sgcd-201, but not Sgcd-202 (Fig. 4A), were reported in the Ensembl database at the time of our previous study [39], we confirmed the presence of the Sgcd-202 transcript in homozygous transgenic, hemizygous transgenic, and wild-type mice by RT-PCR (data not shown). The results suggest that intronic deletion of the Sgcd gene caused by transgene insertion was unlikely to have affected Sgcd-202 gene expression. Therefore, we concluded that DCM in 4C30 mice was caused by altered sialic acid metabolism, induced by overexpression of the mouse ST3GalII gene in the heart [26, 39]. However, it is possible that new intronic control elements may exist in the second intron of the Sgcd-202 gene because intronic microRNA in myosin heavy chain (MHC) genes controls MHC expression [5] and non-coding RNA has been reported to be involved in physiological phenomena, such regulating control factors for expression of various genes (e.g., [9]). The intronic deletion found in 4C30 mice may be a clue for discovering new intronic control elements of genes involved in DCM.
In conclusion, a long-read sequencer is a powerful tool for examining the influence of transgene insertion on the host genome because it can easily determine the entire insertion pattern of transgenes in the genomes of transgenic animals. Recently, target sequencing methods with a CRISPR/Cas9 system [13] and oligo-probes [24] have been proposed. Combining such methods with a long-read sequencer will allow us to more efficiently analyze the effect of the transgene insertion pattern and flanking genome modification on transgenic animals.
Acknowledgments
We wish thank Ms. Yuko Doi and Mr. Masafumi Nakano for assisting with animal care, and Ms. Hiroko Urahama and Ms. Nahoko Kotani for help in preparing the manuscript.
References
- 1.Behringer R., Gertsenstein M., Nagy K.V., Nagy A.2014. Genotyping. pp. 551–570. In: Manipulating the mouse embryo: a laboratory manual, 4th ed., Cold Spring Harbor Press, New York. [Google Scholar]
- 2.Bishop J.O.1996. Chromosomal insertion of foreign DNA. Reprod. Nutr. Dev. 36: 607–618. [PubMed] [Google Scholar]
- 3.Bishop J.O., Smith P.1989. Mechanism of chromosomal integration of microinjected DNA. Mol. Biol. Med. 6: 283–298. [PubMed] [Google Scholar]
- 4.Cain-Hom C., Splinter E., van Min M., Simonis M., van de Heijning M., Martinez M., Asghari V., Cox J.C., Warming S.2017. Efficient mapping of transgene integration sites and local structural changes in Cre transgenic mice using targeted locus amplification. Nucleic Acids Res. 45: e62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Callis T.E., Pandya K., Seok H.Y., Tang R.H., Tatsuguchi M., Huang Z.P., Chen J.F., Deng Z., Gunn B., Shumate J., Willis M.S., Selzman C.H., Wang D.Z.2009. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 119: 2772–2786. doi: 10.1172/JCI36154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camacho C.2019. BLAST+ Release Notes. NCBI, Bethesda. [Google Scholar]
- 7.Collier R., Dasgupta K., Xing Y.P., Hernandez B.T., Shao M., Rohozinski D., Kovak E., Lin J., de Oliveira M.L.P., Stover E., McCue K.F., Harmon F.G., Blechl A., Thomson J.G., Thilmony R.2017. Accurate measurement of transgene copy number in crop plants using droplet digital PCR. Plant J. 90: 1014–1025. doi: 10.1111/tpj.13517 [DOI] [PubMed] [Google Scholar]
- 8.Deamer D., Akeson M., Branton D.2016. Three decades of nanopore sequencing. Nat. Biotechnol. 34: 518–524. doi: 10.1038/nbt.3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Di Mauro V., Barandalla-Sobrados M., Catalucci D.2018. The noncoding-RNA landscape in cardiovascular health and disease. Noncoding RNA Res. 3: 12–19. doi: 10.1016/j.ncrna.2018.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DuBose A.J., Lichtenstein S.T., Narisu N., Bonnycastle L.L., Swift A.J., Chines P.S., Collins F.S.2013. Use of microarray hybrid capture and next-generation sequencing to identify the anatomy of a transgene. Nucleic Acids Res. 41: e70. doi: 10.1093/nar/gks1463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fletcher S.J.2014. qPCR for quantification of transgene expression and determination of transgene copy number. Methods Mol. Biol. 1145: 213–237. doi: 10.1007/978-1-4939-0446-4_17 [DOI] [PubMed] [Google Scholar]
- 12.Gibbs A.J., McIntyre G.A.1970. The diagram, a method for comparing sequences. Its use with amino acid and nucleotide sequences. Eur. J. Biochem. 16: 1–11. doi: 10.1111/j.1432-1033.1970.tb01046.x [DOI] [PubMed] [Google Scholar]
- 13.Gilpatrick T., Lee I., Graham J.E., Raimondeau E., Bowen R., Heron A., Sedlazeck F.J., Timp W.2019. Targeted Nanopore Sequencing with Cas9 for studies of methylation, structural variants, and mutations. bioRxiv: 604173. [Google Scholar]
- 14.Giordano F., Aigrain L., Quail M.A., Coupland P., Bonfield J.K., Davies R.M., Tischler G., Jackson D.K., Keane T.M., Li J., Yue J.X., Liti G., Durbin R., Ning Z.2017. De novo yeast genome assemblies from MinION, PacBio and MiSeq platforms. Sci. Rep. 7: 3935. doi: 10.1038/s41598-017-03996-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goodwin L.O., Splinter E., Davis T.L., Urban R., He H., Braun R.E., Chesler E.J., Kumar V., van Min M., Ndukum J., Philip V.M., Reinholdt L.G., Svenson K., White J.K., Sasner M., Lutz C., Murray S.A.2019. Large-scale discovery of mouse transgenic integration sites reveals frequent structural variation and insertional mutagenesis. Genome Res. 29: 494–505. doi: 10.1101/gr.233866.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartman P.S.1991. Transillumination can profoundly reduce transformation frequencies. Biotechniques 11: 747–748. [PubMed] [Google Scholar]
- 17.Hogan B., Beddington R., Constantini F., Lacy E.1994. Identifying homozygous transgenic mice or embryos. pp. 305–308. In: Manipulating the Mouse Embryo: A Laboratory Manual) Cold Spring Harbor Laboratory Press, Woodbury. [Google Scholar]
- 18.Jain M., Koren S., Miga K.H., Quick J., Rand A.C., Sasani T.A., Tyson J.R., Beggs A.D., Dilthey A.T., Fiddes I.T., Malla S., Marriott H., Nieto T., O’Grady J., Olsen H.E., Pedersen B.S., Rhie A., Richardson H., Quinlan A.R., Snutch T.P., Tee L., Paten B., Phillippy A.M., Simpson J.T., Loman N.J., Loose M.2018. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat. Biotechnol. 36: 338–345. doi: 10.1038/nbt.4060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jain M., Tyson J.R., Loose M., Ip C.L.C., Eccles D.A., O’Grady J., Malla S., Leggett R.M., Wallerman O., Jansen H.J., Zalunin V., Birney E., Brown B.L., Snutch T.P., Olsen H.E., Min I.O.N.A., Reference C., MinION Analysis and Reference Consortium. 2017. MinION Analysis and Reference Consortium: Phase 2 data release and analysis of R9.0 chemistry. F1000 Res. 6: 760. doi: 10.12688/f1000research.11354.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joshi M., Pittman H.K., Haisch C., Verbanac K.2008. Real-time PCR to determine transgene copy number and to quantitate the biolocalization of adoptively transferred cells from EGFP-transgenic mice. Biotechniques 45: 247–258. doi: 10.2144/000112913 [DOI] [PubMed] [Google Scholar]
- 21.Laboulaye M.A., Duan X., Qiao M., Whitney I.E., Sanes J.R.2018. Mapping Transgene Insertion Sites Reveals Complex Interactions Between Mouse Transgenes and Neighboring Endogenous Genes. Front. Mol. Neurosci. 11: 385. doi: 10.3389/fnmol.2018.00385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee Y.C., Kojima N., Wada E., Kurosawa N., Nakaoka T., Hamamoto T., Tsuji S.1994. Cloning and expression of cDNA for a new type of Gal beta 1,3GalNAc alpha 2,3-sialyltransferase. J. Biol. Chem. 269: 10028–10033. [PubMed] [Google Scholar]
- 23.Li H.2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. doi: 10.1093/bioinformatics/bty191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S., Jia S., Hou L., Nguyen H., Sato S., Holding D., Cahoon E., Zhang C., Clemente T., Yu B.2019. Mapping of transgenic alleles in soybean using a nanopore-based sequencing strategy. J. Exp. Bot. 70: 3825–3833. doi: 10.1093/jxb/erz202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang Z., Breman A.M., Grimes B.R., Rosen E.D.2008. Identifying and genotyping transgene integration loci. Transgenic Res. 17: 979–983. doi: 10.1007/s11248-008-9190-7 [DOI] [PubMed] [Google Scholar]
- 26.Nagai-Okatani C., Nishigori M., Sato T., Minamino N., Kaji H., Kuno A.2019. Wisteria floribunda agglutinin staining for the quantitative assessment of cardiac fibrogenic activity in a mouse model of dilated cardiomyopathy. Lab. Invest. 99: 1749–1765. doi: 10.1038/s41374-019-0279-9 [DOI] [PubMed] [Google Scholar]
- 27.Nakanishi T., Kuroiwa A., Yamada S., Isotani A., Yamashita A., Tairaka A., Hayashi T., Takagi T., Ikawa M., Matsuda Y., Okabe M.2002. FISH analysis of 142 EGFP transgene integration sites into the mouse genome. Genomics 80: 564–574. doi: 10.1006/geno.2002.7008 [DOI] [PubMed] [Google Scholar]
- 28.Nicholls P.K., Bellott D.W., Cho T.J., Pyntikova T., Page D.C.2019. Locating and Characterizing a Transgene Integration Site by Nanopore Sequencing. G3 (Bethesda) 9: 1481–1486. doi: 10.1534/g3.119.300582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Niwa H., Yamamura K., Miyazaki J.1991. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108: 193–199. doi: 10.1016/0378-1119(91)90434-D [DOI] [PubMed] [Google Scholar]
- 30.Noguchi A., Takekawa N., Einarsdottir T., Koura M., Noguchi Y., Takano K., Yamamoto Y., Matsuda J., Suzuki O.2004. Chromosomal mapping and zygosity check of transgenes based on flanking genome sequences determined by genomic walking. Exp. Anim. 53: 103–111. doi: 10.1538/expanim.53.103 [DOI] [PubMed] [Google Scholar]
- 31.Palmiter R.D., Norstedt G., Gelinas R.E., Hammer R.E., Brinster R.L.1983. Metallothionein-human GH fusion genes stimulate growth of mice. Science 222: 809–814. doi: 10.1126/science.6356363 [DOI] [PubMed] [Google Scholar]
- 32.Rohan R.M., King D., Frels W.I.1990. Direct sequencing of PCR-amplified junction fragments from tandemly repeated transgenes. Nucleic Acids Res. 18: 6089–6095. doi: 10.1093/nar/18.20.6089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sakamoto A., Ono K., Abe M., Jasmin G., Eki T., Murakami Y., Masaki T., Toyo-oka T., Hanaoka F.1997. Both hypertrophic and dilated cardiomyopathies are caused by mutation of the same gene, delta-sarcoglycan, in hamster: an animal model of disrupted dystrophin-associated glycoprotein complex. Proc. Natl. Acad. Sci. USA 94: 13873–13878. doi: 10.1073/pnas.94.25.13873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwessinger B., Rathjen J.P.2017. Extraction of High Molecular Weight DNA from Fungal Rust Spores for Long Read Sequencing. Methods Mol. Biol. 1659: 49–57. doi: 10.1007/978-1-4939-7249-4_5 [DOI] [PubMed] [Google Scholar]
- 35.Sha H., Xu J., Tang J., Ding J., Gong J., Ge X., Kong D., Gao X.2007. Disruption of a novel regulatory locus results in decreased Bdnf expression, obesity, and type 2 diabetes in mice. Physiol. Genomics 31: 252–263. doi: 10.1152/physiolgenomics.00093.2007 [DOI] [PubMed] [Google Scholar]
- 36.Sharpe J., Lettice L., Hecksher-Sorensen J., Fox M., Hill R., Krumlauf R.1999. Identification of sonic hedgehog as a candidate gene responsible for the polydactylous mouse mutant Sasquatch. Curr. Biol. 9: 97–100. doi: 10.1016/S0960-9822(99)80022-0 [DOI] [PubMed] [Google Scholar]
- 37.Shwed P.S., Crosthwait J., Douglas G.R., Seligy V.L.2010. Characterisation of Muta™Mouse λgt10-lacZ transgene: evidence for in vivo rearrangements. Mutagenesis 25: 609–616. doi: 10.1093/mutage/geq048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki O., Hata T., Takekawa N., Koura M., Takano K., Yamamoto Y., Noguchi Y., Uchio-Yamada K., Matsuda J.2006. Transgene insertion pattern analysis using genomic walking in a transgenic mouse line. Exp. Anim. 55: 65–69. doi: 10.1538/expanim.55.65 [DOI] [PubMed] [Google Scholar]
- 39.Suzuki O., Kanai T., Nishikawa T., Yamamoto Y., Noguchi A., Takimoto K., Koura M., Noguchi Y., Uchio-Yamada K., Tsuji S., Matsuda J.2011. Adult onset cardiac dilatation in a transgenic mouse line with Galβ1,3GalNAc α2,3-sialyltransferase II (ST3Gal-II) transgenes: a new model for dilated cardiomyopathy. Proc. Jpn. Acad., Ser. B, Phys. Biol. Sci. 87: 550–562. doi: 10.2183/pjab.87.550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tosh J.L., Rickman M., Rhymes E., Norona F.E., Clayton E., Mucke L., Isaacs A.M., Fisher E.M.C., Wiseman F.K.2018. The integration site of the APP transgene in the J20 mouse model of Alzheimer’s disease. Wellcome Open Res. 2: 84. doi: 10.12688/wellcomeopenres.12237.2 [DOI] [PMC free article] [PubMed] [Google Scholar]