Abstract
Background
Although there is some evidence that severe acute respiratory syndrome coronavirus 2 can invade the human placenta, limited data exist on the gestational age–dependent expression profile of the severe acute respiratory syndrome coronavirus 2 cell entry mediators, angiotensin-converting enzyme 2 and transmembrane protease serine 2, at the human maternal-fetal interface. There is also no information as to whether the expression of these mediators is altered in pregnancies complicated by preeclampsia or preterm birth. This is important because the expression of decidual and placental angiotensin-converting enzyme 2 and transmembrane protease serine 2 across gestation may affect the susceptibility of pregnancies to vertical transmission of severe acute respiratory syndrome coronavirus 2.
Objective
This study aimed to investigate the expression pattern of specific severe acute respiratory syndrome coronavirus 2 cell entry genes, angiotensin-converting enzyme 2 and transmembrane protease serine 2, in the placenta across human pregnancy and in paired samples of decidua and placenta in pregnancies complicated by preterm birth or preeclampsia compared with those in term uncomplicated pregnancies.
Study Design
In this study, 2 separate cohorts of patients, totaling 87 pregnancies, were included. The first cohort was composed of placentae from first- (7–9 weeks), second- (16–18 weeks), and third-trimester preterm (26–31 weeks) and third-trimester term (38–41 weeks) pregnancies (n=5/group), whereas the second independent cohort included matched decidua and placentae from pregnancies from term uncomplicated pregnancies (37–41 weeks’ gestation; n=14) and pregnancies complicated by preterm birth (26–37 weeks’ gestation; n=11) or preeclampsia (25–37 weeks’ gestation; n=42). Samples were subjected to quantitative polymerase chain reaction and next-generation sequencing or RNA sequencing for next-generation RNA sequencing for angiotensin-converting enzyme 2 and transmembrane protease serine 2 mRNA expression quantification, respectively.
Results
In the first cohort, angiotensin-converting enzyme 2 and transmembrane protease serine 2, exhibited a gestational age–dependent expression profile, that is, angiotensin-converting enzyme 2 and transmembrane protease serine 2 mRNA was higher (P<.05) in the first-trimester placenta than in second-trimester, preterm birth, and term placentae (P<.05) and exhibited a negative correlation with gestational age (P<.05). In the second cohort, RNA sequencing demonstrated very low or undetectable expression levels of angiotensin-converting enzyme 2 in preterm birth, preeclampsia, and term decidua and in placentae from late gestation. In contrast, transmembrane protease serine 2 was expressed in both decidual and placental samples but did not change in pregnancies complicated by either preterm birth or preeclampsia.
Conclusion
The increased expression of these severe acute respiratory syndrome coronavirus 2 cell entry–associated genes in the placenta in the first trimester of pregnancy compared with those in later stages of pregnancy suggests the possibility of differential susceptibility to placental entry to severe acute respiratory syndrome coronavirus 2 across pregnancy. Even though there is some evidence of increased rates of preterm birth associated with severe acute respiratory syndrome coronavirus 2 infection, we found no increase in mRNA expression of angiotensin-converting enzyme 2 or transmembrane protease serine 2 at the maternal-fetal interface.
Key words: angiotensin-converting enzyme 2, coronavirus disease 2019, decidua, gestation age–dependent gene expression, placenta, preeclampsia, preterm birth, severe acute respiratory syndrome coronavirus 2, term pregnancies
AJOG at a Glance.
Why was this study conducted?
This study aimed to investigate the expression pattern of angiotensin-converting enzyme 2 (ACE2) and transmembrane protease serine 2 (TMPRSS2), associated with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) cell entry in the placenta across human pregnancy and at the maternal-fetal interface in pregnancies complicated by preterm birth (PTB) or preeclampsia (PE).
Key findings
ACE2 and TMPRSS2 are highly expressed in the human placenta in early pregnancy, but their expression decreases significantly with advancing gestation. Expression of these genes at the maternal-fetal interface did not change in pregnancies complicated by either PTB or PE.
What does this add to what is known?
The decrease in expression of placental ACE2 and TMPRSS2 with advancing gestational age suggests the potential for differential risk of placental infection across pregnancy. Pregnancies complicated by PTB or PE are not associated with changes in the expression of these SARS-CoV-2 cell entry genes.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection can lead to the life-threatening coronavirus disease 2019 (COVID-19), which emerged in the Wuhan, Hubei Province, China, in December 2019.1 The pathogenesis of COVID-19 is complex, but it may involve cell viral replication and, in severe cases, result in a cytokine storm, a systemic proinflammatory response that seriously harms the brain, heart, kidneys, liver, and lungs and leads to organ failure and ultimately death.1, 2, 3, 4, 5
Currently, the pathogenesis of SARS-CoV-2 infection in pregnancy is poorly understood. However, to date, there has been limited evidence of its maternal to fetal transmission, in contrast to other viruses.6, 7, 8, 9 The most likely path for the virus to access to the fetus would be via the placenta; however, little is known about the tropism of the SARS-CoV-2 for the decidua and placenta. Emerging reports suggest that the SARS-CoV-2 can invade the human placenta10 and can lead to the occurrence of a second-trimester miscarriage in a patient with coronavirus disease symptoms, exhibiting SARS-CoV-2 positivity in the placental submembranes and cotyledons, associated with mixed inflammatory infiltrates and funisitis.11 Furthermore, potential vertical transmission in pregnant women with COVID-19 has been reported. In 1 case of second-trimester preterm delivery, the amniotic fluid and infant tested positive (by polymerase chain reaction [PCR]) for SARS-CoV-2,12 and 1 neonate exhibited elevated immunoglobulin M antibody levels 2 hours after birth.13 In another case, a third-trimester (35 5/7 weeks’ gestation) neonate was born to a mother exhibiting clinical symptoms and testing positive (by PCR) for SARS-CoV-2 genes. The neonate (including blood and nonbronchoscopic bronchoalveolar lavage fluid during first day of life), placenta, and clear amniotic fluid (collected before rupture of membranes during cesarean delivery) also tested positive for SARS-CoV-2 genes. Placental pathologic examination identified diffuse perivillous fibrin deposition with associated infarction and acute and chronic intervillositis. Of importance, SARS-CoV-2 nucleocapsid protein immunostaining in the placenta revealed strong immunosignals concentrated in the syncytial layer.14 This suggests that the syncytial layer is enriched with SARS-CoV-2 cell entry receptors and highlights the need to investigate potential routes and associated mechanisms of placental SARS-CoV-2 infection and vertical transmission.
Compared with the general population, pregnant women are particularly susceptible to specific viral infections, including the cytomegalovirus, herpes simplex virus, and Zika virus, and exhibit greater complications and mortality rates associated with varicella, rubeola, and H1N1 infections. Importantly, the cause of this susceptibility is poorly defined,6 but it may be related to the immunologic adaptations inherent to pregnancy or to the tissue distribution of cell entry viral mediators at the maternal-fetal interface.6 Indeed, it is estimated that approximately one-third of pregnant women died after SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV) infections.15, 16, 17, 18, 19 Recent epidemiologic data suggest that maternal mortality rates and pregnancy complications, such as miscarriage or stillbirth and intrauterine growth restriction, are not as prevalent in cases of COVID-19 compared with cases of SARS-CoV and MERS-CoV infections. However, of importance, emerging evidence points to probable increased rates of preterm birth (PTB) in COVID-19,7 , 17 , 20 with reports showing high percentages of PTB in pregnancies complicated by COVID-19 that range from 23.8% to 39%,17 , 21 or no increased PTB rates.7
The best described mediators of SARS-CoV-2 cell entrance are ACE2 and the TMPRSS2 protein receptors.22, 23, 24 SARS-CoV-2 cell entry is mediated by the binding of the N-terminal portion of its spike (S) protein attached to the SARS-CoV-2–like viral envelope to a pocket of the cell membrane ACE2 receptor. In an important second step, TMPRSS2 cleaves and detaches the S1 from the S2 portion to allow a conformational rearrangement of the viral membrane and subsequent fusion and entry of the virus into the targeted cell.22 , 25 Tissue identification and the expression dynamics of these 2 cell entry–associated proteins are crucial for a better understanding of the SARS-CoV-2 cell tropism and COVID-19 pathogenesis, treatment, and prevention.
Evidence suggests that placental ACE2 expression decreases from early to late pregnancy26; however, even though the expression of TMPRSS2 has been demonstrated in the human placenta,27 , 28 whether the mRNA expression profile of ACE2 and TMPRSS2 in the human placenta across pregnancy (simultaneously comparing first-, second-, and third-trimester pregnancies) and the mRNA expression of these SARS-CoV-2 cell entry–associated proteins are dysregulated in the decidua and placenta from pregnancies complicated by PTB or PE is unclear. We hypothesized that the placenta expresses ACE2 and TMPRSS2 that encode proteins mediating infection of cells within the human maternal-fetal interface in a gestational age–dependent manner and that changes in the expression of these genes at the maternal-fetal interface may be associated with pregnancies complicated by PTB or PE. Information on SARS-CoV-2 expression dynamics at the maternal-fetal interface across pregnancy and in cases of obstetrical complications may provide increased understanding of the potential for placenta and fetal infections and thus support management of patients who present with SARS-CoV-2 infection during pregnancy.
Material and Methods
Sample collection
This is a cross-sectional study involving 2 different cohorts of patients, totaling 87 pregnancies. In the first cohort, we assessed the developmental expression profile of specific SARS-CoV-2 cell entry–associated genes, ACE2 and TMPRSS2, in human placental tissue from (1) first-trimester elective termination (7–9 weeks’ gestation; n=5); (2) second-trimester (16–18 weeks’ gestation; n=5) elective terminations; (3) third-trimester spontaneous onset of PTB (26–35 weeks’ gestation; n=5), 2 delivered vaginally and 3 by cesarean delivery indicated for fetal distress (2) or bleeding from fibroid (1); and (4) term delivery (38–41 weeks’ gestation; n=5) following elective cesarean delivery before labor (4) or following vaginal delivery (1) in otherwise uncomplicated pregnancies. In the second cohort, to evaluate whether the mRNA expression of specific SARS-CoV-2 cell entry–associated genes was altered at the maternal-fetal interface in pregnancies complicated by PTB or PE, a total of 67 patients were recruited. We collected matched decidual and placental tissues from patients experiencing (1) PTB (26–37 weeks’ gestation), 8 patients experienced spontaneous onset of preterm labor, and 3 gave birth by cesarean delivery before labor onset (total n=11); (2) PE (25–37 weeks’ gestation), 11 patients experienced spontaneous labor onset, and 31 gave birth by cesarean delivery before labor onset (total n=42); or (3) term delivery (37–41 weeks’ gestation), 12 patients experienced elective delivery by cesarean delivery, and 2 experienced vaginal delivery following spontaneous labor onset (total n=14) in otherwise uncomplicated pregnancies. PE was defined as new onset of high blood pressure (>140/90 mm Hg) at 20 weeks’ gestation with concurrent proteinuria and end organ dysfunction (renal dysfunction, liver dysfunction, central nervous system disturbances, pulmonary edema, and thrombocytopenia). Tissue samples were collected by the Research Centre for Women’s and Infants’ Health (RCWIH) BioBank at Mount Sinai Hospital (Toronto, Canada) following informed consent. Placental tissue was sampled and processed as previously described.29 Briefly, placental villous tissues from first-trimester pregnancies, following dilation and curettage, were visually identified and dissected from the decidua and other tissues, by specialized RCWIH staff. Second- and third-trimester placental villous tissues and placental fragments from PTB and PE pregnancies were dissected and harvested immediately after birth in a similar way. Placental core sampling was undertaken by dissecting the maternal surface in quadrants in areas 1.5 cm away from the center of the placental disc, the closest placental edge, and the umbilical cord insertion site and from areas of infarcts, thrombosis, or other abnormalities. All cuts were made to exclude the maternal decidua and the chorionic plate, so only placental villous tissues were to be included in the study. For the RNA-sequencing (RNAseq) study, the decidual sample was isolated from the fetal membranes of delivered placenta by scraping and was stored at −80°C until processing. This study was approved by the Institutional Research Ethics Board at Mount Sinai Hospital (Toronto, Canada), under the approval numbers, 20-0006-E and 20-0101-E.
Quantitative polymerase chain reaction analysis in the placental ontogenetic study
Total RNA was isolated from human placental tissue using the RNeasy Plus Universal Mini Kit (73404, Qiagen, Toronto, Ontario, Canada), as described before.30 , 31 RNA quality and concentration were determined using the NanoDrop1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA). RNA integrity was assessed by calculating the RNA integrity number, using the Agilent Bioanalyzer 2100 and RNA 6000 Nano Labchip kit (Agilent Technologies, Santa Clara, CA). In addition, 10 ng/μL of total RNA was reverse transcribed into cDNA using the iScript gDNA Clear cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA). ACE2 and TMPRSS2 mRNA levels were assessed by quantitative polymerase chain reaction (qPCR) using LuminoCt qPCR ReadyMix (Sigma-Aldrich, St. Louis, MO) and the CFX384 Real-Time PCR Detection System (Bio-Rad Laboratories). The cycling conditions were as follows: initial denaturation at 95°C (2 min), followed by 40 cycles of denaturation at 95°C (5 s) and combined annealing and extension at 60°C (20 s). Gene expression was normalized to DNA topoisomerase 1 and succinate-ubiquinone oxidoreductase,32 which presented stable expression among all groups. The primer sequences of all evaluated genes are shown in the Table . Relative expression of target genes was obtained using the 2−ΔΔCT method.33
Table.
Sequence of primers used in this study
| Primer name | Sequence reference |
|---|---|
| ACE2a | Forward: 5'- GGAGTGATAGTGGTTGGCATTGTC -3' |
| Reverse: 5'- GCTAATATCGATGGAGGCATAAGGA -3' | |
| TMPRSS2a | Forward: 5'- AGCTGCAGAAGCCTCTGACTTTC -3' |
| Reverse: 5'- AGCGTTCAGCACTTCTGAGGTC -3' | |
| TOP1 | Forward: 5'-GATGAACCTGAAGATGATGGC -3' |
| Reverse: 5'-TCAGCATCATCCTCATCTCG -3' | |
| SDHA | Forward: 5'- TGGGAACAAGAGGGCATCTG -3' |
| Reverse: 5'- CCACCACTGCATCAAATTCATG -3' | |
ACE2, angiotensin-converting enzyme 2; SDHA, succinate-ubiquinone oxidoreductase; TMPRSS2, transmembrane protease serine 2; TOP1, topoisomerase 1.
Bloise et al. Angiotensin-converting enzyme 2 and transmembrane protease serine 2 in the maternal-fetal interface across pregnancy. Am J Obstet Gynecol 2021.
Gene-specific primers were designed with Primer-BLAST (http://www.ncbi.nlm.nih.gov/tools/primer-blast).
Decidual and placental next-generation sequencing and ribonucleic acid sequencing in patients with preterm birth or preeclampsia
Total RNA was isolated from the decidual and placental samples with RNeasy Plus Universal Mini Kit (Qiagen), as previously described.30 , 31 RNA concentration and quality were assessed with Fragment Analyzer systems (Agilent Technologies). To identify the gene expression signature of ACE2 and TMPRSS2 mRNA levels, RNAseq technology (next-generation sequencing [NGS]) in placental and decidual tissues was undertaken. RNAseq was conducted by an MGISEQ-2000 sequencer at Mount Sinai Hospital, Sinai Health System (Toronto). The RNA libraries were prepared using the MGIEasy RNA Directional Library Prep Set and MGIEasy rRNA Depletion kits (MGI Americas Inc, San Jose, CA). Pair-end RNAseq was conducted at a read length of 100 bp and 50 million reads per library. The workflow was conducted on the same platform by the same research team. We analyzed but found no significant variance owing to batch effect.
Bioinformatics workflow
Sequencing data were analyzed using a high-performance clustering computing system (Galen; Sinai Health System, Toronto, Canada). Data quality of raw fastqc files was examined by FastQC (version 0.11). After trimming off contaminated reads, Bowtie2 (version 2.3), an ultrafast and memory-efficient tool, was used to align sequencing reads to the human reference genome (hg38). Finally, binary alignment and mapping files were built for the downstream analysis. Uniquely mapped reads were summarized to feature counts using GenomicAlignments (version 1.23). Normalized RNA reads for TMPRSS2 expression were extracted by default settings of plotCounts (normalized counts plus a pseudocount of 0.5 as log2 scales) using the outcomes of the DESeqDataSet function provided by DEseq2 package (version 1.27). Transcripts per million (TPM) was calculated by the calculation of TPM function from R package scater (version 1.17) using mapped raw reads and effective gene length of the transcripts. The gene expressions of ACE2 and TMPRSS2 were identified by the ensemble identification ENSG00000130234 and ENSG00000184012, respectively. In this study, we found no evidence of a significant batch effect; therefore, no batch correction was assigned.
Statistical analyses
Statistical analysis was performed using Prism (version 7.0; GraphPad Software, San Diego, CA). qPCR data were assessed for normal distribution using the D’Agostino-Pearson test or the Shapiro-Wilk test; outliers were identified using “QuickCalcs” outlier calculator program (version 7.0; GraphPad Software, San Diego, CA). Gene expression in the ontogenetic study was analyzed using 1-way analysis of variance followed by the Tukey multiple comparison test. NGS statistical analyses were performed by R software (version 3.6) and RStudio (version 1.3). Multiple comparisons of decidual and placental samples from PE, preterm, and term pregnancies were conducted using the Kruskal-Wallis test and followed by pairwise Wilcoxon rank-sum tests. The Spearman correlation was used to evaluate the linear relationship between the gestational age and the given gene. Statistical significance was assumed when P<.05.
Results
The expression of specific severe acute respiratory syndrome coronavirus 2 cell entry–associated genes in human placenta is dependent on gestational age
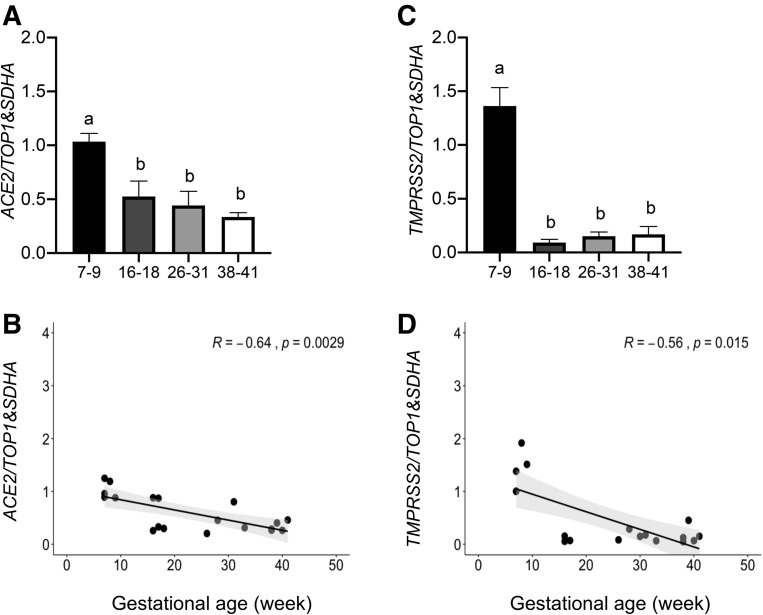
The placental expression of ACE2 and TMPRSS2 mRNA was relatively high in the first trimester of pregnancy (7–9 weeks) but exhibited a significant decrease (P<.05) in samples collected during the second-trimester (16–18 weeks), third-trimester, preterm (26–31 weeks), and term pregnancies (Figure 1 , A and C). Spearman correlation analysis identified a negative correlation between placental ACE2 and TMPRSS2 mRNA expressions (P<.05) with advancing gestational age (Figure 1, B and D).
Figure 1.
The placental expression of ACE2 and TMPRSS2 is gestational age dependent
mRNA levels of ACE2 (A) and TMPRSS2 (C) in the first- (7–9 weeks), second- (16–18 weeks), third-trimester spontaneous onset PTB (26–35 weeks) and third-trimester term (38–41 weeks) pregnancies (n=5/group). A and C, ANOVA followed by Tukey’s multiple comparisons test. B and D, Spearman correlation analysis. Data are presented as mean±SD. Different letters if P<.05.
ACE2, angiotensin-converting enzyme 2; ANOVA, analysis of variance; PTB, preterm birth; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; SD, standard deviation; SDHA, succinate-ubiquinone oxidoreductase; TMPRSS2, transmembrane protease serine 2; TOP1, topoisomerase 1.
Bloise et al. Angiotensin-converting enzyme 2 and transmembrane protease serine 2 in the maternal-fetal interface across pregnancy. Am J Obstet Gynecol 2021.
Angiotensin-converting enzyme 2 and transmembrane protease serine 2 expression at the maternal-fetal interface in term pregnancies and pregnancies complicated by preterm birth or preeclampsia
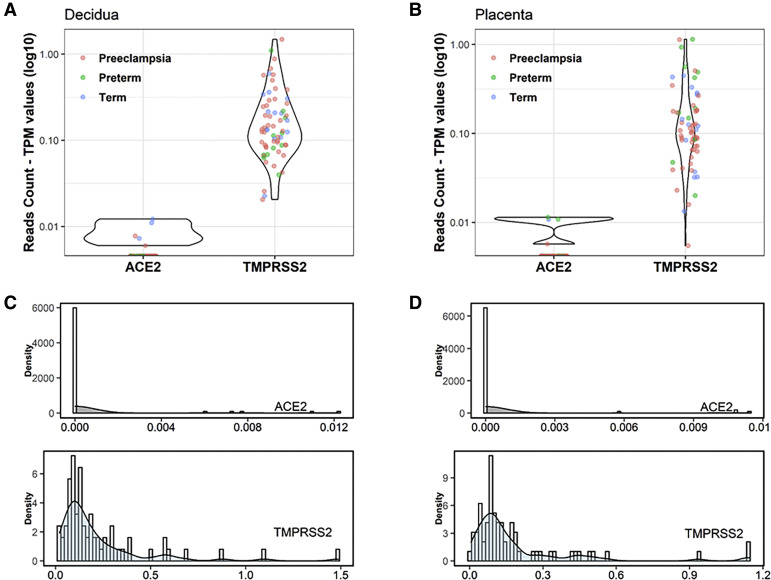
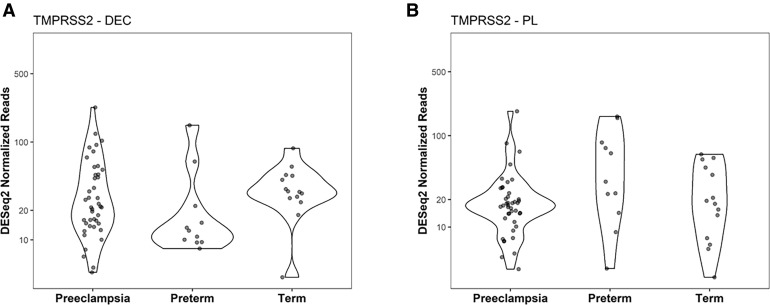
We accessed unpublished RNAseq data generated from paired placental and decidual samples to investigate the expression of ACE2 and TMPRSS2 in a large cohort of patients experiencing a PTB or diagnosed with PE and in patients at term pregnancy undergoing an elective cesarean delivery but otherwise uncomplicated term delivery. All samples from the RNAseq cohort exhibited 0 to 1 ACE2 read count (raw values). Of the 134 samples, only 9 exhibited a single ACE2 read count when the mean coverage of RNAseq ranged around 60 million reads. The remainder of the samples did not exhibit any ACE2 mapping out (data not shown). By evaluating the TPM reads counts (Figure 2 , A and B), we conclude that the expression of ACE2 in the decidua and placenta is undetectable by RNAseq (Figure 2, C and D). In contrast, TMPRSS2 was expressed in both decidual and placental samples (Figure 2, A and B), although the expression of this gene did not change across the patient groups (Figure 3 ) or with labor status (Figure 1, A and C).
Figure 2.
Decidual and placental ACE2 and TMPRSS2 expression in preeclampsia, preterm, and term pregnancies
The values of TPM were calculated by the number of mapped raw reads, the transcript’s length and sequencing depth. The TPM normalized reads count for ACE2 and TMPRSS2 was obtained from decidual (A) and placental (B) samples in preeclampsia (n=42), preterm (n=11), and term (n=14) pregnancies. Density plots (C and D) were constructed to examine the probability of available reads numbers. Histograms were embedded in the graphs to visualize the distribution of read counts.
ACE2, angiotensin-converting enzyme 2; TPM, transcripts per million; TMPRSS2, transmembrane protease serine 2.
Bloise et al. Angiotensin-converting enzyme 2 and transmembrane protease serine 2 in the maternal-fetal interface across pregnancy. Am J Obstet Gynecol 2021.
Figure 3.
Expression of TMPRSS2 in the human decidua and placenta
The expression profile of TMPRSS2 was compared on the basis of Deseq2 normalized RNA reads. Decidua- (A) and placenta-specific (B) expressions were calculated by Kruskal-Wallis test, followed by Wilcoxon rank-sum test. P values were listed when significant differences were detected at P<.05: preeclampsia (n=42), preterm (n=11), and term (n=14), respectively.
TMPRSS2, transmembrane protease serine 2.
Bloise et al. Angiotensin-converting enzyme 2 and transmembrane protease serine 2 in the maternal-fetal interface across pregnancy. Am J Obstet Gynecol 2021.
Comments
Principal findings
In this study, we determined, for the first time, the mRNA expression of 2 key SARS-CoV-2 cell entry–associated proteins in the placentae from first-, second-, and third-trimester pregnancies and in pregnancies complicated by PTB or PE. We found that placental expression of the genes that promote SARS-CoV-2 cell entry, ACE2 and TMPRSS2, is down-regulated as gestation progresses. Expression of ACE2 or TMPRSS2 at the decidual interface (placenta and decidua) did not change in pregnancies complicated by PTB (irrespective of labor status) or PE.
Clinical implications
The higher placental ACE2 mRNA levels in earlier stages of pregnancy raise the possibility of a higher vulnerability to SARS-CoV-2 infection in the first-trimester placenta. SARS-CoV and MERS-CoV infections during pregnancy are associated with increased rates of miscarriage and stillbirth,16 , 17 and there is limited evidence of miscarriage and stillbirth and fetal malformations in pregnancies complicated by COVID-19.7 , 17 The higher levels of placental ACE2 and TMPRSS2 in earlier stages of pregnancy is consistent with evidence showing the placental presence of SARS-COV-210 and the reported case of miscarriage during the second trimester of pregnancy in which the amniotic fluid and infant tested positive (by PCR) for SARS-CoV-2.12 The lower or absence of expression of ACE2 from midpregnancy onward is also consistent with the limited evidence of vertical transmission of SARS-CoV-2 during pregnancy; as noted by Dashraath,17 most reports relate to women who acquired SARS-CoV-2 in the third trimester of pregnancy.
We did not observe altered placental or decidual expression of ACE2 or TMPRSS2 in pregnancies complicated by PTB (whether spontaneous onset or iatrogenic). This is consistent with clinical observations that although rates of PTB are increased, this is largely because of preterm premature rupture of the membranes (PPROM) (there were no cases of PROM in our study) or iatrogenic indications.7 Although PE is often associated with placental inflammation,34 it was not associated with any changes in ACE2 or TMPRSS2 mRNA expression in the decidual or placenta.
Research implications
Placental ACE2 mRNA and protein expression have been previously investigated. ACE2 is an important component of the renin-angiotensin system, where it converts angiotensin II (Ang II) into angiotensin 1-7, an antagonist of Ang II that acts via Mas G protein–coupled receptor regulation.26 , 35 Ang II regulates placental vascular tone and is thought to participate in the pathogenesis of gestational hypertension and PE.36 , 37 In the first-trimester placenta, ACE2 is abundantly immunolocalized to the syncytiotrophoblast and villous stroma, with lower levels in cytotrophoblasts.26 This pattern of localization suggests that SARS-CoV-2 present in the maternal circulation has the potential to enter the maternal blood-bathed syncytiotrophoblast and infect the placenta via ACE2 binding. In fact, a case report depicted SARS-CoV-2 particles being predominantly present in the syncytiotrophoblast of a second-trimester pregnancy complicated with PE and placenta abruption,38 whereas SARS-CoV-2 N protein immunoreactivity was also concentrated in the syncytial layer in a case of SARS-CoV-2 vertical transmission in the third trimester of pregnancy.14 However, ACE2 was not immunolocalized in the fetal vascular endothelium of the villous stroma,26 which theoretically could prevent SARS-CoV-2 penetration via ACE2 binding into the fetal circulation. Furthermore, SARS-CoV-2 syncytiotrophoblast entry has the potential to induce a potent inflammatory response and functionally disrupt the syncytiotrophoblast barrier by negatively impacting nutrient and drug transport efficiency, hormonal output, and cellular turn over. These possibilities clearly require further investigation.
Strengths and limitations
It is possible that there are confounding factors associated with the gestational age differences observed. In our early pregnancy cohort, we were unable (owing to institution ethical policies) to collect further clinical information of the elective pregnancy terminations. As such, confounding factors may include maternal body mass index status, fetal sex, ethnicity, and the presence of unknown maternal infective and/or inflammatory states and/or endocrine and hypertensive disorders.
Using NGS or RNAseq, we were able to concomitantly screen the expression profile of ACE2 and TMPRSS2 in a large number of matched decidua and placentae from pregnancies complicated by PTB or PE. We observed very low levels of decidual and placental ACE2 in all groups investigated in later gestation. This is somewhat similar to our findings using qPCR showing detectable but nevertheless lower placental ACE2 mRNA levels in later stages of pregnancy in healthy patients. The divergent detection of ACE2 mRNA in our cohorts is likely because of the differential sensitivity of techniques. In this context, data extracted from public datasets deposited at ArrayExpress or newly generated single-cell RNAseq identified minimal levels of ACE2 and TMPRSS2 in first-, second-, and third-trimester placentae.28 This is in agreement with our second- and third-trimester findings but in disagreement with our first-trimester results. Differences in the first-trimester findings may be attributed to tissue collection protocols, patient inclusion criteria, and/or differences in the techniques used for gene expression assessment in these studies. Additional studies are clearly required.
In addition, the interpretation of our data requires caution, because there may yet be other undefined mechanisms linking SARS-CoV-2 obstetrical outcomes in normal and in pathologic conditions. Furthermore, future studies should investigate the protein expression pattern, localization, and function of TMPRSS2 in the decidua and placenta and investigate the relationship of gene expression, protein levels, and corresponding function, to better understand the potential routes by which SARS-CoV-2 could gain access to the maternal-fetal interface, especially during early pregnancy and the possibility of vertical transmission at this time.
Conclusion
The gestational age expression pattern of SARS-CoV-2 cell entry–associated proteins suggests a reduced likelihood of placental and decidual cell entries of the virus in later stages of pregnancy. However, earlier stages of pregnancy may be more susceptible to SARS-CoV-2 placental infection. Our data provide no evidence that pregnancies complicated by PTB or PE are at increased risk of placental SARS-CoV-2 infection and vertical transmission.
Acknowledgments
The authors thank the donors, the RCWIH BioBank, the Lunenfeld-Tanenbaum Research Institute, and the Mount Sinai Hospital and University Health Network, Department of Obstetrics and Gynaecology, for the human specimens used in this study (http://biobank.lunenfeld.ca).
Footnotes
The authors report no conflict of interest.
E.B. is supported by the Higher Education Personnel Improvement Coordination (Coordenação de Aperfeiçoamento Pessoal de Nível Superior [CAPES]; finance code 001, CAPES-Print fellowship). S.J.L. is supported by a grant (FDN-143262) from the Canadian Institutes of Health Research (CIHR). S.G.M. is also supported by a grant (FDN-148368) from the CIHR.
The funders had no role in the experimental design, data acquisition, analysis, and interpretation or in the manuscript writing and conception of this study. The corresponding authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Cite this article as: Bloise E, Zhang J, Nakpu J, et al. Expression of severe acute respiratory syndrome coronavirus 2 cell entry genes, angiotensin-converting enzyme 2 and transmembrane protease serine 2, in the placenta across gestation and at the maternal-fetal interface in pregnancies complicated by preterm birth or preeclampsia. Am J Obstet Gynecol 2021;224:298.e1-8.
References
- 1.Huang C., Wang Y., Li X., et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497–506. doi: 10.1016/S0140-6736(20)30183-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen S.F., Ho Y.C. SARS-CoV-2: a storm is raging. J Clin Invest. 2020;130:2202–2205. doi: 10.1172/JCI137647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guzik T.J., Mohiddin S.A., Dimarco A., et al. COVID-19 and the cardiovascular system: implications for risk assessment, diagnosis, and treatment options. Cardiovasc Res. 2020;116:1666–1687. doi: 10.1093/cvr/cvaa106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richardson S., Hirsch J.S., Narasimhan M., et al. Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA. 2020 doi: 10.1001/jama.2020.6775. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Poyiadji N., Shahin G., Noujaim D., Stone M., Patel S., Griffith B. COVID-19-associated acute hemorrhagic necrotizing encephalopathy: imaging features. Radiology. 2020;296:E119–E120. doi: 10.1148/radiol.2020201187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Racicot K., Mor G. Risks associated with viral infections during pregnancy. J Clin Invest. 2017;127:1591–1599. doi: 10.1172/JCI87490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yan J., Guo J., Fan C., et al. Coronavirus disease 2019 in pregnant women: a report based on 116 cases. Am J Obstet Gynecol. 2020;223:111.e1–111.e14. doi: 10.1016/j.ajog.2020.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiao J. What are the risks of COVID-19 infection in pregnant women? Lancet. 2020;395:760–762. doi: 10.1016/S0140-6736(20)30365-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen H., Guo J., Wang C., et al. Clinical characteristics and intrauterine vertical transmission potential of COVID-19 infection in nine pregnant women: a retrospective review of medical records. Lancet. 2020;395:809–815. doi: 10.1016/S0140-6736(20)30360-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Algarroba G.N., Rekawek P., Vahanian S.A., et al. Visualization of severe acute respiratory syndrome coronavirus 2 virus invading the human placenta using electron microscopy. Am J Obstet Gynecol. 2020;223:275–278. doi: 10.1016/j.ajog.2020.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baud D., Greub G., Favre G., et al. Second-trimester miscarriage in a pregnant woman with SARS-CoV-2 infection. JAMA. 2020 doi: 10.1001/jama.2020.7233. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zamaniyan M., Ebadi A., Aghajanpoor Mir S., Rahmani Z., Haghshenas M., Azizi S. Preterm delivery, maternal death, and vertical transmission in a pregnant woman with COVID-19 infection. Prenat Diagn. 2020 doi: 10.1002/pd.5713. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong L., Tian J., He S., et al. Possible vertical transmission of SARS-CoV-2 from an infected mother to her newborn. JAMA. 2020 doi: 10.1001/jama.2020.4621. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vivanti A.J., Vauloup-Fellous C., Prevot S., et al. Transplacental transmission of SARS-CoV-2 infection. Nat Commun. 2020;11:3572. doi: 10.1038/s41467-020-17436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen D., Yang H., Cao Y., et al. Expert consensus for managing pregnant women and neonates born to mothers with suspected or confirmed novel coronavirus (COVID-19) infection. Int J Gynecol Obstet. 2020;149:130–136. doi: 10.1002/ijgo.13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wong S.F., Chow K.M., Leung T.N., et al. Pregnancy and perinatal outcomes of women with severe acute respiratory syndrome. Am J Obstet Gynecol. 2004;191:292–297. doi: 10.1016/j.ajog.2003.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dashraath P., Wong J.L.J., Lim M.X.K., et al. Coronavirus disease 2019 (COVID-19) pandemic and pregnancy. Am J Obstet Gynecol. 2020;222:521–531. doi: 10.1016/j.ajog.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alfaraj S.H., Al-Tawfiq J.A., Memish Z.A. Middle East respiratory syndrome coronavirus (MERS-CoV) infection during pregnancy: report of two cases & review of the literature. J Microbiol Immunol Infect. 2019;52:501–503. doi: 10.1016/j.jmii.2018.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lam C.M., Wong S.F., Leung T.N., et al. A case-controlled study comparing clinical course and outcomes of pregnant and non-pregnant women with severe acute respiratory syndrome. BJOG. 2004;111:771–774. doi: 10.1111/j.1471-0528.2004.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Panahi L., Amiri M., Pouy S. Risks of novel coronavirus disease (COVID-19) in pregnancy; a narrative review. Arch Acad Emerg Med. 2020;8:e34. [PMC free article] [PubMed] [Google Scholar]
- 21.Sentilhes L., De Marcillac F., Jouffrieau C., et al. Coronavirus disease 2019 in pregnancy was associated with maternal morbidity and preterm birth. Am J Obstet Gynecol. 2020 doi: 10.1016/j.ajog.2020.06.022. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann M., Kleine-Weber H., Schroeder S., et al. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell. 2020;181:271–280.e8. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q., Zhang Y., Wu L., et al. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell. 2020;181:894–904.e9. doi: 10.1016/j.cell.2020.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou P., Yang X.L., Wang X.G., et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Verdecchia P., Cavallini C., Spanevello A., Angeli F. The pivotal link between ACE2 deficiency and SARS-CoV-2 infection. Eur J Intern Med. 2020;76:14–20. doi: 10.1016/j.ejim.2020.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pringle K.G., Tadros M.A., Callister R.J., Lumbers E.R. The expression and localization of the human placental prorenin/renin-angiotensin system throughout pregnancy: roles in trophoblast invasion and angiogenesis? Placenta. 2011;32:956–962. doi: 10.1016/j.placenta.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 27.Li M., Chen L., Zhang J., Xiong C., Li X. The SARS-CoV-2 receptor ACE2 expression of maternal-fetal interface and fetal organs by single-cell transcriptome study. PLoS One. 2020;15 doi: 10.1371/journal.pone.0230295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pique-Regi R., Romero R., Tarca A.L., et al. Does the human placenta express the canonical cell entry mediators for SARS-CoV-2? Elife. 2020;9:e58716. doi: 10.7554/eLife.58716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imperio G.E., Javam M., Lye P., et al. Gestational age-dependent gene expression profiling of ATP-binding cassette transporters in the healthy human placenta. J Cell Mol Med. 2019;23:610–618. doi: 10.1111/jcmm.13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lye P., Bloise E., Dunk C., et al. Effect of oxygen on multidrug resistance in the first trimester human placenta. Placenta. 2013;34:817–823. doi: 10.1016/j.placenta.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 31.Lye P., Bloise E., Nadeem L., et al. Breast cancer resistance protein (BCRP/ABCG2) inhibits extra villous trophoblast migration: the impact of bacterial and viral infection. Cells. 2019;8:1150. doi: 10.3390/cells8101150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Drewlo S., Levytska K., Kingdom J. Revisiting the housekeeping genes of human placental development and insufficiency syndromes. Placenta. 2012;33:952–954. doi: 10.1016/j.placenta.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 33.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 34.Tenório M.B., Ferreira R.C., Moura F.A., Bueno N.B., de Oliveira A.C.M., Goulart M.O.F. Cross-talk between oxidative stress and inflammation in preeclampsia. Oxid Med Cell Longev. 2019;2019:8238727. doi: 10.1155/2019/8238727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sampaio W.O., Henrique de Castro C., Santos R.A., Schiffrin E.L., Touyz R.M. Angiotensin-(1-7) counterregulates angiotensin II signaling in human endothelial cells. Hypertension. 2007;50:1093–1098. doi: 10.1161/HYPERTENSIONAHA.106.084848. [DOI] [PubMed] [Google Scholar]
- 36.Gao Q., Tang J., Li N., et al. A novel mechanism of angiotensin II-regulated placental vascular tone in the development of hypertension in preeclampsia. Oncotarget. 2017;8:30734–30741. doi: 10.18632/oncotarget.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ueki N., Takeda S., Koya D., Kanasaki K. The relevance of the renin-angiotensin system in the development of drugs to combat preeclampsia. Int J Endocrinol. 2015;2015:572713. doi: 10.1155/2015/572713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hosier H., Farhadian S.F., Morotti R.A., et al. SARS-CoV-2 infection of the placenta. J Clin Invest. 2020 doi: 10.1172/JCI139569. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]