Abstract
Background
We studied 2 unrelated patients with immune thrombocytopenia and autoimmune hemolytic anemia in the setting of acute infections. One patient developed multisystem inflammatory syndrome in children in the setting of a severe acute respiratory syndrome coronavirus 2 infection.
Objectives
We sought to identify the mechanisms underlying the development of infection-driven autoimmune cytopenias.
Methods
Whole-exome sequencing was performed on both patients, and the impact of the identified variants was validated by functional assays using the patients’ PBMCs.
Results
Each patient was found to have a unique heterozygous truncation variant in suppressor of cytokine signaling 1 (SOCS1). SOCS1 is an essential negative regulator of type I and type II IFN signaling. The patients’ PBMCs showed increased levels of signal transducer and activator of transcription 1 phosphorylation and a transcriptional signature characterized by increased expression of type I and type II IFN-stimulated genes and proapoptotic genes. The enhanced IFN signature exhibited by the patients’ unstimulated PBMCs parallels the hyperinflammatory state associated with multisystem inflammatory syndrome in children, suggesting the contributions of SOCS1 in regulating the inflammatory response characteristic of multisystem inflammatory syndrome in children.
Conclusions
Heterozygous loss-of-function SOCS1 mutations are associated with enhanced IFN signaling and increased immune cell activation, thereby predisposing to infection-associated autoimmune cytopenias.
Key words: SOCS1, Evans syndrome, autoimmune hemolytic anemia, immune thrombocytopenia, COVID-19, MIS-C, SARS-CoV-2
Abbreviations used: AIHA, Autoimmune hemolytic anemia; COVID-19, Coronavirus disease 2019; ES, Evans syndrome; ISG, IFN-stimulated gene; ITP, Immune thrombocytopenia; JAK, Janus kinase; KIR, Kinase inhibitory region; MIS-C, Multisystem inflammatory syndrome in children; SARS-CoV-2, Severe acute respiratory syndrome coronavirus 2; SOCS, Suppressor of cytokine signaling; STAT, Signal transducer and activator of transcription
Background
Suppressor of cytokine signaling (SOCS) 1 is an essential negative regulator of type I and type II IFN signaling. Of the SOCS family members, SOCS1 binds with highest affinity to the substrate-binding pocket of Janus kinase (JAK) 1 and JAK2, thereby inhibiting phosphorylation of signal transducer and activator of transcription (STAT) 1 and STAT2.1 Complete deficiency of Socs1 in mice causes perinatal lethality due to type I and type II IFN-driven inflammatory disease, which can be reduced by neutralizing antibodies to either type I or type II IFNs or genetic deletion of their respective receptors.2 , 3 Furthermore, Socs1-haploinsufficient mice exhibit features of systemic lupus erythematosus, indicating the importance of biallelic Socs1 expression for self-tolerance.2 , 4 In a recent study detailing outcomes of whole-genome sequencing for patients with primary immunodeficiency, SOCS1 haploinsufficiency was briefly described in 2 individuals with recurrent bacterial infections and severe multisystemic autoimmunity.5 However, the immunologic sequelae of SOCS1 haploinsufficiency in humans remain incompletely understood.
Individuals with Evans syndrome (ES) present with immune thrombocytopenia (ITP), autoimmune hemolytic anemia (AIHA), and/or immune neutropenia arising from either primary or secondary causes. The relative risk of ES is significantly higher in children with monogenic disorders of immunity, whereas ES secondary to malignancy is the most common association in adults.6 In addition, viral and bacterial infections have been identified as triggers of ES.6 In this report, we present 2 unrelated children with different heterozygous loss-of-function variants in SOCS1, aberrant IFN signaling, and ES. One patient developed a severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection, precipitating multisystem inflammatory syndrome in children (MIS-C), which responded well to treatment with intravenous immunoglobulins and corticosteroids.
Results and discussion
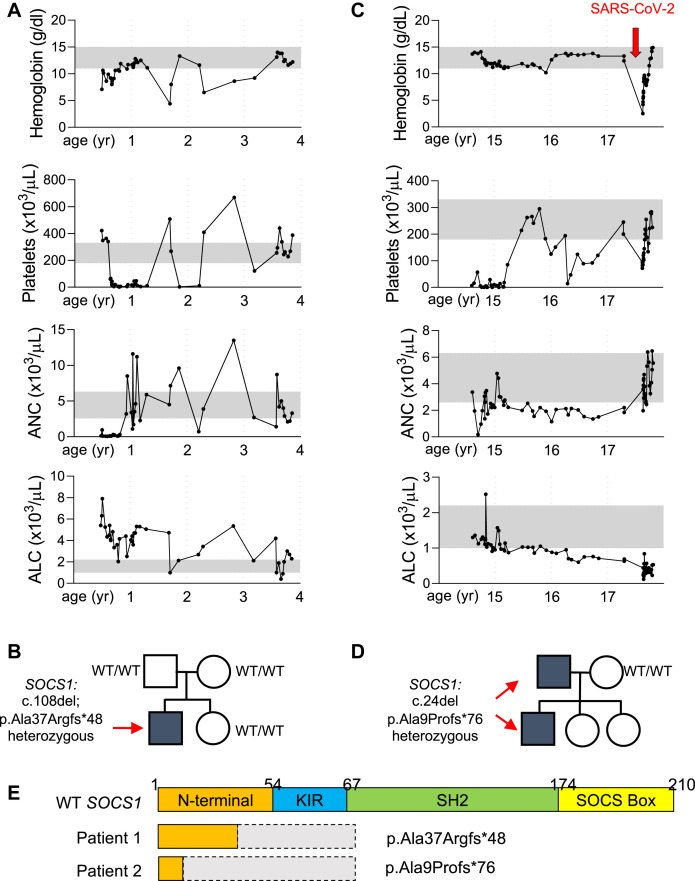
Patient 1 presented at age 5 months with fever, otitis media, oral ulcers, and diarrhea. At that time, he was anemic (hemoglobin, 7.1 g/dL; normal, 10.5-13.0 g/dL) and neutropenic (0.142 × 103 cells/μL; normal, 2.6-7.5 × 103 cells/μL). Several weeks later, he developed ITP (61 × 103 platelets/μL; normal, 215-448 × 103 platelets/μL; Fig 1 , A). At age 2 years, in addition to ITP and immune neutropenia, he developed a warm antibody AIHA, with a hemoglobin level of 4.4 g/dL (normal, 10.5-13.0 g/dL). He was initially treated with corticosteroids and subsequently transitioned to mycophenolate mofetil because of recurrence of AIHA with corticosteroid tapering. In addition to immune cytopenias, he had mild CD8+ T-cell lymphopenia and a predominance of naive IgD+CD27– B cells (Table I ). His percentage of CD4+CD25hiCD127low regulatory T cells was normal. His levels of IgG and IgA were reduced, but tetanus and pneumococcal vaccines titers were protective (Table I), and he had no history of significant infections. Whole-exome sequencing revealed heterozygosity for a truncation variant in SOCS1 (NM 003745.1: c.108delG; p.Ala37Argfs∗48; Fig 1, B). This variant has not been previously reported in the Genome Aggregation Database (gnomAD). The mutation arose de novo because it was absent in both parents.
Fig 1.
Two unique heterozygous truncation SOCS1 variants in 2 unrelated children with ES. A, Indicated laboratory values for patient 1 since initial diagnosis. B, Pedigree of the family of patient 1, who presented at age 5 months with anemia and neutropenia. WES identified a de novo SOCS1 variant. C, Indicated laboratory values for patient 2. The arrow indicates the recent hospital admission for SARS-CoV-2 infection. D, Pedigree of the family of patient 2, who presented with thrombocytopenia at age 14 years and recent development of autoimmune hemolytic anemia. WES identified a paternally inherited SOCS1 variant. E, Schematic of domains for wild-type (WT) SOCS1 and the truncation variants found in the patients. ALC, Absolute lymphocyte count; ANC, absolute neutrophil count; WES, Whole-exome sequencing.
Table I.
Hematologic and Immunologic parameters in patients with SOCS1 mutations
| Parameter | Patient 1 | Patient 2 | Patient 2 |
|---|---|---|---|
| Age (y) | 3 | 14 | 17, during COVID-19 |
| Hemogram | |||
| Hemoglobin (g/dL) | 9.2 (11.0-12.8) | 13.7 (11.0-14.3) | 2.5 (11.0-14.3) |
| WBCs (103 cells/μL) | 5.5 (6.0-10.5) | 5.3 (5.2-9.7) | 4.4 (5.2-9.7) |
| Neutrophils (103 cells/μL) | 2.7 (2.5-6.0) | 3.3 (2.7-6.7) | 3.6 (2.7-6.7) |
| Lymphocytes (103 cells/μL) | 2.1 (1.3-3.5) | 1.3 (1.0-2.2) | 0.4 (1.0-2.2) |
| Monocytes (103 cells/μL) | 0.5 (0.2-0.9) | 0.5 (0.2-0.8) | 0.3 (0.2-0.8) |
| Platelets (103 cells/ μL) | 121 (208-413) | 7 (168-339) | 94 (168-339) |
| Lymphocyte subsets | |||
| CD3+ (103 cells/μL) | 1.24 (1.40-6.20) | 0.81 (1.00-2.60) | 0.34 (1.00-2.60) |
| CD3+CD4+ (cells/μL) | 0.86 (0.7-2.20) | 0.40 (0.53-1.40) | 0.21 (0.53-1.40) |
| CD45RA+CCR7+ (% CD4+) | 65.2 (65.2-84.8) | 33.6 (31.3-69.6) | 7.3 (31.3-69.6) |
| CD45RA+CCR7– (% CD4+) | 1.7 (0.2-3.0) | 1.5 (0.2-2.1) | 0.3 (0.2-2.1) |
| CD45RA–CCR7+ (% CD4+) | 22.7 (10.5-23.2) | 28.7 (21.0-41.3) | 42.5 (21.0-41.3) |
| CD45RA–CCR7– (% CD4+) | 10.3 (2.9-9.8) | 36.2 (7.8-25.9) | 50.0 (7.8-25.9) |
| CD25hiCD127low (% CD4+) | 10.1 (6.3-14.3) | 5.5 (5.9-10.2) | Not done |
| CD3+CD8+ (cells/μL) | 0.32 (0.49-1.30) | 0.32 (0.33-1.10) | 0.09 (0.33-1.10) |
| CD45RA+CCR7+ (% CD8+) | 67.5 (39.0-89.0) | 77.3 (33.1-73.2) | 58.5 (33.1-73.2) |
| CD45RA+CCR7– (% CD8+) | 18.1 (4.8-30.0) | 5.3 (8.7-38.0) | 12.2 (8.7-38.0) |
| CD45RA–CCR7+ (% CD8+) | 2.2 (0.9-5.7) | 4.0 (2.6-8.7) | 8.2 (2.6-8.7) |
| CD45RA–CCR7– (% CD8+) | 12.2 (3.4-28.2) | 13.4 (8.8-44.4) | 21.1 (8.8-44.4) |
| CD19+ (103 cells/μL) | 0.83 (0.39-1.40) | 0.11 (0.11-0.57) | 0.14 (0.11-0.57) |
| IgD−CD27+ (% CD19+) | 4.4 (3.3-7.4) | 2.8 (8.7-25.6) | 1.6 (8.7-25.6) |
| IgD+CD27+ (% CD19+) | 4.3 (2.7-19.8) | 7.6 (4.6-18.2) | 2.9 (4.6-18.2) |
| IgD+CD27− (% CD19+) | 90.0 (54.0-88.4) | 87.7 (51.3-82.5) | 92.1 (51.3-82.5) |
| CD3–CD16+/CD56+ (103 cells/μL) | 134 (130-720) | 0.06 (0.07-0.48) | 0.05 (0.07-0.48) |
| Immunoglobulins | |||
| IgG (mg/dL) | 582 (600-1500) | 463 (639-1344) | 342 (639-1344) |
| IgM (mg/dL) | 40 (22-100) | 20 (34-210) | 42 (34-210) |
| IgA (mg/dL) | 30 (50-150) | 56 (70-312) | 26 (70-312) |
| Tetanus IgG (IU/mL) | 0.34 (0.15-7.00) | 0.26 (0.15-7.00) | Not done |
| Protective antipneumococcal IgG | 12 of 23 serotypes (≥12/23 serotypes) | 14 of 23 serotypes (≥12/23 serotypes) | Not done |
| T-cell activation | |||
| Soluble IL2R (units/mL) | 2,394 (<2,126) | 1,313 (45-1,105) | 15,990 (<1,033) |
WBC, White blood cell.
Age-specific reference ranges in parentheses are derived from healthy controls at Boston Children’s Hospital. Values in boldface are outside the reference range.
Patient 2 was diagnosed with ITP at age 14 years when he presented with diffuse petechiae and a platelet count of 7 × 103 cells/μL (normal, 180-320 × 103 cells/μL; Fig 1, C). Over the course of several years, he developed a weakly positive direct antiglobulin test indicative of erythrocyte autoantibodies without evidence of hemolysis. He subsequently developed neutropenia and lymphopenia (Fig 1, C). His thrombocytopenia improved with the thrombopoietin receptor agonist eltrombopag and mycophenolate mofetil, but he had persistent neutropenia and lymphopenia with reduced numbers of CD4+ and CD8+ T cells and a predominance of naive IgD+CD27+ B cells (Table I). His percentage of CD4+CD25hiCD127low regulatory T cells was minimally reduced. He had reduced levels of IgG, IgA, and IgM, but protective titers to the tetanus and pneumococcal vaccines (Table I). Because he had no history of significant infections, intravenous immunoglobulin was not started. Whole-exome sequencing identified a heterozygous truncation variant in SOCS1 (NM 003745.1: c.24delA; p.Ala9Profs∗76) inherited from his father, who had a history of mucosal bleeding and bruising primarily in his youth (Fig 1, D). This variant is also absent from the gnomAD database.
In March 2020, at the age of 17 years, patient 2 presented with shock-like physiology after having 2 days of fever, recurrent emesis, diarrhea, and dehydration. On presentation, he was tachycardic and hypoxic, with an oxygen saturation of 88%. Chest radiography showed no evidence of acute pneumonia. Laboratory evaluation revealed metabolic acidosis with a bicarbonate level of 15 mmol/L (normal, 20-31 mmol/L) and increased lactate level of 10 mmol/L (normal, 0.5-1 mmol/L). He had a warm IgG autoimmune hemolytic anemia, CD4+ and CD8+ leukopenia, and thrombocytopenia (Table I). He had ongoing hemolysis, as evidenced by his hemoglobin level of 2.5 g/dL, an elevated lactate dehydrogenase level of 1280 units/L (normal, 100-210 units/L), undetectable haptoglobin, and indirect hyperbilirubinemia (indirect bilirubin, 7.9 mg/dL; normal, 0-0.8 mg/dL). His C-reactive protein level was elevated at 48.9 mg/L (normal, <8.0 mg/L), as was his procalcitonin (4.28 mg/dL; normal, <0.08 mg/mL). His prothrombin time was elevated at 24.9 seconds (normal, 12-14.6 seconds). Nasopharyngeal swab was positive for SARS-CoV-2 by RT-PCR. Even after receiving methylprednisolone (1 mg/kg every 6 hours) for 1 day, he had a markedly elevated soluble IL-2 receptor level (15,990 pg/mL), which was increased from his previous level of 1313 pg/mL (normal, 45-1105 pg/mL), reflecting widespread T-cell activation. Although his AIHA was initially thought to be the most prominent feature of his SARS-CoV-2 infection,7 his history of fever, involvement of the gastrointestinal, hematologic, and respiratory systems, and elevated inflammatory markers were consistent with the subsequently described MIS-C.8 He was treated with fresh frozen plasma, intravenous immunoglobulin, and methylprednisolone, and he received multiple transfusions, with eventual resolution of his hypoxia and cytopenias. One month after recovery from COVID-19, while he was still maintained on a high-dose corticosteroid wean, patient 2 was found to have a normal soluble IL-2R level (577 pg/mL) and undetectable levels of all cytokines measured (IL-10, IL-12, IFN-γ, IL-4, IL-5, IL-13, IL-17, IL-1β, IL-6, IL-8, and TNF-α). Corticosteroids were discontinued 2 months after his acute infection and at that time, he was found to have detectable anti–SARS-CoV-2 antibodies (Elecsys anti–SARS-CoV-2 assay; Roche, Basel, Switzerland).
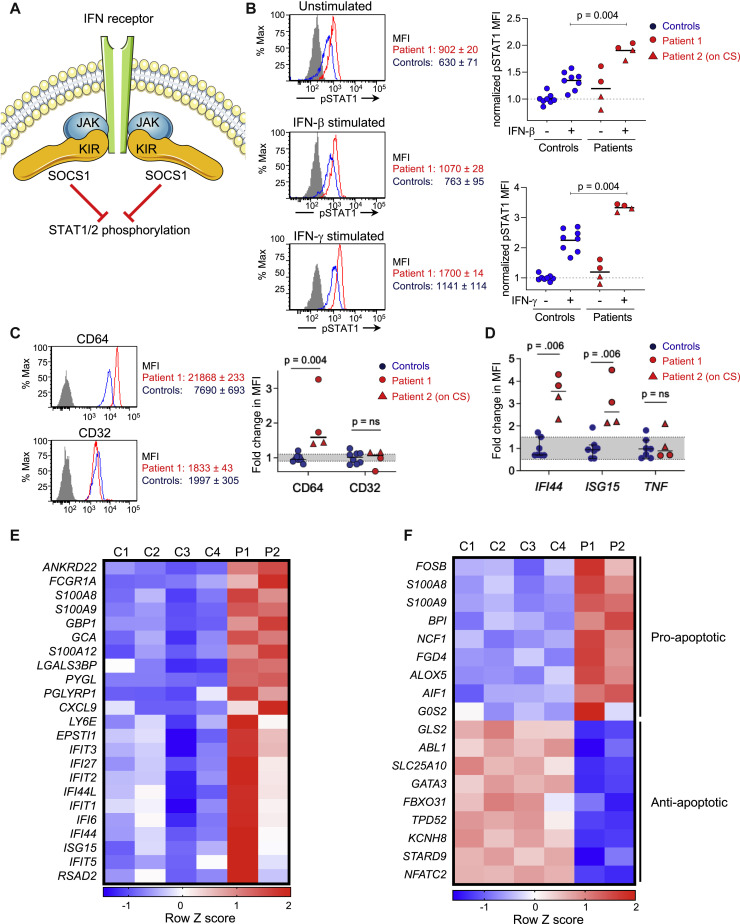
Both patients’ frameshift variants affect the N-terminal domain, upstream of the kinase inhibitory region (KIR), the SH2 dimerization domain, and the SOCS Box domain needed for ubiquitination of target proteins (Fig 1, E). Both variants are thus expected to result in SOCS1 haploinsufficiency. Of the SOCS family members, SOCS1 is the most potent JAK inhibitor, while its ubiquitination activity is 100-fold weaker than that of other family members.1 SOCS1-mediated inhibition of JAKs depends on the binding of the KIR domain to the substrate- binding groove within the JAKs.1 Therefore, both variants are predicted to abrogate the protein’s regulatory effect on IFN signaling. Because SOCS1 inhibits JAK1- and JAK2-mediated phosphorylation of STAT1 (Fig 2 , A),1 we assessed STAT1 phosphorylation in PBMCs from the patients and controls. After stimulation with either IFN-β or IFN-γ, monocytes from both patients demonstrated increased STAT1 phosphorylation compared with controls (Fig 2, B), thus suggesting increased JAK activity in the context of SOCS1 haploinsufficiency.
Fig 2.
Enhanced IFN and proapoptotic signaling associated with loss-of-function SOCS1 variants. Patient 2 was on corticosteroids (CSs) for 2 months at the time of sampling for the assays shown. A, Schematic of the interaction between SOCS1 and the JAKs. B, Quantification of phospho-STAT1 expression in CD14+ monocytes from healthy controls and patients before and after treatment with IFN-β (25 ng/mL) or IFN-γ (25 ng/mL) for 20 minutes. Histograms are representative of 2 independent experiments; the isotype control was comparable between controls and patients. Plotted data are combined from 2 independent experiments, each with 4 controls and 2 patients. C, Expression of the IFN-stimulated gene CD64 compared with CD32, which is independent of IFN signaling, on CD14+ monocytes from healthy controls and patients. The flow cytometry histogram (left) is representative of 2 experiments, which were pooled for quantification (right). The MFI for each sample was normalized to the average expression of healthy controls. Gray shading indicates the 25% to 75% quartiles of control values. D. Quantitative PCR analysis of 2 additional IFN-stimulated genes, IFI44 and ISG15, compared with TNF, which is IFN-independent, in PBMCs from controls and patients. Fold expression for each sample was normalized to the average expression of healthy controls. Gray shading indicates the 25% to 75% quartiles of control values. Data are combined from 2 independent experiments. E and F, Heat-map of type I and type II ISGs (Fig 2, E) as well as proapoptotic vs antiapoptotic genes (Fig 2, F) in 4 controls and patients. MFI, Mean fluorescence intensity.
We next investigated the expression of IFN-stimulated genes (ISGs), which are downstream of IFN-JAK-STAT signaling. Under physiologic conditions, basal IFN signaling primes cells for a rapid response to infectious pathogens.9 Increased tonic expression of ISGs in unstimulated PBMCs is a reliable indicator of disease activity in patients with monogenic interferonopathies as well as systemic lupus erythematosus.10 , 11 To determine whether SOCS1 haploinsufficiency results in increased tonic type I and type II IFN signaling, we assessed expression of ISGs in the patients’ unstimulated PBMCs. Notably, patient 2 had been treated with prednisone and mycophenolic acid for more than 2 months at the time of this assay. CD64, an ISG expressed by monocytes,12 was significantly enhanced on unstimulated monocytes from both patients compared with healthy controls (Fig 2, C). In contrast, the expression of CD32, which is not regulated by IFN signaling, was similar between patients and controls (Fig 2, C). Consistent with these findings, quantitative PCR of unstimulated PBMCs from both patients showed increased expression of 2 additional ISGs, IFI44 and ISG15, compared with 6 healthy controls (Fig 2, D). The specificity of these observations was supported by both patients’ normal expression of TNF-α, a cytokine expressed independently of IFN signaling. Whole-transcriptome analysis of unstimulated PBMCs from the patients and 4 controls identified 114 genes with a greater than or equal to 1.5-fold difference between the 2 groups, of which 42 were upregulated. Differentially expressed genes were enriched in pathways of the inflammatory response (corrected P value = 3.56 × 10−4), with increased expression of genes downstream of type I and type II IFN signaling (Fig 2, E). Differentially expressed genes in the patients’ PBMCs were also enriched in pathways involved in the apoptosis of monocytes and activated T cells (corrected P = 1.42 × 10−2), demonstrated by increased expression of proapoptotic genes and reduced expression of antiapoptosis genes (Fig 2, F). The variability of gene expression in patient 2 likely reflected the variable suppression of inflammatory genes known to be associated with steroid use.13 Collectively, these findings demonstrate that patients with SOCS1 haploinsufficiency have enhanced STAT1 phosphorylation and a transcriptional signature indicative of enhanced IFN signaling and apoptosis.
Increased type I and type II IFN signaling likely have distinct contributions to our patients’ cytopenias. Both our patients developed a warm autoantibody-positive hemolytic anemia. Enhanced type I IFN signaling is associated with the development of autoantibodies and cytopenias in systemic lupus erythematosus and a subset of monogenic interferonopathies, whereas therapeutic use of IFN-α has been reported to cause ES.6 Type I IFNs activate immature myeloid dendritic cells that promote the differentiation and expansion of autoreactive B cells, leading to the generation of autoantibodies.14 Type I IFNs also stimulate the expansion of antigen-specific central memory CD8+ T cells, which may include autoreactive T cells.15 The robust induction of type I IFNs during infections increases apoptosis through multiple mechanisms, including the activation of caspases and inflammasomes.16 Chronic IFN-γ activation is another likely driver of our patients’ cytopenias. IFN-γ impairs multilineage differentiation by inhibiting the proliferation and differentiation of multipotent progenitors cells, thereby leading to pancytopenia.17 In addition, IFN-γ increases T-cell activation, evident in our patients’ consistently elevated soluble CD25 levels, and is required for activation-induced T-cell death.18 By modulating the response to inflammatory cytokines, physiologic levels of SOCS1 thus promote cell survival, which is further underscored by the proapoptotic transcriptional signature of our patients’ PBMCs. Both our patients responded well to treatment with mycophenolate mofetil. Hematopoietic stem cell transplantation is curative for numerous disorders of immune dysregulation, but it has not been investigated yet in mouse models of SOCS1 deficiency. Because SOCS1 is expressed by both stromal and hematopoietic cells, additional studies will be needed to determine whether hematopoietic stem cell transplantation is effective for patients with SOCS1 defects.
To our knowledge, no monogenic risk factors for either COVID-19–associated autoimmunity or MIS-C have been identified to date.19 AIHA, ITP, or ES has been reported in a minority of adults with acute SARS-CoV-2 infections.19 All the patients older than 50 years in these reports had malignancy and/or additional known risk factors for severe COVID-19 (Table II ). In contrast, the patients younger than 50 years had no reported risk factors for severe COVID-19; pre-existing autoimmunity was either absent or remote, and no one was taking immunomodulatory medications (Table II). Patients younger than 50 years with COVID-19–induced autoimmune cytopenias also had pulmonary symptoms and findings commonly seen in adults with SARS-CoV-2 infections, whereas patient 2 in our study had gastrointestinal involvement and coagulopathy, which are more characteristic of MIS-C.8
Table II.
Summary of reported cases of SARS-CoV-2–associated autoimmune cytopenias in adults
| Clinical variable | Patients younger than 50 y |
Patients older than 60 y |
||||
|---|---|---|---|---|---|---|
| This case | Li et al20 | Lopez et al21 | Zagorski et al22 | Capes et al23 | Lazarian et al24 | |
| Age (y) | 17 | 39 | 46 | 46 | 62 | 61-89 (7 patients) |
| Sex | Male | Male | Female | Female | Male | 3 female and 4 male |
| SARS-CoV-2 RT-PCR result | Positive | Positive | Positive | Positive | Positive | Positive (7 of 7) |
| Hemoglobulin (g/dL) | 2.5 | 6.4 | 9.7 | 5.3 | 12, decreased to 6.9 | 3.8-10.8 |
| Platelets (cells/μL) | 94,000 | 3,000 | 43,000 | 318,000 | 101,000 | Not specified |
| Direct antigen test | IgG+ and C3+ Warm agglutinin+ |
Positive, not further specified | IgG+ and C3+ | IgG+ and C3+ Cold agglutinin+ |
C3+ Cold agglutinin+ |
IgG+ (5 of 7) C3+ (4 of 7) Warm agglutinin+ (4 of 7) Cold agglutinin+ (3 of 7) |
| Prior autoimmunity | ITP | None | Congenital thrombocytopenia | ITP during pregnancy | Not reported | Diabetes (2 of 7) |
| Other past medical history | Hypogammaglobulinemia | None | None | None | Arterial hypertension Heavy smoking Oropharyngeal squamous cell carcinoma |
Malignancy (5 of 7) MGUS (1 of 7) Hypertension (4 of 7) Chronic renal failure (3 of 7) Cardiac disease (2 of 7) Obesity (1 of 7) COPD (1 of 7) |
| Other symptoms | Fever Vomiting Diarrhea Dyspnea Coagulopathy |
Fever Dyspnea Cough Hemoptysis, epistaxis |
Fever Pulmonary consolidation |
Fever Pulmonary infiltrates |
Fever Pulmonary infiltrates |
Pulmonary infiltrates (7 of 7) |
| Other infectious disease testing | Negative blood culture | Not reported | Negative: Influenza, RSV, viral respiratory panel Blood cultures Streptococcus pneumoniae and legionella pneumophila urine antigens |
Negative: Hepatitis viral panel Respiratory pathogen panel, including Mycoplasma pneumoniae HIV1/2 antibody |
Negative: Mycoplasma pneumoniae, Legionella pneumophila, Chlamydia pneumoniae, adenovirus, influenza Hepatitis B and C, HIV antibodies |
Not reported |
| Treatment | Corticosteroids IVIG Fresh frozen plasma Transfusions |
IVIG | IVIG Transfusions Corticosteroids |
Transfusions | Transfusions | Corticosteroids (5 of 7) Rituximab (2 of 7) Transfusions (2 of 7) |
| Outcome | Recovered | Recovered | Recovered | Death; acute hemolysis, hypoxia, cardiac arrest | Recovering from respiratory failure | Recovering |
COPD, Chronic obstructive pulmonary disease; IVIG, intravenous immunoglobulin; MGUS, monoclonal gammapathy of unknown significance; RSV, respiratory syncytial virus.
MIS-C is a heterogeneous disease of multisystemic inflammation occurring in the acute or postinfectious phase of SARS-CoV-2 infections.8 In the largest published cohort of patients from the United States with MIS-C, 39% and 31% had either a positive RT-PCR or serologies for SARS-CoV-2, respectively, whereas 30% had exposure to a known case of COVID-19 within 4 weeks of symptom onset.8 Five percent of these patients had pre-existing immune or autoimmune disorders, although none had hemolysis during MIS-C.8 Despite this clinical variability, laboratory evidence of systemic inflammation is an essential and consistent diagnostic feature of MIS-C. IL-2R, an indicator of widespread T-cell activation, and CXCL9, a marker of IFN activation, are among the inflammatory mediators elevated in multiple patients with MIS-C.25 Here, we show that patients with SOCS1 haploinsufficiency exhibit T-cell activation and increased type I and type II IFN signaling, even in the absence of infection. Both patients had elevated soluble IL-2R levels even while on the T-cell–suppressive agent mycophenolate mofetil; during MIS-C, patient 2 developed an even more markedly elevated soluble IL-2R level. In the absence of stimulation or infections, PBMCs from both patients exhibit an enhanced IFN signature. In the general population, loss-of-function variants in SOCS1 are rare, with a minor allelic frequency of 4.27 × 10−6 to 7.52 × 10−6 in gnomAD. Missense variants in the KIR domain, which can impair the inhibitory function of SOCS1 to a lesser extent than loss of the entire KIR domain,1 occur with a minor allelic frequency of 1.15 × 10−5 to 3.21 × 10−5. Future studies are needed to determine whether additional individuals with defects in SOCS1 are susceptible to SARS-CoV-2–triggered autoimmunity.
In addition to ES, both our patients demonstrate a mild humoral defect, because they have hypogammaglobulinemia and elevated percentages of IgD+CD27– naive B cells. Despite this, both patients developed normal antibody titers to the pneumococcal and tetanus vaccines, and patient 2 developed detectable anti–SARS-CoV-2 antibodies even while receiving prednisone for 2 months. Because neither patient developed recurrent or severe bacterial infections, the patients’ hypogammaglobulinemia has thus far not shown a significant biologic impact.
In summary, this study details 2 unrelated patients with SOCS1 haploinsufficiency and ES. We show that both patients have increased phosphorylation of STAT1, elevated T-cell activation, and enhanced IFN signaling, all concordant with reduced SOCS1 activity. For both patients, autoimmunity flared in the setting of infection-driven inflammation. This study highlights the utility of whole-exome sequencing for identifying genetically susceptible individuals at risk for developing autoimmune complications of SARS-CoV-2, including MIS-C.
For detailed methods, please see the Methods section in this article’s Online Repository at www.jacionline.org.
Clinical implications.
Loss-of-function SOCS1 variants are associated with increased immune cell activation, which predisposes to infection-associated autoimmune cytopenias.
Acknowledgments
We thank the patients and their families for their participation in this study. We thank the Turkel family and the Samara Jan Turkel Clinical Center for providing support for the clinical care of these patients.
Footnotes
This work was supported by the National Institutes of Health: National Institute of Arthritis and Musculoskeletal and Skin Diseases (grant no. K08-AR074562 to P.Y.L.), Centers for Disease Control and Prevention (to A.G.R.), Eunice Kennedy Shriver National Institute of Child Health and Human Development (grant no. R21HD095228 to A.G.R.), National Institute of Allergy and Infectious Diseases (grant no. 5K08AI116979 to J.C. and grant no. R01-AI139633 to R.S.G.), the Perkin Fund (to R.S.G.), and the Samara Jan Turkel Center for Autoimmune Diseases (to J.C.).
Disclosure of potential conflict of interest: The authors declare that they have no relevant conflicts of interest.
Methods
Study design and subjects
The studies were approved by the Institutional Review Board at Boston Children’s Hospital. Informed consent was provided by participants or legal guardians. Antibodies to SARS-CoV-2 were tested in the Boston Children’s Hospital clinical laboratory using Elecsys assay (Roche), which detects high-affinity IgG, IgM, and IgA antibodies to SARS-CoV-2.
Whole-exome sequencing
Genomic DNA was isolated from the patients’ PBMCs using the Gentra Puregene blood kit (Qiagen, Hilden, Germany). Library preparation was performed using the Agilent SureSelect V6 kit, followed by sequencing on an Illumina HiSeq-2000 (Illumina, Inc, San Diego, Calif). The average coverage of the exome was 60×. Sequences were aligned for variant calling and annotation with the human genome reference sequence (hg19 build). Minor allelic frequencies for the specified variants were identified using the Genome Aggregation Database (gnomAD).
Flow cytometry
For quantitation of STAT1 and STAT2 phosphorylation, PBMCs were stimulated with recombinant human IFN-β or IFN-γ for 20 minutes before fixation with 4% paraformaldehyde. Antibodies were purchased from Biolegend (Dedham, Mass) unless otherwise stated. Antibodies used in this study include antihuman CD14 (RRID: AB_2565887 and AB_2566791), antihuman CD32 (AB_314337), antihuman CD64 (AB_2561583), antihuman phospho-STAT1Ser727 (AB_2650785), and antihuman phospho-STAT2 Tyr689 (AB_2608580; ThermoFisher Scientific, Waltham, Mass).
Quantitative PCR
Primers used in this study include IFI44 (forward: CTGGGGCTGAGTGAGAAAGA; reverse: AGCGATGGGGAATCAATGTA), ISG15 (forward: GAGGCAGCGAACTCATCTTT; reverse: CCAGCATCTTCACCGTCAG), and TNF (forward: CCAGGGACCTCTCTCTAATCA; reverse: TCAGCTTGAGGGTTTGCTAC).
Transcriptome analysis
RNA was purified from PBMCs using the RNeasy Mini Kit (Qiagen). cDNA was then synthesized from 10 ng of total RNA using SuperScript VILO cDNA Synthesis Kit (ThermoFisher Scientific). Barcoded libraries were prepared using the Ion AmpliSeq Transcriptome Human Gene Expression Kit as per the manufacturer’s protocol and sequenced using an Ion S5 system. Differential gene expression analysis was performed using the AmpliSeqRNA plugin (ThermoFisher). Pathway analysis was done using Ingenuity Pathway Analysis (Qiagen) on genes with at least 1.5-fold difference between controls and patients and a false-discovery rate of less than 0.25, as previously published.
Statistical analysis
For quantitative variables, differences between 2 groups were analyzed by Mann-Whitney U test. All tests were 2-sided, and P less than .05 was considered significant. Statistical analyses were performed using Prism 8.0 software (GraphPad Software, San Diego, Calif).
References
- 1.Liau N.P.D., Laktyushin A., Lucet I.S., Murphy J.M., Yao S., Whitlock E. The molecular basis of JAK/STAT inhibition by SOCS1. Nat Commun. 2018;9:1558. doi: 10.1038/s41467-018-04013-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alexander W.S., Starr R., Fenner J.E., Scott C.L., Handman E., Sprigg N.S. SOCS1 is a critical inhibitor of interferon γ signaling and prevents the potentially fatal neonatal actions of this cytokine. Cell. 1999;98:597–608. doi: 10.1016/s0092-8674(00)80047-1. [DOI] [PubMed] [Google Scholar]
- 3.Fenner J.E., Starr R., Cornish A.L., Zhang J.-G., Metcalf D., Schreiber R.D. Suppressor of cytokine signaling 1 regulates the immune response to infection by a unique inhibition of type I interferon activity. Nat Immunol. 2006;7:33–39. doi: 10.1038/ni1287. [DOI] [PubMed] [Google Scholar]
- 4.Fujimoto M. Inadequate induction of suppressor of cytokine signaling-1 causes systemic autoimmune diseases. Int Immunol. 2004;16:303–314. doi: 10.1093/intimm/dxh030. [DOI] [PubMed] [Google Scholar]
- 5.Thaventhiran J.E.D., Lango Allen H., Burren O.S., Rae W., Greene D., Staples E. Whole-genome sequencing of a sporadic primary immunodeficiency cohort. Nature. 2020;583:90–95. doi: 10.1038/s41586-020-2265-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaime-Pérez J.C., Aguilar-Calderón P.E., Salazar-Cavazos L., Gómez-Almaguer D. Evans syndrome: clinical perspectives, biological insights and treatment modalities. J Blood Med. 2018;9:171–184. doi: 10.2147/JBM.S176144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wahlster L, Weichert-Leahey N, Trissal M, Grace RF, Sankaran VG. COVID-19 presenting with autoimmune hemolytic anemia in the setting of underlying immune dysregulation [published online ahead of print June 3, 2020]. Pediatr Blood Cancer. https://doi.org/10.1002/pbc.28382. [DOI] [PMC free article] [PubMed]
- 8.Feldstein L.R., Rose E.B., Horwitz S.M., Collins J.P., Newhams M.M., Son M.B.F. Multisystem inflammatory syndrome in U.S. children and adolescents. N Engl J Med. 2020;383:334–346. doi: 10.1056/NEJMoa2021680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barrat F.J., Crow M.K., Ivashkiv L.B. Interferon target-gene expression and epigenomic signatures in health and disease. Nat Immunol. 2019;20:1574–1583. doi: 10.1038/s41590-019-0466-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice G.I., del Toro Duany Y., Jenkinson E.M., Forte G.M.A., Anderson B.H., Ariaudo G. Gain-of-function mutations in IFIH1 cause a spectrum of human disease phenotypes associated with upregulated type I interferon signaling. Nat Genet. 2014;46:503–509. doi: 10.1038/ng.2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rice G.I., Melki I., Frémond M.-L., Briggs T.A., Rodero M.P., Kitabayashi N. Assessment of type I interferon signaling in pediatric inflammatory disease. J Clin Immunol. 2017;37:123–132. doi: 10.1007/s10875-016-0359-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y., Lee P.Y., Kellner E.S., Paulus M., Switanek J., Xu Y. Monocyte surface expression of Fcγ receptor RI (CD64), a biomarker reflecting type-I interferon levels in systemic lupus erythematosus. Arthritis Res Ther. 2010;12:R90. doi: 10.1186/ar3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guiducci C., Gong M., Xu Z., Gill M., Chaussabel D., Meeker T. TLR recognition of self nucleic acids hampers glucocorticoid activity in lupus. Nature. 2010;465:937–941. doi: 10.1038/nature09102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kiefer K., Oropallo M.A., Cancro M.P., Marshak-Rothstein A. Role of type I interferons in the activation of autoreactive B cells. Immunol Cell Biol. 2012;90:498–504. doi: 10.1038/icb.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Srivastava S., Koch M.A., Pepper M., Campbell D.J. Type I interferons directly inhibit regulatory T cells to allow optimal antiviral T cell responses during acute LCMV infection. J Exp Med. 2014;211:961–974. doi: 10.1084/jem.20131556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malireddi R.K.S., Kanneganti T.-D. Role of type I interferons in inflammasome activation, cell death, and disease during microbial infection. Front Cell Infect Microbiol. 2013;3:77. doi: 10.3389/fcimb.2013.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lin F., Karwan M., Saleh B., Hodge D.L., Chan T., Boelte K.C. IFN-γ causes aplastic anemia by altering hematopoietic stem/progenitor cell composition and disrupting lineage differentiation. Blood. 2014;124:3699–3708. doi: 10.1182/blood-2014-01-549527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Refaeli Y., Van Parijs L., Alexander S.I., Abbas A.K. Interferon γ is required for activation-induced death of T lymphocytes. J Exp Med. 2002;196:999–1005. doi: 10.1084/jem.20020666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galeotti C., Bayry J. Autoimmune and inflammatory diseases following COVID-19. Nat Rev Rheumatol. 2020;16:413–414. doi: 10.1038/s41584-020-0448-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li M., Nguyen C.B., Yeung Z., Sanchez K., Rosen D., Bushan S. Evans syndrome in a patient with COVID-19. Br J Haematol. 2020;190:e59–e61. doi: 10.1111/bjh.16846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopez C., Kim J., Pandey A., Huang T., DeLoughery T.G. Simultaneous onset of COVID-19 and autoimmune haemolytic anaemia. Br J Haematol. 2020;190:31–32. doi: 10.1111/bjh.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zagorski E., Pawar T., Rahimian S., Forman D. Cold agglutinin autoimmune haemolytic anaemia associated with novel coronavirus (COVID-19) Br J Haematol. 2020;190:e183–e184. doi: 10.1111/bjh.16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capes A., Bailly S., Hantson P., Gerard L., Laterre P.-F. COVID-19 infection associated with autoimmune hemolytic anemia. Ann Hematol. 2020;99:1679–1680. doi: 10.1007/s00277-020-04137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lazarian G., Quinquenel A., Bellal M., Siavellis J., Jacquy C., Re D. Autoimmune haemolytic anaemia associated with COVID-19 infection. Br J Haematol. 2020;190:29–31. doi: 10.1111/bjh.16794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung EW, Zachariah P, Gorelik M, Boneparth A, Kernie SG, Orange JS, et al. Multisystem inflammatory syndrome related to COVID-19 in previously healthy children and adolescents in New York City [published online ahead of print June 8, 2020]. JAMA. https://doi.org/10.1001/jama.2020.10374. [DOI] [PMC free article] [PubMed]
References
- Li M., Nguyen C.B., Yeung Z., Sanchez K., Rosen D., Bushan S. Evans syndrome in a patient with COVID-19. Br J Haematol. 2020;190:e59–e61. doi: 10.1111/bjh.16846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez C., Kim J., Pandey A., Huang T., DeLoughery T.G. Simultaneous onset of COVID-19 and autoimmune haemolytic anaemia. Br J Haematol. 2020;190:31–32. doi: 10.1111/bjh.16786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagorski E., Pawar T., Rahimian S., Forman D. Cold agglutinin autoimmune haemolytic anaemia associated with novel coronavirus (COVID-19) Br J Haematol. 2020;190:e183–e184. doi: 10.1111/bjh.16892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes A., Bailly S., Hantson P., Gerard L., Laterre P.-F. COVID-19 infection associated with autoimmune hemolytic anemia. Ann Hematol. 2020;99:1679–1680. doi: 10.1007/s00277-020-04137-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarian G., Quinquenel A., Bellal M., Siavellis J., Jacquy C., Re D. Autoimmune haemolytic anaemia associated with COVID-19 infection. Br J Haematol. 2020;190:29–31. doi: 10.1111/bjh.16794. [DOI] [PMC free article] [PubMed] [Google Scholar]