Abstract
This is the first genome-wide association study with the Hybrid Mouse Diversity Panel (HDMP) to define the genetic landscape of the variation in the suprathreshold wave 1 amplitude of the auditory brainstem response (ABR) both pre- and post-noise exposure. This measure is correlated with the density of the auditory neurons (AN) and/or the compliment of synaptic ribbons within the inner hair cells of the mouse cochlea. We analyzed suprathreshold ABR for 635 mice from 102 HMDP strains pre- and post-noise exposure (108 dB 10 kHz octave band noise exposure for 2 h) using auditory brainstem response (ABR) wave 1 suprathreshold amplitudes as part of a large survey (Myint et al., Hear Res 332:113–120, 2016). Genome-wide significance levels for pre- and post-exposure wave 1 amplitude across the HMDP were performed using FaST-LMM. Synaptic ribbon counts (Ctbp2 and mGluR2) were analyzed for the extreme strains within the HMDP. ABR wave 1 amplitude varied across all strains of the HMDP with differences ranging between 2.42 and 3.82-fold pre-exposure and between 2.43 and 7.5-fold post-exposure with several tone burst stimuli (4 kHz, 8 kHz, 12 kHz, 16 kHz, 24 kHz, and 32 kHz). Immunolabeling of paired synaptic ribbons and glutamate receptors of strains with the highest and lowest wave 1 values pre- and post-exposure revealed significant differences in functional synaptic ribbon counts. Genome-wide association analysis identified genome-wide significant threshold associations on chromosome 3 (24 kHz; JAX00105429; p < 1.12E-06) and chromosome 16 (16 kHz; JAX00424604; p < 9.02E-07) prior to noise exposure and significant associations on chromosomes 2 (32 kHz; JAX00497967; p < 3.68E-08) and 13 (8 kHz; JAX00049416; 1.07E-06) after noise exposure. In order to prioritize candidate genes, we generated cis-eQTLs from microarray profiling of RNA isolated from whole cochleae in 64 of the tested strains.
This is the first report of a genome-wide association analysis, controlled for population structure, to explore the genetic landscape of suprathreshold wave 1 amplitude measurements of the mouse ABR. We have defined two genomic regions associated with wave 1 amplitude variation prior to noise exposure and an additional two associated with variation after noise exposure.
Keywords: GWAS, HMDP, NIHL, eQTL
Introduction
The sensation of hearing is the result of mechanical impulses (sound waves) being transmitted through the fluid-filled cochlea and coded into neural impulses that travel along the auditory nerve cranial through the brainstem to higher association centers within the cerebral cortex. Each auditory nerve fiber (ANF) contacts a single inner hair cell within the organ of Corti (Eybalin 1993). There is increasing evidence that the ability to hear and understand as we age, in a variety of environments, is dictated by the neural connectivity of the inner hair cells within the cochlea and their contacts with ANFs. Recent data in both mice and humans suggests there exists a “hidden hearing loss” resulting from synaptopathy between inner hair cells (IHCs) and type-I ANFs (Liberman 2017). Disruption of these synapses not only leads to denervation but also a slow degeneration of spiral ganglion neurons (Shi et al. 2016). In humans, this phenomenon has been linked to difficulty understanding speech in complex sound environments (Liberman and Kujawa 2017).
While there are few practical ways of identifying these types of losses in the clinical setting, there is evidence in mice that cochlear synaptopathy can be captured by the amplitude of the first wave (wave I) of the auditory brainstem response (ABR) after a suprathreshold stimulus (Kujawa and Liberman 2015). In several experiments, noise exposure led to a convincing permanent decline in the suprathreshold ABR wave I amplitude despite recovery of otoacoustic emissions and ABR thresholds. It has been determined that the wave I amplitude correlates strongly with the integrity of auditory synapses and/or the compliment AFNs (Kobel et al. 2017).
Recently, evidence for this “hidden hearing loss” has been demonstrated in human temporal bone specimens. (Liberman et al. 2016). It appears that noise exposure over time results in synaptic ribbon loss. In addition, age-related hearing loss (ARHL) has been shown to result from loss of sensory cells and neurons (Schuknecht 1964). It has been shown in mice that ARHL results from a similar synaptic loss between IHCs and ANFs and this cochlear synaptopathy also precedes hair cell loss and permanent threshold shifts in the aging mouse ear (Sergeyenko et al. 2013).
Our laboratory has recently described baseline and noise-induced threshold shifts in 100 strains of the Hybrid Mouse Diversity Panel (HMDP) (Myint et al. 2016) and, with these data, we have begun to define the genetic landscape of these complex traits (Crow et al. 2015) (Lavinsky et al. 2016). The HMDP is a collection of classical inbred (CI) and recombinant inbred (RI) strains whose genomes have been sequenced and/or genotyped at high resolution. In this manuscript we have begun to explore the genetic landscape of cochlear synaptopathy and determine candidate loci and genes. Herein we report the first characterization of suprathreshold wave 1 amplitude of the mouse ABR in over 100 mouse strains and perform the first genome-wide association study (GWAS) in mice corrected for population structure both before and after noise exposure.
Materials and Methods
Ethics Statement
The Institutional Care and Use Committee (IACUC) at University of California San Diego (IACUC 17178) and University of Southern California (IACUC 12033) endorsed the animal protocol for the Hybrid Mouse Diversity Panel (HMDP) inbred strains. Strains and genotypes are accessible from the Jackson Laboratories (www.jax.org). All results required to corroborate the conclusions presented here are provided entirely within this article.
Hybrid Mouse Diversity Panel Strains and Genotypes
The HMDP is a collection of common inbred strains composed of 30 classical inbred strains (CI) providing genetic resolution and allelic diversity, and more than 70 recombinant inbred strains (RI) that enhance resolution and increase power (Bennett et al. 2010). Five-week-old female mice (n = 635) from 102 Hybrid Mouse Diversity Panel strains (n = 6–7/strain) were purchased from the Jackson Laboratories. A detailed characterization of the HMDP is provided in Bennett et al. (2010). Mice were aged until 5 weeks and accommodated in sterilized cages with autoclaved food and water with an average daily ambient noise level of 40 dB. The phenotypes in the HMDP strains were managed using genotypes of 500,000 single nucleotide polymorphisms (SNPs) obtained from the Mouse Diversity Array (minor allele frequencies > 5 %; missing genotype frequencies < 10 %) (Van Nas et al. 2010).
ABR Wave 1 P1-N1 Values for Wave 1 Suprathreshold Amplitude Calculations
In order to analyze pre- and post-noise exposure ABR wave 1 P1-N1 measurements, stainless steel electrodes were placed subcutaneously at the vertex of the head and the right mastoid, with a ground electrode at the base of the tail. Mice were anesthetized with an intraperitoneal injection of ketamine (80 mg/kg body wt) and xylazine (16 mg/kg body wt). Mouse body temperature was maintained through the use of a TCAT-2DF temperature controller and the HP-4 M heating plate (Physitemp Instruments Inc., Clifton, NJ). Artificial tear ointment was applied to the eyes and each mouse was recovered on a heating pad.
Test sounds were presented using a (Intelligent Hearing Systems) speaker attached to an 8-in. long tube that was inserted into the ear canal. Auditory signals were presented as tone pips with a rise and a fall time of 0.5 msec and a total duration of 5 msec at the frequencies 4, 8, 12, 16, 24, and 32 kHz. Tone pips were delivered below threshold and then increased in 5 dB increments until a maximum of 100 dB was reached. Signals were presented at a rate of 30/s. Responses were filtered with a 0.3–3 kHz pass-band (× 10,000 times). For each stimulus intensity 512 waveforms were averaged. Data was stored for offline analysis of ABR peak-to-peak (P1-N1) values for wave 1 amplitudes at 80 dB SPL. Post-exposure thresholds were evaluated by the same method 2 weeks post-exposure.
All hearing tests were performed in a separate (MAC-1 soundproof) chamber in order to eliminate both environmental and electrical noise. An acquisition board (National Instruments Corporation, Austin, TX) was regulated by custom software used to generate the stimuli and to measure the responses. Stimuli were provided by a custom acoustic system (two miniature speakers, and sound pressure was measured by a condenser microphone).
Noise Exposure Protocol
Using a method adapted from Kujawa and Liberman (2009) 6-week-old mice were exposed for 2 h to 10 kHz octave band noise (OBN) at 108 dB SPL. The noise exposure protocol was previously described by White et al. (2009). The cage was arranged in a soundproof chamber (MAC-1) created by Industrial Acoustics (IAC, Bronx, NY) and the sound chamber was lined with soundproofing acoustical foam. Noise recordings were performed with a speaker (Fostex FT17H Tweeter) constructed into the top of the sound chamber. Calibration of the deleterious noise was done with a B&K sound level meter with a variation (1.5 dB) over the cage. For 2 h, mice were positioned in a circular exposure cage with 4 shaped compartments and were capable to move about within the compartment. ABR testing involved the right ear only.
Data Analysis
We performed the association analysis using FaST-LMM (factored spectrally transformed linear mixed model), a method accounts for population structure (https://www.microsoft.com/en-us/download/details.aspx?id=52559 Version 2.0.7 downloaded October 2017). This method set SNPs according to their linear regression p values and then constructs kinship matrices using the SNPs from all other chromosomes to improve power when testing all SNPs on a specific chromosome (Lippert et al. 2011). This procedure includes the SNP being tested for association in the regression equation only once.
Genome-wide significance threshold in the HMDP was determined by the family-wise error rate (FWER) as the probability of observing one or more false positives across all SNPs per phenotype. We performed 100 different sets of permutation tests and parametric bootstrapping of size 1000 and observed that the genome-wide significance threshold at a FWER of 0.05 corresponded to p = 4.1 × 10–6, similar to that used in previous studies with the HMDP (Bennett et al. 2010). This is approximately an order of magnitude larger than the threshold obtained by Bonferroni correction (4.6 × 10–7), which would be an overly conservative estimate of significance because nearby SNPs among inbred mouse strains are highly correlated with each other.
In order to prioritize candidate genes, we used EMMA (Efficient Mixed-Model Association) to perform an association between all SNPs and array probes mapping within each region. EMMA is a statistical test for association mapping correcting for population structure (Kang et al. 2008). An R package implementation of EMMA (version 1.1.2) is available online at www.mouse.cs.ucla.edu/emma.
GWAS Candidate Genes
RefSeq genes were downloaded from the UCSC genome browser (https://genome.ucsc.edu/cgi-bin/hgTracks?db=mm10) using the GRCm38/mm10 genome assembly to characterize genes located within each association. The 95 % confidence interval for the distribution of distances between the most significant and the true causal SNPs, for simulated associations that explain 5 % of the variance in the HMDP, is 2.6 Mb (Bennett et al. 2010). Only SNPs mapping to each associated region were used in this analysis. We selected SNPs that were variants in at least one of the HMDP CI strains.
Cochlear Whole Mount Immunolabeling
We used immunolabeling of synaptic ribbons and glutamate receptors to quantify the functional synapses in strains with the highest and lowest suprathreshold wave 1 amplitudes pre- and post-noise exposure. Animals were euthanized and intracardially perfused after the second hearing measurement and their cochleae were post-fixed with 4 % PFA for 1 h. Fixed samples were rinsed extensively in phosphate-buffered saline (PBS) and dissected under a microscope to three half turns and permeabilized in 1 % Triton X-100 solution for 15 min at room temperature. The specimens were washed three times with PBS and blocked with 10 % goat serum for 1 h at room temperature. Tissues were incubated at 37 °C overnight with the following primary antibodies: monoclonal mouse anti-carboxyl-terminal binding protein 2 (CtBP2) IgG1 at 1:200 (612044; BD Biosciences), monoclonal mouse anti-GluR2 IgG2a at 1:1000 (MAB397; Millipore) and polyclonal rabbit anti-myosin VIIa at 1:200 (25-6790; Proteus Biosciences). The following day, after three 15-min PBS washes, the tissues were incubated with the appropriate Alexa Fluor 594, Alexa Fluor 488 and Alexa Fluor 633-conjugated secondary antibodies at a concentration of 1:1000 for 1 h in darkness at room temperature. Following the final washes after secondary incubations, samples were mounted on slides using ProLong Glass antifade mountant and left to dry for at least 24 h before image acquisition.
Frequency regions corresponding to 16 and 32 kHz were located through their distance from cochlear apex, based on the place-frequency map from Müller et al. (2005) and imaged with a 20 × 1.4NA on a Zeiss 880 LSM airyscan confocal microscope (Carl Zeiss, Oberkochen, Germany). After acquisition, images were Airyscan processed and post-processed and analyzed using Imaris software (BITPLANE) to quantify the number of functional synapses. Briefly, the puncta corresponding to synaptic ribbons and glutamate receptors were reconstructed using the option of spots recognition and limited to cytoplasmic volume through Myo7a staining and a threshold level of 12 voxels to define the minimum size of objects selected was applied to all processed images and the spot colocalization option was number of paired synapses.
Gene Expression Data
Cochleae from each 6-week-old mice were isolated from the 64 HMDP strains (3 mice/strain). The inner ear was microdissected and the surrounding soft tissue and the vestibular labyrinth was removed. The dissected cochleae were then frozen in liquid nitrogen and ground to powder. RNA was extracted and purified by placing cochlea samples in RNA lysis buffer (Ambion). The sample was incubated overnight at 4 °C, centrifuged (12,000×g for 5 min) to pellet insoluble materials, and the RNA isolated following manufacturer’s recommendations. This procedure generated 300 ng of total RNA per mouse.
Gene expression analysis was performed, and gene expression measurements were taken using Illumina’s mouse whole genome expression kit, BeadChips. Amplifications and hybridizations were performed according to Illumina’s protocol (Southern California Genome Consortium microarray core laboratory at University of California, Los Angeles). RNA (100 ng) was reverse transcribed to cDNA using the Ambion cDNA synthesis kit (AMIL1791) and then converted to cRNA and labeled with biotin. Subsequently, 800 ng of biotinylated cRNA product was hybridized to prepare whole genome arrays and was incubated overnight (16–20 h) at 55 °C. Arrays were washed and then stained with Cy3-labeled streptavadin. Excess stain was removed by washing and then arrays were dried and scanned on an Illumina BeadScan confocal laser scanner.
Reagent and Data Availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Baseline and Post-noise Exposure Phenotypic Variation Within the HMDP
We tested 5-week-old female mice (n = 635) from 102 HMDP strains (n = 6–7/strain) at baseline and post-noise exposure using ABR and calculated wave 1 suprathreshold amplitudes (P1-N1) at 8, 12, 16, 24, and 32 kHz tone burst stimuli. A broad range of suprathreshold ABR wave 1 amplitudes were detected across the HMDP with differences between the lowest and the highest strains at specific ABR stimulus frequencies demonstrated Prior and after exposure to damaging noise levels.
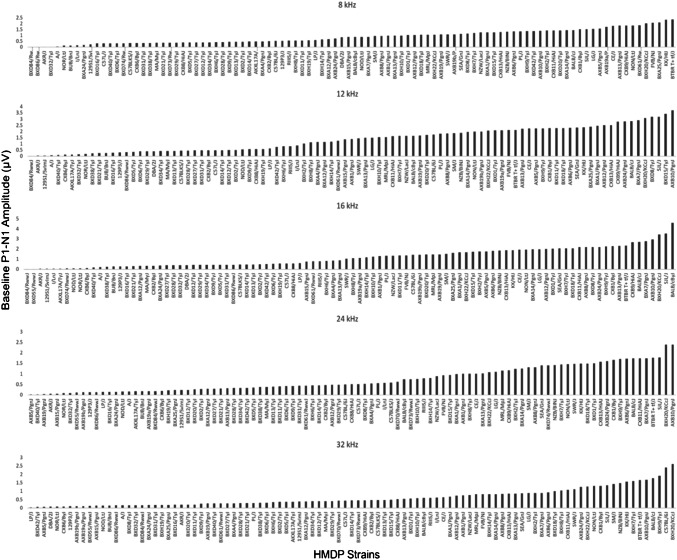
Figure 1 characterizes the variation in ABR wave 1 amplitude at baseline in HMDP strains. Frequencies of 8, 12, 16, 24, and 32 kHz demonstrated differences of 3.82, 2.42, 2.62, 3.75, and 3.43-fold between highest and lowest strains respectively prior to noise (Fig. 1).
Fig. 1.
Characterization of pre-exposure P1-N1 wave 1 amplitudes (at 80 dB SPL) in 102 HMDP inbred mouse strains
At the baseline, several wave 1 amplitude patterns were noticeable by looking to the highest and lowest deciles: three strains among the top ten (BXH20/KCcJ, BTBR T+tf/J and BALB/cByJ) exhibited higher wave 1 amplitude measurement across four consecutive or all frequencies. Other strains showed a particular strength in specific cochlear regions. For example, CXB9/HiAJ had preferentially high wave 1 amplitude values across low frequencies while BXH9/TyJ had stronger measurement at high frequencies. SJL/J and BXA7/PgnJ were more consistent at mid frequencies. A similar pattern was also seen among the worst performer decile, NOR/LtJ and AKR/J recorded lowest wave 1 amplitude across all frequencies. Other strains (BXD84/RwwJ, 129S1/SVlmJ and A/J) had a particular low wave 1 amplitude value at low frequencies while BXD55/RwwJ and AXB19b/PgnJ were more affected at high frequencies.
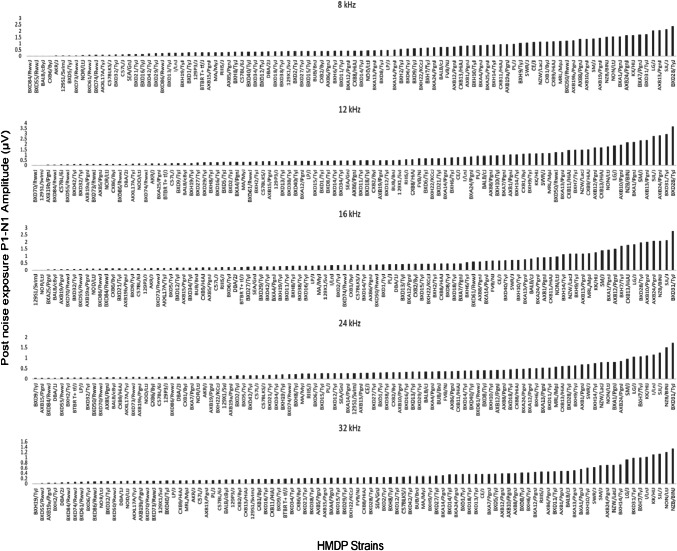
The exposure to noise increased level of strain variation particularly at mid and high frequencies. Frequencies of 8, 12, 16, 24, and 32 kHz demonstrated differences of 3.75, 2.43, 3.23, 5.20, and 7.5-fold post-noise exposure (Fig. 2), respectively.
Fig. 2.
Characterization of post-noise exposure P1-N1 wave 1 amplitudes (at 80 dB SPL) in 102 HMDP inbred mouse strains
An analysis of the highest wave 1 amplitude decile after noise shows that six strains (NZB/BINJ, SJL/J, LJ/J, KK/HiJ, BXD31/TyJ and AXB24/PgnJ) resisted the noise induced decrease in wave 1 amplitude. Another noticeable pattern was the low frequency specific strength in wave 1 amplitude value at BXA1/PgnJ and AXB13/PgnJ. A similar analysis for the lowest wave 1 amplitude deciles shows that four strains BXD84/RwwJ, BXD55/RwwJ, 129S1/SVlmJ, and AXB19b/PgnJ preserved their lower wave 1 amplitude measurement across consecutive or all tested frequencies. This strain associated variation in suprathreshold ABR amplitudes prior and post-exposure to noise support the hypothesis that this phenotype has a genetic basis.
Next, we wanted to explore the relationship between P1-N1 wave 1 amplitudes and their corresponding ABR thresholds in HMDP strains prior and after noise exposure. Scatter plots and Spearman correlation data from Fig. 3 show a small but significant inverse correlation between wave 1 amplitudes and ABR threshold across all tested frequencies prior and post-noise exposure. The degree of correlation increases from low to high frequencies and from pre- to post-noise exposure: 8 kHz frequency pre-noise, (ρ = − 0.353, p < 0.001, Fig. 3a) vs 8 kHz post-noise (ρ = − 0.396, p < 0.001, Fig. 3f). For 12 kHz frequency pre-noise, the Spearman’s ρ = − 0.395 (p < 0.001, Fig. 3b) vs (ρ = 0.476, p < 0.001, Fig. 3g) in post-noise exposure. The Spearman correlation for 16 kHz pre-noise (ρ = − 0.370, p < 0.001, Fig. 3c) vs (ρ = − 0.541, p < 0.001, Fig. 3h) for 16 kHz post-noise. Figure 3d shows (ρ = − 0.442, p < 0.001) for 24 kHz pre-noise vs (ρ = − 0.496, p < 0.001) for 24 kHz post-noise in Fig. 3i; while the highest Spearman’s ρ correlation is attributed to 32 kHz pre-noise (ρ = − 0.570, p < 0.001, Fig. 3e) and 32 kHz post-noise (ρ = − 0.562, p < 0.001, Fig. 3j).
Fig. 3.
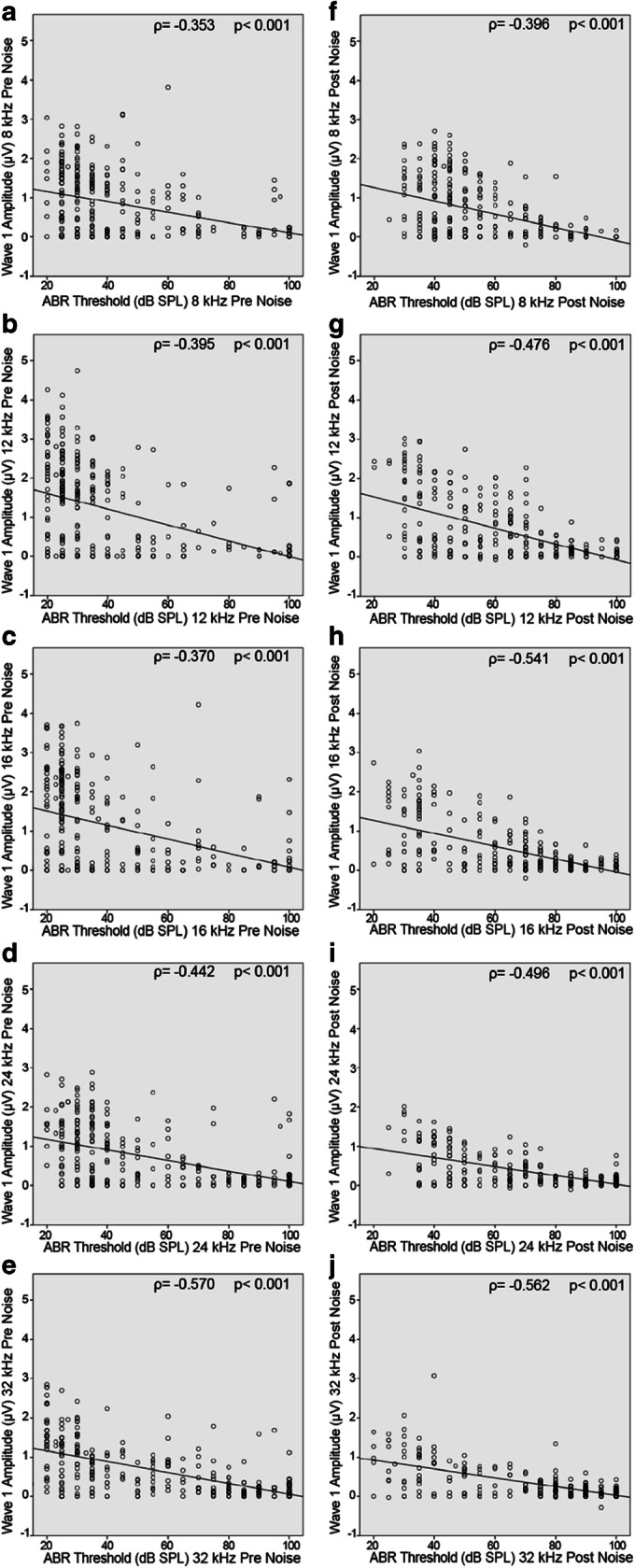
Scatterplot and Spearman correlation between wave 1 amplitude and ABR threshold for individual mice pre- (left) and post- (right) noise across all tested frequencies. There is a consistent negative correlation between ABR threshold and wave 1 amplitude. a 8 kHz pre-noise. b 12 kHz pre-noise. c 16 kHz pre-noise. d 24 kHz pre-noise. e 32 kHz pre-noise. f 8 kHz post-noise. g 12 kHz post-noise. h 16 kHz post-noise. i 24 kHz post-noise. j 32 kHz post-noise
Variation in Wave 1 Amplitudes Correlates with Number of IHC Paired Synapses
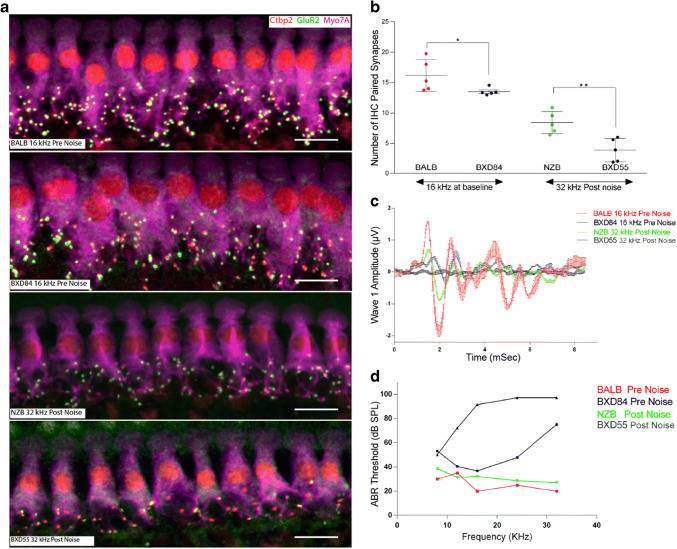
To test the relation between suprathreshold wave 1 amplitudes and the number of functional synapses in the HMDP, we selected the strains with highest and lowest suprathreshold wave 1 amplitudes pre- (BALB/cByJ and BXD84/RwwJ respectively) and post-noise exposure (NZB/BINJ and BXD55/RwwJ respectively) for paired synapse quantification.
Figure 4b shows that the number synapses varied in conjunction with the corresponding wave 1 amplitude values. At the 16 kHz frequency pre-noise, the paired synapses mean for BALB/cByJ was 16.15 ± 1.17 per inner hair cell vs 13.59 ± 0.57 for BXD84/RwwJ (p = 0.044). The difference was higher in the post-noise comparison with the NZB/BINJ strain having a mean value of 8.428 ± 1.82 vs 3.908 ± 1.91 for BXD55/RwwJ (p = 0.005) demonstrating variation in the synapse and/or ANF sensitivity to a noise. Mean ABR representative waves (Fig. 4c) from the strains representing the lowest and the highest ABR wave 1 measurement prior and after noise exposure followed similar pattern to the synaptic count of these strains; the number of functional IHC synapses varied in conjunction with the corresponding wave 1 amplitude values. ABR threshold (Fig. 4d) corresponding of these 4 extreme strains inversely correlated with synapses count in these strains. Mean ABR representative waves (Fig. 4c) from the strains representing the lowest and the highest ABR wave 1 measurement prior and after noise exposure followed similar pattern to the synaptic count of these strains; the number of functional IHC synapses varied in conjunction with the corresponding wave 1 amplitude values. ABR threshold (Fig. 4d) corresponding of these 4 extreme strains inversely correlated with synapses count in these strains.
Fig. 4.
a Representative images of synaptic immunolabeling for the 16 kHz cochlear region pre-noise of highest BALB/cByJ and lowest wave 1 BXD84/RwwJ and the 32 kHz frequency region for the highest and lowest wave 1 values post-noise exposure NZB/BINJ and BXD55/RwwJ respectively. n = 5 for each strain. Scale bar 10 μm. b presents the quantification of paired synapses per IHC from the four conditions presented in a. c Mean ABR trace recorded representing four extreme strains upon 80 dB sound pressure level n = 5 for each strain. d ABR threshold (in dB SPL) plotted as a function of auditory stimulus frequency (in kHz) n = 5 for each strain. All data presented as mean ± SEM. The statistical tests were a priori selected independent samples t test. *p < 0.05; **p < 0.01
GWAS for Suprathreshold ABR P1-N1 Variation at Each Tested Frequency
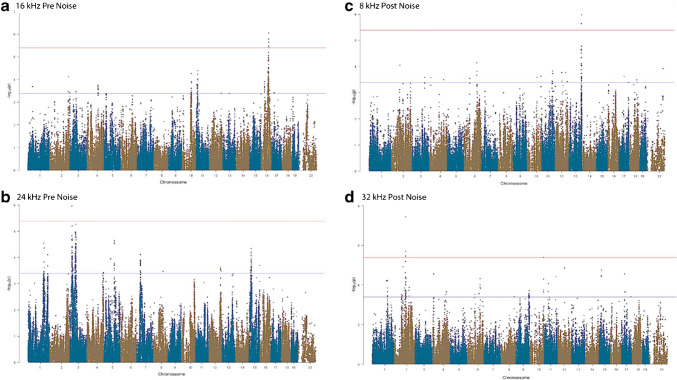
Association analysis was applied to the response to each tone pip stimulus separately to identify genetic associations for the five tone-burst stimuli. We performed a GWAS for both pre- (Fig. 5 a and b) and post-noise exposure ABR P1-N1 suprathreshold (Fig. 5 c and d) in order to identify loci associated with the various frequencies tested. Association p values (adjusted) were calculated for 200,000 SNPs with minor allele frequency of > 5 % (p < 0.05 genome-wide equivalent for GWA using FaST-LMM in the HMDP is p = 4.1 × 10–6, − log10P = 5.39) as described above.
Fig. 5.
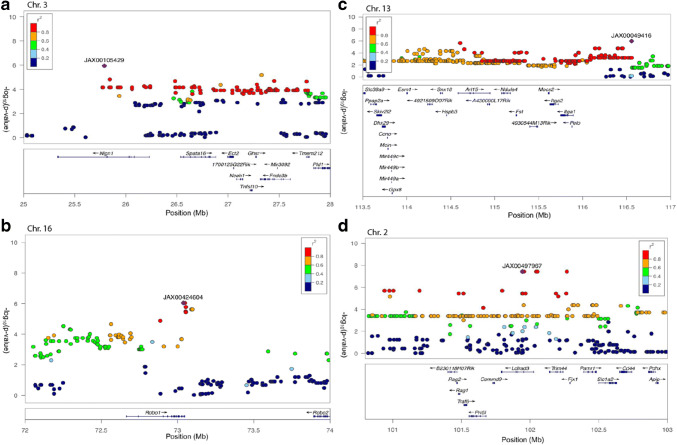
GWAS results for ABR wave 1 P1-N1 suprathreshold amplitude in the HMDP. Manhattan plots showing the association (−log10) p values (−logP) for 16 kHz (a) and 24 kHz (b) at pre-noise and 8 kHz (c) and 32 kHz (d) post-noise exposure in 102 HMDP mouse strains. The analysis was performed using ~ 200,000 SNPs with a minor allele frequency > 5 %. Each chromosome is plotted on the x-axis in alternating brown and blue colors. SNPs on Chr. 16, Chr. 3, Chr. 13, and Chr. 2 exceeded the predetermined genome-wide significance threshold (−logP = 5.39 red line)
At the genome-wide significance threshold (−logP = 5.39), associations on Chr. 3 and Chr. 16 were identified pre-noise exposure (Fig. 5) and on Chr. 2 and Chr. 13 post-noise exposure. The details of each association are provided in Table 1.
Table 1.
Genome-wide association results at baseline and post-noise exposure
| Traita | Chr | SNP | Position (Mb)b | −logP | No. of genesc |
Human region (Chr: start Mb–end Mb) |
|---|---|---|---|---|---|---|
| 16 kHz pre-Exposure | 16 | JAX00424604 | 73.0 | 9.02E-07 | 2 | Chr3:77.5–79.8 |
| 24 kHz pre-exposure | 3 | JAX00105429 | 25.7 | 1.12E-06 | 11 | Chr3:171.3–174.2 |
| 8 kHz post-exposure | 13 | JAX00049416 | 116.5 | 1.07E-06 | 23 | Chr5:50.7–54.9 |
| 32 kHz post-exposure | 2 | JAX00497967 | 101.9 | 3.68E-08 | 14 | Chr11:34.8–37.1 |
aPre- and post-noise exposure suprathreshold ABR wave 1 P1-N1 amplitudes at different stimulus frequencies
bLocations based on genome assembly (NCBI’s Build37)
cNumber of RefSeq genes (NCBI’s Build37 assembly) located in the mouse association confidence interval (2.6 Mb)
Association Analysis
In order to characterize these 4 associated genomic regions, we defined 2.6 Mb as our interval (Table 1). Pre-noise exposure, ABR wave 1 P1-N1 associations exceeding the genome-wide significance threshold were identified on chromosome 3 (Fig. 6a) and 16 (Fig. 6b). For post-noise exposure, associations were significant on chromosome 13 (Fig. 6c) and chromosome 2 (Fig. 6d).
Fig. 6.
Genomic interval of the significant association centered on the lead SNP for 24 kHz (a) and 16 kHz (b) pre-noise exposure and 8 kHz (c) and 32 kHz (d) post-noise exposure in 102 HMDP inbred mouse strains. The blue diamond represents the most significant SNP and SNPs are colored based on their LD with the most significant SNP being red SNPs in LD at r2 > 0.8, orange SNPs in LD at r2 > 0.6 and green SNPs in LD at r2 > 0.4. The positions of all RefSeq genes are plotted using genome locations (NCBI’s Build37 genome assembly)
Exploring Candidate Genes
Quantitative variation in gene expression levels in a specific tissue can act as an intermediate phenotype between genomic sequences variation and phenotypes to explain the remaining “hidden” heritable factors. The heritability of gene expression means it can be subject to the same quantitative trait loci (QTL) analyses as conventional trait data to reveal the so-called expression quantitative trait loci (eQTLs). Transcript abundance of each gene which will be the phenotype analyzed, can therefore be mapped to the genome using standard linkage methods, allowing identification of eQTLs loci that explains fractions of a gene expression phenotype.
Our cochlear expression data allowed us to analyze all 50 identified candidate genes at each association interval. Except the chromosome 16 locus, we identified genes within each of the intervals regulated by a local eQTL. In order to perform eQTL analysis, we generated gene expression microarray profiles using RNA isolated from cochleae in 64 HMDP strains (n = 3 arrays per strain). We then used EMMA to perform an association analysis between all SNPs and array probes mapping within each region. A total of 18,138 genes were represented by at least one probe, after excluding probes that overlapped SNPs present among the classical inbred strains used in the HMDP as describe in the methods.
Loci in which peak SNPs mapped to within 2 Mb of the gene whose expression was regulated were considered “local” or cis-acting eQTLs, while SNPs mapping elsewhere were considered “distal” and presumably trans-acting eQTLs. We calculated the significant p value cutoff (p = 1 × 10–6) for local and distal associations. These genes were prioritized based upon whether they were regulated by a local expression QTL (eQTL). A total of 17 genes (2 genes within Chr. 3 association, 6 genes within Chr. 2 association, and 9 genes within Chr. 13 association) were identified with at least 1 probe whose expression was regulated by a significant cis eQTL in the cochlea (Table 2). Although there were no cochlear cis-eQTLs identified for genes within the interval associated with the pre-noise suprathreshold wave 1 amplitude derived from the 24 kHz tone-burst stimulus, the lead SNP lies within a region containing only two genes, Robo1 and Robo2, both associated with innervation of the mouse cochlea validating our approach (Wang et al. 2013).
Table 2.
Candidate genes within 3 association peaks regulated by cis eQTL in the cochlea
| Gene | RefSeq | Chr | txStart (bp)a | txEnd (bp)b | Local eQTL pc |
|---|---|---|---|---|---|
| Ghsr | ILMN_2872464 | 3 | 27,371,351 | 27,378,010 | 1.42E-06 |
| Fndc3b | ILMN_2862567 | 3 | 27,416,162 | 27,710,439 | 2.35E-06 |
| B230118H07Rik | ILMN_2726101 | 2 | 101,560,781 | 101,649,532 | 3.42E-06 |
| Ldlrad3 | ILMN_1217129 | 2 | 101,950,203 | 102,186,385 | 3.56E-06 |
| Trim44 | ILMN_2471289 | 2 | 102,300,119 | 102,407,828 | 3.6E-06 |
| Cd44 | ILMN_3114585 | 2 | 102,810,241 | 102,901,665 | 3.65E-06 |
| Slc1a2 | ILMN_2642339 | 2 | 102,658,659 | 102,790,784 | 3.82E-06 |
| Traf6 | ILMN_2427560 | 2 | 101,678,429 | 101,701,669 | 3.90E-06 |
| Slc38a9 | ILMN_1242435 | 13 | 112,660,766 | 112,738,743 | 4.01E-06 |
| Esm1 | ILMN_1257574 | 13 | 113,209,659 | 113,218,098 | 2.03E-07 |
| Ndufs4 | ILMN_1234521 | 13 | 114,287,795 | 114,388,094 | 1.84E-06 |
| Ppap2a | ILMN_1222991 | 13 | 112,800,894 | 112,867,881 | 2.46E-06 |
| Itga1 | ILMN_2701778 | 13 | 62,677,826 | 62,783,982 | 9.28E-07 |
| Dhx29 | ILMN_2892376 | 13 | 112,927,730 | 112,969,431 | 2.58E-06 |
| Pelo | ILMN_2853658 | 13 | 115,088,357 | 115,090,188 | 2.62E-06 |
| Ccno | ILMN_2736471 | 13 | 112,987,802 | 112,990,778 | 2.87E-06 |
| Gpx8 | ILMN_2653166 | 13 | 113,042,763 | 113,046,388 | 3.10E-06 |
atxStart, location of transcription (NCBI Build37 genome assembly) start
btxEnd, location of transcription (NCBI Build37 genome assembly) end
cStatistically significant p value ≤ E-06 (Bonferroni corrected for the number of probes tested)
Discussion
This study is the first genome-wide association study in mice using suprathreshold ABR wave 1 amplitude and the first large-scale assessment of this phenotype. The authors acknowledge limitations of this study. First, we measured wave 1 amplitudes at a single stimulus intensity (80 dB SPL) and compared these across strains with varying pre- and post-noise thresholds. A single stimulus level may not activate the same contributing elements across strains and conditions, potentially confounding the phenotypic analysis. Also, because of the tail structure of neural frequency tuning curves (Taberner and Liberman 2005), ABR input/output curves are not frequency-place specific. Thus, damage to inner hair cells or neurons at the basal end of the cochlea may impact the apparent size of wave 1 regardless of stimulus frequency. Conversely, tuning curve tails resist changes that pathology often causes at the tips of tuning curves, so that basal injury need not affect wave I at high sound levels as long as the neurons survive. While it remains possible that differences in wave 1 do not reflect true genetic differences in synaptic operation at the cochlear locations targeted by the frequencies we used, the plausibility of the candidate genes we identified supports our approach. These data will assist researchers in the study of genes and pathways involved in cochlear innervation both at baseline and after noise exposure. The HMDP has been used successfully to examine the genetics of a wide array of phenotypes by us and others including plasma lipids (Bennett et al. 2009), bone density (Farber et al. 2011), blood cell traits (Davis et al. 2013), conditioned fear responses (Park et al. 2011), gene-by-diet interactions in obesity (Parks et al. 2013), inflammatory responses (Orozco et al. 2015), age-related hearing loss (Ohmen et al. 2014), NIHL (Lavinsky et al. 2015), diabetes (Parks et al. 2015), and heart failure (Rau et al. 2015). In many of these studies including ours, genes at the identified loci were validated as causal using engineered mouse models and several of them corresponded to loci identified in human GWAS.
Recent expansion of the depth of genotyping and the number of HMDP strains has led to an increase in the number of significant associations detected (Rau et al. 2015). There exist additional resources for the study of complex traits in mice such as the Collaborative Cross (CC), outbred rodent populations (OS), and Chromosome Substitution Strains (CSS) that have the added advantage of wild strain variation and the potential for identifying additional loci (Buchner and Nadeau 2015). The HMDP has the advantage of genomic homozygosity and complete reproducibility of phenotypic measurements and recent additions to the HMDP including the addition of ~ 75 additional strains and higher density SNP genotyping have substantially increased the power and genomic coverage (Rau et al. 2015). Although the HMDP may not capture SNP variation from wild strains, the limitations of the other resources, including the need for whole genome genotyping of successive generations in the OS and CC and the need for generation of subcongenics in the CSS compelled us to use the HMDP resource.
Our data showed a broad range of suprathreshold ABR wave 1 amplitudes with a discernable pattern detected prior and after noise exposure across HMDP strains and a significant small inverse correlation between ABR wave 1 amplitudes values and their corresponding ABR thresholds. Using ABR measurement, we and others previously identified several distinct patterns of baseline hearing deficits (Zheng et al. 1999) (Myint et al. 2016). We have also characterized several distinct patterns of noise-sensitivity using HMDP strains (Myint et al. 2016). This strain associated variation in ABR amplitudes presented here support the hypothesis that this phenotype has a genetic background.
The analysis of synaptic counts from the 4 extreme strains representing the lowest and the highest ABR wave 1 measurement prior and after noise exposure supported the hypothesis that the number of functional IHC synapses varied in conjunction with the corresponding wave 1 amplitude values and that ABR P1-N1 amplitude can be used as a reliable metric to indirectly analyze the synaptic function and synaptopathy in large populations. Sergeyenko et al. (2013) have shown that loss of IHC synapses correlate with decrease in wave 1 amplitude. Although the significant variation in synapses count between the strains presented in pre-noise condition may not have reflected the full-scale difference in ABR wave 1 amplitude measures, synaptopathy can present itself in the form of abnormal Synaptic function instead of the more common absence of these synapses, particularly at baseline condition. Recently, Clrn1ex4fl/fl Myo15-Cre+/– a mouse model lacking a variant of Clarin1 was shown to have diminished wave I amplitude, defective exocytosis and disorganized distribution of CaV1.3 Ca2+ channels (Dulon et al. 2018). Otof C2C/C2C is another model lacking an otoferlin functional C2C calcium binding site, showed reduced ABR wave I amplitude and disrupted vesicle pool replenishment compared with their wildtype controls (Michalski et al. 2017). These two models presented similar synapses numbers to theirs control despite the apparent synaptopathy.
Our GWAS generated significant associations in two regions for baseline hearing and another two separate regions after exposure to damaging noise generating a total 50 identified candidate genes. Using our cochlear gene expression data and eQTL analysis we were able to narrow down candidate genes substantially. Our findings demonstrate that the genetic architecture of noise induced cochlear synaptopathy is distinct from that of baseline auditory nerve/synapse integrity.
One locus at baseline and two loci at post-exposure to noise had at least two genes within the association peak regulated by a significant local cochlear eQTL (Table 2) and the other locus resides within a region containing only two genes, Robo1 and Robo2. All 17 candidate genes with cis-eQTLs, are expressed within the cells of the cochlear epithelium and spiral ganglion (https://umgear.org/).
Slc1a2 is a strong candidate for the 32-kHz (post-noise exposure) locus based on our cochlear eQTL data. Slc1a2 encodes a member of a family of solute transporter proteins (Arriza et al. 1994). This membrane-bound protein is the key transporter that clears the excitatory neurotransmitter glutamate from the extracellular space at synapses in the nervous system. Glutamate clearance is required for precise synaptic activation and to prevent neuronal damage from excessive activation of glutamate receptors. The synapse is composed of a presynaptic ribbon enclosed by a halo of neurotransmitter-containing vesicles within the inner hair cells (Nouvian et al. 2006) and a postsynaptic active zone on the cochlear nerve terminal with glutamate (AMPA-type) receptors for the released neurotransmitter (Puel et al. 1995). Abnormal regulation of this gene is thought to be related to several neurological disorders (Xu et al. 2016). Puel et al. (1998) has shown that local application of glutamate receptor (GluR) agonists can produce dose-dependent swelling of cochlear nerve terminals contacting IHCs. The dendritic inflammation is observed under inner hair cells, but not outer hair cells, and is prevented by prior intracochlear perfusion of glutamate antagonists (Ruel et al. 2007). Based on conjectures (Kujawa and Liberman 2009), this excitotoxicity would be a primary initial insult in the inflammatory cascade observed after noise. Thus, Slc1a2 could play an important role in the susceptibility to noise-induced cochlear synaptopathy by regulating the glutamate clearance after exposure.
Neuroglin1, a candidate gene at the 24-kHz (pre-noise exposure) locus, is a synaptic cell adhesion molecule that connects pre- and post-synaptic neurons. Neuroglin1 is responsible for signaling across the synapse which regulates synaptic activity and determines the properties of neuronal networks (Gjørlund et al. 2012) but has no known function in the auditory system.
Wang et al., 2013 have previously demonstrated that both Robo1 and Robo2 were actively expressed by spiral ganglion neurons. Also, in Robo1/2 double mutants at E18, spiral ganglion neurons were dislocated in the space dorsal to the cochlear epithelium and did not innervate hair cells. Thus, Robo signaling mediates spatial positioning of spiral ganglion neurons during development of cochlear. Interestingly, there were no significant alterations of the overall cochlear structure in Robo1/2 double mutants in comparison with their heterozygous littermates (hair cells persisted unaffected) (Wang et al. 2013). Thus, our baseline GWAS data on chromosome 16 with the peak SNP within Robo1 validates our approach.
Future studies will be dedicated to the validation of candidate genes through the analyses of the strains with the most extreme phenotypes and the use of transgenic and/or CRISPR models.
Conclusions
We have performed the first comprehensive analysis of suprathreshold ABR wave 1 amplitude in mice and have begun to elucidate the genetic architecture of the auditory hair cell synapse and the susceptibility to noise- induced cochlear synaptopathy. We identified multiple novel loci and, using our cochlear eQTLs, we prioritized positional candidate genes. These findings validate the utility of the HMDP for detecting genes related to auditory function and provide potential candidates for more in-depth analyses.
Acknowledgements
The authors are grateful to the Mills Auditory Foundation.
Funding Information
This work was supported by National Institutes of Health National Institute on Deafness and Other Communication Disorders grants R01DC010856-01 to R.A.F. National Institutes of Health grants R01ES021801, 3R01ES021801-03S1, UL1TR000130, P30ES007048 and Environmental Protection Agency grant RD83544101 grant to H.A.; the funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Compliance with Ethical Standards
The Institutional Care and Use Committee (IACUC) at University of California San Diego (IACUC 17178) and University of Southern California (IACUC 12033) endorsed the animal protocol for the Hybrid Mouse Diversity Panel (HMDP) inbred strains.
Conflict of Interest
The authors declare that they have no conflict of interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Arriza JL, Fairman WA, Wadiche JI, Murdoch GH, Kavanaugh MP, Amara SG. Functional comparisons of three glutamate transporter subtypes cloned from human motor cortex. J Neurosci. 1994;14:5559–5569. doi: 10.1523/JNEUROSCI.14-09-05559.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Wang SS, Wang X, Wu X, Lusis AJ. Genetic regulation of atherosclerotic plaque size and morphology in the innominate artery of hyperlipidemic mice. Arterioscler Thromb Vasc Biol. 2009;29(3):348–355. doi: 10.1161/ATVBAHA.108.176685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BJ, Farber CR, Orozco L, Kang HM, Ghazalpour A, Siemers N, Neubauer M, Neuhaus I, Yordanova R, Guan B, Truong A, Yang WP, He A, Kayne P, Gargalovic P, Kirchgessner T, Pan C, Castellani LW, Kostem E, Furlotte N, Drake TA, Eskin E, Lusis AJ. A high-resolution association mapping panel for the dissection of complex traits in mice. Genome Res. 2010;20(2):281–290. doi: 10.1101/gr.099234.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchner DA, Nadeau JH. Contrasting genetic architectures in different mouse reference populations used for studying complex traits. Genome Res. 2015;25(6):775–791. doi: 10.1101/gr.187450.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow AL, Ohmen J, Wang J, Lavinsky J, Hartiala J, Li Q, Li X, Salehide P, Eskin E, Pan C, Lusis AJ, Allayee H, Friedman RA. The genetic architecture of hearing impairment in mice: evidence for frequency-specific genetic determinants. G3 (Bethesda) 2015;5(11):2329–2339. doi: 10.1534/g3.115.021592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RC, van Nas A, Bennett B, Orozco L, Pan C, Rau CD, Eskin E, Lusis AJ. Genome-wide association mapping of blood cell traits in mice. Mamm Genome. 2013;24(3–4):105–118. doi: 10.1007/s00335-013-9448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulon D, Papal S, Patni P, Cortese M, Vincent PFY, Tertrais M, Emptoz A, et al. Clarin-1 gene transfer rescues auditory synaptopathy in model of Usher syndrome. J Clin Invest. 2018;128(8):3382–3401. doi: 10.1172/JCI94351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eybalin M. Neurotransmitters and neuromodulators of the mammalian cochlea. Physiol Rev. 1993;73:309–373. doi: 10.1152/physrev.1993.73.2.309. [DOI] [PubMed] [Google Scholar]
- Farber CR, Bennett BJ, Orozco L, Zou W, Lira A, Kostem E, Kang HM, Furlotte N, Berberyan A, Ghazalpour A, Suwanwela J, Drake TA, Eskin E, Wang QT, Teitelbaum SL, Lusis AJ. Mouse genome-wide association and systems genetics identify Asxl2 as a regulator of bone mineral density and osteoclastogenesis. PLoS Genet. 2011;7(4):e1002038. doi: 10.1371/journal.pgen.1002038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gjørlund MD, Nielsen J, Pankratova S, Li S, Korshunova I, Bock E, Berezin V. Neuroligin1 induces neurite outgrowth through an interaction with Neurexin-1β and activation of fibroblast growth factor receptor-1. FASEB J. 2012;26(10):4174–4186. doi: 10.1096/fj.11-202242. [DOI] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobel M, Le Prell CG, Liu J, Hawks JW, Bao J. Noise-induced cochlear synaptopathy: past findings and future studies. Hear Res. 2017;349:148–154. doi: 10.1016/j.heares.2016.12.008. [DOI] [PubMed] [Google Scholar]
- Kujawa SG, Liberman MC (2009) Adding insult to injury: cochlear nerve degeneration after temporary” noise-induced hearing loss. J Neurosci 29: 14077–14085 [DOI] [PMC free article] [PubMed]
- Kujawa SG, Liberman MC. Synaptopathy in the noise-exposed and aging cochlea: primary neural degeneration in acquired sensorineural hearing loss. Hear Res. 2015;330:191–199. doi: 10.1016/j.heares.2015.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Crow AL, Pan C, Wang J, Aaron KA, Ho MK, Li Q, Salehide P, Myint A, Monges-Hernadez M, Eskin E, Allayee H, Lusis AJ, Friedman RA. Genome-wide association study identifies nox3 as a critical gene for susceptibility to noise-induced hearing loss. PLoS Genet. 2015;11(4):e1005094. doi: 10.1371/journal.pgen.1005094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavinsky J, Ge M, Crow AL, Pan C, Wang J, Salehi P, Myint A, Eskin E, Allayee H, Lusis AJ, Friedman RA. The genetic architecture of noise-induced hearing loss: evidence for a gene- by-environment interaction. G3 (Bethesda) 2016;6(10):3219–3228. doi: 10.1534/g3.116.032516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC. Noise-induced and age-related hearing loss: new perspectives and potential therapies. F1000Res. 2017;6:927. doi: 10.12688/f1000research.11310.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Kujawa SG. Cochlear synaptopathy in acquired sensorineural hearing loss: manifestations and mechanisms. Hear Res. 2017;349:138–147. doi: 10.1016/j.heares.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman MC, Epstein MJ, Cleveland SS, Wang H, Maison SF. Toward a differential diagnosis of hidden hearing loss in humans. PLoS One. 2016;11(9):e0162726. doi: 10.1371/journal.pone.0162726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippert C, Listgarten J, Liu Y, Kadie CM, Davidson RI, Heckerman D. FaST linear mixed models for genome-wide association studies. Nat Methods. 2011;8(10):833–835. doi: 10.1038/nmeth.1681. [DOI] [PubMed] [Google Scholar]
- Michalski N, Goutman JD, Auclair SM, de Monvel JB, Tertrais M, Emptoz A, Parrin A, et al. Otoferlin acts as a Ca2+ sensor for vesicle fusion and vesicle pool replenishment at auditory hair cell ribbon synapses. Elife. 2017;6:e31013. doi: 10.7554/eLife.31013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M, von Hünerbein K, Hoidis S, Smolders JW. A physiological place-frequency map of the cochlea in the CBA/J mouse. Hear Res. 2005;202:63–73. doi: 10.1016/j.heares.2004.08.011. [DOI] [PubMed] [Google Scholar]
- Myint A, White CH, Ohmen JD, Li X, Wang J, Lavinsky J, Salehi P, Crow AL, Ohyama T, Friedman RA. Large-scale phenotyping of noise-induced hearing loss in 100 strains of mice. Hear Res. 2016;332:113–120. doi: 10.1016/j.heares.2015.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nouvian R, Beutner D, Parsons TD, Moser T. Structure and function of the hair cell ribbon synapse. J Membr Biol. 2006;209(2–3):153–165. doi: 10.1007/s00232-005-0854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmen J, Kang EY, Li X, Joo JW, Hormozdiari F, Zheng QY, Davis RC, Lusis AJ, Eskin E, Friedman RA. Genome-wide association study for age-related hearing loss (AHL) in the mouse: a meta-analysis. J Assoc Res Otolaryngol. 2014;15(3):335–352. doi: 10.1007/s10162-014-0443-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco LD, Morselli M, Rubbi L, Guo W, Go J, Shi H, Lopez D, Furlotte NA, Bennett BJ, Farber CR, Ghazalpour A, Zhang MQ, Bahous R, Rozen R, Lusis AJ, Pellegrini M. Epigenome- wide association of liver methylation patterns and complex metabolic traits in mice. Cell Metab. 2015;21(6):905–917. doi: 10.1016/j.cmet.2015.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park CC, Gale GD, de Jong S, Ghazalpour A, Bennett BJ, Farber CR, Langfelder P, Lin A, Khan AH, Eskin E, Horvath S, Lusis AJ, Ophoff RA, Smith DJ. Gene networks associated with conditional fear in mice identified using a systems genetics approach. BMC Syst Biol. 2011;5:43. doi: 10.1186/1752-0509-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Nam E, Org E, Kostem E, Norheim F, Hui ST, Pan C, Civelek M, Rau CD, Bennett BJ, Mehrabian M, Ursell LK, He A, Castellani LW, Zinker B, Kirby M, Drake TA, Drevon CA, Knight R, Gargalovic P, Kirchgessner T, Eskin E, Lusis AJ. Genetic control of obesity and gut microbiota composition in response to high-fat, high-sucrose diet in mice. Cell Metab. 2013;17(1):141–152. doi: 10.1016/j.cmet.2012.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BW, Sallam T, Mehrabian M, Psychogios N, Hui ST, Norheim F, Castellani LW, Rau CD, Pan C, Phun J, Zhou Z, Yang WP, Neuhaus I, Gargalovic PS, Kirchgessner TG, Graham M, Lee R, Tontonoz P, Gerszten RE, Hevener AL, Lusis AJ. Genetic architecture of insulin resistance in the mouse. Cell Metab. 2015;21(2):334–347. doi: 10.1016/j.cmet.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puel JL, Saffiedine S, Gervais d'Aldin C, Eybalin M, Pujol R. Synaptic regeneration and functional recovery after excitotoxic injury in the guinea pig cochlea. C R Acad Sci III. 1995;318(1):67–75. [PubMed] [Google Scholar]
- Puel J-L, Ruel J, d'Aldin CG, Pujol R. Excitotoxicity and repair of cochlear synapses after noise-trauma induced hearing loss. NeuroReport. 1998;9:2109e2114. doi: 10.1097/00001756-199806220-00037. [DOI] [PubMed] [Google Scholar]
- Rau CD, Parks B, Wang Y, Eskin E, Simecek P, Churchill GA, Lusis AJ. High-density genotypes of inbred mouse strains: improved power and precision of association mapping. G3 (Bethesda) 2015;5(10):2021–2026. doi: 10.1534/g3.115.020784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruel J, Wang J, Rebillard G, Eybalin M, Lloyd R, Pujol R, Puel JL. Physiology, pharmacology and plasticity at the inner hair cell synaptic complex. Hear Res. 2007;227(1–2):19–27. doi: 10.1016/j.heares.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Schuknecht HF. Further observations on the pathology of Presbycusis. Arch Otolaryngol. 1964;80:369–368. doi: 10.1001/archotol.1964.00750040381003. [DOI] [PubMed] [Google Scholar]
- Sergeyenko Y, Lall K, Liberman MC, Kujawa S. Age-related cochlear synaptopathy: an early-onset contributor to auditory functional decline. J Neurosci. 2013;33:13686e13694. doi: 10.1523/JNEUROSCI.1783-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Chang Y, Li X, Aiken S, Liu L, Wang J. Cochlear synaptopathy and noise-induced hidden hearing loss. Neural Plast. 2016;2016:6143164. doi: 10.1155/2016/6143164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taberner AM, Liberman MC. Response properties of single auditory nerve fibers in the mouse. J Neurophysiol. 2005;93(1):557–569. doi: 10.1152/jn.00574.2004. [DOI] [PubMed] [Google Scholar]
- Van Nas A, Ingram-Drake L, Sinsheimer JS, Wang SS, Schadt EE, Drake T, Lusis AJ. Expression quantitative trait loci: replication, tissue- and sex-specificity in mice. Genetics. 2010;185(3):1059–1068. doi: 10.1534/genetics.110.116087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SZ, Ibrahim LA, Kim YJ, Gibson DA, Leung HC, Yuan W, Zhang KK, Tao HW, Ma L, Zhang LI. Slit/Robo signaling mediates spatial positioning of spiral ganglion neurons during development of cochlear innervation. J Neurosci. 2013;33(30):12242–12254. doi: 10.1523/JNEUROSCI.5736-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CH, Ohmen JD, Sheth S, Zebboudj AF, McHugh RK, et al. Genome-wide screening for genetic loci associated with noise-induced hearing loss. Mamm Genome Off J Int Mamm Genome Soc. 2009;20:207–213. doi: 10.1007/s00335-009-9178-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Cao B, Chen Y, Ou R, Wei Q, Yang J, Zhao B, Song W, Shang HF. SLC1A2 rs3794087 are associated with susceptibility to Parkinson’s disease, but not essential tremor, amyotrophic lateral sclerosis or multiple system atrophy in a Chinese population. J Neurol Sci. 2016;365:96–100. doi: 10.1016/j.jns.2016.04.003. [DOI] [PubMed] [Google Scholar]
- Zheng QY, Johnson KR, Erway LC. Assessment of hearing in 80 inbred strains of mice by ABR threshold analyses. Hear Res. 1999;130:94–107. doi: 10.1016/S0378-5955(99)00003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]