Abstract
Exposure to biological fluid envelops a nanoparticle in layers of proteins and biomolecules, which has a profound impact on the nanoparticle’s biological fate. Although the identities and amounts of the proteins in this “corona” have been thoroughly examined, the spatial arrangement of the proteins is unclear, a problem that is compounded on porous nanoparticles due to penetration of proteins within the porous network. To address this problem, we have developed a procedure based on information derived from stochastic optical reconstruction microscopy. We employed a mathematical model to reveal the penetration depth of several proteins within porous nanoparticles. Understanding protein penetration depth provides an explanation for the composition of the protein corona, aiding in the development of safe and effective particle-based therapies.
The formation of the protein corona, that is, the layer of proteins that resides on a material after exposure to biological fluid, has been well documented in recent years.1–4 Particle diameter,5 surface charge,6 surface chemistry,7 and surface area8 have all been shown to influence the formation and composition of the protein corona. Once adsorbed, the protein corona has been shown to affect particle–cell interactions,9–11 inhibit the ability of nanoparticles to target specific cell types,12 influence nanoparticle pathophysiology,1 and mitigate cytotoxicity.13 Although many studies have involved the identification and quantification of the protein corona, very few have investigated the spatial mapping and orientations of the adsorbed proteins,14,15 and of those studies, only dense materials have been used, imposing limits on our understanding of how porous materials interact with biological environments.
We have recently published studies on the composition of the protein corona adsorbed onto mesoporous silica nanoparticles (MSNs).5,16 One feature of these studies was the preferential adsorption of low molecular weight (MW) proteins. As part of these studies, we sought to examine the relationship between MW and penetration depth within MSNs. Although more traditional microscopic techniques may be used to map protein adsorption on dense particles,15 visualizing MSN interiors presents unique challenges. Wide-field fluorescence microscopy methods, such as confocal fluorescence microscopy, have diffraction limits of 250 nm, limiting their abilities to resolve penetration depths within porous nanoparticles (Figure 1). On the other hand, stochastic optical reconstruction microscopy (STORM) exploits the inherent nature of photoswitchable fluorophores to generate an image of high resolution.17
Figure 1.

Microscopic analysis of protein adsorption onto 900 nm MSNs. (a) wide-field, as acquired; (b) wide-field, deconvolved; (c) STORM. Scale bar = 1000 nm.
Under appropriate optical and chemical conditions, photoswitchable fluorophores will stochastically turn on and off, allowing the instrument to record these blinks through many frames. As this process is repeated through numerous cycles, the fluorophores can be mapped with high localization accuracy. Numerous groups have reported an order of magnitude increase in resolution.18–20 A further advantage is that position information about each fluorophore is captured into a database, allowing reconstruction and manipulation. Although STORM has been successfully used in a number of biological applications,19,21 this technique is also an ideal tool for experiments at the topical intersection of biology and materials science.
We chose MSNs with a relatively large diameter for these experiments in order to allow the best determination of penetration depth. Highly monodisperse, spherical MSNs with a diameter of 830 nm as determined by transmission electron microscopy (TEM) were prepared using a previously published procedure.22 Analysis of the porosity of these particles by N2 physisorption showed a surface area of 837 m2 g−1, pore volume of 0.54 cm3 g−1, and pore diameter of 3 nm (Figures S1 and S2). These MSNs are referred to as “small pore” in these experiments. Exposure of this material to 1.0 M NH4OH for 30 min caused etching of the pores, leading to a material with a smaller surface area and pore volume but larger pores (304 m2 g−1, 0.38 cm3 g−1, and 6 nm, respectively). These MSNs are referred to as “large pore” and had a diameter of 913 nm from TEM. To investigate the relationship between pore diameter and protein adsorption, we selected three proteins with different MW: apolipoprotein A-II (17 kDa), albumin (66 kDa), and complement C3c (138 kDa), based on our previous experiments.5 These proteins are commonly identified in the coronas of many types of nanoparticles.23
As observed in Figures 1 and S3, the layer of fluorescently labeled proteins surrounding the silica nanoparticles was readily observed by STORM. However, applying this information to the question of penetration depth was challenging, because the MSNs did not all lie at the same depth within the focal plane of the microscope. Also, assuming the mean of all Cartesian coordinates of the fluorophores to be the center of the MSN was inaccurate due to the asymmetric distribution of proteins on the nanoparticle. To address these issues, individual particles were selected for analysis and all molecules associated with their respective particles were extracted as Cartesian coordinates from the STORM data set for each fluorophore. For a given particle, protein coordinates were used to calculate the sphere of best fit, by minimizing the function:24
| (1) |
Here, I = the number of data points; (xi,yi,zi) = the ith measurement of the origin; (xc, yc, zc) = the center of the sphere; and rc = the radius of the sphere. An example of the processing method is shown in Figure S4 for apolipoprotein A-II adsorbed onto small pore MSN. Once the sphere of best fit was calculated, all data points associated within 100 nm of the center (along the z-axis) were plotted and the opacity was scaled according to each molecule’s intensity. Detailed calculations are provided in the Supporting Information. To allow comparison of protein penetration depths from protein to protein, all calculated radii were normalized to their respective TEM radius, 415.7 nm for small pore and 456.5 nm for large pore MSNs.
This method was employed to analyze the adsorption of apolipoprotein A-II, albumin, and complement C3c on small pore and large pore MSNs (Figure 2). In the case of apolipoprotein A-II, penetration throughout the entire particle was observed for the large pore MSN. Conversely, for the small pore MSN, a clear void near center of the particle indicated the protein was unable to diffuse fully through the particle. Surprisingly, in the case of albumin, localized concentrations of protein were observed around the particle surface for both materials, suggesting that upon adsorption, albumin clogged the pores and inhibited further penetration of the porous scaffold. Only low concentrations of complement C3c were observed in all samples. Given the MW of complement C3c, it is likely that the protein was unable to penetrate into the pores of either material, but instead localized at the nanoparticle surface. These results are consistent with our previous research.5
Figure 2.
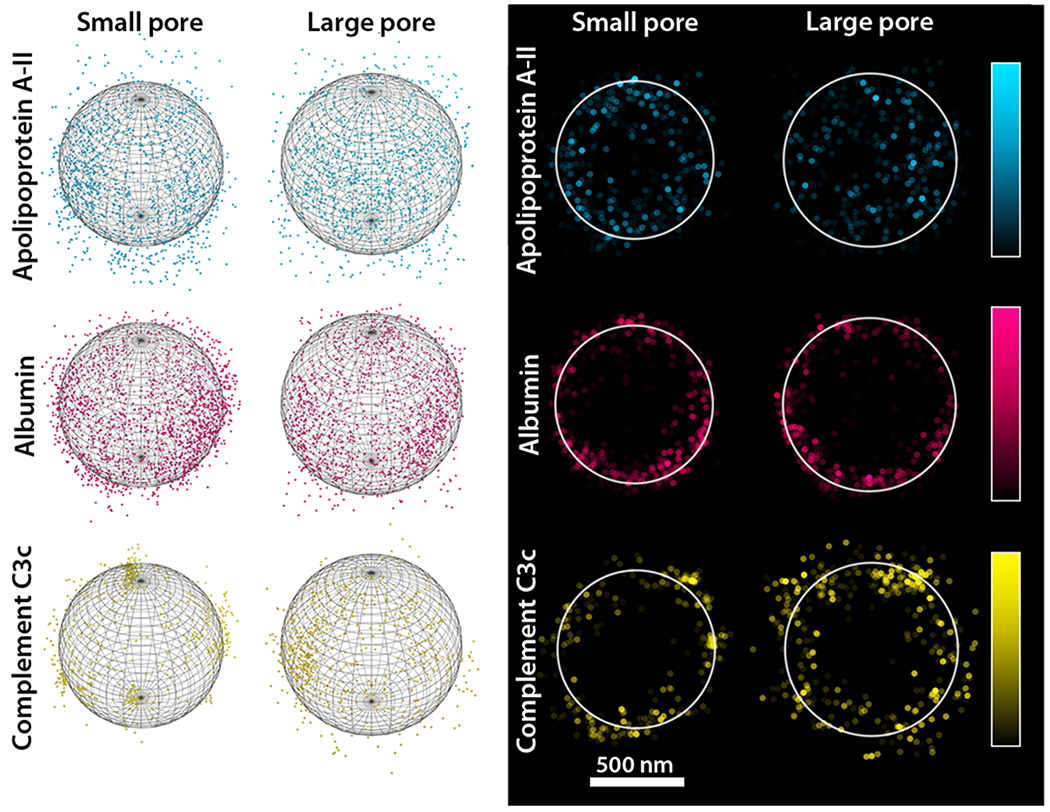
Sphere fitting model and analysis on selected spheres. (Left) The sphere of best fit was determined from STORM data for three fluorescently labeled proteins, represented in dots, adsorbed onto two types of MSNs. (Right) Once the particle origin was calculated, data associated with molecules within 100 nm of the particle center (along the z-axis) were extracted, the data points were flattened and plotted as a function of their fluorescence intensity (indicated in the colored bar).
The distance from the calculated nanoparticle origin to each protein was calculated using the Pythagorean theorem. Comparison among the protein/MSN pairs was evaluated by normalizing all calculated distances to their respective TEM values. Once all distances had been calculated, histograms were plotted and Gaussian fits were applied for both a single sphere and average distances for 10 particles (Figure S5). In a hollow sphere, protein concentrations would scale by r2, as the radius of the particle decreases or increases. This theoretical model exhibits a broad distribution, with a standard deviation of 137 ± 8.1 nm. For proteins capable of penetrating close to the particle center, a broad distribution similar to that of the theoretical model should be obtained. Although apolipoprotein A-II showed this distribution for both MSNs, the larger standard deviation for the distribution in the large pore MSN, 147 ± 7.5 nm vs 108 ± 3.4 nm, indicated that this protein was able to penetrate more deeply into the large pore MSN. This can also be qualitatively observed in Figure 2. Conversely, a narrow distribution would be expected for proteins that localize at or near the surface of the particle; this is observed for both albumin and complement C3c. Although the albumin and complement C3c distributions were similar in both MSNs, the number of albumin molecules identified during analysis was nearly five times that of complement C3c. Thus, there is more error associated with the complement C3c distribution than the albumin distribution, and the plot cannot take the protein clustering observed for the complement C3c data into account. As expected for larger proteins, albumin and complement C3c did not show significant shifts in distribution between the small and large pore MSNs.
The maximum penetration depth across each protein/MSN pair was calculated by arranging the distances from the center of the MSN to each adsorbed protein in ascending order, and then isolating the smallest 2.5% of this population. From this 2.5% subset, a two-sided Grubbs’ Test was applied to remove outliers (based on α = 0.05).25 Penetration depth was then calculated by taking the difference between these distances and the respective TEM radius. Once this process had been applied across all 10 particles, the maximum penetration depth was defined as the average of the largest penetration depth for each particle (Figure 3). Apolipoprotein A-II penetrated on average nearly 75 nm deeper into the large pore MSN compared to the small pore MSN (395 ± 25.0 nm versus 318 ± 27.5 nm), corresponding to nearly complete adsorption within the porous network of the MSN.
Figure 3.

After outliers were removed, the maximum penetration depth was calculated by averaging the greatest distance for each of the 10 particles.
Albumin penetration was similar for both the large pore and small pore MSN (290 ± 18.9 nm versus 280 ± 29.6 nm) supporting the idea that the increase in pore diameter had less of an impact on a protein with a larger MW. Complement C3c penetrated more deeply into the large pore MSN (285 ± 57.5 nm versus 202 ± 58.0 nm). Surprisingly, similar penetration depths for albumin and complement C3c were calculated for the large pore MSN, suggesting that pore diameters >60 Å are large enough to accommodate both of proteins, but it is likely that the initially adsorbed protein began to clog the pores, inhibiting further diffusion. This “bottle-necking” effect was most severe for complement C3c in small pore MSNs but was reduced in large pore MSNs, hence the large change in penetration depth. Because the effect was less severe for albumin in small pore MSNs, the change in penetration depth was not as large. A final compounding factor for complement C3c is that there was much less overall protein adsorbed and this protein tended to cluster on the MSN surface, whereas albumin was more evenly distributed within and across the surface of the particles (Figure 2).
Ultimately, the adsorption process is likely to be more complex when multiple proteins are present. To begin to address this question, we performed binary adsorption experiments using combinations of apolipoprotein A-II with the higher molecular weight proteins. For these experiments, each protein was labeled with a different fluorophore, and independently imaged in their respective channels. The concentrations of the proteins was the same as that used for the single-protein adsorption experiments. Interestingly, different adsorption results were obtained in binary mixtures (Figure 4).
Figure 4.
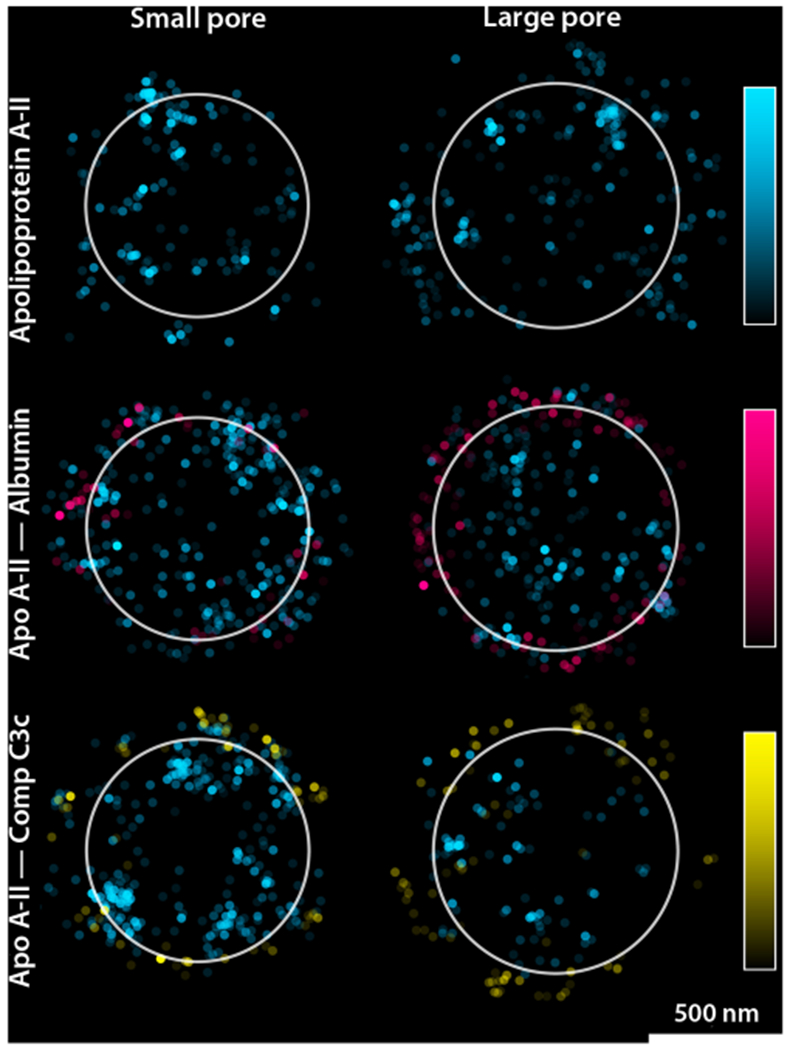
Adsorption of from binary protein mixtures on small and large pore MSNs. STORM data were processed as in Figure 2.
For example, when albumin was the only protein in solution, adsorption took place throughout the porous framework of both small and large pore MSNs (Figure 2). However, in a solution that also contained apolipoprotein A-II, albumin was primarily located on the external surface of the particles (Figure 4), and there was much less albumin present. The protein with the largest molecular weight, complement C3c, was affected by the presence of apolipoprotein A-II even more. In contrast, apolipoprotein A-II penetrated through the particles whether or not the larger proteins were present. This result suggests that in the binary mixture, the binding of apolipoprotein A-II within the pores prevented the larger proteins from accessing the internal pore surfaces.
An additional set of experiments was performed to study protein adsorption from a more complex mixture. The three proteins used above were fluorescently labeled, added to separate solutions of 10% human serum containing MSNs, and STORM was performed as above. Only apolipoprotein A-II could be detected; there was not enough albumin or complement C3c to be observed by STORM. This result is consistent with our previous publications, which showed that the protein corona adsorbed from 10% human serum is largely composed of low MW proteins. Thus, the STORM observations from binary and complex protein mixtures support our previous results and offer conclusive evidence for different protein adsorption patterns on porous and dense nanoparticles. As lower MW proteins compete with higher MW proteins for surface sites, the ability of lower MW proteins to access the pores limits the adsorption of other proteins exclusively to the outer surfaces, reducing the fraction of the corona that is composed of higher MW proteins. Although protein adsorption onto inorganic surfaces is a complex process involving several mechanisms,26 these results and our previous studies indicate that protein size is a predominant factor.
In summary, we have used STORM to develop a quantitative method to study the penetration of proteins within porous silica nanoparticles. The model presented here describes a noninvasive technique for both imaging and analyzing protein and molecular adsorption in a porous material. An important application of STORM is that it may allow a time-resolved study of protein adsorption to be performed, providing important information about the development of the protein corona.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103449. The authors also acknowledge the Microscopy Imaging Center at the University of Vermont.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/jacs.7b01118.
Experimental details, including nanoparticle preparation and characterization, protein labeling and adsorption, imaging conditions, and data analysis (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Tenzer S; Docter D; Kuharev J; Musyanovych A; Fetz V; Hecht R; Schlenk F; Fischer D; Kiouptsi K; Reinhardt C; Landfester K; Schild H; Maskos M; Knauer SK; Stauber RH Nat. Nanotechnol 2013, 8, 772. [DOI] [PubMed] [Google Scholar]
- (2).Lesniak A; Fenaroli F; Monopoli MP; Aberg C; Dawson KA; Salvati A ACS Nano 2012, 6, 5845. [DOI] [PubMed] [Google Scholar]
- (3).Dai Q; Yan Y; Ang C-S; Kempe K; Kamphuis MMJ; Dodds SJ; Caruso F ACS Nano 2015, 9, 2876. [DOI] [PubMed] [Google Scholar]
- (4).Ritz S; Schöttler S; Kotman N; Baier G; Musyanovych A; Kuharev J; Landfester K; Schild H; Jahn O; Tenzer S; Mailander V Biomacromolecules 2015, 16, 1311. [DOI] [PubMed] [Google Scholar]
- (5).Clemments AM; Botella P; Landry CC ACS Appl Mater. Interfaces 2015, 7, 21682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Lundqvist M; Stigler J; Elia G; Lynch I; Cedervall T; Dawson KA Proc. Natl. Acad. Sci. U. S. A 2008, 105, 14265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Monopoli MP; Walczyk D; Campbell A; Elia G; Lynch I; Baldelli Bombelli F; Dawson KA J. Am. Chem. Soc 2011, 133, 2525. [DOI] [PubMed] [Google Scholar]
- (8).Cedervall T; Lynch I; Lindman S; Berggård T; Thulin E; Nilsson H; Dawson KA; Linse S Proc. Natl. Acad. Sci. U. S. A 2007, 104, 2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Fleischer CC; Payne CK Acc. Chem. Res 2014, 47, 2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Treuel L; Brandholt S; Maffre P; Wiegele S; Shang L; Nienhaus GU ACS Nano 2014, 8, 503. [DOI] [PubMed] [Google Scholar]
- (11).Caracciolo G; Callipo L; De Sanctis SC; Cavaliere C; Pozzi D; Laganà A Biochim. Biophys. Acta, Biomembr 2010, 1798, 536. [DOI] [PubMed] [Google Scholar]
- (12).Salvati A; Pitek AS; Monopoli MP; Prapainop K; Bombelli FB; Hristov DR; Kelly PM; Aberg C; Mahon E; Dawson KA Nat. Nanotechnol 2013, 8, 137. [DOI] [PubMed] [Google Scholar]
- (13).Hu W; Peng C; Lv M; Li X; Zhang Y; Chen N; Fan C; Huang Q ACS Nano 2011, 5, 3693. [DOI] [PubMed] [Google Scholar]
- (14).Rocker C; Potzl M; Zhang F; Parak WJ; Nienhaus GU Nat. Nanotechnol 2009, 4, 577. [DOI] [PubMed] [Google Scholar]
- (15).Kelly PM; Åberg C; Polo E; O’Connell A; Cookman J; Fallon J; Krpetić Ž; Dawson KA Nat. Nanotechnol 2015, 10, 472. [DOI] [PubMed] [Google Scholar]
- (16).Clemments AM; Muniesa C; Landry CC; Botella P RSC Adv . 2014, 4, 29134. [Google Scholar]
- (17).Rust MJ; Bates M; Zhuang X Nat. Methods 2006, 3, 793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Szymborska A; de Marco A; Daigle N; Cordes VC; Briggs JAG; Ellenberg J Science 2013, 341, 655. [DOI] [PubMed] [Google Scholar]
- (19).Dudok B; Barna L; Ledri M; Szabo SI; Szabadits E; Pinter B; Woodhams SG; Henstridge CM; Balla GY; Nyilas R; Varga C; Lee S-H; Matolcsi M; Cervenak J; Kacskovics I; Watanabe M; Sagheddu C; Melis M; Pistis M; Soltesz I; Katona I Nat. Neurosci 2015, 18, 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Xu K; Babcock HP; Zhuang X Nat. Methods 2012, 9, 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Huang B; Wang W; Bates M; Zhuang X Science 2008, 319, 810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Nooney RI; Thirunavukkarasu D; Chen Y; Josephs R; Ostafin AE Chem. Mater 2002, 14, 4721. [Google Scholar]
- (23).Cedervall T; Lynch I; Foy M; Berggård T; Donnelly SC; Cagney G; Linse S; Dawson KA Angew. Chem., Int. Ed 2007, 46, 5754. [DOI] [PubMed] [Google Scholar]
- (24).Jennings AL; Black J; Allen C Shock & Vibration 2013, 20, 503. [Google Scholar]
- (25).Grubbs FE Ann. Math. Stat 1950, 21 , 27. [Google Scholar]
- (26).Moerz ST; Huber P Langmuir 2014, 30, 2729. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


