Abstract
Purpose
Genome-wide association studies have identified several genes associated with glaucoma. However, their roles in the pathogenesis of glaucoma remain unclear, particularly their effects on retinal nerve fiber layer (RNFL) thickness. The aim of this study was to investigate the associations between the identified glaucoma risk genes and RNFL thickness.
Methods
A total of 3843 participants (7,020 healthy eyes) were enrolled from the Singapore Epidemiology of Eye Diseases (SEED) study, a population-based study composing of three major ethnic groups—Malay, Indian, and Chinese—in Singapore. Ocular examinations were performed, and spectral-domain optical coherence tomography (SD-OCT) was used to measure circumpapillary RNFL thickness. We selected 35 independent glaucoma-associated genetic loci for analysis. An linear regression model was conducted to determine the association of these variants with circumpapillary RNFL, assuming an additive genetic model. We conducted association analysis in each of the three ethnic groups, followed by a meta-analysis of them.
Results
The mean age of the included participants was 59.4 ± 8.9 years, and the mean RFNL thickesss is 92.3 ± 11.2 µm. In the meta-analyses, of the 35 glacuoma loci, we found that only SIX6 was significantly associated with reduction in global RNFL thickness (rs33912345; β = −1.116 um per risk allele, P = 1.64E-05), and the effect size was larger in the inferior RNFL quadrant (β = −2.015 µm, P = 2.9E-6), and superior RNFL quadrant (β = −1.646 µm, P = 6.54E-5). The SIX6 association were consistently observed across all three ethnic groups. Other than RNFL, we also found several genetic varaints associated with vertical cuo-to-disc ratio (ATOH7, CDKN2B-AS1, and TGFBR3-CDC7), rim area (SIX6 and CDKN2B-AS1), and disc area (SIX6, ATOH7, and TGFBR3-CDC7). The association of SIX6 rs33912345 with NRFL thickness remained similar after further adjusting for disc area and 3 other disc parameter associated SNPs (ATOH7, CDKN2B-AS1, and TGFBR3-CDC7).
Conclusions
Of the 35 glaucoma identified risk loci, only SIX6 is significantly and independently associated with thinner RNFL. Our study further supports the involvement of SIX6 with RNFL thickness and pathogensis of glaucoma.
Keywords: glaucoma, RNFL, genetic diseases, Asian, meta-analysis
Glaucoma is one of the leading causes of blindness, affecting about 64 million people worldwide, and the number is projected to increase to 111.8 million by 2040.1 It is a disease characterized by the progressive loss of retinal ganglion cells with structural changes to optic nerve, as well as retinal nerve fiber layers (RNFL). Of the various glaucoma subtypes, primary open angle glaucoma (POAG) is the most common form of glaucoma.1
Understanding of the complex genetics underlying POAG has been aided by genome-wide association studies (GWAS), which identified several genes associated with the disease in the past decade.2–9 More recently, seven novel susceptibility loci were identified through a GWAS in Japan, highlighting the importance of ethnic difference in the development of the disease.10 Furthermore, GWAS meta-analyses also identified risk genes associated with glaucoma-related endophenotypes, such as intraocular pressure (IOP) and optic disc parameters.2,8,11,12 Taking together, these studies contributed to a better understanding of the underlying genetics of glaucoma.
In addition to optic disc parameters, another important structural endophenotype of POAG is the circumpapillary RNFL thickness. Loss of RNFL may precede change in optic disc, in particular in early stages of glaucoma. RNFL thickness is a heritable trait,13,14 and SIX6 and CDKN2B-AS1 are the only loci consistently reported to be associated with reduction of RNFL thickness15–17; moreover only a few studies have investigated the effect of glaucoma risk genes on RNFL thickness.17–19 Therefore the purpose of this study was to investigate the effects of known glaucoma risk variants on circumpapillary RNFL thickness.
Methods
Study Population
Study subjects were enrolled from the Singapore Epidemiology of Eye Disease (SEED) Study, a population-based cohort comprising three major ethnic groups (Malays, Indians, and Chinese) in Singapore. The details of the study design and methodology of the SEED study were previously reported.20,21 In brief, the study was conducted in the southwestern part of Singapore, using a standardized study protocol. An age-stratified random sampling strategy was adopted in each ethnic group to select adults aged 40 or older. Optical coherence tomography (OCT) was performed at the the baseline examination of the Chinese particiants (year 2009 to 2011), and at the six-year follow-up visit of the Malay (year 2011 to 2013), and Indian participants (year 2014 to 2016). The study adhered to the tenets of the Declaration of Helsinki, and ethics committee approval was obtained from the SingHealth Centralized Institutional Review Board. Institutional Review Board (IRB)/Ethics Committee approval was obtained for all participants.
Ocular Examination
All participants underwent a standardized and comprehensive ocular examination.21,22 In brief, IOP was measured using the Goldmann applanation tonometer (GAT; Haag-Streit, Bern, Switzerland) before pupil dilation. One reading was taken from each eye. If the IOP reading was greater than 21 mm Hg, a repeat reading was taken, and the second reading was used for analysis. Central corneal thickness (CCT) was measured using an ultrasound pachymeter (Advent; Mentor O & O, Inc., Norwell, MA, USA); the mean of five measurements was used for analysis. Axial length was measured using noncontact partial coherence interferometry (IOL Master V3.01; Carl Zeiss Meditec AG, Jena, Germany); the mean of five measurements was used for analysis.
Static automated perimetry (Swedish Interactive Threshold Algorithm standard 24-2, Humphrey Field Analyzer II; Carl Zeiss Meditec, Dublin, CA, USA) was performed on one in five participants and in all glaucoma subjects and glaucoma suspects (defined as below). A visual field was defined as reliable when fixation losses were less than 20%, and false-positive and false-negative rates were less than 33%. A glaucomatous visual field defect was defined as the presence of three or more significant (P <0.05) non-edge contiguous points with at least one at the P < 0.01 level on the same side of the horizontal meridian in the pattern deviation plot, and classified as “outside normal limits” on the Glaucoma Hemifield Test, confirmed on two consecutive visual field examinations. After pupil dilation using tropicamide 1% and phenylephrine hydrochloride 2.5%, the optic disc was evaluated using a 78-diopter (D) lens at 16 times magnification during slit-lamp fundoscopy (Haag-Streit model BQ-900), and vertical cup-to-disc ration (VCDR) was measured with an eyepiece measuring graticule.
Spectral-Domain OCT Imaging
Optic nerve head (ONH) and circumpapillary RNFL imaging was performed using spectral-domain optical coherence tomography (SD-OCT; Cirrus HD-OCT; Carl Zeiss Meditec) after pupil dilation. Details of the Cirrus HD-OCT optic disc scan protocol have been described in detail elsewhere.23 Briefly, optic nerve head and RNFL scan acquisitions were performed for each participant using an optic disc cube 200 × 200 scan protocol, which generated a cube of data in a 6- × 6-mm2 grid with 200 × 200 scans. The peripapillary measurement circle (3.46-mm diameter) was centered on the optic disc before capturing the image. After each capture, motion artifact was checked, and rescanning was performed if motion artifacts (indicated by discontinuity of blood vessels) or saccades through the peripapillary measurement circle were detected. The ONH and RNFL algorithms native to Cirrus HD-OCT (software version 6.0.2) were then used to measure the disc area, neural retinal rim area, and peripapillary RNFL thicknesses (average and quadrants) automatically. Detailed descriptions of the ONH and RNFL measurement algorithms have been previously published.24 Study eyes with OCT scans showing retinal layer segmentation errors, signal strength of less than six, or artifacts due to eye movements or blinking were excluded from the analysis.
Definition of Glaucoma
In principla, glaucoma was defined according to the International Society of Geographical and Epidemiological Ophthalmology (ISGEO) criteria based on three categories.14 In brief, category 1 cases were defined as optic disc abnormality (VCDR or VCDR asymmetry ≥ 97.5th percentile) with a corresponding glaucomatous visual field defect. Category 2 cases were defined as having a severely damaged optic disc (VCDR or VCDR asymmetry ≥ 99.5th percentile) in the absence of reliable visual field test results. Category 3 cases were defined for subjects who were blind (corrected visual acuity < 3/60), without visual field or optic disc data, and previous glaucoma surgery or IOP > 99.5th percentile.
A glaucoma suspect was defined as meeting any of the following criteria: (1) IOP > 21 mm Hg, (2) VCDR > 0.6 or VCDR asymmetry > 0.2, (3) signs consistent with pseudoexfoliation or pigment dispersion syndrome, (4) narrow angles (posterior trabecular meshwork visible for <1808 during static gonioscopy), (5) peripheral anterior synechiae, (6) other findings consistent with secondary glaucoma, and (7) known history of glaucoma.
Genotyping
Genotyping was performed using the Illumina Infinium OmniExpress BeadChips (llumina, Inc., San Diego, CA, USA). Stringent quality control filters were used to remove poorly performing samples and genetic variants. with a call rate < 99%, minor allele frequency (MAF) <0.1%, or showing deviation from Hardy-Weinberg equilibrium (P < 10−6) were removed. Routine quality control criteria on a per sample basis were carried out, and poorly performing samples were removed from further analysis. The remaining samples were then subjected to biologic relationship verification by using the principle of variability in allele sharing according to the degree of relationship. Identity-by-state information was derived using the PLINK software.25 For those pairs of individuals who showed evidence of cryptic relatedness, we removed the sample with the lower call rate before performing principal component (PC) analysis. Principal component analysis was undertaken using EIGENSTRAT26 to account for spurious associations resulting from ancestral differences of individual SNPs.
We conducted an extensive literature review on published GWAS of POAG and identified loci reported to be genome-wide significant (P <5 × 10−8) to date.2,4–10,16,27–31 We identified a total of 52 genetic variants from 35 POAG genetic loci (Supplementary Table S1).
Statistical Analysis
A linear regression model was fitted to determine the association between the the risk alleles and RNFL thickness, as well as VCDR, rim area and disc area. We assumed an additive genetic model where the genotype of the genetic variant was a variable varying from 0, 1, or 2, representing the number of copies of the risk allele carried. We conducted association analysis in each ethnic group, followed by a meta-analysis combining all three ethnic groups. Analysis for the association was adjusted for age, sex, and the first three PCs in each ethnic groups.
Data from two eyes of the individuals were included in the analysis when data from both eyes were available. If one eye was excluded or not eligible, data from the other eye were used. Generalized estimating equations with exchangeable correlation structures were applied to account for the correlation between pairs of eyes for each individual. In the meta-analysis, significance level of per-variant analysis was defined as P < 0.0014 after Bonferroni correction (0.05/35 for testing 35 loci). We performed conditional analayis to assess the independent association of SIX6 SNPs with RNFL thickness. Here we used nominal P < 0.05 as the significance cutoff instead of the one corrected for multiple testing because only one association test was performed for SIX6 SNP rs33912345. All association analyses were done using R package “geepack.”
Results
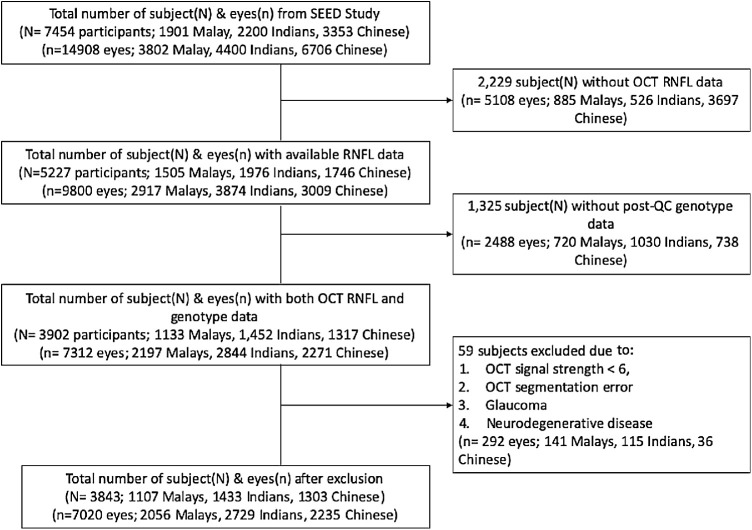
Of the 7454 SEED subjects who participants in the ocular examination, 5227 had available OCT RNFL data. We further excluded 1325 individuals without post-quality control genotype, and 59 with major ocular dieases or unacceptable RNFL data. This left a total of 7020 healthy eyes from 3843 participants (1303 Chinese, 1433 Indians, and 1107 Malays) with OCT, clinical and genotype data for this analysis (Fig.). The clinical and demographic characteristics of the study subjects are shown in Table 1. The mean (standard deviation [SD]) age was 55.0 (7.3) years for Chinese, 61.5 (8.4) years for Indians, and 61.8 (9.2) years for Malays in our study. The overall mean (SD) RNFL thickness in the study was 92.3 (11.2) µm.
Figure.
Consort diagram of study population.
Table 1.
Demographic and Clinical Characteristic of the Study Subjects
| Characteristics | Overall | Chinese | Indian | Malay | P Value* |
|---|---|---|---|---|---|
| Number of subjects | 3843 | 1303 | 1433 | 1107 | |
| Number of Eyes | 7020 | 2235 | 2729 | 2056 | |
| Age, yr | 59.4 (8.9) | 55.0 (7.3) | 61.5 (8.4) | 61.8 (9.2) | <0.001 |
| Gender, female | 1928 (50.2) | 644 (49.4) | 711 (49.6) | 573 (51.8) | 0.452 |
| Intraocular pressure, mm Hg | 14.8 (3.0) | 14.3 (3.0) | 15.3 (2.8) | 14.5 (3.0) | <0.001 |
| Vertical cup-to-disc ratio | 0.5 (0.1) | 0.5 (0.1) | 0.6 (0.1) | 0.5 (0.1) | < 0.001 |
| Disc area, mm2 | 2.0 (0.4) | 2.0 (0.4) | 2.0 (0.4) | 2.1 (0.4) | < 0.001 |
| RNFL thickness, µm | |||||
| Global | 92.3 (11.2) | 96.5 (9.6) | 87.1 (10.6) | 94.6 (10.8) | < 0.001 |
| Temporal sector | 65.5 (12.8) | 71.7 (12.2) | 58.8 (10.9) | 67.8 (11.4) | < 0.001 |
| Superior sector | 115.4 (17.7) | 121.3 (16.6) | 108.6 (16.4) | 118.0 (17.5) | < 0.001 |
| Nasal sector | 69.3 (11.0) | 68.1 (10.7) | 69.2 (11.0) | 70.8 (11.0) | < 0.001 |
| Inferior sector | 118.8 (18.4) | 124.6 (16.8) | 111.9 (17.2) | 121.8 (18.6) | < 0.001 |
Data are presented as mean (standard deviation) or number (%), as appropriate by eye.
In the single cohort analysis, SIX6 single nucleotide polymorphisms (SNP) rs33912345 was significantly associated with superior RNFL in Chinese individuals (β = −2.479, P = 0.001) (Supplementary Table S3), but not significantly associated with global RNFL and inferior RNFL in all 3 populations (Supplementary Tables S2, S4). In addition, the risk allele of ABCA1 SNP (rs2472494) was significantly associated with thinner inferior RFNL thickness in Malays (β = −2.378 µm per risk allele, P = 0.001; Supplementary Table S4).
Table 2 shows the association of the glaucoma loci with RNFL thickness in the meta-analysis of all three ethnic groups. The lead variant within a locus, defined by the variant with the most significant P value from the meta analysis of global RNFL, was reported. Of the 35 loci included in the analysis, only SIX6 SNP (rs33912345) was significantly associated with global (P = 1.64E-05), superior (P = 6.54E-05) and inferior (P = 2.9E-06) RNFL thickness. The effect size of rs33912345 was largest in the inferior RNFL (β = −2.015) followed by the superior (β = −1.646) and global RNFL quadrant (β = −1.116). Other seven loci reached nominal significant level (P < 0.05), including CDKN1A, CDKN2B-AS1 and 8q22 in the global and inferior RNFL thickness, TMCO1 in the global RNFL thickness, ARHGEF12, EXOC2 in the inferior RNFL thickness, and PMM2 in the superior RNFL thickness.
Table 2.
Associations of Lead SNPs With RNFL Thickness Parameters From Combined Analyses of All Three Ethnic Groups
| Global RNFL | Superior Quadrant | Inferior Quadrant | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | rs ID | Risk Allele | β* | P Value | β* | P Value | β* | P Value |
| 8q22 | rs284489 | A | 0.454 | 0.045 | 0.693 | 0.056 | 0.799 | 0.036 |
| ABCA1 | rs2472494 | T | −0.127 | 0.574 | −0.070 | 0.847 | −0.523 | 0.169 |
| AFAP1 | rs4619890 | G | −0.194 | 0.401 | −0.533 | 0.141 | −0.232 | 0.556 |
| ANKH | rs76325372 | C | 0.205 | 0.410 | 0.225 | 0.568 | 0.272 | 0.510 |
| ANKRD55 | rs61275591 | A | −0.229 | 0.418 | −0.177 | 0.698 | −0.091 | 0.850 |
| ARHGEF12 | 11:120357425 | AT/A | −0.468 | 0.061 | −0.331 | 0.400 | −0.919 | 0.031 |
| ATOH7 | rs1900004 | T | −0.202 | 0.427 | 0.471 | 0.252 | −0.367 | 0.395 |
| ATXN2 | rs7137828 | C | −0.128 | 0.877 | 0.277 | 0.826 | −0.721 | 0.607 |
| CADM2 | rs34201102 | G | 0.445 | 0.072 | 0.516 | 0.185 | 0.581 | 0.158 |
| CAV1/2 | rs10281637 | C | −0.349 | 0.446 | −0.199 | 0.772 | −0.521 | 0.481 |
| CDKN1A | 6:36592986 | TCA/T | −0.540 | 0.039 | −0.472 | 0.254 | −0.922 | 0.042 |
| CDKN2B-AS1 | rs1360589 | T | −0.749 | 0.019 | −0.979 | 0.052 | −1.269 | 0.015 |
| DGKG | rs9853115 | T | 0.132 | 0.573 | 0.308 | 0.409 | 0.327 | 0.400 |
| EXOC2 | rs2073006 | T | −0.627 | 0.057 | −0.651 | 0.213 | −1.178 | 0.033 |
| FLNB | rs12494328 | A | 0.018 | 0.935 | −0.092 | 0.798 | −0.298 | 0.425 |
| FMNL2 | rs56117902 | C | 0.241 | 0.457 | 0.087 | 0.861 | 0.276 | 0.602 |
| FNDC3B | rs7636836 | T | −0.557 | 0.350 | −1.680 | 0.110 | −0.813 | 0.460 |
| FOXC1 | rs2745572 | A | −0.090 | 0.720 | −0.163 | 0.680 | −0.224 | 0.597 |
| GAS7 | rs9913991 | A | −0.365 | 0.409 | −0.408 | 0.583 | −1.135 | 0.129 |
| GMDS | rs11969985 | G | −0.128 | 0.679 | −0.076 | 0.880 | −0.190 | 0.713 |
| HMGA2 | rs343093 | G | −0.128 | 0.597 | −0.165 | 0.665 | −0.392 | 0.333 |
| IKZF2 | rs56335522 | C | 0.301 | 0.756 | 1.021 | 0.492 | 1.455 | 0.355 |
| LHPP | rs12262706 | G | −0.314 | 0.214 | −0.680 | 0.094 | −0.590 | 0.167 |
| LMO7 | rs9530458 | T | −0.181 | 0.570 | −0.204 | 0.675 | −0.212 | 0.685 |
| LMX1B | rs10819187 | G | 0.207 | 0.447 | 0.142 | 0.738 | 0.173 | 0.707 |
| LOXL1 | rs1048661 | T | 0.158 | 0.489 | 0.077 | 0.834 | 0.152 | 0.693 |
| MEIS2 | rs28480457 | C | 0.402 | 0.424 | −0.103 | 0.899 | −0.065 | 0.940 |
| PDE7B | rs9494457 | A | 0.155 | 0.505 | 0.366 | 0.313 | 0.176 | 0.644 |
| PMM2 | rs3785176 | G | 0.475 | 0.094 | 0.891 | 0.043 | 0.679 | 0.155 |
| SIX6 | rs33912345 | C | −1.116 | 1.64E-5 | −1.646 | 6.54E-5 | −2.015 | 2.9E-6 |
| TAP2 | rs241430 | C | 0.153 | 0.478 | 0.056 | 0.873 | 0.421 | 0.246 |
| TGFBR3-CDC7 | rs1192415 | G | −0.048 | 0.865 | −0.033 | 0.939 | 0.099 | 0.830 |
| TMCO1 | rs4656461 | G | −1.362 | 0.026 | −1.873 | 0.052 | −1.122 | 0.285 |
| TMTC2 | rs324794 | G | −0.117 | 0.686 | −0.103 | 0.821 | −0.301 | 0.531 |
| TXNRD2 | rs35934224 | T | 0.650 | 0.107 | 0.869 | 0.148 | 1.065 | 0.108 |
In bold, Bonferroni-corrected significance value P < 0.0014 (0.05/35).
β, Changes in RNFL thickness (in micrometers) per risk allele count. Model adjusted for age, gender, top three genotype PCs.
In addition to RNFL thickness, we also conducted association tests for other glaucoma endophenotypes including VCDR, rim area, and disc area to check whether the same set of genetic variants are responsible for the change in these glaucoma-related structure phenotypes. SIX6 was significantly associated with both rim area (β = −0.052, P < 1E-8) and disc area (β = −0.055, P = 3.42E-8), but not VCDR (P = 0.006). CDKN2B-AS1 was associated with both VCDR (β = 0.016, P = 1.42E-4) and rim area (β = −0.026, P = 3.87E-4), but not disc area (P = 0.405). ATOH7 was associated with both VCDR (β = −0.018, P = 1.29E-6) and disc area (β = −0.087, P < 1E-8), but not rim area (P = 0.014). And TGFBR3-CDC7 was associated with VCDR (β = 0.014, P = 2.08E-4) only (Table 3).
Table 3.
Association of SNPs With Vertical Cup-to-Disc Ratio, Rim Area, and Disc Area From Combined Analyses of Three Etnic Groups
| Vertical Cup-to-Disc Ratio | Rim Area, Mm2 | Disc Area, Mm2 | ||||||
|---|---|---|---|---|---|---|---|---|
| Gene | rs ID | Risk Allele | β* | P Value | β* | P Value | β* | P Value |
| 8q22 | rs284489 | A | 0.002 | 0.451 | 0.003 | 0.529 | 0.012 | 0.182 |
| ABCA1 | rs2472494 | T | 0.004 | 0.211 | −0.010 | 0.069 | −0.002 | 0.797 |
| AFAP1 | rs4619890 | G | −0.003 | 0.313 | 0.001 | 0.896 | −0.012 | 0.168 |
| ANKH | rs76325372 | C | −0.005 | 0.125 | 0.012 | 0.041 | 0.003 | 0.769 |
| ANKRD55 | rs61275591 | A | 0.009 | 0.032 | −0.017 | 0.014 | 0.005 | 0.652 |
| ARHGEF12 | 11:120357425 | AT/A | 0.006 | 0.096 | −0.012 | 0.041 | −0.001 | 0.912 |
| ATOH7 | rs1900004 | T | −0.018 | 1.29E-6 | −0.015 | 0.014 | −0.087 | <1E-8 |
| ATXN2 | rs7137828 | C | −0.003 | 0.812 | 0.002 | 0.918 | −0.011 | 0.718 |
| CADM2 | rs34201102 | G | −0.001 | 0.806 | −0.002 | 0.781 | −0.006 | 0.499 |
| CAV1/2 | rs10281637 | C | 0.002 | 0.733 | −0.004 | 0.691 | 0.002 | 0.900 |
| CDKN1A | 6:36592986 | TCA/T | 0.005 | 0.133 | −0.020 | 0.002 | −0.008 | 0.420 |
| CDKN2B-AS1 | rs1360589 | T | 0.016 | 1.42E-4 | −0.026 | 3.87E-4 | 0.010 | 0.405 |
| DGKG | rs9853115 | T | 0.006 | 0.072 | −0.005 | 0.415 | 0.003 | 0.762 |
| EXOC2 | rs2073006 | T | 0.007 | 0.088 | −0.012 | 0.126 | −0.003 | 0.798 |
| FLNB | rs12494328 | A | 0.007 | 0.027 | −0.014 | 0.011 | 0.001 | 0.953 |
| FMNL2 | rs56117902 | C | 0.003 | 0.473 | −0.006 | 0.438 | 0.007 | 0.552 |
| FNDC3B | rs7636836 | T | 0.005 | 0.554 | −0.018 | 0.237 | −0.007 | 0.792 |
| FOXC1 | rs2745572 | A | 0.005 | 0.114 | −0.005 | 0.387 | 0.008 | 0.397 |
| GAS7 | rs9913991 | A | −0.004 | 0.557 | 0.000 | 0.974 | −0.019 | 0.304 |
| GMDS | rs11969985 | G | 0.006 | 0.167 | −0.013 | 0.073 | −0.002 | 0.896 |
| HMGA2 | rs343093 | G | 0.002 | 0.454 | −0.011 | 0.047 | −0.011 | 0.248 |
| IKZF2 | rs56335522 | C | 0.007 | 0.571 | −0.027 | 0.200 | −0.039 | 0.292 |
| LHPP | rs12262706 | G | 0.000 | 0.980 | −0.009 | 0.170 | −0.011 | 0.257 |
| LMO7 | rs9530458 | T | 0.001 | 0.813 | −0.006 | 0.409 | −0.004 | 0.742 |
| LMX1B | rs10819187 | G | 0.001 | 0.892 | 0.004 | 0.541 | 0.006 | 0.606 |
| LOXL1 | rs1048661 | T | 0.002 | 0.542 | −0.007 | 0.221 | −0.001 | 0.936 |
| MEIS2 | rs28480457 | C | −0.004 | 0.520 | −0.006 | 0.591 | −0.031 | 0.112 |
| PDE7B | rs9494457 | A | −0.004 | 0.182 | 0.007 | 0.225 | −0.001 | 0.950 |
| PMM2 | rs3785176 | G | 0.002 | 0.529 | −0.001 | 0.825 | 0.008 | 0.470 |
| SIX6 | rs33912345 | C | 0.010 | 0.006 | −0.052 | <1E-8 | −0.055 | 3.42E-8 |
| TAP2 | rs241430 | C | 0.000 | 0.900 | 0.001 | 0.913 | 0.002 | 0.844 |
| TGFBR3-CDC7 | rs1192415 | G | 0.014 | 2.08E-4 | 0.012 | 0.078 | 0.072 | <1E-8 |
| TMCO1 | rs4656461 | G | 0.014 | 0.095 | −0.030 | 0.034 | 0.003 | 0.905 |
| TMTC2 | rs324794 | G | −0.003 | 0.380 | -0.007 | 0.298 | −0.024 | 0.028 |
| TXNRD2 | rs35934224 | T | −0.013 | 0.012 | 0.025 | 0.007 | −0.001 | 0.919 |
In bold, Bonferroni-corrected significance value P < 0.0014 (0.05/35).
β denotes per unit changes in respective outcome parameter, per risk allele count. Model adjusted for age, gender, top three genotype PCs.
Because SIX6 is associated with several glaucoma endophenotypes (RNFL, rim area and disc area), we further included disc area as one of the covariates when testing the association between SIX6 and RNFL thickness. After adjusting for disc area, SIX6 was still significantly associated with global RNFL (β = −0.882, P = 4.9E-4), superior RNFL (β = −1.2162, P = 2.6E-3), and inferior RNFL (β = −1.7115, P = 5.7E-5) (Table 4) in the meta-analysis of three populations. In addiation, after further adjusting for ATOH7 rs1900004, CDKN2B-AS1 rs1360589, and TGFBR3-CDC7 rs1192415, the effect size and significance level still remained similar (Table 4) in the meta-analysis of three populations. These sensitivity analyses suggest that SIX6 SNP is independently associated with RNFL thickness.
Table 4.
Association Between SIX6 (rs33912345) and Peripapillary Retinal Nerve Fiber Thickness Parameters From Combined Analyses of Three Etnic Groups
| Global RNFL | Superior Quandrant | Inferior Quandrant | ||||
|---|---|---|---|---|---|---|
| β* | P Value | β* | P Value | β* | P Value | |
| Model 1† | −0.882 | 4.9E-4 | −1.216 | 2.6E-3 | −1.712 | 5.7E-5 |
| Model 2‡ | −0.847 | 8.8E-4 | −1.172 | 4.0E-3 | −1.682 | 8.4E-5 |
Changes in RNFL thickness (in µm) per risk allele C of rs33912345.
Model 1 adjusted for age, gender, top three genotype PCs, and disc area.
Model 2 adjusted for age, gender, top three genotype PCs, disc area, rs1900004 (ATOH7 gene), rs1360589 (CDKN2B-AS1), rs1192415 (TGFBR3-CDC7).
On the other hand, for ATOH7 rs1900004 and TGFBR3-CDC7 rs1192415, after further adusting for disc area, the associations with VCDR became non-significant (data not shown), suggesting that the original observed associations with VCDR of the two genes were mainly driven by their associations with disc area, also given the known correlation between disc area and VCDR.
We also conducted two more sensitivity analyses for all the variants by including axial length and disc area as additional covariates in the original model (Supplementary Tables S5, S6, respectively). The associations between all variants (including SIX6) and RNFL remained similar in these sensitivity analyses.
Discussion
Of the 35 known POAG risk loci included in our study, we found that only SIX6 rs33912345 was significantly associated with thinner RNFL in normal healthy eyes. The association was more apparent in the superior and inferior quadrant, consistent with previous reports.18,32 In our study, we also noticed that the association between SIX6 and RNFL remains similar after further adjusting for disc area and the other three disc parameters associated SNPs suggesting that this association is independent of disc size as well as other disc related genotypes. Our findings provided a more comprehensive insight into the complex genetic architecture underlying glaucoma and the role of SIX6 in determining RNFL thinning. This suggests that subjects with the SIX6 genetic risk variant are at higher risk for glaucoma due to thinner RNFL, while the other glaucoma risk genes may lead to glaucoma through mechanisms.
The human SIX gene consists of SIX1-SIX6, was initially discovered through homology to Drosophila melanogaster (Drosophila) sine oculis (so) gene, and its function is important for ectodermal differentiation and ocular development.33 Carnes et al.32 first reported their association with RNFL thinning and hypothesized that SIX6 causes glaucoma by disrupting neural retinal tissue development leading to loss of retinal ganglion cells eventually leading to glaucomatous visual loss. The association of SIX6 with thinning of RNFL was later confirmed by several other studies.17,18,34,35 These associations were also found to apply to nonglaucomatous eyes by one of our earlier studies with smaller smaple size of 1243 Chinese individuals,16 which strongly supported SIX6's contribution to RNFL thickness.16 In the article, each copy of C allele of rs33912345 was associated with 1.44-µm decrease in global RNFL thickness. Similarly, Kuo et al.17 reported the association between RNFL thickness and another SNP re10483727 in a populaton of 231 individuals of European descent; each T allele of rs10483727 was associated with 0.16 um decrease in global RNFL thickness. Khawaja et al.34 have also reported association of rs33912345 C allele with smaller rim area (0.030-mm decrease, P = 5.4E-9), a larger VCDR (0.025 increase, P = 3.3E-10) and thinner RNFL (0.39-µm decrease, P = 0.001). This is also an European based study. They used 3699 samples to estimate the association between the SIX6 SNP and RNFL, and 5433 samples to estimate the associations between the SIX6 SNP and rim/VCDR parameters. As such, our finding of association of SIX6 with thinner RNFL further reinforces these previous findings in a larger cohort.
The risk allele of SIX6 (rs33912345) was associated with thinner global RNFL (β = −1.116, P = 2.9E-6 per risk allele) in our analysis, and was previously reported to be associated with higher POAG risk.36 In addition, CDKN1A, CDKN2B-AS1, 8q22, TMCO1, ARHGEF12, EXOC2, PMM2 were associated with thinner RNFL thickness parameters (either global, inferior, or superior) but with nominal significance (P < 0.05). Of these seven loci, the risk alleles of CDKN1A, CDKN2B-AS1, TMCO1, ARHGEF12, EXOC2 were also associated with increased risk of POAG, based on previous reports.37–39 On the other hand, the risk allele of 8q22 (rs284489), was previously reported to be associated with normal tension glaucoma40 but was shown to be associated with thicker global RNFL (β = 0.454, P = 0.045) in our meta-analysis. Similarly, the risk allele of PMM2 (rs3785176) was previously reported to be associated with higher POAG risk41 but was shown to be associated with thicker superior RNFL (β = 0.891, P = 0.043) in our current analysis. Nevertheless, it should be noted that the associations RNFL thickness in these two loci were only marginally significant. Thus further investigation is needed to further confirm whether a differing direction with RNFL and glaucoma risk was indeed present in these two loci.
Based on Tables 2 and 3, we observed that the risk allele of SIX6 was significantly associated with thinner RNFL thickness (P = 1.64E-5), and consistently showed to be associated with smaller rim area (P = 1E-8). In addition, its association with vertical cup-to-disc ratio (VCDR) was nominally significant (P = 0.006) but with consistent direction (per risk allele, larger VCDR). The less significant association with VCDR may be partly owing to the reason that thinning of RNFL and rim area are better proxy markers for RGC compared to VCDR. Furthermore, it should be noted that SIX6 was also significantly associated with smaller disc area (P = 3.42E-8). In light of this and given that VCDR is an indirect measure of rim loss that is calculated from the cup length (which can also be thought of as “disc length minus the rim length”) and disc length—it is therefore possible that the association with VCDR was slightly negated compared to associations with rim area and disc area.
Other than SIX6, of the 35 lcoi only one of the other previously identified POAG risk genes, ABCA1, were significantly associated with reduction in inferior RNFL thickness in our Malay group, despite that there loci (including ATOH7, CDKN2B-AS and TGFBR3-CDC7 SNPs) are significantly associated with glaucoma-related disc parameters. We therefore postulated that glaucoma risk genes may have different effects on RNFL thinning and disc alteration, and RNFL thinning in glaucoma is most likely a manifestation of various ocular insults rather than a direct effect of the gene products and their associated pathway on the integrity of RNFL. Further studies are needed to evaluate the mechanisms in which these genes predispose individuals to glaucoma.
The strength of our study includes a multiethnic population-based sample with a standardized protocol for RFNL assessment. Nevertheles, our study has a few limiattions. First, our study is limited by its cross-sectional design, and thus we cannot determine whether individuals carrying the SIX6 risk variants have an accelerated rate of RNFL thinning, or that the risk variants only have an effect during the embryological ocular development phase which results in these individuals having a thinner RNFL. A prospective study might be useful to distinguish between these mechanisms via which the variant may have effect. On the other hand, based on our power calculation, given a type 1 error rate of 0.0014, the current sample size of 7020 would have 80% power to minimally detect an effect size of 1.01 µm in RNFL thickness per risk allele (for allele frequency of 0.2), and 0.78 µm per risk allele (for allele frequency of 0.4). Thus our current sample size may be limited in identifying new loci especially one with lower allele frequency.
In conclusion, this is the first study comparing effect of glaucoma risk genes on RNFL across multiple ethnic groups in Asians. Of the 35 glaucoma risk loci we investigated in our population, only SIX6 is significantly and independently associated with thinner RNFL in the global, superior and inferior RNFL quadrants. Our study further supports the involvement of SIX6 with RNFL thickness and pathogensis of glaucoma. These findings may provide further insights into the genetic variants associated with pathogenesis of glaucoma.
Supplementary Material
Acknowledgments
Supported by the National Medical Research Council (NMRC/1249/2010, NMRC/CIRG/1371/2013, NMRC/CIRG/1417/2015, and NMRC/CIRG/1488/2018), Singapore. C-YC is supported by National Medical Research Council (NMRC/CSA-SI/0012/2017).
Disclosure: X. Chai, None; K.Y. Low, None; Y.C. Tham, None; M.L. Chee, None; S. Thakur, None; L. Zhang, None; N.Y. Tan, None; C.C. Khor, None; T. Aung, None; T.Y. Wong, None; C.-Y. Cheng, None
References
- 1. Tham Y-C, Li X, Wong TY, et al.. Global prevalence of glaucoma and projections of glaucoma burden through 2040: a systematic review and meta-analysis. Ophthalmology. 2014; 121: 2081–2090. [DOI] [PubMed] [Google Scholar]
- 2. Nakano M, Ikeda Y, Tokuda Y, et al.. Common variants in CDKN2B-AS1 associated with optic-nerve vulnerability of glaucoma identified by genome-wide association studies in Japanese. PloS One. 2012; 7: e33389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ramdas WD, van Koolwijk LM, Lemij HG, et al.. Common genetic variants associated with open-angle glaucoma. Hum Mol Genet. 2011; 20: 2464–2471. [DOI] [PubMed] [Google Scholar]
- 4. Osman W, Low S-K, Takahashi A, et al.. A genome-wide association study in the Japanese population confirms 9p21 and 14q23 as susceptibility loci for primary open angle glaucoma. Hum Mol Genet. 2012; 21: 2836–2842. [DOI] [PubMed] [Google Scholar]
- 5. Thorleifsson G, Walters GB, Hewitt AW, et al.. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010; 42: 906–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burdon KP, Macgregor S, Hewitt AW, et al.. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011; 43: 574–578. [DOI] [PubMed] [Google Scholar]
- 7. Chen Y, Lin Y, Vithana EN, et al.. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014; 46: 1115–1119. [DOI] [PubMed] [Google Scholar]
- 8. Springelkamp H, Iglesias AI, Mishra A, et al.. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet. 2017; 26: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Choquet H, Paylakhi S, Kneeland SC, et al.. A multiethnic genome-wide association study of primary open-angle glaucoma identifies novel risk loci. Nat Commun. 2018; 9: 2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiga Y, Akiyama M, Nishiguchi KM, et al.. Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum Mol Genet. 2018; 27: 1486–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hysi PG, Cheng C-Y, Springelkamp H, et al.. Genome-wide analysis of multi-ancestry cohorts identifies new loci influencing intraocular pressure and susceptibility to glaucoma. Nat Genet. 2014; 46: 1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wiggs JL, Yaspan BL, Hauser MA, et al.. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012; 8: e1002654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. van Koolwijk LM, Despriet DD, van Duijn CM, et al.. Genetic contributions to glaucoma: heritability of intraocular pressure, retinal nerve fiber layer thickness, and optic disc morphology. Invest Ophthalmol Vis Sci. 2007; 48: 3669–3676. [DOI] [PubMed] [Google Scholar]
- 14. Hougaard JL, Kessel L, Sander B, et al.. Evaluation of heredity as a determinant of retinal nerve fiber layer thickness as measured by optical coherence tomography. Invest Ophthalmol Vis Sci. 2003; 44: 3011–3016. [DOI] [PubMed] [Google Scholar]
- 15. Yoshikawa M, Nakanishi H, Yamashiro K, et al.. Association of glaucoma-susceptible genes to regional circumpapillary retinal nerve fiber layer thickness and visual field defects. Invest Ophthalmol Vis Sci. 2017; 58: 2510–2519. [DOI] [PubMed] [Google Scholar]
- 16. Cheng C-Y, Allingham RR, Aung T, et al.. Association of common SIX6 polymorphisms with peripapillary retinal nerve fiber layer thickness: the Singapore Chinese Eye StudyAssociation of SIX6 Variant With RNFL Thickness. Invest Ophthalmol Vis Sci. 2015; 56: 478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kuo JZ, Zangwill LM, Medeiros FA, et al.. Quantitative trait locus analysis of SIX1-SIX6 with retinal nerve fiber layer thickness in individuals of European descent. Am J Ophthalmol. 2015; 160: 123–130. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng C-Y, Allingham RR, Aung T, et al.. Association of common SIX6 polymorphisms with peripapillary retinal nerve fiber layer thickness: the Singapore Chinese Eye Study. Invest Ophthalmol Vis Sci 2015; 56: 478–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mydel P, Wang Z, Brisslert M, et al.. Carbamylation-dependent activation of T cells: a novel mechanism in the pathogenesis of autoimmune arthritis. J Immunol. 2010; 184: 6882–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Foong AW, Saw S-M, Loo J-L, et al.. Rationale and methodology for a population-based study of eye diseases in Malay people: The Singapore Malay Eye Study (SiMES). Ophthalmic Epidemiol. 2007; 14: 25–35. [DOI] [PubMed] [Google Scholar]
- 21. Lavanya R, Jeganathan VSE, Zheng Y, et al.. Methodology of the Singapore Indian Chinese Cohort (SICC) eye study: quantifying ethnic variations in the epidemiology of eye diseases in Asians. Ophthalmic Epidemiol. 2009; 16: 325–336. [DOI] [PubMed] [Google Scholar]
- 22. Chua J, Tham YC, Liao J, et al.. Ethnic differences of intraocular pressure and central corneal thickness: the Singapore Epidemiology of Eye Diseases study. Ophthalmology. 2014; 121: 2013–2022. [DOI] [PubMed] [Google Scholar]
- 23. Cheung CY, Chen D, Wong TY, et al.. Determinants of quantitative optic nerve measurements using spectral domain optical coherence tomography in a population-based sample of non-glaucomatous subjects. Invest Ophthalmol Vis Sci. 2011; 52: 9629–9635. [DOI] [PubMed] [Google Scholar]
- 24. Tham Y-C, Cheung CY, Koh VT, et al.. Relationship between ganglion cell-inner plexiform layer and optic disc/retinal nerve fibre layer parameters in non-glaucomatous eyes. Br J Ophthalmol. 2013; 97: 1592–1597. [DOI] [PubMed] [Google Scholar]
- 25. Purcell S, Neale B, Todd-Brown K, et al.. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007; 81: 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price AL, Patterson NJ, Plenge RM, et al.. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006; 38: 904–909. [DOI] [PubMed] [Google Scholar]
- 27. Gharahkhani P, Burdon KP, Fogarty R, et al.. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014; 46: 1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bailey JNC, Loomis SJ, Kang JH, et al.. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016; 48: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gharahkhani P, Burdon KP, Bailey JNC, et al.. Analysis combining correlated glaucoma traits identifies five new risk loci for open-angle glaucoma. Sci Rep. 2018; 8: 3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Shiga Y, Akiyama M, Nishiguchi KM, et al.. Genome-wide association study identifies seven novel susceptibility loci for primary open-angle glaucoma. Hum Mol Genet. 2018; 27: 1486–1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Micheal S, Ayub H, Khan MI, et al.. Association of known common genetic variants with primary open angle, primary angle closure, and pseudoexfoliation glaucoma in Pakistani cohorts. Mol Vis. 2014; 20: 1471–1479. [PMC free article] [PubMed] [Google Scholar]
- 32. Carnes MU, Liu YP, Allingham RR, et al.. Discovery and functional annotation of SIX6 variants in primary open-angle glaucoma. PLoS Genet. 2014; 10: e1004372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kumar J. The sine oculis homeobox (SIX) family of transcription factors as regulators of development and disease. Cell Mol Life Sci. 2009; 66: 565–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Khawaja AP, Chan MP, Yip JL, et al.. A common glaucoma-risk variant of SIX6 alters retinal nerve fiber layer and optic disc measures in a European population: the EPIC-Norfolk Eye Study. J Glaucoma. 2018; 27: 743–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Shiga Y, Nishiguchi KM, Kawai Y, et al.. Genetic analysis of Japanese primary open-angle glaucoma patients and clinical characterization of risk alleles near CDKN2B-AS1, SIX6 and GAS7. PloS One. 2017; 12: e0186678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bailey JN, Loomis SJ, Kang JH, et al.. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016; 48: 189–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Springelkamp H, Iglesias AI, Mishra A, et al.. New insights into the genetics of primary open-angle glaucoma based on meta-analyses of intraocular pressure and optic disc characteristics. Hum Mol Genet 2017; 26: 438–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gharahkhani P, Burdon KP, Fogarty R, et al.. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014; 46: 1120–1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. MacGregor S, Ong JS, An J, et al.. Genome-wide association study of intraocular pressure uncovers new pathways to glaucoma. Nat Genet. 2018; 50: 1067–1071. [DOI] [PubMed] [Google Scholar]
- 40. Nordberg A, Hartvig P, Lilja A, et al.. Decreased uptake and binding of 11C-nicotine in brain of Alzheimer patients as visualized by positron emission tomography. J Neural Transm Park Dis Dement Sect. 1990; 2: 215–224. [DOI] [PubMed] [Google Scholar]
- 41. Chen Y, Lin Y, Vithana EN, et al.. Common variants near ABCA1 and in PMM2 are associated with primary open-angle glaucoma. Nat Genet. 2014; 46: 1115–1119. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.