Abstract
The gateway for invasion by the novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) into human host cells is via the angiotensin-converting enzyme 2 (ACE2) transmembrane receptor expressed in multiple immune and nonimmune cell types. SARS-CoV-2, that causes coronavirus disease 2019 (COVID-19; CoV-19) has the unusual capacity to attack many different types of human host cells simultaneously via novel clathrin- and caveolae-independent endocytic pathways, becoming injurious to diverse cells, tissues and organ systems and exploiting any immune weakness in the host. The elicitation of this multipronged attack explains in part the severity and extensive variety of signs and symptoms observed in CoV-19 patients. To further our understanding of the mechanism and pathways of SARS-CoV-2 infection and susceptibility of specific cell- and tissue-types and organ systems to SARS-CoV-2 attack in this communication we analyzed ACE2 expression in 85 human tissues including 21 different brain regions, 7 fetal tissues and 8 controls. Besides strong ACE2 expression in respiratory, digestive, renal-excretory and reproductive cells, high ACE2 expression was also found in the amygdala, cerebral cortex and brainstem. The highest ACE2 expression level was found in the pons and medulla oblongata in the human brainstem, containing the medullary respiratory centers of the brain, and may in part explain the susceptibility of many CoV-19 patients to severe respiratory distress.
Keywords: Alzheimer’s disease, Angiotensin-converting enzyme 2 (ACE2) receptor, Coronavirus, COVID-19, CoV-19, Hartnup's disease, microRNA, miRNA-5197, SARS-CoV-2, single stranded RNA (ssRNA)
Introduction
Coronavirus disease 2019 (COVID-19; CoV-19) first emerged about ~ mid-December 2019 in an open seafood, poultry and wild game wet market in the city of Wuhan in east-central China. Circumstantial evidence suggested the possibility of an initial zoonotic emergence of SARS-CoV-2 and the role of intermediate hosts such as the fruit bat (Pteropus scapulatus), pangolin (Manis javanica), feral chicken (Gallus gallus) and other wild animals, via ‘zoonotic spillover’, in SARS-CoV-2 interspecies transmission (Rodriguez-Morales et al. 2020; Tiwari et al. 2020; Ye et al. 2020). Not previously observed in humans, SARS-CoV-2 is now known to be caused by a single-stranded ribonucleic acid (ssRNA) beta-coronavirus known as ‘severe acute respiratory syndrome coronavirus 2’ (SARS-CoV-2; Bourgonje et al. 2020; Guan et al. 2020; Wackett 2020). Highly transmissible, widespread infection by SARS-CoV-2 was declared a global pandemic by the World Health Organization (WHO) by mid-March 2020 (https://genome.ucsc.edu/covid19.html). This novel human viral pathogen primarily induces a dangerous and acute pulmonary disruption with fever, cough, shortness of breath and severe respiratory dysfunction and distress often requiring assisted ventilation. The clinical symptoms of CoV-19 are similar to those reported for ‘severe acute respiratory syndrome coronavirus’ (SARS-CoV) and Middle Eastern respiratory syndrome coronavirus (MERS-CoV; Du et al. 2017; Evans et al. 2020; Ivashchenko et al. 2020). When established and severe, fulminant SARS-CoV-2 infection can rapidly evolve into a systemic illness characterized by hyper-inflammation, cytokine storm and rapid increase in systemic inflammatory biomarkers, elevations in cardiovascular malfunction and severe respiratory failure that is lethal in about 3% of all cases. Multiple neurological symptoms include loss of smell (anosmia), altered taste (ageusia), loss of the coordination of movement (ataxia), headache and nausea, dizziness, intermittent loss or impairment of consciousness, acceleration or aggravation of pre-existing cognitive deficits, direct negative effects of the heightened immune reaction, unusual hyper-inflammatory responses, increased vascular inflammation, inflammation of the brain and spinal cord and atherogenesis, exacerbation or de novo induction of inflammatory neurodegeneration, loss of respiratory control and progressive disturbances in cognition (Baig and Sanders 2020; Zhang et al. 2020) including encephalitis (Efe et al. 2020), stroke, seizures, encephalopathy (Zubair et al. 2019) and acute disseminated encephalomyelitis (ADEM; Panupattanapong and Brooks 2020; Parsons et al. 2020). One worrisome auxiliary sequela of CoV-19 infection called ‘multisystem inflammatory syndrome in children’ (MIS-C) has been observed in which children who have recovered from CoV-19 infection subsequently develop a severe inflammatory syndrome with Kawasaki disease-like features including persistent fever, conjunctivitis, hyper-inflammatory responses, elevated inflammatory markers and immunodeficiency, and a constellation of symptoms including hypotension, cardiac, gastrointestinal (GI), renal, hematologic, dermatologic and neurological involvement (Panupattanapong and Brooks 2020; Ronconi et al. 2020). The WHO reports that globally there are currently about ~ 17 million cases CoV-19 along with about ~ 700,000 total deaths as of late July 2020 (https://www.worldometers.info/coronavirus/; last accessed 29 July 2020).
SARS-CoV-2 possesses an enveloped, positive-stranded ssRNA genome of 29,811 nucleotides (nt) and shares highly homologous RNA sequence with SARS-CoV and MERS-CoV (and like SARS-CoV and MERS-CoV causes severe and acute respiratory distress and a highly lethal pneumonia). SARS-CoV-2 encodes at least six membrane proteins: envelope (E), membrane (M), spike (S1) and nucleocapsid (N) proteins N3-to-N5 (Fehr and Perlman 2015; Sah et al. 2020; Shang et al. 2020). With regard to pathogenicity, spike (S) proteins are essential for viral entry into host cells (Du et al. 2017; Guan et al. 2020; Li et al. 2020a, b; Zubair et al. 2019). SARS-CoV-2 appears to bind exclusively to the angiotensin-converting enzyme (ACE2) protein, a single-pass type 1 transmembrane receptor with its enzymatically active domain exposed on the surface of multiple cell types, such as type II alveolar cells of the respiratory system, enterocytes and intestinal epithelial cells, endothelial cells, epithelial cells of the conjunctival epithelium, kidney cells (renal tubules) and certain immune cells, such as alveolar monocytes/macrophages and certain cells of the CNS including those of the cerebral cortex and brainstem (Guan et al. 2020; Kabbani et al. 2020; Li et al. 2020a, b; Zhou et al. 2020; Zubair et al. 2019). More specifically, the binding of the spike S1 protein of both SARS-CoV and SARS-CoV-2 to the enzymatic domain of ACE2 on the surface of cells results in the endocytosis and translocation of both SARS-CoV-2 and the ACE2 enzyme into endosomes of infected cells (Wang et al. 2008; Millet and Whittaker 2018; Lewis 2020). The cell surface associated transmembrane serine protease 2 TMPRSS2 (epitheliasin; EC 3.4.21.109) also facilitates CoV-19 entry into cells following binding of the viral spike protein S1 to ACE2 (https://www.genecards.org/cgi-bin/carddisp.pl?gene=TMPRSS2; Zhou et al. 2020). The inhibition of SARS-CoV-2 and ACE2 recognition, TMPRSS2 signaling and SARS-CoV-2 translocation into host cells is currently under extremely intense research investigation as a cornerstone to the implementation of prospective and efficacious anti-SARS-CoV-2 therapeutic strategies (Akhmerov and Marbán 2020; Bourgonje et al. 2020; Datta et al. 2020; Saavedra 2020; Shang et al. 2020).
Because of the singular importance of ACE2 metalloprotein in the recognition, attachment and entry of SARS-CoV-2 into cells, in the current communication we investigated the expression of ACE2 in 85 human tissues and controls including 21 different anatomical regions of the human brain using multiple tissue expression (MTE) arrays. Interestingly, ACE2 expression was easily detected in multiple brain regions. The highest levels of ACE2 expression in the brain were found in the pons and medulla oblongata, the breathing centers of the brain, which may in part explain SARS-CoV-2 unusually strong ability to disrupt normal respiration and pulmonary manifestations including shortness of breath, impaired breathing and severe respiratory distress.
Methods
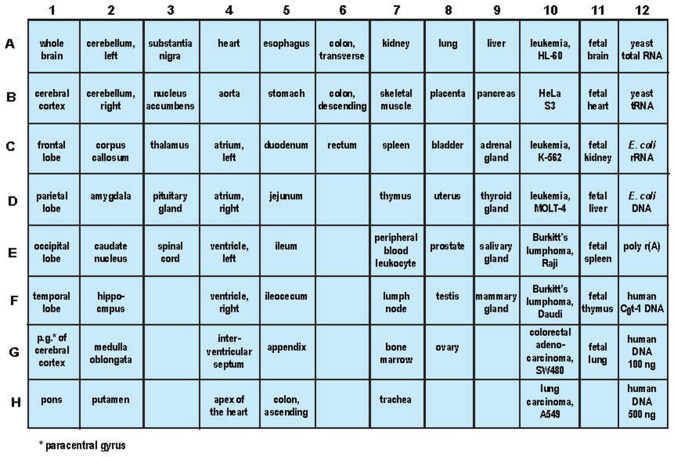
Human MTE arrays (Invitrogen-Clontech, Palo Alto, CA; cat no. 7775‐1) were used at minimum in triplicate to analyze tissue- or brain-specific expression of ACE2 mRNA; this MTE array contains polyA + mRNA samples (each sector containing ~ 2.0 µg polyA+ RNA) enriched from 77 human tissues with 8 independent DNA and RNA controls (Fig. 1). A unique human-specific 26 nt ACE2-specific DNA probe 5′-CTTGCAGCTACACCAGTTCCCAGGCA-3′ (US NIH/NLM Sequence ID: AY217547.1; https://blast.ncbi.nlm.nih.gov/Blast.cgi#27978647; see also accession AB046569.1 https://www.ncbi.nlm.nih.gov/nuccore/AB046569.1) and a human-specific 35 nt ACE2 DNA probe (5′-GGGCTCGAGTTTTTCTAAAAGGAGGTCTGAACATC-3′; derived from exon 18 of the ~ 3.4 knt ACE2 mRNA; Genbank Accession AF291820; NM_021804.2; https://www.genscript.com/gene/homo-sapiens/59272/ace2.html; https://www.ncbi.nlm.nih.gov/gene/59272; last accessed 29 July 2020) and exhibiting no homology to ACE1; (Tipnis et al. 2005; Hofmann et al. 2004) were generous gifts from Dr. PN Alexandrov; both the 26 nt and 35 nt ACE2 DNA probes gave similar hybridization signals; data for the 26 nt ACE2 DNA probe is presented here; both of these DNA probes were specifically designed so that they crossed two exons, thereby detecting only ACE2 mRNA and not ACE2 genomic DNA; several other ACE2-specific radiolabeled cDNA probes were used that gave similar hybridization signals; DNA probes were used because RNA–DNA hybrids elicit a more accurate and stable hybridization with a higher energy of association (EA) between the ACE2 DNA probe and the membrane-bound polyA + mRNA target (see Lukiw et al. 1992; Goulter et al. 2004; https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=59,272); ACE2 DNA probes were radiolabeled (i) using Random Primer Labeling and the direct incorporation of [α-32P]dCTP (3000 Ci/mmol, Amersham, Chicago, IL) or (ii) were end-labeled using [γ-32P]dATP (3000 Ci/mmol, Amersham redivue); glyceraldehyde 3-phosphate dehydrogenase (G3PDH) was used for control normalization (see https://www.takara.co.kr/file/manual/pdf/PT3307-1.pdf; Lukiw et al. 1990,1992; Goulter et al 2004); to obtain maximal signal quantitation accuracy hybridized and washed membranes were overlaid on the MTE template (Fig. 1) excised and each sector counted separately in 10 ml liquid scintillation fluid (Ultima Gold, PerkinElmer, Waltham, MA) in a Liquid Scintillation Counter (LS600016 LSC, Beckman, Fullerton, CA); relative signal intensities (Figs. 2, 3) were expressed by comparing the ACE2 hybridization signal to the G3PDH signal for each polyA + mRNA on the MTE array (see Lukiw et al. 1990, 1992; Zhao et al. 2001; Jaber et al. 2017; Piras et al. 2020). Statistical significance was analyzed using a two-way factorial analysis of variance (p, ANOVA; SAS Institute, Cary, NC). A p < 0.01 (ANOVA) was deemed as high statistical significance; experimental values are expressed as the means ± SD.
Fig. 1.
Organization of human MTE blot for Northern blot analysis; ACE2 signals were compared to G3PDH signals in the same sample and expressed as ‘relative signal intensity’; these are shown bar graph format in Figs. 2 and 3; a minimum of N = 6 MTE arrays (3 for ACE2 and 3 for G3PDH) were used in each type-of-tissue determination
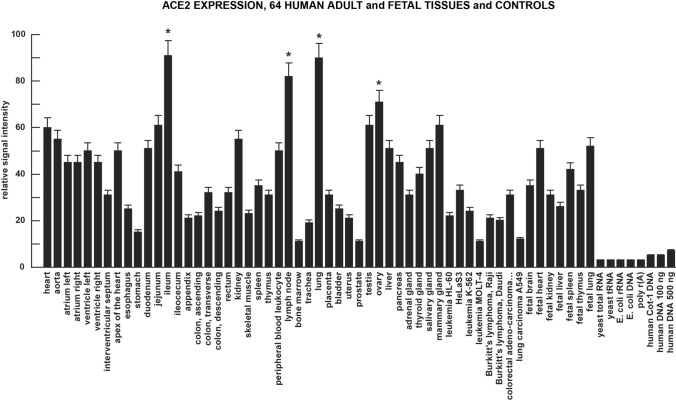
Fig. 2.
Tissue-specific patterns of ACE2 expression are a strong indicator of gene function and susceptibility to SARS-CoV-2 invasion and the development of CoV-19. High ACE2 expression in vascular, gastrointestinal (GI) tract, respiratory, excretory and reproductive tissues and some relevant cancers and controls; the highest values of expression were found in the heart (both adult and fetal), ileum, kidney, lung (both adult and fetal), testes, ovary and mammary gland; ACE2 is a member of the angiotensin-converting enzyme family of dipeptidyl carboxydipeptidases and has considerable homology to human angiotensin 1 converting enzyme (ACE1), however the ACE2 DNA probe here was unique showing no homology with ACE1; ACE2, expressed as both a secreted protein and trans-membrane form catalyzes the cleavage of angiotensin I (ANG) into ANG1–9, and ANG II into the vasodilator ANG1–7; ACE-2 is also a receptor to the spike (S1) glycoprotein of the human coronavirus HCoV-NL63 and the human SARS-CoV and SARS-CoV-2 virus that causes CoV-19 (see: https://www.youtube.com/watch?v=W1k1sUoLPlA; https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=59,272; last accessed 29 July 2020); at least N = 6 MTE arrays (at minimum 3 for ACE2 and 3 for G3PDH) were used in each tissue determination; *p < 0.01 (ANOVA); error bars represent one standard deviation of the mean
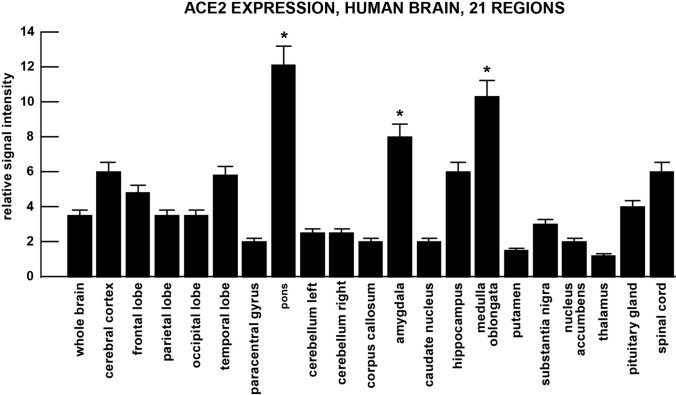
Fig. 3.
ACE2 expression in 21 anatomical regions of the human brain. The highest expression of the SARS-CoV-2 receptor ACE-2 was found in the amygdala, pons and medulla oblongata; the later two regions contain the respiratory control centers of the brain; considerable expression of ACE2 in the brain, temporal lobe and hippocampus may in part explain neurological disruption and cognitive dysfunction associated with SARS-CoV-2 infection; note difference of scale in the Y-axis (ordinate) compared to Fig. 2; to obtain maximal signal quantitation hybridized and washed membranes were overlaid on the MTE template (Fig. 1) excised and counted in liquid scintillation fluid as previously described; relative signal intensities (Figs. 2, 3) were expressed by comparing the ACE2 hybridization signal to the G3PDH signal for each polyA + mRNA on the same MTE array sector (see Lukiw et al. 1990, 1992; Zhao et al. 2001; Jaber et al. 2017); at least N = 6 MTE arrays (3 for ACE2 and 3 for G3PDH) were used in each brain tissue determination; *p < 0.01 (ANOVA); error bars represent one standard deviation of the mean
Results
At the mRNA level ACE2 is very widely expressed in multiple human cell- and tissue-types and organ systems. The most consistent expression was found in all regions of the heart sampled and was highest in the adult lung, ileum, lymph node, testis and ovaries. This pattern of high ACE2 expression is reciprocated in both adult and fetal heart and lung (Fig. 2). In the brain there was a consistent ACE2 expression across the cerebral cortex and the highest ACE2 expression was found in the pons and medulla oblongata (Fig. 3). Patterns of cell-, tissue- and organ-specific expression of ACE2 suggests that this common dipeptidyl carboxydipeptidase (E.C.3.4.17.23) plays an important role in the regulation of respiratory, cardiovascular, GI-tract and renal-excretory system functions, as well as in a wide range of brain and CNS anatomical regions with multiple neurological functions (see Figs. 2, 3 and legends). In general, the data indicate that ACE2 represents a very common and widespread receptor type and that the SARS-CoV-2 virus has taken advantage of this wide range of potential cellular entry points and a remarkably efficient viral invasion strategy to ensure its survival and proliferation in multiple human cell types (see below).
Discussion
The major established biological function for the 805 amino acid, 92.5 kDa, zinc-containing ACE2 membrane-integral metalloprotein (E.C.3.4.17.23) is catalyzing the hydrolysis of the vasoconstrictor peptide angiotensin II (ANG II) into the vasodilator ANG1–7. Reducing the amount of ANG II while increasing ANG1–7 results in a relaxation of vascular smooth muscle, a lowering of both local and systemic vascular resistance to blood flow and hence elicits a decrease in blood pressure (Wang et al. 2016; Datta et al. 2020; Li et al. 2020a, b). ACE2 therefore plays a key regulatory role in the classical axis of the renin-angiotensin system (RAS), a hormonal system that regulates not only systemic vascular resistance to blood flow and blood pressure but also fluid and electrolyte balance (Kehoe et al. 2016; Wang et al. 2016; Datta et al. 2020). ACE2 has also been reported to have ancillary physiological roles: (i) in the regulation of membrane trafficking of the sodium-dependent neutral amino acid transporter SLC6A19 that has been implicated in Hartnup's disease (a disorder of amino acid homeostasis and translocation; Gheblawi et al. 2020); (ii) in the interaction with other vasoactive peptides such as Apelin that regulate blood pressure and myocardial contractility (Kuba et al. 2013); and (iii) in GI-tract inflammation and the regulation of the complexity and speciation of the GI-tract microbiome (Kuba et al. 2013; Gheblawi et al. 2020; Lukiw 2020; Zuo et al. 2020; unpublished observation). Other important contributory factors associated with variable ACE2 receptor expression (and hence potential for SARS-CoV-2 infection and CoV-19 severity) include age, sex, ethnicity, medication and several co-morbidities, such as cardiovascular disease, metabolic syndrome and obesity, and dementia and cognitive decline (Baig and Sanders 2020; Gheblawi et al. 2020). As might be expected the blockage or downregulation of ACE2 leads to ‘unopposed’ ANG II accumulation resulting in pleiotropic effects including vasoconstriction, aldosterone secretion, endothelial dysfunction and altered inflammatory signaling which may impact the progress of CoV-19 infection all via increased activity of the RAS (Du et al. 2017; Datta et al. 2020; Saavedra 2020). Indeed molecular-based rational design of angiotensin receptor blockers (ARBs) with enhanced affinities for ACE2 or the spike (S1) protein of the coronaviruses may further advance the development of neutralizing antibodies, decoy ligands and perhaps advance more effective pharmacological strategies for the suppression of SARS-CoV-2 infection, but these therapeutic approaches are currently in the very earliest stages of CoV-19 research investigation and their clinical efficacies, if any, remains to be established (Du et al. 2017; de Vries 2020; Evans et al. 2020; Lewis 2020).
Besides considerable ACE2 expression in cells and tissues of the respiratory, cardiovascular, GI-tract, renal-excretory and reproductive systems (Harmer et al. 2002; Datta et al. 2020) there is in addition significant ACE2 expression in different anatomical regions of the brain and CNS (Fig. 3). This is in concordance with the multiple neurological anomalies associated with CoV-19 infection including headache, anosmia, ageusia, lethargy, confusion, cognitive impairment and loss of autonomic respiratory control (Baig and Sanders 2020; Bourgonje et al. 2020; Datta et al. 2020; Efe et al. 2020; Heneka et al. 2020; Kabbani and Olds 2020). Significant neuro-invasion involving SARS-CoV has been reported from both patients and experimental animals where the brainstem was heavily infected from apparent spreading via a synapse-connected route to the medullary cardiorespiratory centers (Zubair et al. 2019; Chigr et al. 2020). Indeed SARS-CoV-2-mediated neuro-invasion may be achieved by several routes, including blood-borne transport to highly vascularized brain tissues, by trans-synaptic transferal across infected neurons, via entry through the olfactory nerve, infection via the ocular epithelium or vascular endothelium, or leukocyte migration across the blood–brain barrier (Kabbani and Olds 2020; Zhou et al. 2020). Interestingly ACE2 receptor expression was also easily detected in retinal cells of the eye and visual system (manuscript in preparation), suggesting that the eye and/or visual system may provide additional entry points for SARS-CoV-2 invasion (and suggesting that eyeglasses or face shields may be as important as face masks in reducing SARS-CoV-2 transmission). Co-localization studies have suggested that SARS-CoV-2 viral entry may be mediated by a clathrin- and caveolae-independent mechanism and that cholesterol- and sphingolipid-rich lipid raft micro-domains assist in SARS-CoV-2-ACE2-interaction and subsequent SARS-CoV-2 entry into susceptible cells via the ubiquitous ACE2 receptor (Simmons et al. 2004; Wang et al. 2008; Gheblawi et al. 2020).
Interestingly (i) ACE2 expression is significantly decreased in Alzheimer’s disease (AD) affected brain tissues and in amyloid over-expressing transgenic murine models of AD (Kehoe et al. 2016; Rocha et al. 2018; Royea and Hamel 2020), (ii) this deficit is associated with increased amyloid-beta (Aβ) peptide load and tau pathology, the two pathophysiological hallmarks of AD (Kehoe et al. 2016; Rocha et al. 2018; Evans et al. 2020), and (iii) hyperactivity of the classical axis of the renin-angiotensin system (RAS), mediated by ANG II activation of the ANG II type 1 receptor (AT1R), is further implicated in biofluid imbalance in the neurovascular and neuro-immune pathology associated with AD (Kehoe et al. 2016; Evans et al. 2020).
Lastly, the ssRNA SARS-CoV-2 virus (at about ~ 30,000 nt), considerably larger than the size of the average cellular messenger RNA (mRNA), may be intrinsically susceptible to complementary base-pair interaction with other ssRNAs including microRNAs (miRNAs) and the RNA binding proteins associated with pre-miRNA processing into mature miRNA and translocation. For example the 23 nt human miRNA hsa-miRNA-5197 is well over 90% homologous to a 3′ down-stream region of the SARS-CoV-2 RNA sequence (Ivashchenko et al. 2020; unpublished observations). Targeting and inactivation of SARS-CoV-2 by pre-existing or artificially synthesized and stabilized miRNAs (such as hsa-miRNA-5197): (i) may provide a natural protection mechanism against SARS-CoV-2 invasion; (ii) may help explain individual natural immunity to SARS-CoV-2 infection of some persons who naturally express hsa-miRNA-5197 or other miRNAs or ssRNAs that target pathological viruses such as SARS-CoV, SARS-CoV-2or MERS-CoV; and (iii) may be of therapeutic value in the accelerated depolymerization and destruction of the invading SARS-CoV-2 viral sequence itself, resulting in SARS-CoV-2 degradation and the rapid cessation and termination of SARS-CoV-2 s lethal effects.
Conclusions
The omnipresence of ACE2 expression in multiple human cell- and tissue-types is noteworthy. SARS-CoV-2 has evolved a remarkable evolutionary strategy to ensure its replication, survival and spread by exploiting the presence of a very common, abundant and susceptible transmembrane ACE2 receptor making many different neural and extra-neural human cell- and tissue-types at high risk and susceptibility to SARS-CoV-2 invasion. Many research gaps remain, and additional basic investigative research is urgently required to more fully understand the etiology, epidemiology, pathological mechanisms and neurobiology of CoV-19 disease. A real danger of SARS-CoV-2 infection is not only its highly transmissible and contagious nature and lethality but also: (i) SARS-CoV-2′s simultaneous and multipronged attack on many human cell- and tissue-types involving vital and critical respiratory, immunological, vascular, renal-excretory and neural systems; and (ii) an unprecedented coordinated disruption of the complex neurophysiology, neurochemistry and neurology of the cells of the brain that normally regulate these multiple neurobiological systems.
Acknowledgements
Sincere thanks are extended to Drs. Jian-Guo Cui, Christopher Eicken, Elizabeth Head, Christopher Hebel, Wayne Poon and Piotr N. Alexandrov for short post-mortem interval (PMI) human brain and/or retinal tissues or extracts, ACE2 DNA and cDNA probes, miRNA array work and initial data interpretation, and to Darlene Guillot for expert technical assistance and medical artwork. Dr. James M Hill (now deceased) had a life-long interest in virology and in ACE2 expression in the aging CNS and in neurological disease and the renin-angiotensin system (RAS) in AD. This communication is dedicated to his many years of scientific work in this fascinating research area. Thanks are further extended to the many neuropathologists, physicians and researchers of Canada and the USA who have provided high quality, short post-mortem interval (PMI) human CNS, retinal tissues or extracted total brain and retinal RNA for scientific study and quantitative analysis. Research on human brain and host cell transcriptomics in the Lukiw laboratory involving total human RNA transcription and gene expression, the innate-immune response in AD and in other forms of neurological or retinal disease, amyloidogenesis and neuro-inflammation was supported through an unrestricted grant to the LSU Eye Center from Research to Prevent Blindness (RPB); the Louisiana Biotechnology Research Network (LBRN) and NIH grants NEI EY006311, NIA AG18031 and NIA AG038834 (WJL).
Author Contributions
JMH and WJL conceived and wrote the paper; AP participated in literature search and data collection and analysis; all authors read and approved the manuscript.
Footnotes
For interested readers and ACE2-CoV-19 researchers key updated website links, excellently presented, regarding multiple aspects of ACE2 expression, SARS-CoV-2 invasion and CoV-19 disease can be found at: https://genome.ucsc.edu/covid19.html (COVID-19 Pandemic Resources at UCSC); https://www.rndsystems.com/resources/articles/ace-2-sars-receptor-identified (ACE2: The Receptor for SARS-CoV-2); https://academic.oup.com/eurheartj/article/26/4/322/439241 (ACE and ACE2: a tale of two enzymes, from the European Hearth Journal 2005); and https://medicalxpress.com/news/2020–05-ace2-receptor-coronavirus.html (What is the ACE2 receptor, and how is it connected to coronavirus? From the Medical Express, 2020).
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Akhmerov A, Marbán E. COVID-19 and the heart. Circ Res. 2020;126:1443–1455. doi: 10.1161/circresaha.120.317055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baig AM, Sanders EC. Potential neuroinvasive pathways of SARS-CoV-2: deciphering the spectrum of neurological deficit seen in coronavirus disease 2019 (COVID-19) J Med Virol. 2020 doi: 10.1002/jmv.26105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgonje AR, Abdulle AE, Timens W, et al. Angiotensin-converting enzyme-2 (ACE2), SARS-CoV-2 and pathophysiology of coronavirus disease 2019 (COVID-19) J Pathol. 2020 doi: 10.1002/path.5471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigr F, Merzouki M, Najimi M. Autonomic brain centers and pathophysiology of COVID-19. ACS Chem Neurosci. 2020;11(11):1520–1522. doi: 10.1021/acschemneuro.0c00265. [DOI] [PubMed] [Google Scholar]
- Datta PK, Liu F, Fischer T, Rappaport J, Qin X. SARS-CoV-2 pandemic and research gaps: understanding SARS-CoV-2 interaction with the ACE2 receptor and implications for therapy. Theranostics. 2020;10(16):7448–7464. doi: 10.7150/thno.48076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries AAF. Renin-angiotensin system inhibition in COVID-19 patients. Neth Heart J. 2020;28(7–8):396–405. doi: 10.1007/s12471-020-01439-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du L, Yang Y, Zhou Y, Lu L, Li F, Jiang S. MERS-CoV spike protein: a key target for antivirals. Expert Opin Ther Targets. 2017;21:131–143. doi: 10.1080/14728222.2017.1271415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efe IE, Aydin OU, Alabulut A, Celik O, Aydin K. COVID-19-associated encephalitis mimicking glial tumor: a case report. World Neurosurg. 2020;140:46–48. doi: 10.1016/j.wneu.2020.05.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans CE, Miners JS, Piva G, et al. ACE2 activation protects against cognitive decline and reduces amyloid pathology in the Tg2576 mouse model of Alzheimer's disease. Acta Neuropathol. 2020;139:485–502. doi: 10.1007/s00401-019-02098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. 2015;1282:1–23. doi: 10.1007/978-1-4939-2438-7_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gheblawi M, Wang K, Viveiros A, et al. Angiotensin-converting enzyme 2: SARS-CoV-2 receptor and regulator of the renin-angiotensin system: celebrating the 20th anniversary of the discovery of ACE2. Circ Res. 2020;126(10):1456–1474. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulter AB, Goddard MJ, Allen JC, Clark KL. ACE2 gene expression is up-regulated in the human failing heart. BMC Med. 2004;2:19. doi: 10.1186/1741-7015-2-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan WJ, Ni ZY, Hu Y, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer D, Gilbert M, Borman R, et al. Quantitative mRNA expression profiling of ACE 2, a novel homologue of angiotensin converting enzyme. FEBS Lett. 2002;532:107–110. doi: 10.1016/S0014-5793(02)03640-2. [DOI] [PubMed] [Google Scholar]
- Heneka MT, Golenbock D, Latz E, Morgan D, Brown R. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimers Res Ther. 2020;12:69. doi: 10.1186/s13195-020-00640-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H, Geier M, Marzi A, et al. Susceptibility to SARS coronavirus S protein-driven infection correlates with expression of angiotensin converting enzyme 2 and infection can be blocked by soluble receptor. Biochem Biophys Res Commun. 2004;319:1216–1221. doi: 10.1016/j.bbrc.2004.05.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivashchenko A, Rakhmetullina A, Aisina D. How miRNAs can protect humans from coronaviruses COVID-19, SARS-CoV and MERS-CoV. Bioinformatics. 2020 doi: 10.21203/rs.3.rs-16264/v1. [DOI] [Google Scholar]
- Jaber V, Zhao Y, Lukiw WJ. Alterations in micro RNA-messenger RNA (miRNA-mRNA) coupled signaling networks in sporadic Alzheimer's disease (AD) hippocampal CA1. J Alzheimers Dis Parkinsonism. 2017;7:312. doi: 10.4172/2161-0460.1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabbani N, Olds JL. Does COVID19 infect the brain? Mol Pharmacol. 2020;97(5):351–353. doi: 10.1124/molpharm.120.000014.PMID32238438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehoe PG, Wong S, Al Mulhim N, Palmer LE, Miners JS. Angiotensin-converting enzyme 2 is reduced in Alzheimer's disease in association with increasing amyloid-β and tau pathology. Alzheimers Res Ther. 2016;8:50. doi: 10.1186/s13195-016-0217-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuba K, Imai Y, Penninger JM. Multiple functions of angiotensin-converting enzyme 2 and its relevance in cardiovascular diseases. Circ J. 2013;77:301–308. doi: 10.1253/circj.cj-12-1544. [DOI] [PubMed] [Google Scholar]
- Lewis R (2020). COVID-19 vaccine will close in on the spikes. DNA Science Blog. Public Library of Science. Archived from the original on 2020‐02‐22.
- Li MY, Li L, Zhang Y, Wang XS. Expression of the SARS-CoV-2 cell receptor gene ACE2 in a wide variety of human tissues. Infect Dis Poverty. 2020;9:45. doi: 10.1186/s40249-020-00662-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YC, Bai WZ, Hashikawa T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J Med Virol. 2020 doi: 10.1002/jmv.25728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ. Gastrointestinal (GI) tract microbiome-derived neurotoxins-potent neuro-inflammatory signals from the GI tract via the systemic circulation into the brain. Front Cell Infect Microbiol. 2020;10:22. doi: 10.3389/fcimb.2020.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukiw WJ, Wong L, McLachlan DR. Cytoskeletal messenger RNA stability in human neocortex: studies in normal aging and in Alzheimer's disease. Int J Neurosci. 1990;55:81–88. doi: 10.3109/00207459008985953. [DOI] [PubMed] [Google Scholar]
- Lukiw WJ, Handley P, Wong L, Crapper McLachlan DR. BC200 RNA in normal human neocortex, non-Alzheimer dementia (NAD), and senile dementia of the Alzheimer type (AD) Neurochem Res. 1992;17:591–597. doi: 10.1007/BF00968788. [DOI] [PubMed] [Google Scholar]
- Millet JK, Whittaker GR. Physiological and molecular triggers for SARS-CoV membrane fusion and entry into host cells. Virology. 2018;517:3–8. doi: 10.1016/j.virol.2017.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panupattanapong S, Brooks EB. New spectrum of COVID-19 manifestations in children: Kawasaki-like syndrome and hyperinflammatory response Cleve. Clin J Med. 2020 doi: 10.3949/ccjm.87a.ccc039. [DOI] [PubMed] [Google Scholar]
- Parsons T, Banks S, Bae C, Gelber J, Alahmadi H, Tichauer M. COVID-19-associated acute disseminated encephalomyelitis (ADEM) J Neurol. 2020 doi: 10.1007/s00415-020-09951-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piras IS, Bleul C, Schrauwen I, et al. Transcriptional profiling of multiple system atrophy cerebellar tissue highlights differences between the Parkinsonian and cerebellar sub-types of the disease. Acta Neuropathol Commun. 2020;8:76. doi: 10.1186/s40478-020-00950-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha NP, Toledo A, Corgosinho LTS, et al. Cerebrospinal fluid levels of angiotensin-converting enzyme are associated with amyloid-β42 burden in Alzheimer's disease. J Alzheimers Dis. 2018;64:1085–1090. doi: 10.3233/JAD-180282. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Morales AJ, Bonilla-Aldana DK, Balbin-Ramon GJ, et al. History is repeating itself: probable zoonotic spillover as the cause of the 2019 novel coronavirus epidemic. Infez Med. 2020;28:3–5. [PubMed] [Google Scholar]
- Ronconi G, Teté G, Kritas SK, et al. SARS-CoV-2, which induces COVID-19, causes Kawasaki-like disease in children: role of pro-inflammatory and anti-inflammatory cytokines. J Biol Regul Homeost Agents. 2020 doi: 10.23812/editorial-ronconi-e-59. [DOI] [PubMed] [Google Scholar]
- Royea J, Hamel E. Brain angiotensin II and angiotensin IV receptors as potential Alzheimer's disease therapeutic targets. Geroscience. 2020 doi: 10.1007/s11357-020-00231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra JM. Angiotensin receptor blockers and COVID-19. Pharmacol Res. 2020;156:104832. doi: 10.1016/j.phrs.2020.104832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sah R, Rodriguez-Morales AJ, Jha R, et al. Complete genome sequence of a 2019 novel coronavirus (SARS-CoV-2) strain isolated in Nepal. Microbiol Resour Announc. 2020;9(11):e00169–e220. doi: 10.1128/MRA.00169-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi K, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons G, Reeves JD, Rennekamp AJ, Amberg SM, Piefer AJ, Bates P. Characterization of severe acute respiratory syndrome-associated coronavirus (SARS-CoV) spike glycoprotein-mediated viral entry. Proc Natl Acad Sci USA. 2004;101(12):4240–4245. doi: 10.1073/pnas.0306446101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipnis SR, Hooper NM, Hyde R, Karran E, Christie G, Turner AJ. A human homolog of angiotensin-converting enzyme. Cloning and functional expression as a captopril-insensitive carboxypeptidase. J Biol Chem. 2005;275:33238–33243. doi: 10.1074/jbc.M002615200. [DOI] [PubMed] [Google Scholar]
- Tiwari R, Dhama K, Sharun K, et al. COVID-19: animals, veterinary and zoonotic links. Vet Q. 2020;40:169–182. doi: 10.1080/01652176.2020.1766725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wackett LP. SARS-CoV-2: environment and spread: an annotated selection of World Wide Web sites relevant to the topics in environmental microbiology. Environ Microbiol. 2020;22:2443–2444. doi: 10.1111/1462-2920.15089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang P, Liu K, Guo F, Zhang Y, Zhang G, Jiang C. SARS coronavirus entry into host cells through a novel clathrin- and caveolae-independent endocytic pathway. Cell Res. 2008;18:290–301. doi: 10.1038/cr.2008.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, McKinnie SM, Farhan M, Paul M, McDonald T, McLean B, et al. Angiotensin-converting enzyme 2 metabolizes and partially inactivates pyr-apelin-13 and apelin-17: physiological effects in the cardiovascular system". Hypertension. 2016;68:365–377. doi: 10.1161/hypertensionaha.115.06892. [DOI] [PubMed] [Google Scholar]
- Ye ZW, Yuan S, Yuen KS, Fung SY, Chan CP, Jin DY. Zoonotic origins of human coronaviruses. Int J Biol Sci. 2020;16:1686–1697. doi: 10.7150/ijbs.45472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shan KS, Abdollahi S, Nace T. Anosmia and ageusia as the only indicators of coronavirus disease 2019 (COVID-19) Cureus. 2020;12(5):e7918. doi: 10.7759/cureus.7918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Alvarado D, Rainier S, et al. Mutations in a newly identified GTPase gene cause autosomal dominant hereditary spastic paraplegia. Nat Genet. 2001;29:326–331. doi: 10.1038/ng758. [DOI] [PubMed] [Google Scholar]
- Zhou L, Xu Z, Castiglione GM, Soiberman US, Eberhart CG, Duh EJ. ACE2 and TMPRSS2 are expressed on the human ocular surface, suggesting susceptibility to SARS-CoV-2 infection. Ocul Surf. 2020 doi: 10.1101/2020.05.09.086165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubair AS, McAlpine LS, Gardin T, Farhadian S, Kuruvilla DE, Spudich S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: a review. JAMA Neurol. 2019 doi: 10.1001/jamaneurol.2020.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T, Zhang F, Lui GCY, et al. Alterations in gut microbiota of patients with COVID-19 during time of hospitalization. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]