Summary
Dendritic cells (DCs) are antigen-presenting cells controlling T cell activation. In humans, the diversity, ontogeny, and functional capabilities of DC subsets are not fully understood. Here, we identified circulating CD88−CD1c+CD163+ DCs (called DC3s) as immediate precursors of inflammatory CD88−CD14+CD1c+CD163+FcεRI+ DCs. DC3s develop via a specific pathway activated by GM-CSF, independent of cDC-restricted (CDP) and monocyte-restricted (cMoP) progenitors. Like classical DCs but unlike monocytes, DC3s drove activation of naive T cells. In vitro, DC3s displayed a distinctive ability to prime CD8+ T cells expressing a tissue homing signature and the epithelial homing alpha-E integrin (CD103) through transforming growth factor β (TGF-β) signaling. In vivo, DC3s infiltrated luminal breast cancer primary tumors, and DC3 infiltration correlated positively with CD8+CD103+CD69+ tissue-resident memory T cells. Together, these findings define DC3s as a lineage of inflammatory DCs endowed with a strong potential to regulate tumor immunity.
Keywords: conventional DCs, DC3s, cDC2s, mononuclear phagocytes, inflammatory DCs, monocytes, DC progenitors, TRM
Graphical Abstract

Highlights
-
•
DC3s are phenotypic and functional intermediates between cDC2s and monocytes
-
•
GM-CSF alone, but not FLT3L, supports efficient differentiation of DC3s
-
•
DC3s do not differentiate via cDC (CDP)- or monocyte-restricted (cMoP) progenitors
-
•
DC3s prime TRM cells in vitro and correlate with TRM expansion in primary breast cancer
Bourdely et al. identify human CD88−CD1c+CD163+ DC3s as a pro-inflammatory phagocyte lineage sharing features with monocytes and conventional DCs. DC3s efficiently induce differentiation of CD103+CD8+ T cells in vitro, and their infiltration correlates with CD8+CD69+CD103+ TRM accumulation in breast tumors.
Introduction
Human dendritic cells (DCs) are sentinel cells of the immune system specialized in controlling T cell function (Banchereau and Steinman, 1998; Palucka and Banchereau, 2013; Steinman et al., 2003). The mouse model has brought important concepts to our understanding of DCs and suggests that multiple DC subsets arising from specialized ontogenetic pathways are endowed with specific immune functions (Briseño et al., 2016; Guermonprez et al., 2019; Merad et al., 2013; Murphy et al., 2016).
Definition of human DC subsets is a prerequisite to understanding the division of labor underpinning induction of various types of immune responses. At homeostasis, conventional DCs (cDCs) include cDC1s (CD141+XCR1+CLEC9A+IRF8+) and cDC2s (CD1c+CD11c+CD172a+IRF4+) (Bachem et al., 2010; Crozat et al., 2010; Heidkamp et al., 2016; Jongbloed et al., 2010; Schlitzer et al., 2013). cDC1s and cDC2s arise through a specialized ontogenetic pathway from a common DC precursor (CDP) (Lee et al., 2015b) or from early IRF8+ multipotent lympho-myeloid progenitors (MLPs) (Helft et al., 2017; Lee et al., 2017). Bone marrow progenitors for cDCs generate a common circulating precursor that progressively diverges into pre-cDC1 and pre-cDC2 (Breton et al., 2015, 2016; See et al., 2017). This is further complicated by inclusion of AXL+SIGLEC6+CD11c+CD1c+ cells (AS-DCs also called type 5 DCs), which have been proposed to act as precursors for cDCs (pre-cDCs) or a lineage on its own (See et al., 2017; Villani et al., 2017).
An additional layer of complexity in the DC network lies in its responsiveness to perturbations. For instance, inflammation affects hematopoiesis and phagocyte trafficking, resulting in leukocyte mobilization and tissue infiltration. Specifically, inflammation affects DC diversity and triggers mobilization of CD14+CD1c+ DCs, called inflammatory DCs (iDCs) (Binnewies et al., 2019; Granot et al., 2017; Segura et al., 2012, 2013; Wollenberg et al., 1996; Zaba et al., 2009). The expression of CD1c lectin is shared between iDCs and cDCs. However, CD1c+ iDCs also express multiple monocytic markers, such as CD14, CCR2, and FcγRI/CD64. iDCs have been reported in inflamed skin, synovial fluid, ovarian cancer ascites, solid tumor infiltrates, and lymph nodes (Bakdash et al., 2016; Binnewies et al., 2019; Granot et al., 2017; Lavin et al., 2017; Segura et al., 2012, 2013; Wollenberg et al., 1996; Zaba et al., 2009). The developmental pathway of human CD1c+CD14+ iDCs is poorly understood. In vitro studies suggest that iDCs obtained after differentiation of CD14+ monocytes in granulocyte macrophage colony-stimulating factor (GM-CSF) and interleukin-4 (IL-4) (Sallusto and Lanzavecchia, 1994) might correspond to in vivo iDCs (Granot et al., 2017; Segura et al., 2012, 2013). In this context, IL-4 acts through induction of the transcriptional regulator NCOR2 (Sander et al., 2017). In addition, triggering the aryl hydrocarbon receptor in monocytes supports activation of IRF4-dependent differentiation of iDCs (Goudot et al., 2017). Together, these studies support the prevailing notion that CD14+ monocytes act as immediate precursors for iDCs.
Re-evaluation of circulating mononuclear phagocyte diversity has been enabled by single-cell RNA sequencing (scRNA-seq). Recent studies have revealed that a subset of DC-like cells, called DC3s, express mRNA for the CD14 and CD1c genes (Villani et al., 2017). However, this analysis was performed after excluding cells expressing the highest amount of CD14 (Villani et al., 2017). As a consequence, this approach renders a problematic distinction between DC3s and bona fide CD14+ monocytes (Villani et al., 2017). This discrimination is further complicated by previous reports of CD14+CD1c+ “inflammatory” DCs recruited at inflammatory sites (Binnewies et al., 2019; Granot et al., 2017; Segura et al., 2012, 2013; Wollenberg et al., 1996; Zaba et al., 2009).
Here we intended to re-evaluate the definition of DC3s using unbiased scRNA-seq and high-dimensional flow cytometry by exploring the full spectrum of CD14 and CD1c expression. In addition, we identify DC3 growth factor requirements and developmental pathways. Finally, we show that DC3s activate CD103+ T cells and that DC3 infiltration in human breast tumors correlates with the abundance of CD8+CD103+CD69+ tissue-resident memory (TRM) T cells.
Results
DC3s Represent a Discrete Subset of CD88−CD1c+CD163+ Cells in Human Peripheral Blood
To probe the diversity of CD16−CD141−CD123− blood mononuclear phagocytes, we developed a sorting strategy including all phenotypic intermediates between CD14hiCD1clo and CD14loCD1chi cells. The proportions between cell populations were compensated to enrich in less abundant CD14loCD1chi cells (Figure S1A). Flow cytometry-sorted cells isolated from blood were analyzed using a droplet-based scRNA-seq approach (Figure 1A; Figure S1A). We found that cells expressing CD14 and/or CD1c could be separated into four CD33+ clusters (A, B, C, and D) (Figure 1A; Figure S1B). Contaminating clusters containing B and T lymphocytes and neutrophils were excluded from the analysis (Figure S1B). Hierarchical clustering performed on averaged single cell expression data within clusters showed that A and B were closer to each other than any of the other subsets (Figures 1B–1D). Cluster D fell between the group of clusters A and B and cluster C (Figure 1B). Classical cDC2 markers, such as CLEC10A, FCER1A, and major histocompatibility complex (MHC) class II genes (HLA-DQA, HLA-DPA, HLA-DRA, and the MHC class II-associated invariant chain CD74) were expressed prominently in clusters A, B, and D (Figures 1D and 1E; Figure S1C; Heidkamp et al., 2016; Lavin et al., 2017; Segura et al., 2013). In contrast, monocytic markers such as CD14, S100A8, S100A9, S100A12, and VCAN were more expressed in clusters C and D, with higher expression in C compared with D (Figures 1D and 1E). Finally, expression of the C5 receptor C5AR1 (CD88) was found to be restricted to cluster C together with SOD2 and RBP7 (Figures 1D and 1E).
Figure 1.
DC3s Are a Discrete Subset of CD88−CD1c+CD163+ Cells in Human Peripheral Blood
(A) Gating strategy used to define mononuclear phagocytes expressing CD14 and/or CD1c. Cells expressing CD14 and/or CD1c were sorted by flow cytometry from 3 healthy donors and pooled before scRNA-seq analysis. To improve the resolution of CD1c+ subsets, the cellular input was enriched in CD1high cells (Figure S1A). Single cells were isolated using a droplet-based approach and sequenced. Dimensionality reduction of scRNA-seq data was performed using dimensionality reduction (t-distributed stochastic neighbor embedding [tSNE]). Clusters A, B, C, and D were identified using the shared nearest neighbor (SNN) clustering algorithm. Each dot represents an individual cell (n = 1,622).
(B) Hierarchal clustering of groups A, B, C, and D based on average gene expression (14,933 genes).
(C) Absolute number of differentially expressed genes (DEGs) for pairwise comparisons between groups A, B, and D.
(D) Heatmaps displaying relative expression of up to 20 DEGs defining each cluster.
(E) Violin plots illustrating expression probability distributions across clusters of representative DEGs (226 total DEGs). Feature plots display the average expression of groups of genes (identified in violin plots) in each cell of the tSNE plot defined in (A).
(F) Expression distribution across clusters A, B, C, and D of gene signatures identified by Villani et al. (2017) and Yin et al. (2017). (∗∗p < 0.01, ∗∗∗∗p < 0.0001, one-way ANOVA test)
(G) Identification of 4 subsets within CD14lo to hi CD1clo to hi cells by unsupervised clustering of flow cytometry data using the FlowSOM algorithm. tSNE and unsupervised clustering were performed using the following markers: CD88, CD1c, FcεRI, CD14, CD163, BTLA, CD123, and CD5. tSNE plots (right) display the relative expression of each marker among the subsets. Dot plots (below) show the expression of specific markers in clusters 1, 2, and 3 when combined in 2-dimensional analysis.
(H) Improved gating strategy for identification of cDC2s, DC3s, and CD14+ monocytes in circulating PBMCs and histograms showing expression of S100A8/9, FcεRI, CD5, CD14, CD116, and CD206.
(I) Principal-component analysis (PCA) for bulk-sequenced mononuclear phagocyte populations as defined in (H).
(J) Cluster dendrogram of the different cell types using the 2,000 most variable genes.
(K) Heatmaps comparing the relative expression of markers discriminating clusters in scRNA-seq analysis (A, B, C, and D, left) and in bulk RNA-seq analysis on sorted subsets based on the gating strategy defined in (H) (right).
To determine whether our clustering reflects previously published data, we decided to evaluate the expression of gene signatures obtained from Villani et al. (2017). We found that signature genes discriminating cDC2 within CD14lo cells were mostly represented in clusters A and B, confirming their identity (Figure 1F). Likewise, signature genes defining DC3s within CD14lo cells (Villani et al., 2017) and CD14+ monocytes were significantly enriched in cluster D and cluster C, respectively (Figure 1F). Of note, genes enriched in DC3s compared with cDC2s (S100A8, S100A9, and CD14) were highly expressed in clusters C and D (Figure 1E; Figure S1E).This underlines the need to integrate a monocyte reference in any comparison aiming to define DC3s. We conclude that scRNA-seq analysis identifies DC3s (cluster D) as a specific subset sharing transcriptional features of cDC2s (clusters A and B) and monocytes (cluster C).
Next we sought to define a flow cytometry-based strategy enabling analysis and prospective isolation of DC3s in blood. To this end, we performed an unsupervised flow cytometry data analysis based on genes identified by scRNA-seq (e.g., CD88, CD14, FcεRI, and CD1c) as well as markers previously associated with cDC2s (e.g., BTLA and CD5) (Yin et al., 2017) and DC3s (e.g., CD163) (Villani et al., 2017), even though they were not detected in the scRNA-seq analysis. We identified three main clusters (1, 2, and 3) together with a rarer cluster (4) of CD5hi cells (Figure 1G; Figure S1D). Cluster 4 appeared to be CD123hi contamination of AS-DCs (Figure S1D). Cluster 1 highly expressed CD88, aligning with monocyte cluster C identified by scRNA-seq. Cluster 2 did not express the monocyte-associated markers CD14 and CD88 but was characterized by expression of CD1c, FcεRI, and B and T lymphocyte attenuator (BTLA) (Figure 1G). In addition, cluster 2 showed heterogeneous expression of CD5 (Figure 1G). Similar to cluster 2, cluster 3 did not express the monocytic marker CD88 and displayed higher amounts of CD1c and FcεRI. However, cluster 3 could be distinguished from cluster 2 by higher expression of CD163 and heterogeneous expression of CD14 (Figure 1G). The unsupervised flow cytometry analysis allowed us to define a simple gating strategy enabling prospective isolation of monocytes (CD88+CD14+), cDC2s (CD5+ and CD5−CD88−CD1c+BTLA+CD163−), and DC3s (CD88−CD1c+BTLA−CD163+) (Figure 1H).
To further validate the flow cytometry-based identification of cell subsets, we performed bulk RNA-seq analysis of sorted monocytes, cDC2s (CD5+ and CD5−), DC3s, plasmacytoid DCs (pDCs), and AS-DCs (Figure S1E). Principal-component analysis (PCA) indicated that, overall, CD5+ and CD5− cDC2s, DC3s, and monocytes separated from pDCs and AS-DCs along the PC1 axis, accounting for 61% variance (Figure 1I). CD5+ and CD5− cDC2 clustered closely together, and DC3s separated clearly from cDC2s and monocytes (Figure 1I; Figure S1F). Hierarchical clustering (HC) and differentially expressed gene analysis led to the same conclusion, with DC3s sitting between monocytes and cDC2s (CD5+ and CD5−) (Figure 1J; Figure S1G). Overall, DC3s are closer to cDC2s than monocytes (Figures 1I and 1J).
Together, this validates that cellular clusters isolated by a flow cytometry-based approach align to clusters identified by unbiased scRNA-seq (Figure 1K).
We conclude that DC3s are a separable entity within CD1c+ cells, defined by a distinct gene expression profile, and that they can be prospectively isolated using CD88, CD1c, CD163, and BTLA (Figure S1H). scRNA-seq and bulk gene expression profiling identify markers shared between DC3s and cDC2s (e.g., CLEC10A and FCER1A) and markers shared between DC3s and monocytes (such as S100A8, S100A9, CD14, and CD163).
DC3s Give Rise to CD14+CD1c+ DCs Infiltrating Tumors
Tumor-infiltrating CD14+CD1c+ DCs have been reported in multiple instances, including ovarian cancer ascites (Segura et al., 2013), breast cancer (Michea et al., 2018), and melanoma (Bakdash et al., 2016; Binnewies et al., 2019). Therefore, we asked whether CD14+CD1c+ cells would align with DC3s. To this end, we analyzed mononuclear phagocytes infiltrating luminal breast cancer primary tumors. Using the gating strategy described in Figure 1, we found that, after exclusion of CD88+ monocytes and macrophages, the remaining CD45+HLA-DR+CD123−CD88− fraction contained cDC1s, CD14−CD1c+CD163−CD5+ cDC2s (CD5+ cDC2s), and CD14+CD1c+CD163+CD5− DC3s (CD14+ DC3s) (Figure 2A; Figure S2A). At this stage, we observed that CD163 and BTLA were particularly susceptible to enzymatic digestion of solid tissue, preventing consistent and reliable quantification of cells throughout the cohort of samples. Hence, we adopted a more restrictive definition of cDC2s and DC3s as CD1c+CD14−CD5+ and CD1c+CD14+CD5− cells, respectively (Figure 2A). This strategy enabled isolation of CD5+ cDC2s and CD14+ DC3s in peripheral tissues even though it might result in underestimation of their absolute numbers. Nevertheless, both subsets aligned phenotypically with their blood counterparts (Figures S2B and S2C) and expressed markers reported previously for CD1c+CD14+ iDCs, such as CD11c and FcεRI (Figure 2A; Segura et al., 2013).
Figure 2.
DC3s Infiltrate Human Breast Tumors
(A) Representative gating strategy used to define macrophages, CD5+ cDC2s, and CD14+ DC3s and histograms showing the expression of CD163, FcεRI, BTLA, and CD11c in human breast cancer primary tumors.
(B) Violin plot quantifying cDC1, CD5+cDC2, CD14+ DC3, and CD14+CD88+ macrophage subsets identified in (A) in human breast cancer primary tumors (n = 25; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA test).
(C) Pearson correlations of the frequencies of macrophages, cDC1s, CD5+cDC2s, and CD14+ DC3s within HLA-DR+ cells in human breast cancer primary tumors (red, significantly correlated p < 0.05; black, not correlated).
(D) HC showing the relative expression of markers used for subset identification in Figure 1 in CD1c+, CD1c+CD14+, and CD14+ cells from invaded lymph nodes draining human breast cancer primary tumors.
(E) GSEA of pairwise comparisons of CD1c+CD14+ cells with CD1c+ or CD14+ from invaded lymph nodes draining human breast cancer primary tumors. Gene signatures of blood DC3s compared with cDC2s (DC3 > cDC2) or CD14+ monocytes (DC3 > Mono) and, vice versa, of blood cDC2s (cDC2 > DC3) or CD14+ monocytes (Mono > DC3) compared with DC3s were used (Villani et al., 2017).
DC3s were consistently identified in 25 samples of primary tumors of clinical stages I, II, and III (Figure S2D). Macrophages represented by far the most abundant population. DC3s outnumbered cDC1s but were on par with bona fide CD5+ cDC2s (Figure 2B). The relative abundance of DC3s did not correlate with disease progression (Figure S2D) or with macrophages or cDCs (Figure 2C). In contrast, cDC1 infiltration correlated with cDC2s (Figure 2C).
Bulk RNA-seq analysis of CD1c+CD14+ cells sorted from tumor-invaded lymph nodes indicated that they displayed a similar expression profile as blood DC3s (high expression of cDC2 markers such as CLEC10A and FCER1A combined with low expression of monocyte-associated markers such as C5AR1 and SOD2) (Figure 2D). Gene set enrichment analysis (GSEA) revealed that CD14+CD1c+ were more enriched for the DC3 > cDC2 signature compared with CD1c+ cells (normalized enrichment score [NES], 2.06; p = 0.0; Subramanian et al., 2005; Villani et al., 2017). In addition, CD14+CD1c+ were more enriched for the DC3 > monocyte (Mono) signature compared with CD14+ cells (NES, 1.42; p = 0.02; Villani et al., 2017; Figure 2E). Conversely, compared with CD1c+CD14+, the cDC2 > DC3 and Mono > DC3 gene signatures were enriched in CD1c+ (NES, −1.57; p = 0.004) and CD14+ (NES, −2.07; p = 0.0) cells, respectively (Figure 2E).
We conclude that CD11c+FcεRI +CD14+CD1c+ iDCs infiltrating breast cancer align with DC3s.
Inflammatory cues promoting mobilization of CD14+CD1c+ cells at the site of inflammation are not fully defined. Mouse studies define GM-CSF as a likely candidate (Mach et al., 2000; Menezes et al., 2016). For this reason, we decided to test whether GM-CSF was sufficient to mobilize human CD14+CD1c+ DCs in a humanized mouse metastatic lung model. We generated B16 mouse melanoma engineered to overexpress human GM-CSF (B16_huGM) or FLT3L (B16_huFLT3L) (Figure S3A). Immunodeficient NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were injected intravenously with B16 control (CTRL), B16_huGM, or B16_huFLT3L (Figure 3A). Lung metastasis-bearing mice were engrafted with human peripheral blood mononuclear cells (PBMCs). Two days later, human CD45+ leukocytes were found in lung tumor foci and juxta-tumor areas (Figure 3B; Figures S3B and S3C). Flow cytometry analysis of metastatic lungs showed that FLT3L, but not GM-CSF, promotes expansion of CD1c+CD5+ cells aligning with blood cDC2s (Figure 3C, blue). In contrast, GM-CSF, but not FLT3L, led to accumulation of CD1c+CD14+ cells aligning with blood DC3s (Figure 3C, orange; Figure S3D, orange). As shown for circulating peripheral blood subsets in Figure 1, FLT3L-dependent cDC2s and GM-CSF-dependent DC3s shared expression of Clec10A, CD11c, and FcεRI (Figure 3C; Figure S3E). All tumor-bearing lungs contained some monocytes and/or macrophages (CD14+CD88+) (Figure 3C; Figure S3B). To further establish alignment of GM-CSF-mobilized DC3s, we performed an unbiased scRNA-seq on human CD45+HLA-DR+ cells expressing CD14 and/or CD1c (Figures S3F and S3G). Two major clusters could be identified. Cluster 0 was characterized by expression of genes associated with DC3s and cDC2s, such as CLEC10A, FCER1A, and CD74 (Figures S3H and S3I), and was enriched in DC3 differentially expressed transcripts (Villani et al., 2017; Figure S3J). Conversely, cluster 1 expressed markers defining monocytes (SOD2, C5AR1, and G0S2) and CXCL2, CXCL3, and CXCL8 chemokines (Figures S3H and S3I). We conclude that GM-CSF drives mobilization of CD14+CD1c+ cells aligning phenotypically with circulating DC3s.
Figure 3.
DC3s Give Rise to CD14+CD1c+ DCs at Inflammatory Sites
(A) Experimental model. NSG mice were injected intravenously (i.v.) with B16_CTRL, B16_huFLT3L, or B16_huGM on day 0. On days 7 and 8, 108 human PBMCs were injected i.v. Metastatic lungs were collected on day 9.
(B) Pseudocolor images of B16_huGM (green) metastatic lung on day 9 post-injection, stained for human CD45 (red). Nuclei were stained with Hoechst (blue). Scale bar, 100 μm.
(C) Gating strategy for cDC2 and DC3 identification in B16_huGM and B16_huFLT3L metastatic mouse lung and histograms showing the expression of CD163, CD206, and Clec10A. The bar graph summarizes the frequency of cDC2s and DC3s among total HLA-DR+ cells in metastatic B16_CTRL, B16_huFLT3L, or B16_huGM mouse lungs (n = 3 independent mice; ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA test).
(D) Flow cytometry analysis of flow cytometry-sorted cDC2s, DC3s, and CD14+ monocytes after 2 days of culture with MS5 stromal cells expressing human GM-CSF (MS5_GM). Bar graphs show the frequency of output cells among total huCD45+ cells (n = 4–5 healthy donors).
(E) Histograms showing CD14 expression on cDC2s, DC3s, and CD14+ monocytes before and after 2 days of coculture with MS5_GM and bar graphs summarizing the frequency of CD14 expression within each cell type (n = 5 healthy donors, ∗∗p < 0.01, Mann-Whitney two-tailed t test).
See also Figures S3 and S4 and Table S1.
We next wondered whether GM-CSF could induce trans-differentiation of circulating cDC2s or monocytes into CD14+CD1c+ DC3s. To test this hypothesis, cDC2s, DC3s, and monocytes were sorted by flow cytometry from blood and cocultured in vitro in the presence of GM-CSF-expressing stromal cells (MS5_GM). After 2 days, cDC2s did not acquire CD163 or CD14 expression (Figure 3D). In contrast, DC3s did upregulate CD14 (Figures 3D and 3E). We found that monocytes differentiated into CD16+ and CD16− macrophages (Figure 3D). Importantly, CD88+CD1c−CD14+ monocytes did not give rise to CD1chiCD163hi DC3s (Figure 3D). Addition of IL-4 to GM-CSF culture did not affect the outcome (Figure S4A). The lack of trans-differentiation of cDC2s or monocytes into DC3s was also confirmed in vivo upon adoptive transfer into immunodeficient NSG mice carrying GM-CSF-expressing tumors (Figures S4B and S4C). Overall, we conclude that CD88−CD14+CD1c+ cells can differentiate from DC3s independent of cDC or monocytic lineages.
GM-CSF Stimulates Differentiation of CD34+ Hematopoietic Progenitors into DC3s In Vitro
We next wondered how DC3s would differentiate from bone marrow progenitors. To this end, we sought to define a culture system capable of generating DC3s together with cDC2s and macrophages. We cocultured human cord blood-derived CD34+ hematopoietic stem and progenitor cells (HSPCs) with stromal cells engineered to overexpress cDC-promoting factors (membrane-bound FLT3L together with stem cell factor (SCF) and CXCL12 [MS5_FS12]; Anselmi et al., 2020) in the presence or absence of GM-CSF (Figure 4A; Figure S5A). We found that GM-CSF increased differentiation of CD1c+CD14+CD163+ cells (orange) phenotypically aligning with blood DC3s (Figures 4A and 4B). CD1c+CD14− cells did not express CD163, suggesting that they mostly align with cDC2s. In addition, CD163 expression was restricted to CD1c+CD14+ cells (Figure 4B). GM-CSF alone (MS5_GM), but not FLT3L, was sufficient to induce differentiation of DC3s in vitro (Figure 4C). In contrast, FLT3L (MS5_FL) was sufficient to induce differentiation of cDC2s (Figure 4C). We conclude that cDC2s and DC3s have distinct growth factor requirements.
Figure 4.
DC3s Differentiate from Hematopoietic Progenitors upon GM-CSF Exposure Independent of Mono-Committed Progenitors (cMoPs) or cDC-Committed Progenitors (CDPs)
(A) Flow cytometry analysis of cord blood-derived CD34+ HSPCs cultured on stromal cells expressing human FLT3L, SCF, and CXCL12 (MS5_FS12) with or without human recombinant GM-CSF (MS5_FS12+recGM-CSF) for 14 days.
(B) Flow cytometry plots of BTLA and CD163 expression within CD1c+CD14− and CD1c+CD14+ cells identified in (A). Bar graphs summarize the absolute numbers of differentiated CD1c+CD14− cDC2s and CD1c+CD14+ DC3s (a line represents the median; n = 6 independent cord blood donors, ∗p < 0.05, Wilcoxon test).
(C) Bar graphs summarizing the absolute numbers of CD1c+CD14−CD163− cDC2s and CD1c+CD14+CD163+ DC3s differentiated from cord blood-derived CD34+ HSPCs cocultured for 14 days with stromal cells expressing human FLT3L (MS5_FL), GM-CSF (MS5_GM), or neither (MS5_CTRL) (a line represents the median; n = 5–7 independent cord blood donors, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, Wilcoxon test).
(D) HC based on 19,791 protein-coding genes of in-vitro-generated subsets differentiated from CD34+ HSPCs cultured with MS5_FS12 supplemented with human recombinant GM-CSF (MS5_FS12+recGM-CSF). Each dot represents an average of three donors.
(E) Volcano plots showing the DEGs between in-vitro-generated DC3s cells (orange) compared with cDC2s (blue and turquoise, left plot) or macrophages (gray and brown, right plot). Genes with Log2(fold change, FC) > ±2 and a false discovery rate (FDR)-adjusted p value of less than 0.05 were considered significant.
(F) GSEA of pairwise comparisons of DC3s with cDC2s or macrophages generated in vitro. Gene signatures (gene set) defining genes upregulated in blood DC3s compared with cDC2s (DC3 > cDC2) or blood DC3s compared with CD14+ monocytes (DC3 > CD14 Mono) were used (Villani et al., 2017) (NES, normalized enrichment score).
(G) BubbleMap summarizing the enrichment of defined gene sets in pairwise comparisons of in-vitro-differentiated DC3s versus in vitro cDC2s or in vitro macrophages. Gene signatures (gene sets) of blood DC3s compared with cDC2s (DC3 > cDC2) or CD14+ monocytes (DC3 > Mono) and, vice versa, of blood cDC2s (cDC2 > DC3) or CD14+ monocytes (Mono > DC3) compared with DC3s were used (Villani et al., 2017).
(H) Flow cytometry analysis of cord blood-derived CDPs, cMoPs, and GMDPs cultured for 7 days with MS5_FS12 or MS5_GM. Bar graphs summarize the absolute number of differentiated cells from each progenitor (a line represents the median, n = 4–7 independent cord blood donors).
We next intended to determine whether the transcriptional landscape of in-vitro-generated DC3s from stromal cell cocultures aligned with their in vivo counterparts. Bulk RNA-seq analysis of in-vitro-generated cells showed that CD14+CD1c+ DC3s (orange) sat between cDC2s (blue and turquoise) and macrophages (gray and brown) (Figure 4D). Of note, CD1c+CD14− cells generated in vitro were heterogenous for CD206 expression and were therefore analyzed as two independent subsets (CD1c+CD206− [blue] and CD1c+CD206+ [turquoise]; Figure S5B). However, both subsets displayed very similar transcriptomes regardless of CD206 expression, and the CD1c+CD14−CD206+ and CD1c+CD14−CD206− fractions strongly resembled circulating cDC2s (Anselmi et al., 2020).
Further analysis of differentially expressed genes (DEGs) showed that in vitro DC3s differed from in vitro cDC2s by expression of monocyte-associated markers such as S100A8, S100A9, S100A12, and CD14 (Figure 4E; Figure S5C). Conversely, in vitro cDC2s displayed higher expression of CD1C, LAMP3, CD52, and LTB, as reported for lung cDC2s (Lavin et al., 2017; Figure 4E; Figure S5C). Using the GSEA methodology (Subramanian et al., 2005), we found that the set of genes upregulated in primary DC3s compared with cDC2s (DC3 > cDC2 from Villani et al., 2017) was enriched in GM-CSF-dependent in vitro DC3s compared with in vitro cDC2s (Figure 4F). Of note, GM-CSF-exposed cDC2s did not convert to DC3s (Figures S5D and S5E). We found that markers common for primary cDC2s and DC3s (CLEC10A, Figure 1) were higher in in vitro DC3s compared with in vitro macrophages (Figure 4E; Heidkamp et al., 2016). Conversely, in vitro macrophages expressed more FCGR1A, C5AR1 (CD88), CXCL8, CXCL1, CXCL2, CXCL3, CCL2, CCL3, and CCL7 compared with in vitro DC3s (Ruffell et al., 2009; Figure 4E; Figures S5C and S5F). Genes upregulated in primary DC3s compared with CD14+ monocytes (DC3 > CD14+ Mono from Villani et al., 2017) were significantly enriched in in vitro DC3s compared with macrophages (Figures 4F and 4G). We conclude that GM-CSF drives, in vitro, the differentiation of DC3s aligning to primary blood DC3s.
DC3s Develop via a Differentiation Pathway Independent of CDPs and cMoPs
The developmental relationship between DC3s and cDCs or monocyte lineages is not known. A classical view defines the development of phagocytes as a stepwise and ordered loss of developmental potential concomitant with lineage commitment. Historically, this process has been identified using prospective isolation of progenitor populations of decreasing potential. Specifically, early granulocyte-monocyte and DC progenitors (GMDPs) carries a tri-lineage potential (Lee et al., 2015b). Loss of neutrophil potential defines monocyte and DC progenitors (MDPs) (Lee et al., 2015b). Loss of monocyte potential defines CDPs, which generate cDCs via a pre-cDC intermediate (Breton et al., 2015, 2016; Lee et al., 2015b; Naik et al., 2007; Onai et al., 2007; See et al., 2017). Finally, loss of DC and neutrophil potential defines monocyte-committed progenitors (cMoPs) (Kawamura et al., 2017). Having established the growth factor requirements for DC3 development, we decided to test the contribution of CDPs and cMoPs to generation of DC3s. Flow cytometry-sorted CDPs and cMoPs and the remaining GMDP-containing fraction (CD34+CD38+CD45RA+CD123−CD64−) isolated from cord blood-derived HSPCs (see the gating strategy in Figure S5G) were cocultured with stromal cells supporting cDC (MS5_FS12) or DC3 (MS5_GM) differentiation.
Flow cytometry analysis after 7 days of culture showed that CDP gave rise exclusively to cDC1s and cDC2s in MS5_FS12 cocultures, as described previously (Figure 4H; Figure S5H; Lee et al., 2015b). cMoP cultures gave rise solely to CD14+CD1c− cells, as described previously (Figure 4H; Figure S5H; Kawamura et al., 2017). As expected, the GMDP-containing fraction gave rise to granulocytes, macrophages, and cDC2s. Importantly, the GMDP-containing fraction also gave rise to DC3s in MS5_GM coculture (Figure 4H; Figure S5H). Therefore, we asked whether DC3s would arise directly from a multipotent progenitor or via formation of an intermediate DC3-committed progenitor devoid of any other lineage potential.
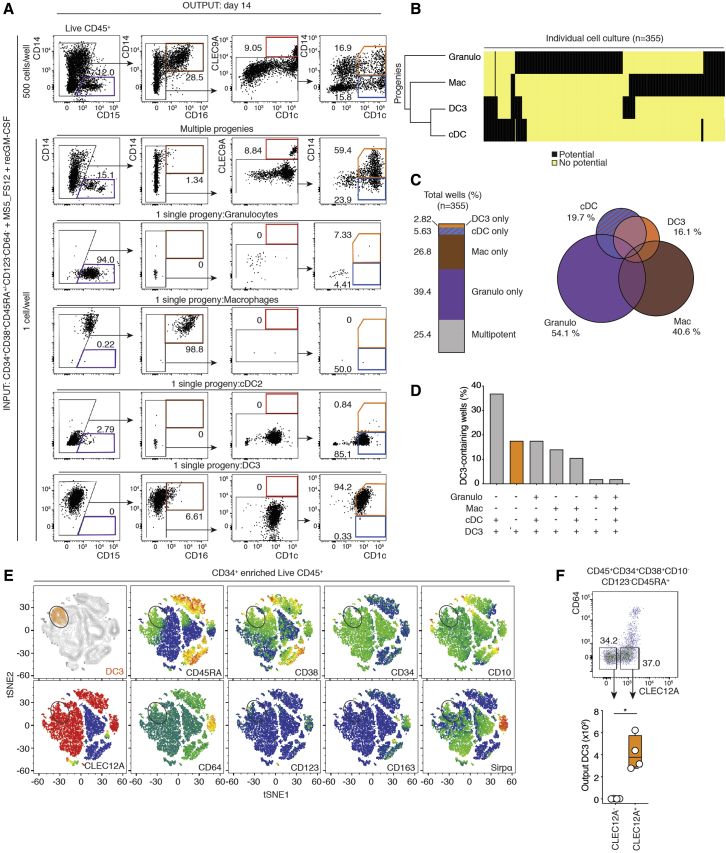
To address this question in unbiased settings, we developed a single cell culture of CD34+CD38+CD123−CD64− progenitors distinct from CDPs or cMoPs (Figure 5A). We chose to combine MS5_FS12 with soluble GM-CSF for two reasons: (1) GM-CSF alone did not support growth of individual progenitors (Figure S5I), and (2) MS5_FS12 coculture was found to more efficiently support cDC and monocyte differentiation (Figure 4G).
Figure 5.
Single-Cell Analysis of DC3 Commitment
(A) Flow cytometry analysis of bulk (500 cells) or single CD34+CD38+CD123−CD64− progenitor cells cocultured for 14 days with MS5_FS12 supplemented with recombinant human GM-CSF (MS5_FS12+recGM-CSF). Flow cytometry plots resulting from single CD34+CD38+CD123−CD64− progenitor cells with different potentials are shown as representative examples (n = 355 cells from 2 independent experiments).
(B) HC of lineage potential from single CD34+CD38+CD123−CD64− progenitor cells (n = 355).
(C) Bar graph and Venn diagram summarizing the frequency of the potential of mono-, bi-, tri-, or multipotent individual CD34+CD38+CD123−CD64− cells within the total wells analyzed (n = 355).
(D) Bar graphs summarizing the frequency of mono-, bi-, tri-, or multipotent individual CD34+CD38+CD123−CD64− cells among DC3-generating progenitors only. An orange bar represents the frequency of DC3-restricted progenitors.
(E) Cell surface phenotype of DC3-restricted progenitors before differentiation cultures inferred by index flow cytometry sorting. tSNE plots display an overlay of total CD45+ cells (gray) and DC3-restricted progenitor cells (orange) (top left). Shown is relative expression of the markers CD45RA, CD38, CD34, CD10, Clec12A, CD64, CD123, CD163, and SIRPα.
(F) Validation experiment for identification of Clec12A as a marker for DC3-committed progenitors. Shown is flow cytometry analysis of bulk-sorted CD34+CD38+CD45RA+CD123−CD64−Clec12A− and Clec12A+ cells. 500 cells were cocultured with MS5_GM for 7 days. The bar graph summarizes the number of differentiated DC3s from each bulk population (n = 4 healthy donors, ∗p < 0.05, Mann-Whitney two-tailed t test).
See also Figure S5.
Flow cytometry analysis of 14-day progeny of individual progenitor cultures revealed multiple patterns of developmental potential (Figure 5A). Overall, the granulocyte potential was segregated from the potential for mononuclear phagocytes (Figure 5B). We found that only 0.3% of progenitors could differentiate into all four lineages corresponding to the GMDP functional definition (Figure 5D). Most individual progenitors gave rise to single lineage progeny (Figures 5C and 5D). Unipotent wells containing only neutrophils represented the most abundant outcome (39.4%), followed by macrophage-only (26.8%) and cDC-only (5.63%) wells (Figure 5C). Most individual progenitors endowed with DC3 potential had multi-lineage potential. DC3 potential was more associated with mononuclear phagocytes rather than granulocyte potential (61.4% and 19.3% of DC3-containing wells, respectively) (Figure 5D). Importantly, we also identified a minor fraction of individual progenitors giving rise exclusively to DC3s (Figures 5B–5D).
We next aimed to further define the cell surface phenotype of progenitors endowed with DC3 potential. To this end, we attempted to establish correlations between the cell surface phenotype of single sorted cells (inferred from index flow cytometry sorting) and their developmental potential. A posteriori identification of the cell surface phenotype of DC3-committed progenitors revealed that they had a CD34+CD38+CD45RAintCD123−CD64−SIRPαloCD10−Clec12A+ phenotype (Figure 5E). As validation, we showed that the potential for DC3s lay in the Clec12A+ population of the GMDP-containing fraction (Figure 5F). We conclude that DC3s can develop via a DC3-restricted intermediate distinct from cDC-restricted CDPs or monocyte-restricted cMoPs. Even though the existence of a DC3-commited unipotent progenitor is not formally proven, our data are compatible with the notion that DC3 specification arises downstream of MDP.
Activated DC3s Induce Priming of Naive T Cells and Differentiation of CD103+ T Cells
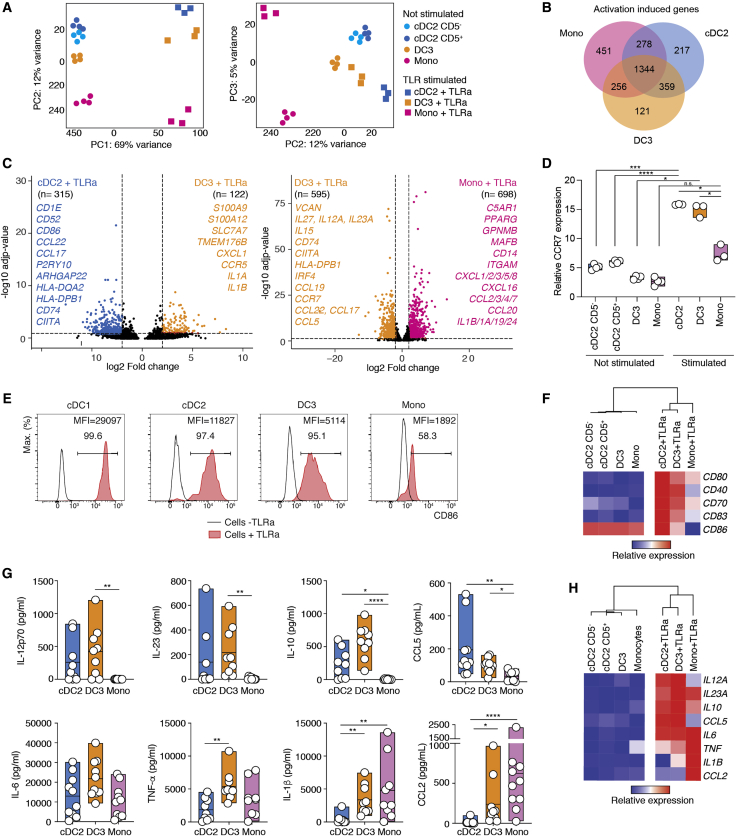
We next aimed to understand the immunological function of DC3s and to compare it with cDC2s and monocytes. First, we decided to test the responsiveness of DC3s, cDC2s, and monocytes to a cocktail of Toll-like receptor (TLR) agonists. cDC2s, DC3s, and monocytes were sorted by flow cytometry from blood and stimulated overnight ex vivo. PCA analysis of the total transcriptome of unstimulated and stimulated populations evidenced that all subsets underwent a certain degree of convergence in their transcriptome (Figure 6A). In support of this, we found an important overlap in the set of activation-induced genes defined for each subset (1,344 genes; Figure 6B). Despite the relative convergence of activated cells, we found that overnight activation did not compromise cell surface discrimination of DC3s from cDC2s and monocytes (Figure S6A). Indeed, TLR-activated DC3s could still be discriminated from TLR-activated cDC2s by 437 DEGs or from TLR-activated monocytes by 1,293 genes (Figure 6C). The same was true for the pairwise comparison of activated cDC2s and circulating DC3s (Figure S6B). In sum, we conclude that innate activation does not trigger conversion of cDC2s or monocytes into DC3s despite induction of a common transcriptional response to TLR stimulation.
Figure 6.
DC3s Respond to TLR Stimulation
We performed bulk RNA-seq analysis of BTLA+CD5+ and BTLA+CD5− cDC2s, DC3s, and monocytes sorted as shown in Figure 1H and stimulated overnight (16 h, 3 donors) or not (4 donors) with a TLR agonist cocktail (25 μg/mL poly(I:C), 1 μg/mL R848, and 10 ng/mL LPS). For activation of cDC2s, BTLA+CD5+ and BTLA+CD5− were pooled.
(A) PCA analysis for all genes.
(B) Venn diagram summarizing the number of activation-induced DEGs upregulated in stimulated compared with unstimulated cells within each cell population.
(C) Volcano plots showing DEGs between TLR agonist-stimulated DC3s compared with TLR agonist-stimulated cDC2s or TLR agonist-stimulated monocytes. Genes with Log2(FC) > ±2 and a FDR-adjusted p value of less than 0.05 were considered significant.
(D) Bar graph summarizing relative CCR7 gene expression within TLR-agonist stimulated or unstimulated mononuclear phagocyte populations (n = 3–4; a line represents the median; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA test).
(E) Histograms showing the frequency and median of fluorescence intensity (MFI) of CD86 on TLR agonist-stimulated or unstimulated mononuclear phagocyte populations.
(F) Heatmap showing the relative gene expression of selected costimulatory molecules on TLR agonist-stimulated or unstimulated mononuclear phagocyte populations.
(G) Quantification of cytokines and chemokines secreted by cDC2s, DC3s, and CD14+ monocytes in response to overnight stimulation with a cocktail of TLR agonists (n = 9 healthy donors; a line represents the median; ∗p < 0.05, ∗∗p < 0,01, ∗∗∗∗p < 0,0001, one-way ANOVA test).
(H) Heatmap showing the relative gene expression of cytokines and chemokines analyzed in (G) within TLR-agonist stimulated or unstimulated mononuclear phagocyte populations.
From the perspective of adaptive immunity, activated DC3s shared a lot of common features with activated cDC2s but less with activated monocytes: (1) stimulated DC3s and cDC2s upregulated CCR7 upon activation (Figure 6D), potentially enabling their ability to migrate toward T cell zones; (2) activated DC3s and cDC2s upregulated cell surface co-stimulatory molecules (CD80, CD86, CD70, and CD40; Figure 6E and 6F); (3) activated DC3s and cDC2s efficiently increased the expression of T cell-attracting chemokines such as CCL5 (Figures 6G and 6H), CCL19, CCL17, CCL22, CXCL9, CXCL10, CXCL11, and CXCL13 (Figure S6C); and (4) activated DC3s and cDC2s produced higher amounts of IL-12p70, IL-23, IL27, and IL-10 (Figures 6G and 6H; Figure S6E).
In addition, activated DC3s shared some common features with activated monocytes that are less pronounced in activated cDC2s: (i) activated DC3s and monocytes secreted more inflammatory cytokines, such as tumor necrosis factor alpha (TNF-α) and IL-1β (Figures 6G and 6H); and (2) activated DC3s and activated monocytes upregulated inflammatory chemokines, such as CCL2 (Figures 6G and 6H), CCL1, and CCL3 (Figure S6C) or granulocytes attracting CXCL1, CXCL3, and CXCL5 (Figures S6C and S6D).
Collectively, our data suggest that the transcriptome and secretome of activated DC3s, unlike the ones of activated monocytes, are consistent with a function in priming of naive T cells.
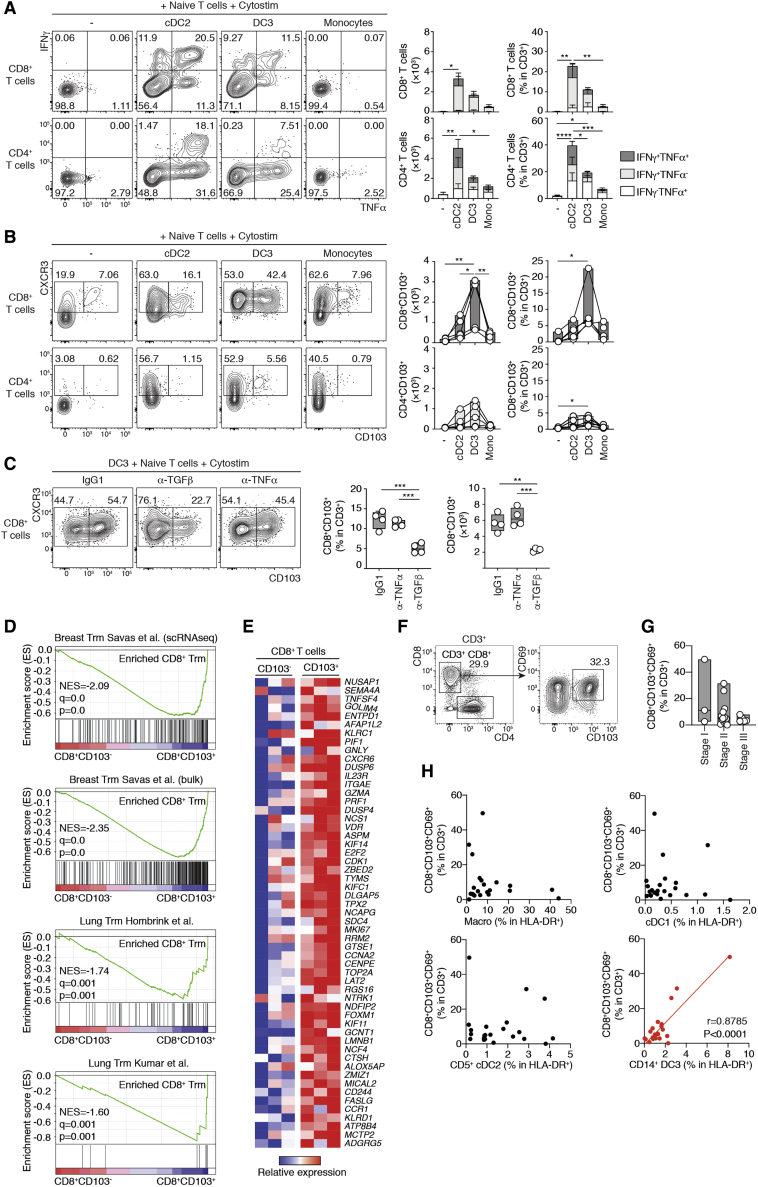
To directly test the T cell priming capabilities of DC3s, we performed 5-day cocultures of flow cytometry-sorted, activated DC3s, cDC2s, and monocytes (see the cell sorting strategy in Figure S7A) with allogenic CD45RA+ naive T cells in the presence of a synthetic superantigen. We found that activated DC3s and cDC2s, unlike monocytes, triggered proliferative expansion and effector differentiation in CD4+ and CD8+ T cells probed by CD45RO acquisition (Figure S7B). The same results were also obtained using in-vitro-generated DC3s (Figure S7C). In contrast to monocytes, activated DC3s and cDC2s induced interferon γ (IFN-γ)- and TNF-α-producing CD4+ and CD8+ T cells but not IL- 17A (Figure 7A; Figure S7D). We found that DC3s had a specific ability to efficiently trigger CD103 expression in CD8+ T cells (Figure 7B), even without the presence of superantigen (Figure S7E). CD103 expression is a hallmark of tissue-resident memory T (TRM) cells because of its interaction with E-cadherin (Mueller and Mackay, 2016). Mechanistically, multiple factors, including transforming growth factor β (TGF-β), have been proposed to induce CD103 expression on T cells (Mueller and Mackay, 2016; Rihs et al., 1996; Yu et al., 2013). Here we showed that anti-TGF-β neutralizing antibodies, but not others, blocked the expression of CD103 on CD8+ T cells (Figure 7C; Figure S7F).
Figure 7.
DC3s Prime Naive T Cells and Drive Acquisition of the CD103+ TRM Phenotype
(A and B) Representative flow cytometry plots and quantification of CD4+ and CD8+ naive T cells cultured for 5 days with flow cytometry-sorted blood cDC2s, DC3s, or CD14+ monocytes after overnight activation with TLR agonists (25 μg/mL poly(I:C), 1 μg/mL R848, and 10 ng/mL LPS) in the presence of a synthetic superantigen (Cytostim). Absolute numbers and frequencies of cytokine-producing and other activated T cells (A) and CD103+ T cells (B) are shown (n = 5 healthy donors in 5 independent experiments; a line represents the median; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way ANOVA test).
(C) Representative flow cytometry plots and quantification showing CD103 expression on CD8+ naive T cells cocultured with blood DC3s sorted by fluorescence-activated cell sorting (FACS) in the presence of 10 μg/mL of neutralizing antibodies against TNF-α or TGF-β or an isotype CTRL (n = 4 healthy donors in 3 independent experiments; a line represents the median; ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001, one-way paired ANOVA test).
(D and E) Bulk RNA-seq analysis of CD8+CD103− T cells (n = 3) and CD8+CD103+ T cells (n = 3) sorted by flow cytometry after 5 days in vitro coculture of naive blood CD8+ T cells with blood DC3s activated overnight by TLR agonists.
(D) GSEA of pairwise comparisons of CD8+CD103+ T cells with CD8+CD103− T cells. Gene signatures (gene set) defining genes upregulated in breast or lung CD103+CD69+CD8+ TRM cells were used (Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018).
(E) Heatmap displaying 56 representative genes significantly upregulated in CD8+CD103+ cells compared with CD8+CD103− induced by blood DC3s in vitro (of 205 DEGs). Selected genes are shared with at least one of the previously reported gene signatures defining human TRM cells (Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018).
(F–H) Correlative analysis of TRM cell content in luminal breast cancer primary tumors.
(F) Representative flow cytometry plots showing the gating strategy for CD103+CD69+CD8+ T cells in 21 human luminal breast cancer primary tumors.
(G) Quantification of CD103+CD69+CD8+ T cells in different stages of human breast tumors (stage I, n = 3; stage II, n = 13; stage III, n = 5).
(H) Pearson correlation of the frequencies of the macrophages and cDC1, CD1c+ CD14−, and CD1c+ CD14− cells and the frequencies of CD103+CD69+CD8+ T cells in human breast cancer primary tumors (red, significantly correlated p < 0.05; black, not correlated).
We next wondered whether CD103+ T cells aligned with bona fide TRM cells isolated from human tissue. To this end, we evaluated the gene expression profile of CD103− and CD103+ CD8+ T cells obtained after coculture of naive T cells with activated DC3s (Figure S7G). Using the GSEA methodology (Subramanian et al., 2005), we found that the signatures obtained for breast cancer or lung tissue CD103+ TRM cells (Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018) were enriched in CD103+ T cells compared with CD103− T cells induced by DC3s (Figure 7D). DEG analysis revealed that human TRM cell markers such as NUSAP1, DUSP4, CXCR6, and FASLG (Figure 7E; Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018) were upregulated in CD103+ compared with CD103− CD8+ T cells. In addition, DC3-activated CD103+ CD8+ T cells expressed core components of the cytotoxic machinery (PRF1 and GZMA), as reported earlier for breast cancer-invading TRM cells (Figure 7E; Savas et al., 2018).
CD8+CD103+ TRM cell infiltration has a protective prognosis value in breast cancer (Savas et al., 2018; Wang et al., 2016). To test the physiological relevance of DC3-dependent induction of CD8+CD103+ T cells, we analyzed CD103 and CD69 expression in CD3+CD8+ T cells from 18 samples of primary luminal breast cancer (Figure 7F). We found that CD8+CD103+CD69+ T cells were present across different cancer stages (Figure 7G). The frequency of CD8+CD103+CD69+ T cells was positively correlated with DC3 infiltration but not with other mononuclear phagocytes (Figure 7H). Despite the statistical significance of this correlation, it is important to underline that this was mainly driven by a subset of the samples. This suggests that further stratification of patients could help improve our understanding of the relationship between TRM cells and DC3s in breast cancer.
Together, our results anchor DC3 function within the DC lineage; DC3s, just like cDC2s but unlike monocytes, are competent for priming and polarization of CD45RA+ naive T cells. In addition, we define induction of bona fide TRM cell-like CD103+CD8+ T cells (Yu et al., 2013) as a specific privilege of DC3s but not cDC2s.
Discussion
Using scRNA-seq and high-dimensional flow cytometry, we provided evidence that DC3s represent a DC subset that can be separated and isolated from other DC subtypes. Regarding gene expression, our results are largely congruent with findings reported in previous studies (Dutertre et al., 2019; Villani et al., 2017). In addition, we provide a robust flow cytometry strategy to identify and purify DC3s by taking in account CD14+ monocytes and cDC2s. In agreement with previous studies, our results challenge the widely accepted notion that CD14 is a specific marker for monocytes (Dutertre et al., 2019; Villani et al., 2017). Indeed, we clearly demonstrate that a large fraction of DC3s expresses cell surface CD14. We identified CD88 (encoded by the C5AR1 gene) as a proper monocyte marker, enabling prospective purification of monocytes devoid of CD14+CD1c+ DC3s. We found that ex vivo GM-CSF cultures of pure CD88+CD14+ monocytes did not give rise to CD88-CD14+CD1c+ iDCs. Therefore, our findings provide an incentive to carefully revisit the prevailing notion that CD14+CD1c+ iDCs arise exclusively from monocytes in vivo.
From the DC perspective, we refined the strategy to analyze the functional heterogeneity of CD1c+ DCs, including cDC2s and DC3s. Indeed, previous studies have reported heterogeneous expression of CD5 in CD1c+ DCs (Dutertre et al., 2019; Yin et al., 2017). However, our findings highlight that CD5−CD1c+ DCs contain CD163− cDC2s and CD163+ DC3s, which are transcriptionally distinct. Overall, our scRNA-seq and bulk RNA-seq analyses cluster together CD5+ and CD5− cDC2s. However, the developmental relationship between CD5− cDC2s, CD5+ cDC2s, and AS-DCs remains to be clarified (Dutertre et al., 2019; See et al., 2017; Villani et al., 2017).
Because of CD1c expression and their relative similarity to cDC2s, DC3s have been embedded in the group of cDCs. Here we have provided substantial evidence arguing against this notion. Indeed, a conservative definition of cDCs includes (1) dependence on the FLT3L growth factor (Breton et al., 2015; Guermonprez et al., 2013, 2019; McKenna et al., 2000; Pulendran et al., 1998; Waskow et al., 2008) and (2) reliance on the CDP and pre-cDC developmental pathway (Breton et al., 2015, 2016; Guermonprez et al., 2019; Lee et al., 2015b; Naik et al., 2007; Onai et al., 2007). Here we have shown that DC3s do not meet any of these criteria. First, FLT3L alone was poorly active in stimulating the production of DC3s from CD34+ HSPCs in a controlled setting in vitro. Conversely, GM-CSF drives the commitment of DC3s under the same conditions. Moreover, we have shown that CDP did not give rise to DC3s although they are competent to generate cDC1s and cDC2s (Lee et al., 2015b). Taking in account the heterogeneity of defined progenitor populations (Paul et al., 2015), we developed single-cell cultures enabling analysis of multiple lineage populations. Our single-cell cultures demonstrated that DC3s develop from CLEC12A+ DC3-restricted progenitors. In addition, the most frequent progenies differentiating along with DC3s from a bi-potent progenitor were cDCs and macrophages. This strongly suggest that DC3 progenitors diverge downstream of the MDP stage (Fogg et al., 2006; Lee et al., 2015a). Further studies will define more precisely the cell surface and molecular phenotype of the DC3-restricted progenitor. In support of a distinct regulation of cDC2s and DC3s, we have shown that cDC2 infiltration in breast cancer is correlated with cDC1s but not DC3s. Also, Dutertre et al. (2019) have shown recently that DC3s, but not cDC2s, expand in the blood of systemic lupus erythematosus patients (Dutertre et al., 2019). Further delineation of the inflammatory cues and transcription factors underpinning the development of the DC3 lineage is needed.
Identification of DC3 as a cellular entity arising from a specific lineage brings forward the question of their specific immune function. We have shown that activated DC3s, just as cDC2s but unlike monocytes, secrete high amounts of T cell-polarizing cytokines (IL-12p70 and IL-23) and T cell-attracting chemokines (CXCL9, CXCL10, CXCL11, and CCL5). In addition, DC3s also secrete other cytokines (IL-10, IL-6, and TNF-α), some of which were poorly secreted by cDC2s (e.g., TNF-α). DC3s are polyvalent phagocytes with a cytokine pattern encompassing T cell and inflammation cues.
A quintessential defining feature of DCs is their capacity to activate naive T cells. For instance, infiltration of CD1c+ DCs is associated with priming of T cell effectors when regulatory T (Treg) cell infiltration is low or Treg cell-mediated suppression is alleviated by checkpoint blockade (Binnewies et al., 2019). However, even when the heterogeneity of CD1c+ cells is appreciated, including recruitment of CD14+CD1c+ DCs in tumor-draining lymph nodes (Binnewies et al., 2019), little is known about the function of CD1c+ subtypes. Therefore, we asked whether the functions attributed to CD1c+ DCs would be carried out by cDC2s and/or DC3s. We found that DC3s, like cDC2s but unlike monocytes, primed and drove robust activation of naive T cells into IFN-γ- and TNF-α-secreting polyfunctional effectors (Acosta-Rodriguez et al., 2007; Leal Rojas et al., 2017; Napolitani et al., 2005; Nizzoli et al., 2013; Schlitzer et al., 2013; Yin et al., 2017).
αE integrin (CD103) is a key marker of TRM cells through its interaction with E-cadherin, participating in retention of T cells at epithelial and mucosal sites (Mueller and Mackay, 2016). Yu et al. (2013) have identified a feature of total CD1c+ DCs in their ability to drive acquisition of CD103 in CD8+ T cells. Here we have shown that DC3s, but not cDC2s, induced expression of CD103 on CD8+ and CD4+ T cells. In addition, we have shown that CD103+ T cells primed by DC3s ex vivo align with bona fide TRM cells isolated from lung or breast cancer (Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018). This finding is consistent with the existence of early imprinting of the TRM cell program at the level of T cell priming by DC3s. This notion is supported by a recent in vivo study in a mouse model, evidencing early imprinting of the TRM cell program during T cell priming in the lymph nodes by αV integrin-expressing migratory DCs (Mani et al., 2019). Further supporting this view, we found that (1) DC3s upregulated CCR7 when activated by TLR agonists, potentially acting as migratory DCs, and (2) blocking studies indicated that TGF-β was required to prime CD103+ T cells. This role of priming in lymph nodes does not exclude that tumor-infiltrating DC3s might also provide TGF-β and other signals important for maintenance of TRM cells, as demonstrated in a mouse model (Mani et al., 2019). In support of this view, we found that infiltration of DC3s was selectively associated with the abundance of CD8+CD103+ T cells in luminal breast cancer primary tumors. Further studies are needed to delineate the full molecular mechanisms of CD103 induction in T cells by DC3s and their physiological in vivo relevance during immune responses. Indeed, CD103+CD8+ T cells are a protective biomarker in triple-negative breast cancer (Savas et al., 2018; Wang et al., 2016), and lung CD103+CD8+ T cells are a hallmark of protective immunity afforded by influenza vaccination (Yu et al., 2013). This underlines the potential of DC3s to regulate tissue immunity and defines them as targets for vaccines and immunotherapeutic interventions.
Limitations of Study
We identify DC3s as CD88-CD1c+CD163+CD14+/-. Further studies will be needed to determine if this phenotype corresponds to all CD1c+CD14+ cells that had been reported in various inflammatory settings.
We showed that DC3 differentiation is driven by GM-CSF in humanized mouse metastatic lung model. However, human circulating PBMCs were used in these experiments, preventing the assessment of GM-CSF impact on bone marrow DC3 progenitors in vivo. Hence, we cannot exclude a role of GM-CSF on DC3s survival instead of differentiation. In addition, we showed that GM-CSF drives DC3 differentiation from CD34+ umbilical cord HSPCs in vitro. This finding does not exclude that other growth factors control DC3 development in vivo.
We aligned CD1c+CD14+ cells infiltrating breast tumor-draining lymph node with blood DC3s compared to cDC2s and monocytes. Due to limited availability of healthy human lymph nodes, we did not investigate if CD1c+CD14+ cells are infiltrating secondary lymphoid organs in homeostatic conditions. Blood DC3s upregulate CCR7 upon TLR stimulation suggesting they have a migratory potential. However, we did not investigate whether lymph nodes DC3s originate from non-lymphoid peripheral tissues via lymphatic vessels or directly from blood.
A defining feature of cDCs lies in their ability to prime naïve T cells. We showed that DC3s can induce the proliferation of naïve T cell in a non-autologous priming context, i.e. the mixed leukocyte reaction. The capability of DC3s to uptake, process and present antigens by MHCI and MHCII molecules remains to be addressed.
Our in vitro experiments showed that i) DC3s induce efficiently TRM differentiation from naïve CD8+T cells and, ii) DC3 infiltration correlates with TRM abundance in vivo. However, this does not provide a direct evidence that DC3s control TRM specification within tumor draining lymph nodes where they are likely to interact with naïve T cells. Indeed, we cannot rule out that, in vivo, DC3s act selectively in the tissue to maintain TRM populations primed in lymph nodes by a different DC subtype. Further studies will be needed to address how DC3s control CD8+ TRM populations in vivo.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Biotin anti-human CD335 (NKp46) antibody (clone 9E2) | BioLegend | Cat#331906; RRID: AB_1027671 |
| Biotin anti-human CD3 antibody (clone OKT3) | BioLegend | Cat# 317320; RRID: AB_10916519 |
| Biotin anti-human CD19 antibody (clone HIB19) | BioLegend | Cat# 302204; RRID: AB_314234 |
| Biotin anti-human CD20 antibody (clone 2H7) | BioLegend | Cat# 302350; RRID: AB_2565524 |
| Biotin anti-human CD56 (NCAM) antibody (clone HCD56) | BioLegend | Cat# 318320; RRID: AB_893390 |
| Biotin anti-human CD203c (E-NPP3) antibody (clone NP4D6) | BioLegend | Cat# 324604; RRID: AB_756042 |
| Biotin anti-human CD66b antibody (clone G10F5) | BioLegend | Cat# 305120; RRID: AB_2566608 |
| Brilliant Violet 421 anti-human CD16 antibody (clone 3G8) | BioLegend | Cat# 302038; RRID: AB_2561578 |
| Brilliant Violet 510 anti-human CD45RA antibody (clone HI100) | BioLegend | Cat# 304142; RRID: AB_2561947 |
| Brilliant Violet 510 anti-human HLA-DR antibody (clone L243) | BioLegend | Cat# 307646; RRID: AB_2561948 |
| Brilliant Violet 510 anti-human CD15 (SSEA-1) antibody (clone W6D3) | BioLegend | Cat# 323028; RRID: AB_2563400 |
| FITC anti-human CD163 antibody (clone GHI/61) | BioLegend | Cat# 333618; RRID: AB_2563094 |
| Brilliant Violet 711 anti-human CD163 antibody (clone GHI/61) | BioLegend | Cat# 333630; RRID: AB_2650972 |
| Brilliant Violet 421 anti-human CD163 antibody (clone GHI/61) | BioLegend | Cat# 333612; RRID:AB_2562463 |
| Brilliant Violet 785 anti-human CD14 antibody (clone M5E2) | BioLegend | Cat# 301840; RRID: AB_2563425 |
| PE/Cy7 anti-human CD14 antibody (clone HCD14) | BioLegend | Cat# 325618; RRID: AB_830691 |
| FITC anti-human CD14 antibody (clone M5E2) | BioLegend | Cat# 301804; RRID: AB_314186) |
| Brilliant Violet 785 anti-human CD3 antibody (clone UCHT1) | BioLegend | Cat# 300472; RRID: AB_2687178 |
| APC anti-human CD34 antibody (clone 561) | BioLegend | Cat# 343608; RRID: AB_2228972 |
| FITC anti-human CD34 antibody (clone 561) | BioLegend | Cat# 343604; RRID: AB_1732005 |
| PerCP/Cyanine5.5 anti-human CD303 (BDCA-2) antibody (clone 201A) | BioLegend | Cat# 354210; RRID: AB_11219604 |
| PerCP/Cyanine5.5 anti-human CD141 (Thrombomodulin) antibody (clone M80) | BioLegend | Cat# 344112; RRID: AB_2561625 |
| PerCP/Cyanine5.5 anti-human CD371 (CLEC12A) antibody (clone 50C1) | BioLegend | Cat# 353612; RRID: AB_2565544 |
| PE anti-human CD370 (CLEC9A/DNGR1) antibody (clone 8F9) | BioLegend | Cat# 353804; RRID: AB_10965546 |
| PE anti-human CD5 antibody (clone L17F12) | BioLegend | Cat# 364014; RRID: AB_2565284 |
| PE anti-human CD116 antibody (clone 4H1) | BioLegend | Cat# 305908; RRID: AB_2085686 |
| PE anti-human CD206 (MMR) antibody (clone 15-2) | BioLegend | Cat# 321106; RRID: AB_571911 |
| PE anti-human CD88 (C5aR) antibody (clone S5/1) | BioLegend | Cat# 344304; RRID: AB_2067175 |
| PE/Dazzle 594 anti-human CD123 antibody (clone 6H6) | BioLegend | Cat# 306034; RRID: AB_2566450 |
| PerCP/Cyanine5.5 anti-human CD123 antibody (clone 6H6) | BioLegend | Cat# 306016; RRID: AB_2264693 |
| FITC anti-human CD1c antibody (clone L161) | BioLegend | Cat# 331518; RRID: AB_2073403 |
| PE/Cy7 anti-human CD1c antibody (clone L161) | BioLegend | Cat# 331516; RRID: AB_2275574 |
| APC anti-human CD272 (BTLA) antibody (clone MIH26) | BioLegend | Cat# 344510; RRID: AB_10613101 |
| APC anti-human CD64 antibody (clone 10.1) | BioLegend | Cat# 305014; RRID: AB_1595428 |
| APC/Cyanine7 anti-human CD45 antibody (clone HI30) | BioLegend | Cat# 304014; RRID: AB_314402 |
| APC/Cy7 Streptavidin | BioLegend | Cat# 405208 |
| Brilliant Violet 785 anti-human CD95 (Fas) antibody (clone DX2) | BioLegend | Cat# 305646; RRID: AB_2629742 |
| Brilliant Violet 711 anti-human CD8 antibody (clone SK1) | BioLegend | Cat# 344734; RRID: AB_2565243 |
| Brilliant Violet 605 anti-human CD45 antibody (clone HI30) | BioLegend | Cat# 304042; RRID: AB_2562106 |
| Brilliant Violet 650 anti-human CD45RO antibody (clone UCHL1) | BioLegend | Cat# 304232; RRID: AB_2563462 |
| Brilliant Violet 421 anti-human CD103 (Integrin E) antibody (clone Ber-ACT8) | BioLegend | Cat# 350214; RRID: AB_2563514 |
| Brilliant Violet 785 anti-human CD103 (Integrin E) antibody (clone Ber-ACT8) | BioLegend | Cat# 350230; RRID: AB_2734364 |
| FITC anti-human CD223 (LAG-3) antibody (clone 7H2C65) | BioLegend | Cat# 369210; RRID: AB_2716129 |
| FITC anti-human CD3 antibody (clone UCHT1) | BioLegend | Cat# 300406; RRID: AB_314060 |
| PE/Cy7 anti-human CD183 (CXCR3) antibody (clone G025H7) | BioLegend | Cat# 353720; RRID: AB_11219383 |
| PE/Dazzle 594 anti-human CD28 antibody (clone CD28.2) | BioLegend | Cat# 302942; RRID: AB_2564235 |
| PE anti-human CD197 (CCR7) antibody (clone G043H7) | BioLegend | Cat# 353204; RRID: AB_10913813 |
| APC/Cyanine7 anti-human CD4 antibody (clone SK3) | BioLegend | Cat# 344616; RRID: AB_2028483 |
| Alexa Fluor® 700 anti-human CD127 (IL-7Ra) antibody (clone A019D5) | BioLegend | Cat# 351344; RRID: AB_2566200 |
| APC anti-human CD279 (PD-1) antibody (clone EH12.2H7) | BioLegend | Cat# 329908; RRID: AB_940475 |
| PE anti-human CD115 antibody (clone 9-4D2-1D4) | BioLegend | Cat# 347310; RRID: AB_2565491 |
| FITC anti-human CD116 antibody (clone 4H1) | BioLegend | Cat# 305906; RRID: AB_2085687 |
| FITC anti-human CD86 antibody (clone BU63) | BioLegend | Cat# 374204; RRID: AB_2721574 |
| Human Axl PE antibody (clone 108724) | R&D | Cat# FAB154P; no RRID |
| PerCP/Cyanine5.5 anti-mouse CD45 antibody (clone 30-F11) | BioLegend | Cat# 103132; RRID: AB_893340 |
| APC-eFluor 780 anti-human HLA-DR antibody (clone LN3) | eBioscience | Cat# 47-9956; RRID:AB_1963603 |
| PE/Cy7 anti-human CD11c antibody (clone Bu15) | Biolegend | Cat# 337216; RRID:AB_2129790 |
| FITC anti-human CD14 antibody (clone 61D3) | eBioscience | Cat# 11-0149; RRID:AB_464951 |
| PerCP-eFluor 710 anti-human CD1c antibody (clone L161) | eBioscience | Cat# 46-0015; RRID:AB_10548936 |
| PE anti-human CD304 antibody (clone AD5-17F6) | Miltenyi | Cat# 130-113-517; RRID:AB_2751124 |
| PE-Cy5 Mouse anti-Human CD1a antibody (clone HI149) | BD | Cat# 555808; RRID:AB_396142 |
| Alexa Fluor® 647 anti-human CD206 (MMR) antibody (clone 15-2) | Biolegend | Cat# 321116; RRID:AB_571881 |
| VioBlue anti-human CD141 antibody (clone AD5-14H12) | Miltenyi | Cat# 130-113-882; RRID:AB_2726374 |
| Purified anti-human CD45 antibody (clone HI30) | BioLegend | Cat# 304002; RRID: AB_314390 |
| Purified anti-human TGF 1-2-3 antibody (clone 1d11) | R&D | Cat# MAB1835; RRID:AB_357931 |
| Purified anti-human TNFα antibody (clone 1825) | R&D | Cat# MAB210; RRID:AB_2240620 |
| Purified anti-human IL-15 antibody (clone 34593) | R&D | Cat# MAB247; RRID:AB_2124578 |
| Purified anti-human Lymphotoxin α antibody (clone 5802) | R&D | Cat# MAB211; RRID:AB_2138622 |
| Purified polyclonal anti-human OX40L antibody | R&D | Cat# AF1236; RRID:AB_354686 |
| Purified polyclonal anti-human IL-33 antibody | R&D | Cat# AF3625; RRID:AB_1151900 |
| Purified anti-human TNFSF8 antibody (clone 116614) | R&D | Cat# MAB1028; RRID:AB_2303710 |
| Purified Mouse IgG1 (clone 11711) | R&D | Cat# MAB002; RRID:AB_357344 |
| Purified polyclonal Goat IgG | R&D | Cat# AB-108-C; RRID:AB_354267 |
| Purified Mouse IgG2b (clone 20116) | R&D | Cat# MAB004; RRID:AB_357346 |
| Bacterial and Virus Strains | ||
| pMX-IRES-GFP | Cell biolabs | Cat# RTV-0133 |
| pMX-huFLT3L-IRES-GFP | This paper | N/A |
| pMX-huGMCSF-IRES-GFP | This paper | N/A |
| Biological Samples | ||
| Leukocyte cones for bulk RNaseq | NHS | N/A |
| Leukocytes cones for functionnal experiments | NHS | N/A |
| umbilical cord-blood for in vitro diffentiation cells | Anthony Nolan Cell Therapy Centre | N/A |
| PBMC from whole blood for blood scRNaseq | Institut Curie Hospital (Paris, France) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Liberase TL | Sigma | Cat# 5401020001 |
| Collagenase D | Sigma | Cat# 11088866001 |
| DNase I | Sigma | Cat# 10104159001 |
| Dispase | Sigma | Cat# D4693 |
| CD34 MicroBead Kit UltraPure, human | Miltenyi | Cat# 130-100-453 |
| Accucheck counting beads | Thermo Fischer | Cat# PCB100 |
| Matrigel® | BD | Cat# 354230 |
| Poly I:C | InvivoGen | Cat# 31852-29-6 |
| R848 | InvivoGen | Cat# 144875-48-9 |
| lipopolysaccharide (LPS) | Sigma-Aldrich | Cat# L2630 |
| Cell Trace Violet (CTV) | Thermo Fischer | Cat# C34557 |
| Naive Pan T cells isolation Kit | Miltenyi | Cat# 130-097-095 |
| Cytostim | Miltenyi | Cat# 130-092-172 |
| Cytofix/ cytoperm Kit | BD Bioscience | Cat# 554714 |
| Critical Commercial Assays | ||
| Human Flt-3 Ligand/FLT3L Quantikine ELISA Kit | R&D | Cat# DFK00 |
| ELISA MAX Deluxe Set Human GM-CSF | Biolegend | Cat# 432004 |
| Human Proinflammatory Chemokine Panel (13-plex) | Biolegend | Cat# 740003 |
| Human Macrophage/Microglia Panel (13-plex) | Biolegend | Cat# 740503 |
| Deposited Data | ||
| RNaseq data raw reads and processed data | This paper | GEO: SuperSeries GSE151095 |
| Experimental Models: Cell Lines | ||
| B16_Control | This paper | N/A |
| B16_human FLT3L | This paper | N/A |
| B16_human GM-CSF | This paper | N/A |
| MS5_FLT3L | Anselmi et al., 2020 | N/A |
| MS5_GM-CSF | Anselmi et al., 2020 | N/A |
| MS5_FS12 | Anselmi et al., 2020 | N/A |
| Experimental Models: Organisms/Strains | ||
| mouse: NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ | The Jakson Laboratory | Stock#005557 |
| Software and Algorithms | ||
| Genomics Suite | Partek | N/A |
| FlowJo V10 | BD | https://www.flowjo.com |
| GraphPad Prism 8 | GraphPad Software | https://www.graphpad.com |
| R4.4 | The R Foundation | https://www.r-project.org |
| Fiji | open source | https://imagej.net/Fiji |
Resource Availability
Lead Contact
Additional information and request for resources and reagents should be directed to and will be made available by the Lead Contact, Pierre Guermonprez (pierre.guermonprez@kcl.ac.uk).
Materials Availability
The reagents generated in this study will be made available on request, but we may require a payment and/or a completed Materials Transfer Agreement if there is potential for commercial application.
Data and Code Availability
The scRNA-seq and the bulk RNA-seq datasets are deposited in the Genome Expression Omnibus under the SuperSeries accession numbers GSE151095.
Experimental Model and Subject Details
Human umbilical cord and adult blood
Human umbilical cord blood units were obtained from Anthony Nolan Cell Therapy Centre (ANCTC). Leukophoretic adult blood (buffy coats or leukocyte cones) were obtained from healthy volunteers through NHS.
Clinical samples
Tumor-invaded lymph nodes (tdLN) and primary tumors were collected from luminal breast cancer submitted to surgical resection at the Institut Curie Hospital (Paris, France), in accordance with institutional ethical guidelines. Patients’ clinical and pathologic characteristics are summarized in Table S2.
Mice
NSG (NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ) mice were bred and maintained in specific pathogen-free animal facility in accordance with institutional KCL guidelines. All procedures involving animals were conducted according to requirements of UK Animals (Scientific Procedures) Act 1986.
Method Details
Human blood and CD34+ progenitors
Peripheral blood mononuclear cells (PMBCs) were obtained by gradient centrifugation using Ficoll-Paque (GE Healthcare). Progenitor cells were enriched using CD34+ microbead isolation kit (Miltenyi).
Tumor cell lines
B16_CTRL, B16_huGM-CSF and B16_huFLT3L were generated by retroviral transduction of B16-F10 (C57BL/6 melanoma cell line) with an empty pMX-IRES-GFP vector or coding for human GM-CSF and human Flt3L respectively. Tumor cell lines were cultured in RPMI 1640 medium (Thermo Fisher) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher), penicillin-streptomycin (Thermo Fisher) and b-mercaptoethanol (Thermo Fisher) (complete RPMI) and maintained at 37°C and 5% CO2.
Metastasis model
Engineered B16-F10 cells were counted and resuspended in RPMI 1640 medium. NSG mice (8-12 weeks, males and females) were injected intravenously with 106 B16-F10 at day 0. 108 human blood PBMCs were injected intravenously at day 7 and 8. Mice were culled at day 9 and lungs were harvested.
Histology
Mouse lungs were fixed with 1% PFA (Alfa Aesar) for 1hr at 4°C, washed and incubated in 34% sucrose solution (Sigma-Aldrich) overnight at 4°C. Lungs were embedded in Cryomatrix (Thermo Fischer) and frozen for cryostat sectioning (9 μm-thick). Sections were permeabilized using 0.5% saponin (Sigma-Aldrich), 2% BSA (Sigma-Aldrich), 1% FBS (Life Technologies) for 30 minutes at room temperature. Sections were labeled overnight at 4°C with mouse anti-human purified CD45 (HI30, Biolegend) followed by incubation for 1hr at room temperature with goat anti-mouse Cy3 (Jackson laboratory). All sections were labeled with Hoechst (Molecular Probes, Thermo Fisher) for nuclei staining 5 minutes at room temperature and mounted with Prolong diamond (Thermo scientific). Slides were imaged using a SP5 (Leica) and analyzed with Fiji software.
Preparation of cell suspensions from lung mouse
Mouse lungs were harvested and transferred to 3ml digestion buffer (Hank’s Balanced Salt Solution (HBSS) with calcium and magnesium (Thermo Fisher) and with 0.1 mg/ml of Liberase TL (Roche) and 0.02μg/ml DNase I (Thermo Fisher). Lungs were dissociated using gentleMACS Octo Dissociator (Miltenyi) and incubated at 37°C for 45 minutes. The cell suspension was passed through a cell strainer (70μm, Corning) and red blood cells were lysed using ACK lysing buffer (Thermo Fischer). The absolute number of cells in the resulting cell suspension was assessed using AccuCheck Counting Beads (Thermo Fisher) on BD FACSCanto II (BD Biosciences).
Stromal cell line maintenance
Mouse bone marrow-derived MS5 stromal cells engineered to express human membrane bound FLT3L alone (MS5_FL) or in combination with SCF and soluble CXCL12 (MS5_FS12) or human GM-CSF alone (MS5_GM) generated as previously described (Anselmi et al., 2020). Stomal cell lines were cultured in IMDM medium supplemented with 10% heat-inactivated FBS, penicillin/streptomycin, 50 μM β-mercaptoethanol (complete IMDM) and maintained at 37°C and 5% CO2.
In vitro differentiation from CD34+ progenitors or blood phagocytes
For in vitro co-culture experiments with stromal cells, MS5_FL, MS5_FS12 and MS5_GM feeders were seeded at 104 cells/well density in a 96-well plate (Thermo Fischer) and maintained overnight at 37°C and 5% CO2.
Total enriched-CD34+ progenitors and flow cytrometry-sorted progenitor subsets (GMDP, CDP and cMoP) were plated on top of stromal cells, supplemented or not with 100ng/ml of recombinant human GM-CSF (Peprotech), for 14 days and 7 days, respectively. Blood phagocytes subsets were flow cytometry-sorted and plated on MS5_GM for 2 days.
For bulk RNA sequencing analysis, total enriched-CD34+ progenitors were plated on top of MS5_FS12 feeders and with or without 100ng/ml of recombinant human GM-CSF (Peprotech). On day 5 and 10 of differentiation, half the medium was replaced with fresh complete IMDM or complete IMDM containing 100ng/ml of recombinant human GM-CSF. Cells were collected at day 15.
All cells were collected with a solution of phosphate buffered saline (PBS) (GIBCO) 5mM EDTA (Thermo Fisher) at 4°C for 10min.
In vivo conversion assay
PBMCs from healthy donors were stained and sorted as described previously. Blood cDC2 (105) and monocytes (3 × 105) were flow cytometry-sorted and injected subcutaneously along with B16_huGM (105) in 200 μL of ice-cold Matrigel® (BD Biosciences). Mice were sacrificed at day 4 by cervical dislocation and Matrigel® plugs were collected. Subcutaneous Matrigel® plugs were recovered, cut in pieces and incubated in HBSS (Life Technologies) 1% FBS, 0.37 U/ml Collagenase D (Roche), 10 μg/ml DNaseI (Roche) and 1 mg/ml Dispase (Sigma-Aldrich) for 30 minutes at 37°C. After digestion, plugs were smashed on a 70 μm strainer (Corning) and cells were collected and resuspended in flow cytometry buffer for flow cytometry analysis.
Flow cytometry analysis
Cells were stained in flow cytometry buffer (PBS 5mM EDTA 1% BSA) according to antibody panels (Key Resources Table) for 30 min. Dead cells were stained using DAPI or Live/Dead Blue staining (Thermo Fischer). Lineage (Lin) included CD3, CD19, CD20, NKp46, CD56, CD203c and CD66b, all conjugated with biotin. Multiparameter analysis was performed on LSRFortessa and Symphony (BD Biosciences) flow cytometers and analyzed using FlowJo software (Tree Star). The flow cytometry-sort was performed on BD FACS Aria II or BD FACS Aria Fusion at the Biomedical Research Centre (BRC) Flow Core Facility (Guy’s and St Thomas’ NHS Foundation Trust and King’s College London).
Unsupervised clustering of flow cytometry data (Figure 1F) was performed using Self-Organizing Map clustering algorithm FlowSOM (Cytofkit R package). Data was then extracted as FCS files and further analyzed in FlowJo software.
Bulk RNA sequencing
For bulk sequencing of in vitro differentiated subsets, up to 100 flow cytometry-sorted cells from three individual donors were collected directly in Lysis buffer (Takara Clontech, Cat# 635015) containing RNase inhibitors. RNaseq libraries were prepared on the contactless liquid handling system Labcyte Echo 525 (Labcyte Inc). In brief, ERCC was added to each sample and first strand full length cDNA was generated with a modified protocol of the SMARTseq v4 Ultra Low Input RNA Kit (Takara Clontech, Cat# 634891) using poly dT primers and a template switching oligo. Full length cDNA was amplified using SeqAmp DNA Polymerase (Takara Clontech, Cat# 638509). 12 ng of amplified cDNA from each sample was used to generate non-stranded RNA libraries using a modified protocol of the Ovation Ultralow System V2 1-96 kit (NuGEN, Cat# 0347-A01). In brief, amplified cDNA was fragmented through sonication on Covaris E220 (Covaris Inc), repaired and polished followed by ligation of indexed adapters. Adaptor ligated cDNA were pooled before final amplification to add flow cell primers. Libraries were sequenced on HiSeq2500 (Illumina Cambridge) for 100 cycles PE in Rapid mode.
Bulk RNA sequencing data processing
The raw sequencing data was initially processed using open source, web-based platform Galaxy (version 18.05.rc1) (https://usegalaxy.org). Reads were filtered for quality with more than 80% of the sequence having quality score > 33 using FastQC tool. Mapping against reference genome was performed with Hisat2 to the hg38 human genome. Adaptor sequences were detected automatically with TrimGalore!. Reads under 20bp were discarded. All processed sequencing files were imported in Partek® Genomics Suite software®, version 7.0©; 2017 (PGS), where they were processed further.
Primary data analysis and visualization
mRNA was quantified using PGS built in RNA-seq workflow. Normalization method used was Reads Per Kilobase per Million mapped reads (RPKM) and mRNA was quantified against RefSeq Transcripts 2018-11-20 database. Hierarchal clustering on average expression within the group was performed on all identified protein coding genes (19791 genes). Based on the CD14-CD1c+CD206+/− two blue cluster similarity, they were both considered as cDC2-like cells and their datasets were merged for further analysis. The same was done for the two macrophage-like cells (gray and brown). Differentially expressed genes (Fold-Change ≤ −2 or ≥ 2 and p value < 0.05) were determined using one-way ANOVA in all pairwise comparisons with three donors grouped and visualized as Volcano plots in PGS. Individual samples were visualized via principal component analysis (PCA) using 500 most variable genes, which were determined based on median absolute deviation (MAD). The expression patterns of selected gene lists were displayed in the form of heatmaps, where rows and/or columns were ordered based on hierarchical clustering using Euclidean distance and average linkage in Morpheus (Broad Institute; https://software.broadinstitute.org/morpheus/). A few gene names are depicted next to the heatmap.
Single-cell RNA sequencing
PBMCs from three healthy donors or human CD45+ cells isolated from metastatic mouse lungs were isolated as previously described. Sorted cells from each donor were pooled together and CD1c+ and CD14+ cells were mixed at 80:20 ratio. 3 × 103 cells from the resulting cell suspension were partitioned into an emulsion of nanoliter-sized droplets using a 10X Genomics Chromium Single Cell Controller and RNA sequencing libraries were constructed using the Chromium Single Cell 3′ Library & Gel Bead Kit v2 (10X Genomics, Cat# PN-120237). Briefly, droplets containing individual cells, reverse transcription reagents and a gel bead were loaded with poly(dT) primers that include a 16 base cell barcode and a 10 base unique molecular index (UMI). Reverse transcription reactions were engaged to generate barcoded full-length cDNA followed by the disruption of emulsions using the recovery agent and cDNA clean up with DynaBeads MyOne Silane Beads (Thermo Fisher Scientific, Cat# 37002D). Bulk cDNA was amplified, and indexed sequencing libraries were constructed using the reagents from the Chromium Single Cell 3′ v2 Reagent Kit. Libraries were sequenced on NovaSeq 6000 Sequencing System (Illumina Cambridge).
Single-cell RNaseq data processing and analysis
Cell Ranger (version 2.1.1) (from 10x genomics) was used to process Chromium single cell 3′ v2 RNA-seq output files. First, we generated fastq files for the Read1 for cell barcode and UMI and Read2 for transcript applying cellranger mkfastq (with default parameters). Second, we aligned the Read2 to the human reference genome GRCh38 using STAR (version 2.5.1) with cell ranger count (with default parameters) (https://support.10xgenomics.com/single-cell-gene-expression/software/pipelines/latest/using/mkfastq). Further analysis was performed using Seurat package (version 2.3.4) in R (version 3.4.0) (Butler et al., 2018). Before performed analysis, we applied the following filtering step: only genes expressed in 3 or more cells have been preserved and cells with less 200 unique genes and more than 4000 unique expressed genes were discarded (as they are potentially cells doublets). After filtering step, we used an expression matrix resulting in 14933 genes across 1622 cells (among 1625 cells) for the rest of the analysis. The matrix was normalized using genes expression values for each cell were divided by the total number of transcripts and multiplied by median of UMI counts. Then, these values were natural log-transformed before downstream analysis. For dimensionality reduction analysis, we first identified 3288 genes as highly variable genes across the single cells (cutoff value for dispersion = 0.5; cutoff value for average expression = 0). PCA performed using the variable genes as input and determined 10 PCs as significant PCs. These principal components were used as input for t-Distributed Stochastic Neighbor Embedding (tSNE) (van der Maaten, 2008). We used the shared nearest neighbor (SNN) modularity optimization-based clustering algorithm from the Seurat package (FindClusters function with default parameters) to identify the clusters of cells, following by Clustree analysis (clustree R package, version 0.2.2) by changing the resolution parameters from 0 to 2. Finally, we kept a resolution parameter at 0.8 and defined distinct 9 clusters. After controlling expression of some quality control genes, we excluded clusters E, F, G, H and I as contamination. Finally, we identified 4 relevant clusters. We identified cell specific marker by comparing cells in a specific cluster with cells in all other clusters using FindAllMarkers from Seurat package (MAST; logFC threshold = 0.5; only positive markers). Heatmap, feature plots and violin plots were performed using Seurat package.
Signature expression analysis
Single-cell RNA sequencing matrix for clusters A, B, C and D was created with gene signatures from Villani et al. (2017). Signatures were defined as mean expression of discriminative markers for cDC2s and DC3s among lin-CD14- cells (cDC2 enriched and DC3 enriched, respectively) and of discriminative markers for CD14+ monocytes within lin-CD14+/− and/or CD16+/− monocytes (CD14+ mono enriched). The average expression of signature genes for each cell was calculated and plotted as a violin plot using R package ggplot2 (version 3.1.0).
Gene set enrichment analysis
To statistically evaluate the enrichment of previously reported gene signatures (Gene Sets) (Hombrink et al., 2016; Kumar et al., 2017; Savas et al., 2018; Villani et al., 2017) in our dataset, we used pairwise comparisons using the gene set enrichment analysis (GSEA) (Subramanian et al., 2005) method from the Massachussets Institute of Technology (https://www.broadinstitute.org/gsea). GSEA tests the relative position of a collection of genes (Gene Set) within an independent, ranked dataset (GeneList). Statistical analysis was performed by evaluation of nominal p value and false discovery rate (q value) based on 1,000 random permutations. Results were considered significant when the p value was below 0.05 and when the q value was below 0.25 (false discovery rate below 25%) accordingly to the recommendation from the software developers. For each pairwise comparison, the GSEA output can be represented as a bar code where each bar corresponds to the projection of one Probe Sets of the Gene Sets on the list of all the Probe Sets of the gene chips ranked from those having a high signal in one cell population (represented in red) to those having a high signal in the other cell population (represented in blue). Each bar code can be characterized by two parameters. The normalized enrichment score (NES) represents the number and differential expression intensity of the genes enriched in the corresponding cell subset. The NES is positive if the Gene Set is enriched in the first cell population and negative if it is enriched in the second cell population. The false discovery rate (FDR) statistical value (q) represents the likelihood that the enrichment of the Gene Set represents a false positive finding (for example if q = 0.25, 25% of the Gene Sets found enriched can be false positives). An absolute value of the NES below or around 1 means no enrichment as confirmed with associated q-values above 0.25.
To simultaneously visualize pairwise comparisons of transcriptomes from cord blood-derived DC2s (light and dark blue) DC3s (orange) and macrophages (gray and brown), the BubbleMap module of BubbleGum (Spinelli et al., 2015) was used. Results were considered significant when the p value was below 0.05 and the FDR (false discovery rate, q) value was below 0.25. The BubbleGum was performed using previously published gene signatures of pairwise comparison between DC2s and DC3s and DC3s and CD14+ monocytes (Villani et al., 2017).
Human tissue processing and cell suspension
Tumor-draining lymph nodes (tdLN) and primary tumors were collected in CO2 independent medium (GIBCO; Cat# 18045-054) within few hours after the primary surgery. Tissue were cut into small fragments and submitted to enzymatic digestion using 0.1 mg/ml of Liberase TL (Roche) and 0.1 mg/ml of DNase (Roche) for 30 min. Cells were filtered on 40-μm cell strainer (BD), washed using CO2 independent medium (GIBCO; Cat# 18045-054) containing 0.4g/ml of human albumin and resuspended for cell counting.
Cell sorting of myeloid subsets from patients’ tdLN for RNA sequencing
After tissue processing, cells obtained from tdLN were submitted to myeloid cells enrichment accordingly to (Durand and Segura, 2016) prior flow cytometry-sorting. In brief, T and B lymphocytes, NK cells, erythrocytes and myelomonocytic cells were depleted using monoclonal antibodies against: CD3, CD19, CD56, CD235a and CD15, respectively. Subsequently, cell suspensions were stained for 30 min with antibody-conjugated as the following: HLA-DR, CD11c, CD14, CD1c, CD304, CD1a, CD206, CD141. Around 1,000 cells of each DC subset were sorted by flow cytometry using BD FACS ARIA II cell sorter, (purity > 98%). Cells were centrifuged and lysed with TCL buffer (QIAGEN) containing 1% of beta-mercaptoethanol before storage at −80°C. RNA were extracted and isolated using the Single Cell RNA purification kit (Norgen, Cat#51800) according to the manufacturer’s instructions and the RNA integrity number was evaluated with an Agilent RNA 6000 pico kit.
LEGENDplex™ assay
PBMCs and in vitro generated cells were stained and sorted as described previously. In total 3 × 105 of blood cell subsets (cDC2, DC3 and Mono) or 105 in vitro generated cells (cDC2-, DC3 and Macro-like) were flow cytometry-sorted and cultured with TLR agonists cocktail containing 25 μg/ml Poly I:C (InvivoGen, Cat# 31852-29-6), 1 μg/ml R848 (InvivoGen, Cat# 144875-48-9) and 10ng/ml LPS (Sigma-Aldrich, Cat# L2630) for 16 h. Culturing supernatants were collected and stored at −20°C until the LEGENDplex™ assay execution day. LEGENDplex Human Macrophage/Microglia Panel (13-plex) with V-bottom Plate (Biolegend Cat# 740503) and Human Proinflammatory Chemokine Panel (13-plex) with V-bottom Plate (Biolegend Cat# 740003) was used according to manufacturer’s instructions. In short, samples and standards were thawed and plated with capture beads and incubated for 2 h. Plate was then washed, and Detection Antibodies were added. After 1 h incubation SA-PE was added and incubated for 30 min. Samples were acquired on BD FACSCanto II. Samples were analyzed using LEGENDplex™ Data Analysis Software.
In vitro T cell assay
PBMCs and in vitro generated cells were stained and flow cytometry-sorted as described previously. 104 mononuclear phagocytes were cultured with TLR agonists cocktail containing 25 μg/ml PolyI:C, 1 μg/ml R848 and 10 ng/ml LPS for 16 h. T cells were isolated from fresh or frozen PBMCs using Naive Pan T Cell Isolation Kit (Miltenyi, Cat# 130-097-095) and 105 cells were plated on top of mononuclear phagocytes in presence of CytoStim (Miltenyi, Cat# 130-092-172), according to manufacturer’s instructions. For mixed leukocyte reaction (MLR) experiments, isolated Naive Pan T cells were labeled with Cell Tracer Violet (CTV) (Thermo Fischer, Cat# C34557) as per manufacturer’s instructions and cultured with flow cytometry-sorted DCs without CytoStim. At day 5, cells were collected and stained for extracellular and intracellular marker expression and analyzed using BD LSRFortessa. For intracellular staining, fixation and permeabilization were performed using BD Cytofix/Cytoperm solution (BD Biosciences, Cat# 554714) according to manufacturer’s instructions.
Quantification and Statistical Analysis
Statistical analysis was performed using Prism 8.3 (GraphPad Software Inc., USA). When two experimental groups were compared, non-parametric Mann-Whitney test was used. When three or more groups were compared, statistically significant differences between means were determined using the one-way or two-way analysis of variance (ANOVA) method. A p value of less than 0.05 was considered as significant.
Acknowledgments
P.G. is a CNRS investigator. J.H. is an INSERM investigator. We thank the Anthony Nolan Cord Blood Bank and Cell Therapy Centre for providing the cord blood units used in this study, the flow core and NGS platform from Institut Curie, and Daniel Chung for technical help. This research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy's and St Thomas' NHS Foundation Trust and King’s College London and/or the NIHR Clinical Research Facility. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health. The research was supported by the BBSRC, NC3RS, CRUK, Rosetrees Trust, CRUK King’s Health Partners Cancer Centre in United Kingdom. The research was supported by IDEX (ANR-18-IDEX-0001) and INCA (PL-BIO-ICR1), Institut Curie, Institut National de la Santé et de la Recherche Médicale, Labex DCBIOL (ANR-10-IDEX-0001-02 PSL), ANR-11-LABX0043, SIRIC INCa-DGOS-INSERM_12554, in France and the Programa de Apoyo a Centros con Financiamiento Basal (AFB 170004), and FONDECYT 1171703 in Chile.
Author Contributions
Conceptualization, P.G., P.B., G.A., K.V., and J.H.; Software, K.V., W.R., and Y.M.K.; Methodology, G.A., P.B., K.V., E.P., S.H., and A.L.; Formal Analysis, G.A., P.B., K.V., W.R., J.H., and P.G.; Investigation, G.A., K.V., P.B., S.H., R.N.R., K.W., and A.S.; Resources, E.P., J.H., and P.G.; Writing, P.G., G.A., K.V., P.B., and J.H.; Visualization, G.A., K.V., and P.B.; Supervision, A.L., E.P., J.H., and P.G.; Funding acquisition, A.L., E.P., J.H., and P.G.
Declaration of Interests
The authors declare no competing interests.
Published: June 30, 2020
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.immuni.2020.06.002.
Supplemental Information
References
- Acosta-Rodriguez E.V., Napolitani G., Lanzavecchia A., Sallusto F. Interleukins 1beta and 6 but not transforming growth factor-beta are essential for the differentiation of interleukin 17-producing human T helper cells. Nat. Immunol. 2007;8:942–949. doi: 10.1038/ni1496. [DOI] [PubMed] [Google Scholar]
- Anselmi G., Vaivode K., Dutertre C.A., Bourdely P., Missolo-Koussou Y., Newell E., Hickman O., Wood K., Saxena A., Helft J. Engineered niches support the development of human dendritic cells in humanized mice. Nat. Commun. 2020;11:2054. doi: 10.1038/s41467-020-15937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachem A., Güttler S., Hartung E., Ebstein F., Schaefer M., Tannert A., Salama A., Movassaghi K., Opitz C., Mages H.W. Superior antigen cross-presentation and XCR1 expression define human CD11c+CD141+ cells as homologues of mouse CD8+ dendritic cells. J. Exp. Med. 2010;207:1273–1281. doi: 10.1084/jem.20100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakdash G., Buschow S.I., Gorris M.A., Halilovic A., Hato S.V., Sköld A.E., Schreibelt G., Sittig S.P., Torensma R., Duiveman-de Boer T. Expansion of a BDCA1+CD14+ Myeloid Cell Population in Melanoma Patients May Attenuate the Efficacy of Dendritic Cell Vaccines. Cancer Res. 2016;76:4332–4346. doi: 10.1158/0008-5472.CAN-15-1695. [DOI] [PubMed] [Google Scholar]
- Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- Binnewies M., Mujal A.M., Pollack J.L., Combes A.J., Hardison E.A., Barry K.C., Tsui J., Ruhland M.K., Kersten K., Abushawish M.A. Unleashing Type-2 Dendritic Cells to Drive Protective Antitumor CD4(+) T Cell Immunity. Cell. 2019;177:556–571.e16. doi: 10.1016/j.cell.2019.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Lee J., Zhou Y.J., Schreiber J.J., Keler T., Puhr S., Anandasabapathy N., Schlesinger S., Caskey M., Liu K., Nussenzweig M.C. Circulating precursors of human CD1c+ and CD141+ dendritic cells. J. Exp. Med. 2015;212:401–413. doi: 10.1084/jem.20141441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breton G., Zheng S., Valieris R., Tojal da Silva I., Satija R., Nussenzweig M.C. Human dendritic cells (DCs) are derived from distinct circulating precursors that are precommitted to become CD1c+ or CD141+ DCs. J. Exp. Med. 2016;213:2861–2870. doi: 10.1084/jem.20161135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briseño C.G., Haldar M., Kretzer N.M., Wu X., Theisen D.J., Kc W., Durai V., Grajales-Reyes G.E., Iwata A., Bagadia P. Distinct Transcriptional Programs Control Cross-Priming in Classical and Monocyte-Derived Dendritic Cells. Cell Rep. 2016;15:2462–2474. doi: 10.1016/j.celrep.2016.05.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler A., Hoffman P., Smibert P., Papalexi E., Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Contreras V., Feuillet V., Dutertre C.A., Ventre E., Vu Manh T.P., Baranek T., Storset A.K., Marvel J. The XC chemokine receptor 1 is a conserved selective marker of mammalian cells homologous to mouse CD8alpha+ dendritic cells. J. Exp. Med. 2010;207:1283–1292. doi: 10.1084/jem.20100223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand M., Segura E. Dendritic Cell Subset Purification from Human Tonsils and Lymph Nodes. Methods Mol. Biol. 2016;1423:89–99. doi: 10.1007/978-1-4939-3606-9_6. [DOI] [PubMed] [Google Scholar]
- Dutertre C.A., Becht E., Irac S.E., Khalilnezhad A., Narang V., Khalilnezhad S., Ng P.Y., van den Hoogen L.L., Leong J.Y., Lee B. Single-Cell Analysis of Human Mononuclear Phagocytes Reveals Subset-Defining Markers and Identifies Circulating Inflammatory Dendritic Cells. Immunity. 2019;51:573–589.e8. doi: 10.1016/j.immuni.2019.08.008. [DOI] [PubMed] [Google Scholar]
- Fogg D.K., Sibon C., Miled C., Jung S., Aucouturier P., Littman D.R., Cumano A., Geissmann F. A clonogenic bone marrow progenitor specific for macrophages and dendritic cells. Science. 2006;311:83–87. doi: 10.1126/science.1117729. [DOI] [PubMed] [Google Scholar]
- Goudot C., Coillard A., Villani A.C., Gueguen P., Cros A., Sarkizova S., Tang-Huau T.L., Bohec M., Baulande S., Hacohen N. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. 2017;47:582–596.e6. doi: 10.1016/j.immuni.2017.08.016. [DOI] [PubMed] [Google Scholar]
- Granot T., Senda T., Carpenter D.J., Matsuoka N., Weiner J., Gordon C.L., Miron M., Kumar B.V., Griesemer A., Ho S.H. Dendritic Cells Display Subset and Tissue-Specific Maturation Dynamics over Human Life. Immunity. 2017;46:504–515. doi: 10.1016/j.immuni.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Helft J., Claser C., Deroubaix S., Karanje H., Gazumyan A., Darasse-Jèze G., Telerman S.B., Breton G., Schreiber H.A. Inflammatory Flt3l is essential to mobilize dendritic cells and for T cell responses during Plasmodium infection. Nat. Med. 2013;19:730–738. doi: 10.1038/nm.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guermonprez P., Gerber-Ferder Y., Vaivode K., Bourdely P., Helft J. Origin and development of classical dendritic cells. Int. Rev. Cell Mol. Biol. 2019;349:1–54. doi: 10.1016/bs.ircmb.2019.08.002. [DOI] [PubMed] [Google Scholar]
- Heidkamp G.F., Sander J., Lehmann C.H.K., Heger L., Eissing N., Baranska A., Lühr J.J., Hoffmann A., Reimer K.C., Lux A. Human lymphoid organ dendritic cell identity is predominantly dictated by ontogeny, not tissue microenvironment. Sci. Immunol. 2016;1 doi: 10.1126/sciimmunol.aai7677. eaai7677. [DOI] [PubMed] [Google Scholar]
- Helft J., Anjos-Afonso F., van der Veen A.G., Chakravarty P., Bonnet D., Reis e Sousa C. Dendritic Cell Lineage Potential in Human Early Hematopoietic Progenitors. Cell Rep. 2017;20:529–537. doi: 10.1016/j.celrep.2017.06.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hombrink P., Helbig C., Backer R.A., Piet B., Oja A.E., Stark R., Brasser G., Jongejan A., Jonkers R.E., Nota B. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 2016;17:1467–1478. doi: 10.1038/ni.3589. [DOI] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J., Dunbar P.R., Wadley R.B., Jeet V. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 2010;207:1247–1260. doi: 10.1084/jem.20092140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura S., Onai N., Miya F., Sato T., Tsunoda T., Kurabayashi K., Yotsumoto S., Kuroda S., Takenaka K., Akashi K., Ohteki T. Identification of a Human Clonogenic Progenitor with Strict Monocyte Differentiation Potential: A Counterpart of Mouse cMoPs. Immunity. 2017;46:835–848 e4. doi: 10.1016/j.immuni.2017.04.019. [DOI] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.H., Lerner H. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 2017;20:2921–2934. doi: 10.1016/j.celrep.2017.08.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Kobayashi S., Leader A., Amir E.D., Elefant N., Bigenwald C., Remark R., Sweeney R., Becker C.D., Levine J.H. Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell. 2017;169:750–765.e17. doi: 10.1016/j.cell.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal Rojas I.M., Mok W.H., Pearson F.E., Minoda Y., Kenna T.J., Barnard R.T., Radford K.J. Human Blood CD1c+ Dendritic Cells Promote Th1 and Th17 Effector Function in Memory CD4+ T Cells. Front. Immunol. 2017;8:971. doi: 10.3389/fimmu.2017.00971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Breton G., Aljoufi A., Zhou Y.J., Puhr S., Nussenzweig M.C., Liu K. Clonal analysis of human dendritic cell progenitor using a stromal cell culture. J. Immunol. Methods. 2015;425:21–26. doi: 10.1016/j.jim.2015.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Breton G., Oliveira T.Y., Zhou Y.J., Aljoufi A., Puhr S., Cameron M.J., Sékaly R.P., Nussenzweig M.C., Liu K. Restricted dendritic cell and monocyte progenitors in human cord blood and bone marrow. J. Exp. Med. 2015;212:385–399. doi: 10.1084/jem.20141442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Zhou Y.J., Ma W., Zhang W., Aljoufi A., Luh T., Lucero K., Liang D., Thomsen M., Bhagat G. Lineage specification of human dendritic cells is marked by IRF8 expression in hematopoietic stem cells and multipotent progenitors. Nat. Immunol. 2017;18:877–888. doi: 10.1038/ni.3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mach N., Gillessen S., Wilson S.B., Sheehan C., Mihm M., Dranoff G. Differences in dendritic cells stimulated in vivo by tumors engineered to secrete granulocyte-macrophage colony-stimulating factor or Flt3-ligand. Cancer Res. 2000;60:3239–3246. [PubMed] [Google Scholar]
- Mani V., Bromley S.K., Äijö T., Mora-Buch R., Carrizosa E., Warner R.D., Hamze M., Sen D.R., Chasse A.Y., Lorant A. Migratory DCs activate TGF-β to precondition naïve CD8+ T cells for tissue-resident memory fate. Science. 2019;366:eaav5728. doi: 10.1126/science.aav5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna H.J., Stocking K.L., Miller R.E., Brasel K., De Smedt T., Maraskovsky E., Maliszewski C.R., Lynch D.H., Smith J., Pulendran B. Mice lacking flt3 ligand have deficient hematopoiesis affecting hematopoietic progenitor cells, dendritic cells, and natural killer cells. Blood. 2000;95:3489–3497. [PubMed] [Google Scholar]
- Menezes S., Melandri D., Anselmi G., Perchet T., Loschko J., Dubrot J., Patel R., Gautier E.L., Hugues S., Longhi M.P. The Heterogeneity of Ly6Chi Monocytes Controls Their Differentiation into iNOS+ Macrophages or Monocyte-Derived Dendritic Cells. Immunity. 2016;45:1205–1218. doi: 10.1016/j.immuni.2016.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., Mortha A. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 2013;31:563–604. doi: 10.1146/annurev-immunol-020711-074950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michea P., Noël F., Zakine E., Czerwinska U., Sirven P., Abouzid O., Goudot C., Scholer-Dahirel A., Vincent-Salomon A., Reyal F. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat. Immunol. 2018;19:885–897. doi: 10.1038/s41590-018-0145-8. [DOI] [PubMed] [Google Scholar]
- Mueller S.N., Mackay L.K. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 2016;16:79–89. doi: 10.1038/nri.2015.3. [DOI] [PubMed] [Google Scholar]
- Murphy T.L., Grajales-Reyes G.E., Wu X., Tussiwand R., Briseño C.G., Iwata A., Kretzer N.M., Durai V., Murphy K.M. Transcriptional Control of Dendritic Cell Development. Annu. Rev. Immunol. 2016;34:93–119. doi: 10.1146/annurev-immunol-032713-120204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naik S.H., Sathe P., Park H.Y., Metcalf D., Proietto A.I., Dakic A., Carotta S., O’Keeffe M., Bahlo M., Papenfuss A. Development of plasmacytoid and conventional dendritic cell subtypes from single precursor cells derived in vitro and in vivo. Nat. Immunol. 2007;8:1217–1226. doi: 10.1038/ni1522. [DOI] [PubMed] [Google Scholar]
- Napolitani G., Rinaldi A., Bertoni F., Sallusto F., Lanzavecchia A. Selected Toll-like receptor agonist combinations synergistically trigger a T helper type 1-polarizing program in dendritic cells. Nat. Immunol. 2005;6:769–776. doi: 10.1038/ni1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nizzoli G., Krietsch J., Weick A., Steinfelder S., Facciotti F., Gruarin P., Bianco A., Steckel B., Moro M., Crosti M. Human CD1c+ dendritic cells secrete high levels of IL-12 and potently prime cytotoxic T-cell responses. Blood. 2013;122:932–942. doi: 10.1182/blood-2013-04-495424. [DOI] [PubMed] [Google Scholar]
- Onai N., Obata-Onai A., Schmid M.A., Manz M.G. Flt3 in regulation of type I interferon-producing cell and dendritic cell development. Ann. N Y Acad. Sci. 2007;1106:253–261. doi: 10.1196/annals.1392.015. [DOI] [PubMed] [Google Scholar]
- Palucka K., Banchereau J. Dendritic-cell-based therapeutic cancer vaccines. Immunity. 2013;39:38–48. doi: 10.1016/j.immuni.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul F., Arkin Y., Giladi A., Jaitin D.A., Kenigsberg E., Keren-Shaul H., Winter D., Lara-Astiaso D., Gury M., Weiner A. Transcriptional Heterogeneity and Lineage Commitment in Myeloid Progenitors. Cell. 2015;163:1663–1677. doi: 10.1016/j.cell.2015.11.013. [DOI] [PubMed] [Google Scholar]
- Pulendran B., Smith J.L., Jenkins M., Schoenborn M., Maraskovsky E., Maliszewski C.R. Prevention of peripheral tolerance by a dendritic cell growth factor: flt3 ligand as an adjuvant. J. Exp. Med. 1998;188:2075–2082. doi: 10.1084/jem.188.11.2075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rihs S., Walker C., Virchow J.C., Jr., Boer C., Kroegel C., Giri S.N., Braun R.K. Differential expression of alpha E beta 7 integrins on bronchoalveolar lavage T lymphocyte subsets: regulation by alpha 4 beta 1-integrin crosslinking and TGF-beta. Am. J. Respir. Cell Mol. Biol. 1996;15:600–610. doi: 10.1165/ajrcmb.15.5.8918367. [DOI] [PubMed] [Google Scholar]
- Ruffell D., Mourkioti F., Gambardella A., Kirstetter P., Lopez R.G., Rosenthal N., Nerlov C. A CREB-C/EBPbeta cascade induces M2 macrophage-specific gene expression and promotes muscle injury repair. Proc. Natl. Acad. Sci. USA. 2009;106:17475–17480. doi: 10.1073/pnas.0908641106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lanzavecchia A. Efficient presentation of soluble antigen by cultured human dendritic cells is maintained by granulocyte/macrophage colony-stimulating factor plus interleukin 4 and downregulated by tumor necrosis factor alpha. J. Exp. Med. 1994;179:1109–1118. doi: 10.1084/jem.179.4.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander J., Schmidt S.V., Cirovic B., McGovern N., Papantonopoulou O., Hardt A.L., Aschenbrenner A.C., Kreer C., Quast T., Xu A.M. Cellular Differentiation of Human Monocytes Is Regulated by Time-Dependent Interleukin-4 Signaling and the Transcriptional Regulator NCOR2. Immunity. 2017;47:1051–1066.e12. doi: 10.1016/j.immuni.2017.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savas P., Virassamy B., Ye C., Salim A., Mintoff C.P., Caramia F., Salgado R., Byrne D.J., Teo Z.L., Dushyanthen S., Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 2018;24:986–993. doi: 10.1038/s41591-018-0078-7. [DOI] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W., See P., Shin A., Wasan P.S. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 2013;38:970–983. doi: 10.1016/j.immuni.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- See P., Dutertre C.A., Chen J., Günther P., McGovern N., Irac S.E., Gunawan M., Beyer M., Händler K., Duan K. Mapping the human DC lineage through the integration of high-dimensional techniques. Science. 2017;356 doi: 10.1126/science.aag3009. eaag3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Valladeau-Guilemond J., Donnadieu M.H., Sastre-Garau X., Soumelis V., Amigorena S. Characterization of resident and migratory dendritic cells in human lymph nodes. J. Exp. Med. 2012;209:653–660. doi: 10.1084/jem.20111457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Touzot M., Bohineust A., Cappuccio A., Chiocchia G., Hosmalin A., Dalod M., Soumelis V., Amigorena S. Human inflammatory dendritic cells induce Th17 cell differentiation. Immunity. 2013;38:336–348. doi: 10.1016/j.immuni.2012.10.018. [DOI] [PubMed] [Google Scholar]
- Spinelli L., Carpentier S., Montañana Sanchis F., Dalod M., Vu Manh T.P. BubbleGUM: automatic extraction of phenotype molecular signatures and comprehensive visualization of multiple Gene Set Enrichment Analyses. BMC Genomics. 2015;16:814. doi: 10.1186/s12864-015-2012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M., Hawiger D., Nussenzweig M.C. Tolerogenic dendritic cells. Annu. Rev. Immunol. 2003;21:685–711. doi: 10.1146/annurev.immunol.21.120601.141040. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Maaten J. Visualizing Data Using t-SNE. J. Mach. Learn. Res. 2008;9:2579–2605. [Google Scholar]
- Villani A.C., Satija R., Reynolds G., Sarkizova S., Shekhar K., Fletcher J., Griesbeck M., Butler A., Zheng S., Lazo S. Single-cell RNA-seq reveals new types of human blood dendritic cells, monocytes, and progenitors. Science. 2017;356 doi: 10.1126/science.aah4573. eaah4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Milne K., Derocher H., Webb J.R., Nelson B.H., Watson P.H. CD103 and Intratumoral Immune Response in Breast Cancer. Clin. Cancer Res. 2016;22:6290–6297. doi: 10.1158/1078-0432.CCR-16-0732. [DOI] [PubMed] [Google Scholar]
- Waskow C., Liu K., Darrasse-Jèze G., Guermonprez P., Ginhoux F., Merad M., Shengelia T., Yao K., Nussenzweig M. The receptor tyrosine kinase Flt3 is required for dendritic cell development in peripheral lymphoid tissues. Nat. Immunol. 2008;9:676–683. doi: 10.1038/ni.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollenberg A., Kraft S., Hanau D., Bieber T. Immunomorphological and ultrastructural characterization of Langerhans cells and a novel, inflammatory dendritic epidermal cell (IDEC) population in lesional skin of atopic eczema. J. Invest. Dermatol. 1996;106:446–453. doi: 10.1111/1523-1747.ep12343596. [DOI] [PubMed] [Google Scholar]
- Yin X., Yu H., Jin X., Li J., Guo H., Shi Q., Yin Z., Xu Y., Wang X., Liu R. Human Blood CD1c+ Dendritic Cells Encompass CD5high and CD5low Subsets That Differ Significantly in Phenotype, Gene Expression, and Functions. J. Immunol. 2017;198:1553–1564. doi: 10.4049/jimmunol.1600193. [DOI] [PubMed] [Google Scholar]
- Yu C.I., Becker C., Wang Y., Marches F., Helft J., Leboeuf M., Anguiano E., Pourpe S., Goller K., Pascual V. Human CD1c+ dendritic cells drive the differentiation of CD103+ CD8+ mucosal effector T cells via the cytokine TGF-β. Immunity. 2013;38:818–830. doi: 10.1016/j.immuni.2013.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaba L.C., Krueger J.G., Lowes M.A. Resident and “inflammatory” dendritic cells in human skin. J. Invest. Dermatol. 2009;129:302–308. doi: 10.1038/jid.2008.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNA-seq and the bulk RNA-seq datasets are deposited in the Genome Expression Omnibus under the SuperSeries accession numbers GSE151095.