Abstract
Background
Exosomes are small membrane vesicles that are secreted by most cell types. Circular RNAs (circRNAs) have recently been identified in exosomes, and exosomal circRNAs exert important biological activities in human cancers, including oral squamous cell carcinoma (OSCC). The purpose of this study was to investigate whether circRNA GDP dissociation inhibitor 2 (circGDI2) was transferred by exosomes and how exosomal circGDI2 regulated OSCC cell malignant behaviors.
Methods
The levels of circGDI2, miR-424-5p and suppressor of cancer cell invasion (SCAI) were detected by quantitative real-time polymerase chain reaction (qRT-PCR) or Western blot. Cell proliferation was evaluated using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay. Transwell assays were performed to detect cell migration and invasion. Measurement of glucose consumption and lactate production were conducted using a corresponding assay kit. Targeted correlations among circGDI2, miR-424-5p and SCAI were confirmed by dual-luciferase reporter assays. Xenograft assays were used to observe the role of circGDI2 in tumor growth in vivo.
Results
Our data indicated that circGDI2 was down-regulated in OSCC, and it could be transferred by the exosomes in OSCC cells. The up-regulation of exosomal circGDI2 weakened OSCC cell proliferation, migration, invasion and glycolysis. CircGDI2 functioned as a molecular sponge of miR-424-5p, and SCAI was a direct target of miR-424-5p. MiR-424-5p mediated the repression of exosomal circGDI2 overexpression on OSCC cell malignant behaviors, and miR-424-5p silencing mitigated OSCC cell progression by up-regulating SCAI. Moreover, exosomal circGDI2 regulated SCAI expression through sponging miR-424-5p. Additionally, the overexpression of exosomal circGDI2 inhibited tumor growth in vivo.
Conclusion
The present study had led to the identification of exosomal circGDI2 that regulated OSCC cell malignant behaviors through targeting the miR-424-5p/SCAI axis, highlighting circGDI2 as a novel exosome-based cancer biomarker and therapeutic agent for OSCC treatment.
Keywords: OSCC, exosome, circGDI2, miR-424-5p, SCAI
Introduction
Oral squamous cell carcinoma (OSCC) is one of the most common malignancies, resulting in millions of deaths all over the world.1 Traditional therapies, including surgery, chemotherapy and radiotherapy, have been widely used for OSCC management and significantly improved the outcomes. However, they also lead to a series of associated problems and morbidities.2,3 Thus, identifying novel therapeutic biomarkers and targets is very vital for the design of innovative treatments against OSCC.
Circular RNAs (circRNAs) are highly conserved and stable covalently closed RNA circles that have no gene-coding potential.4 It has recently become apparent that some circRNAs have important gene-regulatory function in cancer biology through acting as efficient microRNA (miRNA) sponges.5 A growing number of researches have demonstrated that groups of circRNAs are involved in the hallmarks of OSCC.6 For instance, Ouyang et al reported that has_circ_0109291 was up-regulated in OSCC, and its deficiency suppressed OSCC progression.7 Wang et al underscored that circDOCK1 modulated OSCC cell apoptosis by mediating BIRC3 expression via sponging miR-196a-5p.8 As for circRNA GDP dissociation inhibitor 2 (circGDI2, hsa_circ_0005379), derived from the back-spliced exons of the GDI2 gene, it was found to be down-regulated in OSCC, and the elevated expression of circGDI2 repressed OSCC cell malignant behaviors and promoted cell sensitivity to chemotherapy.9
Exosomes are small (40–100 nm) membrane vesicles that are secreted by most cell types and can be horizontally transferred to recipient cells.10 CircRNAs have recently been identified in exosomes, and exosomal circRNAs exert crucial biological activities in human cancers.11 For example, Li et al reported that exosomal circRNA phosphodiesterase 8A (circ-PDE8A) from metastatic tumor cells was correlated with the development and prognosis of the patients with pancreatic ductal adenocarcinoma through acting as a miR-338 sponge.12 Li et al underscored that circRNA transverse aeolian ridges (circ-TARS) was transferred by exosomes and exosomal circ-TARS enhanced pancreatic cancer invasion and metastasis.13 However, whether circGDI2 is transferred by the exosomes derived from OSCC cells remains undefined.
The main objective of this study was to explore whether circGDI2 was transferred by exosomes and how exosomal circGDI2 regulated OSCC cell malignant behaviors. By combining the in vitro and in vivo studies, the present work had led to the identification of exosomal circGDI2 that regulated OSCC cell proliferation, migration, invasion and glycolysis through targeting miR-424-5p/suppressor of cancer cell invasion (SCAI) axis.
Materials and Methods
Clinical Samples and Ethics Statement
In this research, 30 OSCC patients were recruited from The Second Affiliated Hospital of Jinzhou Medical University. The clinicopathological features of these patients were presented in Table 1. The clinical samples, including tumor tissues and adjacent healthy tissues, were snap-frozen in liquid nitrogen and stored at −80°C. All patients provided written informed consent, and this study was approved by the Ethics Committee of The Second Affiliated Hospital of Jinzhou Medical University.
Table 1.
The Clinicopathological Features of These Patients (n = 30)
| Variables | Clinicopathological Features | |
|---|---|---|
| Gender | Female | Male |
| 19 | 11 | |
| Age (years) | ≥60 | <60 |
| 13 | 17 | |
| Tumor size (cm) | ≥5 | <5 |
| 16 | 14 | |
| Tumor stage | I–II | III–IV |
| 20 | 10 | |
| Lymph node status | Negative | Positive |
| 18 | 12 | |
Cell Culture and Transfection
Two OSCC cell lines (CAL-27 and SCC-15) and human normal oral keratinocyte cell line HOK (all from Cosmo Bio, Tokyo, Japan) were used in this study. CAL-27 and SCC-15 cells were maintained at 37°C in RPMI-1640 medium (Sigma-Aldrich, Toyko, Japan) containing 10% fetal bovine serum (Gibco, Cergy Pontoise, France) under 5% CO2. HOK cells were cultured in oral keratinocyte growth supplement (OKGS, CliniSciences, Nanterre, France).
The sequence of circGDI2 (hsa_circ_0005379) was synthesized by GeneWiz (Suzhou, China) and then cloned in a pcDNA3.1 vector (GenePharma, Shanghai, China) with Hind III and BamH I sites to generate circGDI2 overexpression vector (pcDNA-circGDI2). For in vitro circGDI2 up-regulation studies, CAL-27 and SCC-15 cells were transfected with 50 ng of pcDNA-circGDI2 or negative vector (pcDNA-NC) using Lipofectamine 3000 reagent (Thermo Fisher Scientific, Basingstoke, UK) based on the protocols of the manufacturers. For in vitro knockdown studies, the two cells were introduced with siRNA against circGDI2 (si-circGDI2, 25 nM, 5ʹ-UUCAAACAAUCCCAUUAGGCU-3ʹ) or SCAI (si-SCAI, 25 nM, 5ʹ-UUUUCGAGGCGGUUUCUCCAC-3ʹ), and nontarget siRNA (si-NC, 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ) was used as a negative control. MiR-424-5p up-regulation or knockdown cells were generated using commercial miR-424-5p mimic (25 nM) or miR-424-5p inhibitor (anti-miR-424-5p, 25 nM), with a scrambled sequence (miR-NC mimic or anti-miR-NC) as the negative control. To in vivo circGDI2 overexpression studies, lentiviral vectors encoding circGDI2 (lenti-circGDI2) or negative control sequence (lenti-NC) were obtained from GenePharma and used to transduce CAL-27 cells in 8 μg/mL polybrene as recommended by the manufacturers. Vector-transduced cells were selected with puromycin in a final concentration of 1 μg/mL over 72 h.
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA from tissues, cells and exosomes was extracted by homogenization in TRIzol reagent (Thermo Fisher Scientific) based on the recommendations of the manufacturers. For circGDI2 and mRNA expression quantification, cDNA was generated using the RevertAid Reverse Transcription Kit (Thermo Fisher Scientific) and subsequently subjected to qRT-PCR using TaqMan Universal PCR Master Mix (Applied Biosystems, Cheshire, UK). The level of miR-424-5p was detected using the mirVana qRT-PCR miRNA Detection Kit (Applied Biosystems) as recommended by the manufacturers. The glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 snRNA was used as an internal control. The primers sequences for qRT-PCR were shown in Table 2. The qRT-PCR results were calculated by the 2−ΔΔCt formula.14
Table 2.
Primers for PCR
| Primers for PCR (5ʹ-3ʹ) | ||
|---|---|---|
| CircGDI2 | Forward | CATGGTTCCCGCTACTGTGT |
| Reverse | TAGGCCTCGAATCCACCGAT | |
| GDI2 mRNA | Forward | TCAATGGGGAGAGGAAGAGA |
| Reverse | CTGCTTCAGTGGAAGGAACC | |
| SCAI | Forward | GGCAAAGACCACTTCAGGCA |
| Reverse | CCTCCACGCTCAAAGAACCA | |
| GAPDH | Forward | CCTGGCCAAGGTCATCCATG |
| Reverse | GGAAGGCCATGCCAGTGAGC | |
| MiR-424-5p | Forward | CAGCAGCAATTCATGT |
| Reverse | TGGTGTCGTGGAGTCG | |
| U6 snRNA | Forward | CTCGCTTCGGCAGCACA |
| Reverse | AACGCTTCACGAATTTGCGT | |
Ribonuclease (RNase) R Digestion
Total RNAs (10 µg) were incubated in 20 µL of reactions containing 10 U of RNase R (Geneseed Biotech, Guangzhou, China) for 30 min at 37°C. Then, the measurement of circGDI2 and GDI2 linear mRNA levels were conducted by qRT-PCR.
Subcellular Fractionation
Total RNA from cytoplasmic fraction and nuclear of CAL-27 and SCC-15 cells was obtained using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, ON, Canada) as recommended by the manufacturers, followed by the detection of circGDI2 level by qRT-PCR.
Isolation and Characterization of Exosomes
The ExoQuick Exosome Precipitation reagent (System Biosciences, Palo Alto, CA, USA) was used in the experiments for exosomes isolation, following the suggestions of manufacturers. Then, the exosomes were observed by a transmission electron microscopy (TEM) JEM-1200 EX microscope as previously described.15 The exosomal markers CD63 and CD915 were determined by Western blot.
Western Blot
The isolated exosomes and treated cells were subjected to Western blot analysis as previously reported.16 Antibodies against SCAI (ab124688), lactate dehydrogenase A (LDHA, ab101562), glucose transporter 1 (GLUT1, ab652), CD63 (ab8219), CD9 (ab223052), lamin B (ab122919), α-tubulin (ab52866) and GAPDH (ab9485, all from Abcam, Cambridge, UK) were used as primary antibodies. As for secondary antibody, we used anti-rabbit IgG (ab205718, Abcam) antibody which was conjugated with horseradish peroxidase.
Determination of Cell Proliferation, Migration and Invasion
CAL-25 and SCC-15 cells were transfected with or without the indicated oligonucleotides and treated with 30 µg/mL of the exosomes from pcDNA-NC- or pcDNA-circGDI2-transfected Donor cells. The assessment of cell proliferation was carried out using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay as previously expressed.17 The transwell assays were performed to assess cell migration and invasion using 24-Transwell chambers (8 μm insert, BD Biosciences, Heidelberg, Germany) and invasion chambers with Matrigel-coated membrane (BD Biosciences), respectively, following the instructions of the manufacturers.18
Measurement of Glucose Consumption, Lactate Production and Extracellular Acidification Rate (ECAR)
Determination of glucose consumption and lactate production were conducted using a Glucose Assay Kit (Thermo Fisher Scientific) and Lactate Assay Kit (Sigma-Aldrich), respectively, referring to the manufacturer’s recommendations. ECAR detection of CAL-27 and SCC-15 cells after various treatments was implemented using the Seahorse Extracellular Flux analyzer (XFe 96, Seahorse Bioscience, Billerica, MA, USA) based on the guidance of the manufacturers. Briefly, the cells were sequentially exposed to glucose, the oxidative phosphorylation inhibitor oligomycin (OM) and the glycolytic inhibitor 2-DG, which were followed by the analysis of ECAR using the Seahorse XF-96 Wave software.
Dual-Luciferase Reporter Assay
Bioinformatic analyses for the interacted miRNAs of circGDI2 and the molecular targets of miR-424-5p were done using the starBase v.2 software at http://starbase.sysu.edu.cn/. CircGDI2 luciferase reporter vector (circGDI2 WT) containing the miR-424-5p-binding sites and the site-directed mutant in the target sites (circGDI2 MUT), SCAI 3ʹUTR reporter vector (SCAI 3ʹUTR WT) and the mutation of the seed sequence (SCAI 3ʹUTR MUT) were established by Ribobio (Guangzhou, China). Each reporter construct (100 ng) and 25 nM of miR-424-5p mimic or miR-NC mimic were cotransfected into CAL-27 and SCC-15 cells. The cells were harvested 48 h post-transfection and analyzed for luciferase activities using the Dual-luciferase Reporter Assay System (Promega, Southampton, UK).
Animal Studies
The protocol of animal studies was approved by the Ethics Committee of The Second Affiliated Hospital of Jinzhou Medical University, and all animal processes were done based on the National Standard of the Care and Use of Laboratory Animals. Twelve 6-old-week BALB/c mice (Henan Research Center of Laboratory Animal, Zhengzhou, China) were used for the tumor growth xenograft assays. 5 × 106 CAL-27 cells were subcutaneously injected into the nude mice (n= 6 each group). After 7 days cell implantation, we performed the intratumor injection for the exosomes (30 µg per mouse) from lenti-NC- or lenti-circGDI2-transduced CAL-27 cells for four times every week. Tumor volume measurement began on 7 days after implantation and was conducted every week. After 35 days cell implantation, all mice were euthanized, and tumor tissues were removed.
Statistical Analysis
Experimental results were the mean ± standard deviation (SD). A Student’s t-test, Mann–Whitney U-test or one-way analysis of variance (ANOVA) was used to compare the differences in two groups or multiple groups. Correlations among circGDI2, miR-424-5p and SCAI expression in OSCC tissues were analyzed using the Spearman test. Statistical significance was set as P < 0.05.
Results
CircGDI2 Level Was Down-Regulated in OSCC Tissues and Cells
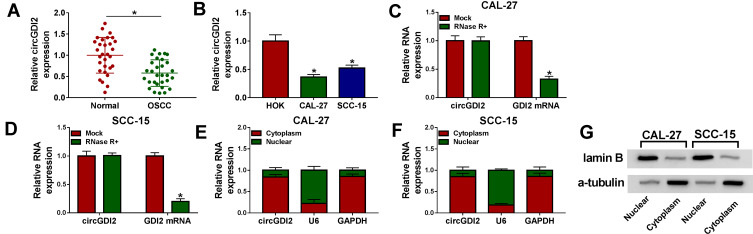
The data of qRT-PCR revealed that in comparison to the corresponding negative control, circGDI2 level was significantly down-regulated in OSCC tissues and cells (Figure 1A and B). The incubation with RNase R led to a striking reduction in the level of GDI2 linear mRNA, and circGDI2 was resistant to RNase R (Figure 1C and D), indicating the stability of circGDI2. Moreover, subcellular localization analysis showed that circGDI2 was mainly localized in the cytoplasm of CAL-27 and SCC-15 cells (Figure 1E and F). The nice separation of cytoplasmic and nuclear fractionations was confirmed by the expression of lamin B (mainly localized in the nucleus) and α-tubulin (mainly localized in the cytoplasm), respectively (Figure 1G).
Figure 1.
CircGDI2 expression was decreased in OSCC tissues and cells. (A and B) CircGDI2 expression by qRT-PCR in 30 pairs of OSCC tissues and matched normal tissues, HOK, CAL-27 and SCC-15 cells. Blots were representative of n = 3. (C and D) The levels of circGDI2 and GDI2 linear mRNA by qRT-PCR in RNase R-treated RNA extracts from CAL-27 and SCC-15 cells. Blots were representative of n = 6. (E and F) The subcellular localization of circGDI2 in both CAL-27 and SCC-15 cells. Error bars indicated SD of triplicate experiments. (G) The levels of lamin B and α-tubulin by Western blot in cytoplasmic and nuclear fractionations of CAL-27 and SCC-15 cells. A representative experiment was shown in triplicate. *P < 0.05.
CircGDI2 Was Transferred by Incorporation into Exosomes
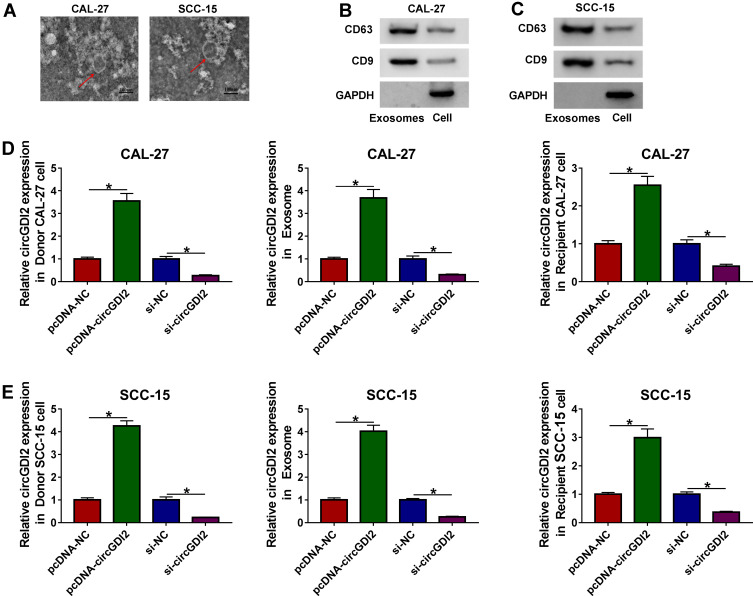
Then, we determined whether circGDI2 was transferred by exosomes in OSCC cells. The morphological characteristics by TEM revealed that the vesicles contained a round or oval membrane (Figure 2A). Western blot analyses showed that exosomal markers CD9 and CD63 levels were significantly increased in the vesicles derived from CAL-27 and SCC-15 cells (Figure 2B and C). After that, the exosomes derived from the transfected OSCC cells (Donor cells) were isolated and used to treat the corresponding Recipient cells. The results of qRT-PCR revealed that in contrast to their counterparts, circGDI2 expression was prominently elevated by the transfection of pcDNA-circGDI2, while it was remarkably reduced by si-circGDI2 introduction in the two Donor cells (Figure 2D and E). Moreover, circGDI2 level was higher in the exosomes derived from circGDI2-overexpressing OSCC cells than that of control, and the exosomes from si-circGDI2-transfected Donor cells had a lower circGDI2 level when comparing to the negative group (Figure 2D and E). More interestingly, circGDI2 expression level in corresponding Recipient cells was consistent with the Donor cells and the corresponding exosomes (Figure 2D and E). These results together suggested that OSCC cells might transmit circGDI2 to surrounding cancer cells by exosomes.
Figure 2.
CircGDI2 was transferred by exosomes in OSCC cells. (A) The representative micrograph of the exosomes derived from CAL-27 and SCC-15 cells by TEM (scale bars=100 nm). Red arrows pointed the exosomes. (B and C) The levels of CD63 and CD9 by Western blot in the exosomes and cell lysates. GAPDH was used as a negative control. A representative experiment was shown in triplicate. (D) CAL-27 cells were treated with 30 µg/mL of the exosomes derived from CAL-27 cells transfected with pcDNA-NC, pcDNA-circGDI2, si-NC or si-circGDI2, and then circGDI2 expression was assessed by qRT-PCR in the Donor cells, the exosomes and Recipient cells. Blots were representative of n = 6. (E) SCC-15 cells were treated with 30 µg/mL of the exosomes derived from SCC-15 cells transfected with pcDNA-NC, pcDNA-circGDI2, si-NC or si-circGDI2, followed by the determination of circGDI2 level in the Donor cells, the exosomes and Recipient cells. Blots were representative of n = 6. *P < 0.05.
Overexpression of Exosomal CircGDI2 Suppressed OSCC Cell Proliferation, Migration, Invasion and Glycolysis
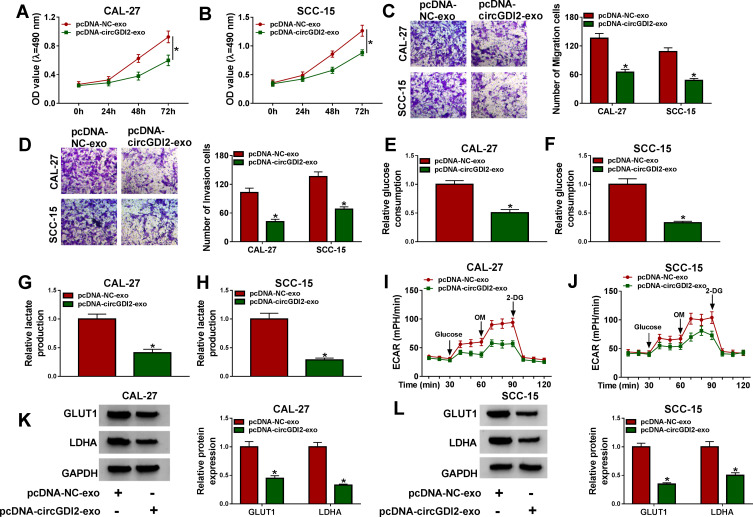
To observe the role of exosomal circGDI2 in OSCC progression, we used the exosomes (pcDNA-NC-exo and pcDNA-circGDI2-exo) derived from Donor cells to treat the Recipient cells, followed by the assessment of cell malignant behaviors. In comparison to the negative group, the elevated expression of exosomal circGDI2 led to a significant inhibition in cell proliferation (Figure 3A and B), migration (Figure 3C) and invasion (Figure 3D) in the corresponding Recipient cells. Moreover, the up-regulation of exosomal circGDI2 triggered a striking reduction in glucose consumption (Figure 3E and F) and lactate production (Figure 3G and H), as well as a diminishment in ECAR (Figure 3I and J) which reflects overall glycolytic flux, demonstrating the suppression of exosomal circGDI2 overexpression on OSCC cell glycolysis. Additionally, the elevated expression of exosomal circGDI2 resulted in decreased levels of glycolysis-related LDHA and GLUT1 proteins in the Recipient cells (Figure 3K and L).
Figure 3.
The elevated expression of exosomal circGDI2 hampered OSCC cell proliferation, migration, invasion and glycolysis. CAL-27 and SCC-15 cells (Recipient cells) were treated with 30 µg/mL of the exosomes from pcDNA-NC- or pcDNA-circGDI2-transfected Donor cells. (A and B) MTT assay for cell proliferation. (C and D) Transwell assay for cell migration and invasion. (E–H) A corresponding assay kit for glucose consumption and lactate production. (I and J) Measurement of ECAR in treated Recipient cells. (K and L) Western blot for GLUT1 and LDHA levels in treated Recipient cells. Error bars indicated SD of triplicate experiments. *P < 0.05.
Abbreviations: pcDNA-NC-exo, exosomes derived from OSCC cells transfected with pcDNA; pcDNA-circGDI2-exo, exosomes derived from OSCC cells transfected with pcDNA-circGDI2.
CircGDI2 Functioned as a Molecular Sponge of MiR-424-5p
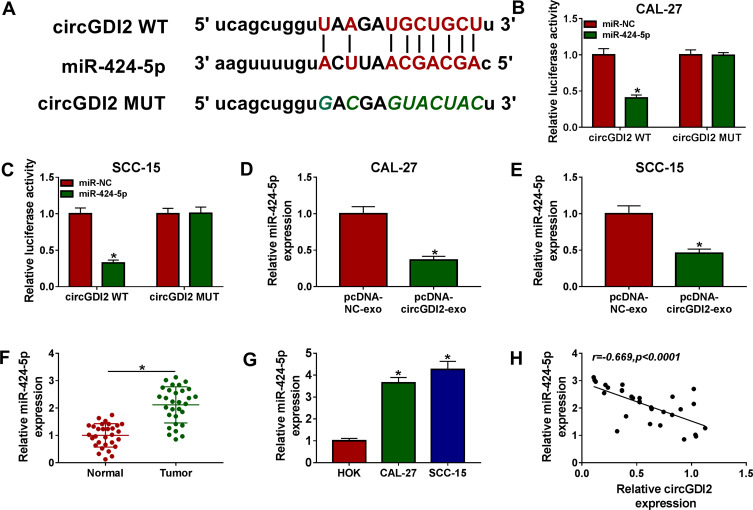
To understand the mechanism by which exosomal circGDI2 repressed OSCC cell malignant behaviors, we used the starBase v.2 software to help identify the targeted miRNAs of circGDI2. The predicted data revealed that circGDI2 harbored a potential miR-424-5p-binding sequence (Figure 4A). To validate this, circGDI2 luciferase reporter vector containing the miR-424-5p-binding sites or a site-directed mutant was transfected into the OSCC cells with miR-424-5p mimic or miR-NC mimic. With the wild-type reporter and miR-424-5p overexpression caused a prominent reduction in luciferase activity (Figure 4B and C). Nevertheless, no reduction in luciferase of the mutant-type reporter was observed with miR-424-5p overexpression (Figure 4B and C). Moreover, in contrast to the negative group, miR-424-5p expression was prominently decreased by exosomal circGDI2 overexpression in the two OSCC cells (Figure 4D and E).19,20 Additionally, the data of qRT-PCR showed that miR-424-5p level was significantly up-regulated in OSCC tissues and cells (Figure 4F and G). Besides, the expression of miR-424-5p was inversely correlated with circGDI2 level in OSCC tissues (Figure 4H).
Figure 4.
CircGDI2 acted as a sponge of miR-424-5p. (A) Schematic model of the miR-424-5p-binding sequence within circGDI2 and the mutant of the binding sequence. (B and C) The luciferase activity in CAL-27 and SCC-15 cells cotransfected with circGDI2 luciferase reporter vector (circGDI2 WT) or the site-directed mutant (circGDI2 MUT) and miR-424-5p mimic or miR-NC mimic. Blots were representative of n = 6. The expression of miR-424-5p by qRT-PCR in CAL-27 and SCC-15 cells treated with pcDNA-NC-exo or pcDNA-circGDI2-exo (D and E), in 30 pairs of OSCC tissues and matched normal tissues (F), HOK, CAL-27 and SCC-15 cells (G). Blots were representative of n = 3. (H) Correlation between circGDI2 level and miR-424-5p expression in OSCC tissues using the Spearman test. *P < 0.05.
Abbreviations: pcDNA-NC-exo, exosomes derived from OSCC cells transfected with pcDNA; pcDNA-circGDI2-exo, exosomes derived from OSCC cells transfected with pcDNA-circGDI2.
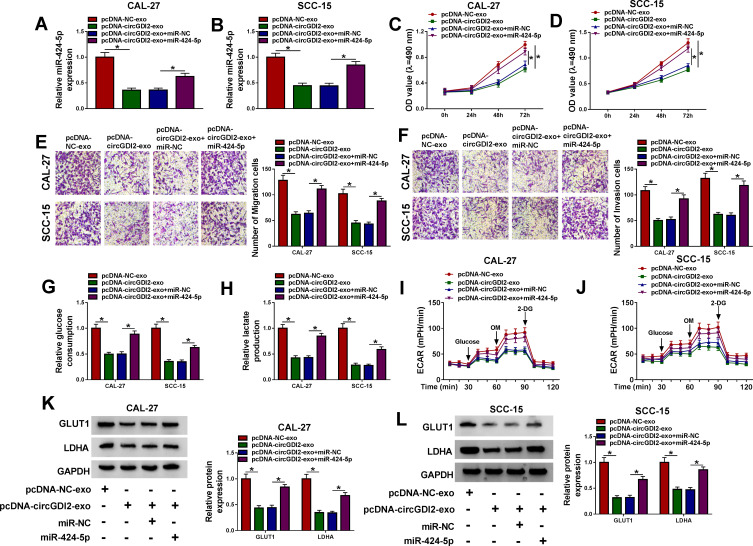
The Inhibition of Exosomal CircGDI2 Overexpression on OSCC Cell Proliferation, Migration, Invasion and Glycolysis Was Mediated by MiR-424-5p
Next, we determined whether exosomal circGDI2 hindered OSCC cell malignant behaviors by miR-424-5p. qRT-PCR results revealed that in comparison to their counterparts, the elevated expression of exosomal circGDI2 resulted in a significant down-regulation in miR-424-5p expression, and this effect was dramatically abolished by the introduction of miR-424-5p mimic (Figure 5A and B). MTT and transwell assays showed that exosomal circGDI2 overexpression-mediated anti-proliferation (Figure 5C and D), anti-migration (Figure 5E) and anti-invasion (Figure 5F) were strongly abrogated by miR-424-5p level restoration. Moreover, the decreased impact of exosomal circGDI2 overexpression on glucose consumption (Figure 5G), lactate production (Figure 5H), ECAR (Figure 5I and J) and LDHA and GLUT1 levels (Figure 5K and L) were significantly reversed by the restored level of miR-424-5p.
Figure 5.
The suppressive impact of exosomal circGDI2 overexpression on OSCC cell malignant behaviors was reversed by the restored level of miR-424-5p. CAL-27 and SCC-15 cells (Recipient cells) were transfected with miR-424-5p mimic or miR-NC mimic and then treated with 30 µg/mL of the exosomes from pcDNA-NC- or pcDNA-circGDI2-transfected Donor cells, followed by the detection of miR-424-5p expression by qRT-PCR (A and B), cell proliferation by MTT assay (C and D), cell migration and invasion by transwell assay (E and F), glucose consumption and lactate production using a corresponding assay kit (G and H), ECAR using a standard assay (I and J), LDHA and GLUT1 levels by Western blot (K and L). A representative experiment was shown in triplicate. *P < 0.05.
Abbreviations: pcDNA-NC-exo, exosomes derived from OSCC cells transfected with pcDNA; pcDNA-circGDI2-exo, exosomes derived from OSCC cells transfected with pcDNA-circGDI2.
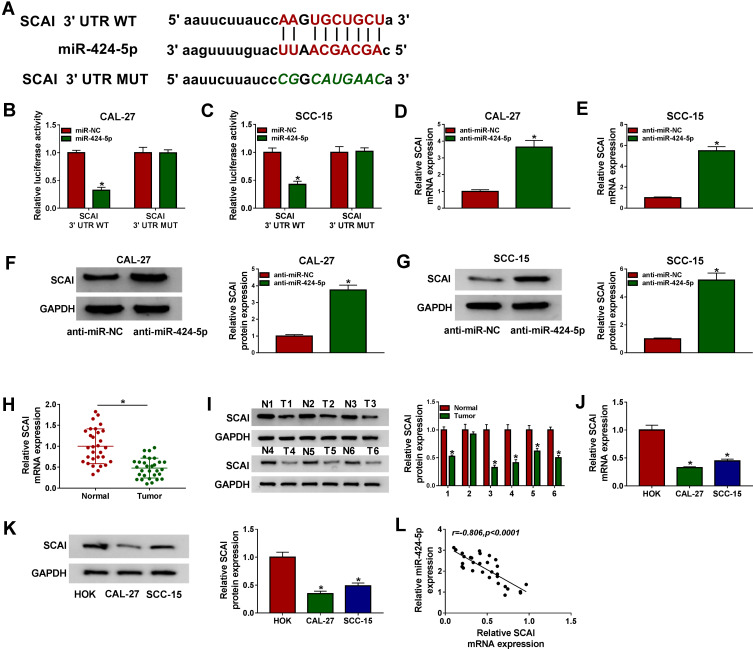
SCAI Was a Direct Target of MiR-424-5p in OSCC Cells
Using the online software starBase v.2, a putative target sequence for miR-424-5p was predicted within the 3ʹ-UTR of SCAI (Figure 6A). Transient introduction of miR-424-5p mimic, but not the scrambled miR-NC control, led to a striking reduction in the luciferase activity of SCAI 3ʹUTR reporter vector (Figure 6B and C). However, the down-regulated effect of miR-424-5p overexpression on reporter gene expression was notably abolished by the site-directed mutant of the miR-424-5p-binding sites (Figure 6B and C). Moreover, in contrast to the negative control, SCAI mRNA and protein levels were markedly elevated by anti-miR-424-5p in both CAL-27 and SCC-15 cells (Figure 6D–G). Furthermore, SCAI expression was significantly reduced in OSCC tissues and cells when comparing to the corresponding negative control (Figure 6H–K). Additionally, an inverse correlation between SCAI mRNA level and miR-424-5p expression was discovered in OSCC tissues (Figure 6L).
Figure 6.
SCAI was a direct target of miR-424-5p. (A) Schematic of the miR-424-5p-binding sites within SCAI 3ʹUTR identified by the starBase v.2 software and mutated miR-424-5p-binding sites. (B and C) The luciferase activity in CAL-27 and SCC-15 cells cotransfected with SCAI 3ʹUTR WT or SCAI 3ʹUTR MUT and miR-424-5p mimic or miR-NC mimic. Blots were representative of n = 6. SCAI mRNA expression by qRT-PCR and protein level by Western blot in CAL-27 and SCC-15 cells transfected with anti-miR-NC or anti-miR-424-5p (D–G), OSCC tissues and matched normal tissues (H and I), HOK, CAL-27 and SCC-15 cells (J and K). Blots were representative of n = 3. (L) Correlation between SCAI mRNA level and miR-424-5p expression in OSCC tissues using the Spearman test. *P < 0.05.
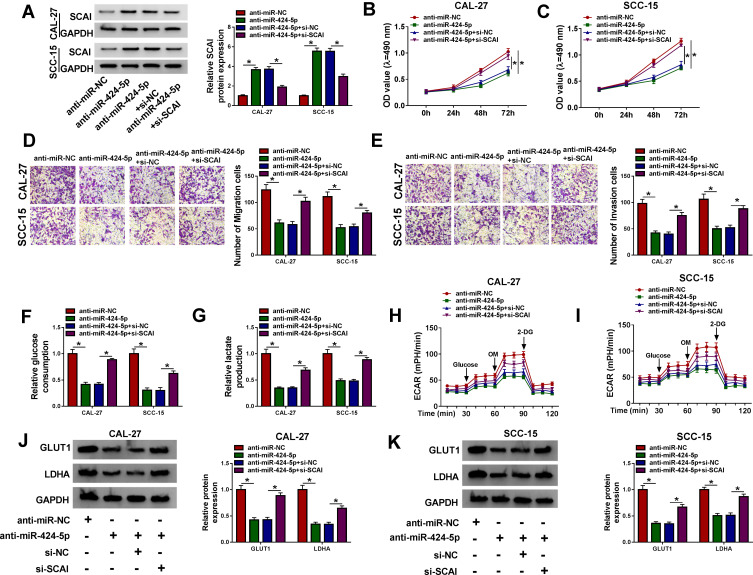
MiR-424-5p Knockdown Mitigated OSCC Cell Proliferation, Migration, Invasion and Glycolysis via Up-Regulating SCAI
We then validated whether SCAI was a functional target of miR-424-5p in regulating OSCC malignant behaviors. Western blot analyses showed that in comparison to the negative control, anti-miR-424-5p-mediated SCAI up-regulation was notably abolished by si-SCAI cotransfection in the two OSCC cells (Figure 7A). Functional experiments revealed that the knockdown of miR-424-5p led to a remarkable repression in cell proliferation (Figure 7B and C), migration (Figure 7D), invasion (Figure 7E), and a distinct reduction in glucose consumption (Figure 7F), lactate production (Figure 7G) and ECAR (Figure 7H and I), as well as a clear decrease in LDHA and GLUT1 expression (Figure 7J and K). Nevertheless, these effects of miR-424-5p knockdown were prominently abrogated by the restored expression of SCAI (Figure 7B–K).
Figure 7.
The mitigative effects of miR-424-5p knockdown on OSCC cell malignant behaviors were mediated by SCAI. CAL-27 and SCC-15 cells were transfected with anti-miR-NC, anti-miR-424-5p, anti-miR-424-5p+si-NC or anti-miR-424-5p+si-SCAI. (A) SCAI expression by Western blot in transfected cells. (B and C) Cell proliferation by MTT assay. (D and E) Cell migration and invasion by transwell assay. (F and G) Glucose consumption and lactate production using a corresponding assay kit. (H and I) ECAR using a standard assay. (J and K) LDHA and GLUT1 levels by Western blot in transfected cells. A representative experiment was shown in triplicate. *P < 0.05.
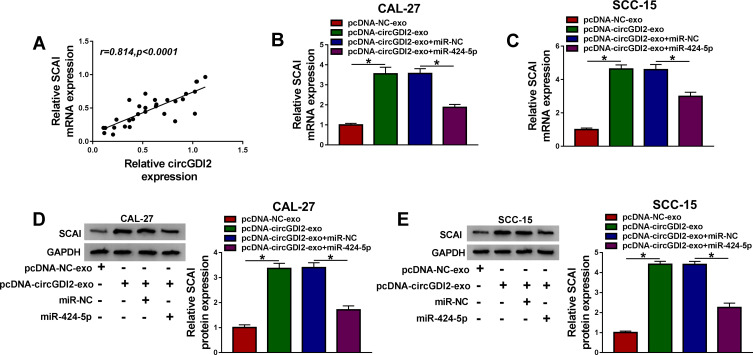
Exosomal CircGDI2 Mediated SCAI Expression Through Sponging MiR-424-5p
Interestingly, SCAI mRNA expression was positively correlated with circGDI2 level in OSCC tissues (Figure 8A). We further determined whether exosomal circGDI2 regulated SCAI expression in the two OSCC cells. In comparison to the negative group, SCAI mRNA and protein levels were notably increased by exosomal circGDI2 overexpression, and this effect was remarkably reversed by miR-424-5p mimic (Figure 8B–E).
Figure 8.
Exosomal circGDI2 regulated SCAI expression through sponging miR-424-5p. (A) Correlation between circGDI2 expression and SCAI mRNA level in OSCC tissues using the Spearman test. CAL-27 and SCC-15 cells (Recipient cells) were transfected with miR-424-5p mimic or miR-NC mimic and then treated with 30 µg/mL of the exosomes from pcDNA-NC- or pcDNA-circGDI2-transfected Donor cells, followed by the measurement of SCAI mRNA expression by qRT-PCR (B and C), SCAI protein level by Western blot (D and E). Blots were representative of n = 6. *P < 0.05.
Abbreviations: pcDNA-NC-exo, exosomes derived from OSCC cells transfected with pcDNA; pcDNA-circGDI2-exo, exosomes derived from OSCC cells transfected with pcDNA-circGDI2.
Overexpression of Exosomal CircGDI2 Hindered Tumor Growth in vivo
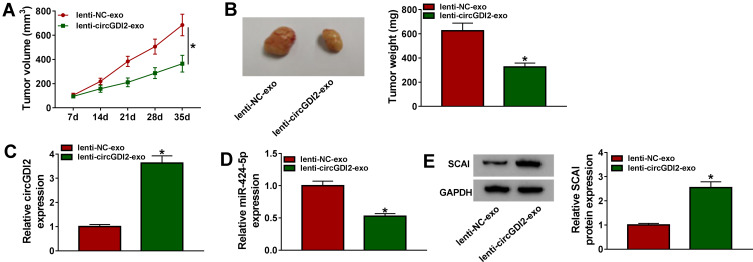
Lastly, we observed whether exosomal circGDI2 affected tumor growth in vivo using the xenograft mice model. After 7 days cell implantation, we performed the intratumor injection for the exosomes from lenti-NC- or lenti-circGDI2-transduced CAL-27 cells. In comparison to the negative control, the treatment of the exosomes from lenti-circGDI2-transduced cells resulted in a striking repression in tumor growth (Figure 9A and B). Moreover, circGDI2 and SCAI levels were significantly increased and miR-424-5p expression was highly decreased in the tumor tissues when the treatment with exosomes derived from lenti-circGDI2-transduced cells (Figure 9C–E).
Figure 9.
Overexpression of exosomal circGDI2 hampered tumor growth in vivo. CAL-27 cells were subcutaneously injected into the nude mice (n= 6 each group). One week later, intratumor injection for the exosomes from lenti-NC- or lenti-circGDI2-transduced CAL-27 cells was performed. After 35 days cell implantation, all mice were euthanized. (A) Tumor volume measurement began on 7 days after implantation and was conducted every week. (B) Representative pictures and average weight of the xenograft tumors. (C–E) The levels of circGDI2, miR-424-5p and SCAI were assessed by qRT-PCR or Western blot in excised tumor tissues. A representative experiment was shown in triplicate. *P < 0.05.
Discussion
Recently, circRNAs have been discovered in exosomes, and exosomal circRNAs can be taken up by adjacent or distant cells and then regulate recipient cells.21 Conveying information via exosomal circRNAs has been proposed to play essential roles in cancer biology, highlighting a possibility for exosomal circRNAs as diagnostic biomarkers and therapeutic targets in further personalized medicine.11 Considering the tumor-suppressive role of circGDI2 in OSCC,9 we undertook to identify whether circGDI2 could be transferred by exosomes and how circGDI2 suppressed OSCC progression.
Our data validated that circGDI2 was down-regulated in OSCC, in agreement with a recent study.9 As previously reported for other circRNAs,22,23 we confirmed that circGDI2 was stable and resistant to RNase R, owing to the lack of 3ʹ poly(A) tails and 5ʹ caps.24 To determine whether circGDI2 was transferred by exosomes, we isolated the vesicles from the cell culture medium. For the first time, we highlighted that circGDI2 was transmitted by the OSCC cells to surrounding cancer cells by incorporation into exosomes. Glycolysis is remarkably elevated in cancer cells, which contributes to tumor cell malignant progression.25 GLUT1 is a key factor for glucose transport and LDHA is a glycolytic enzyme in the final step of glycolysis, which exert important function in tumorigenesis and tumor progression.26–28 We were first to uncover that the overexpression of exosomal circGDI2 weakened OSCC cell proliferation, migration, invasion and glycolysis in vitro. Our data also demonstrated that circGDI2 was mainly localized in the cytoplasm of the two OSCC cells, which provided the possibility for the interplay between circGDI2 and miRNAs.
Then, we used the starBase v.2 software to help identify the miRNAs that potentially bind to circGDI2. Among these candidates, we selected miR-424-5p for further research and confirmed that circGDI2 directly interacted with miR-424-5p through sequestration. MiR-424-5p has been demonstrated to regulate tumor progression in a number of human cancers, such as thyroid cancer, basal-like breast cancer and cervical cancer.29–31 MiR-424-5p was also reported to be involved in the progression of tongue squamous cell carcinoma.32 Moreover, miR-424-5p was highly expressed in OSCC, and it accelerated OSCC progression via targeting suppressor of cytokine signaling (SOCS2).33 We observed that miR-424-5p knockdown suppressed cell proliferation, migration, invasion and glycolysis of OSCC cells. More interestingly, for the first time, we illuminated that miR-424-5p mediated the inhibition of exosomal circGDI2 overexpression on OSCC cell malignant behaviors. Zhang et al reported that circRNA La-related RNA-binding protein 4 (circ-LARP4) hindered gastric cancer cell growth and invasion through controlling large tumor suppressor 1 (LATS1) expression via functioning as a miR-424-5p sponge.34
SCAI has been identified to be down-regulated in many human cancers, and it functions as a suppressor of tumor cell invasion through regulating beta1-integrin.35 Moreover, it was reported that miR-424-5p was underexpressed in OSCC and it participated in the regulation of OSCC cell migration.36 Using the starBase v.2 software, we found that SCAI 3ʹ-UTR harbored a miR-424-5p-binding sequence, and we validated that SCAI in OSCC cells was directly targeted and inhibited by miR-424-5p. Furthermore, for the first time, we uncovered that miR-424-5p silencing repressed OSCC cell malignant behaviors through up-regulating SCAI. Xu et al described that miR-625-3p enhanced OSCC cell migration by targeting SCAI.36 Previous studies had also reported that some other miRNAs, such as miR-1228 and miR-1270, regulated tumor progression through targeting SCAI.37,38 More interestingly, we found that exosomal circGDI2 controlled SCAI expression through sponging miR-424-5p. Additionally, our data showed a strong inverse correlation between miR-424-5p expression and SCAI mRNA level in OSCC tissues. However, using online TCGA database, there was no significant correlation between miR-424-5p and SCAI mRNA in over 200 OSCC samples. These controversial conclusions may attribute to different clinicopathological feature of these patients or different sample size, which is expected to be investigated in further work.
Lastly, we also confirmed the suppressive effect of exosomal circGDI2 overexpression on tumor growth in vivo. CircGDI2 overexpression also triggered a decrease in miR-424-5p expression and an increase in SCAI level in the tumor tissues, providing evidence for the mediator role of the miR-424-5p/SCAI axis in exosomal circGDI2-mediated regulation on OSCC progression. However, the in vivo research about the novel mechanism is incomplete in the present work, which should be investigated in further work. Besides, circGDI2 influenced the expression of GLUT1 and LDHA in the two OSCC cells, which possibly was due to the miR-424-5p/SCAI axis. Further studies will build on the findings by determining precisely how the novel mechanism affects GLUT1 and LDHA levels.
In conclusion, the current work had led to the identification of exosomal circGDI2 regulated OSCC cell malignant behaviors via controlling SCAI expression by acting as a miR-424-5p sponge. Our findings provided the evidence for the circGDI2 as a novel exosome-based cancer biomarker and demonstrated the potential biological effect of exosomal circGDI2.
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
- 1.Ng JH, Iyer NG, Tan M-H, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: a global study. Head Neck. 2017;39(2):297–304. doi: 10.1002/hed.24589 [DOI] [PubMed] [Google Scholar]
- 2.Wang C, Liu XQ, Hou JS, Wang JN, Huang HZ. Molecular mechanisms of chemoresistance in oral cancer. Chin J Dent Res. 2016;19(1):25–33. doi: 10.3290/j.cjdr.a35694 [DOI] [PubMed] [Google Scholar]
- 3.Perri F, Pacelli R, Della Vittoria Scarpati G, et al. Radioresistance in head and neck squamous cell carcinoma: biological bases and therapeutic implications. Head Neck. 2015;37(5):763–770. doi: 10.1002/hed.23837 [DOI] [PubMed] [Google Scholar]
- 4.Chen -L-L, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi: 10.1080/15476286.2015.1020271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi: 10.1038/onc.2017.361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y-F, Li B-W, Sun S, et al. Circular RNA expression in oral squamous cell carcinoma. Front Oncol. 2018;8:398. doi: 10.3389/fonc.2018.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ouyang S-B, Wang J, Zhao S-Y, Zhang X-H, Liao L. CircRNA_0109291 regulates cell growth and migration in oral squamous cell carcinoma and its clinical significance. Iran J Basic Med Sci. 2018;21(11):1186–1191. doi: 10.22038/IJBMS.2018.30347.7313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang L, Wei Y, Yan Y, et al. CircDOCK1 suppresses cell apoptosis via inhibition of miR‑196a‑5p by targeting BIRC3 in OSCC. Oncol Rep. 2018;39(3):951–966. doi: 10.3892/or.2017.6174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Su W, Wang Y, Wang F, et al. Hsa_circ_0005379 regulates malignant behavior of oral squamous cell carcinoma through the EGFR pathway. BMC Cancer. 2019;19(1):400. doi: 10.1186/s12885-019-5593-5 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Ruivo CF, Adem B, Silva M, Melo SA. The biology of cancer exosomes: insights and new perspectives. Cancer Res. 2017;77(23):6480–6488. doi: 10.1158/0008-5472.CAN-17-0994 [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, Yanfang W, Li J, et al. Tumor-released exosomal circular RNA PDE8A promotes invasive growth via the miR-338/MACC1/MET pathway in pancreatic cancer. Cancer Lett. 2018;432:(237–250. doi: 10.1016/j.canlet.2018.04.035 [DOI] [PubMed] [Google Scholar]
- 13.Li J, Li Z, Jiang P, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi: 10.1186/s13046-018-0822-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L-F, Lou J-T, Lu M-H, et al. Suppression of miR-199a maturation by HuR is crucial for hypoxia-induced glycolytic switch in hepatocellular carcinoma. EMBO J. 2015;34(21):2671–2685. doi: 10.15252/embj.201591803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu X, Lu Y, Xu Y, et al. Exosomal transfer of miR-501 confers doxorubicin resistance and tumorigenesis via targeting of BLID in gastric cancer. Cancer Lett. 2019;459:122–134. doi: 10.1016/j.canlet.2019.05.035 [DOI] [PubMed] [Google Scholar]
- 16.Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol. 2019;862:172615. doi: 10.1016/j.ejphar.2019.172615 [DOI] [PubMed] [Google Scholar]
- 17.Sylvester PW. Optimization of the tetrazolium dye (MTT) colorimetric assay for cellular growth and viability. Methods Mol Biol. 2011;716:157–168. doi: 10.1007/978-1-61779-012-6_9 [DOI] [PubMed] [Google Scholar]
- 18.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449(7163):682–688. doi: 10.1038/nature06174 [DOI] [PubMed] [Google Scholar]
- 19.Ding C, Yi X, Wu X, et al. Exosome-mediated transfer of circRNA CircNFIX enhances temozolomide resistance in glioma. Cancer Lett. 2020;479:1–12. doi: 10.1016/j.canlet.2020.03.002 [DOI] [PubMed] [Google Scholar]
- 20.Zhu X, Shao P, Tang Y, Shu M, Hu -W-W, Zhang Y. hsa_circRNA_100533 regulates GNAS by sponging hsa_miR_933 to prevent oral squamous cell carcinoma. J Cell Biochem. 2019;120(11):19159–19171. doi: 10.1002/jcb.29245 [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, Liu J, Ma J, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi: 10.1186/s12943-019-1041-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang J, Li X, Lu L, He L, Hu H, Xu Z. Circular RNA hsa_circ_0000567 can be used as a promising diagnostic biomarker for human colorectal cancer. J Clin Lab Anal. 2018;32(5):e22379–e22379. doi: 10.1002/jcla.22379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li F, Zhang L, Li W, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget. 2015;6(8):6001–6013. doi: 10.18632/oncotarget.3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hsiao K-Y, Sun HS, Tsai S-J. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi: 10.1177/1535370217708978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akram M. Mini-review on glycolysis and cancer. J Cancer Educ. 2013;28(3):454–457. doi: 10.1007/s13187-013-0486-9 [DOI] [PubMed] [Google Scholar]
- 26.Amann T, Hellerbrand C. GLUT1 as a therapeutic target in hepatocellular carcinoma. Expert Opin Ther Targets. 2009;13(12):1411–1427. doi: 10.1517/14728220903307509 [DOI] [PubMed] [Google Scholar]
- 27.Yeh C-S, Wang J-Y, Chung F-Y, et al. Significance of the glycolytic pathway and glycolysis related-genes in tumorigenesis of human colorectal cancers. Oncol Rep. 2008;19(1):81–91. [PubMed] [Google Scholar]
- 28.Jin L, Chun J, Pan C, et al. Phosphorylation-mediated activation of LDHA promotes cancer cell invasion and tumour metastasis. Oncogene. 2017;36(27):3797–3806. doi: 10.1038/onc.2017.6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu X, Fu Y, Zhang G, et al. miR-424-5p promotes anoikis resistance and lung metastasis by inactivating hippo signaling in thyroid cancer. Mol Ther Oncolytics. 2019;15:248–260. doi: 10.1016/j.omto.2019.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang J, Wang S, Zhou J, Qian Q. miR-424-5p regulates cell proliferation, migration and invasion by targeting doublecortin-like kinase 1 in basal-like breast cancer. Biomed Pharmacother. 2018;102:147–152. doi: 10.1016/j.biopha.2018.03.018 [DOI] [PubMed] [Google Scholar]
- 31.Dong J, Wang Q, Li L, Xiao-Jin Z. Upregulation of long non-coding RNA small nucleolar RNA host gene 12 contributes to cell growth and invasion in cervical cancer by acting as a sponge for miR-424-5p. Cell Physiol Biochem. 2018;45(5):2086–2094. doi: 10.1159/000488045 [DOI] [PubMed] [Google Scholar]
- 32.Li D, Liu K, Li Z, Wang J, Wang X. miR-19a and miR-424 target TGFBR3 to promote epithelial-to-mesenchymal transition and migration of tongue squamous cell carcinoma cells. Cell Adh Migr. 2018;12(3):236–246. doi: 10.1080/19336918.2017.1365992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng H-Y, Jiang -S-S, Hsiao J-R, et al. IL-8 induces miR-424-5p expression and modulates SOCS2/STAT5 signaling pathway in oral squamous cell carcinoma. Mol Oncol. 2016;10(6):895–909. doi: 10.1016/j.molonc.2016.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang J, Liu H, Hou L, et al. Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol Cancer. 2017;16(1):151. doi: 10.1186/s12943-017-0719-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brandt DT, Baarlink C, Kitzing TM, et al. SCAI acts as a suppressor of cancer cell invasion through the transcriptional control of beta1-integrin. Nat Cell Biol. 2009;11(5):557–568. doi: 10.1038/ncb1862 [DOI] [PubMed] [Google Scholar]
- 36.Xu D, Gu M, Liu HL. MicroRNA-625-3p promotes cell migration of oral squamous cell carcinoma by regulating SCAI expression. Eur Rev Med Pharmacol Sci. 2019;23(2):641–648. doi: 10.26355/eurrev_201901_16878 [DOI] [PubMed] [Google Scholar]
- 37.Lin L, Liu D, Liang H, Xue L, Su C, Liu M. MiR-1228 promotes breast cancer cell growth and metastasis through targeting SCAI protein. Int J Clin Exp Pathol. 2015;8(6):6646–6655. [PMC free article] [PubMed] [Google Scholar]
- 38.Yi T, Zhou X, Sang K, Zhou J, Ge L. MicroRNA-1270 modulates papillary thyroid cancer cell development by regulating SCAI. Biomed Pharmacother. 2019;109:2357–2364. doi: 10.1016/j.biopha.2018.08.150 [DOI] [PubMed] [Google Scholar]