Abstract
Background
Colon cancer (CRC) is a common type of tumour, and IQGAP family proteins play an important role in many tumours. However, their roles in CRC remain unclear.
Methods
First, we searched many public databases to comprehensively analyze expression of IQGAPs in CRC. Next, real-time PCR, immunohistochemical (IHC), transwell, siRNA transfection and Western blot assays were used to evaluate relationships among IQGAP3 expression, clinical pathological parameters and CRC prognosis, and the underlying molecular mechanism was investigated.
Results
IQGAP3 was elevated in CRC tissues, whereas there was no significant change in expression of IQGAP1 or IQGAP2. Additionally, IQGAP3 expression in CRC tissues was associated with tumour progression, invasion and poor prognosis. In mechanistic studies, we found that IQGAP3 was positively coexpressed with PIK3C2B. In an in vitro assay, the PIK3C2B expression level was increased after exogenous overexpression of IQGAP3, resulting in the promotion of cell invasion, which was blocked by pretransfecting cells with PIK3C2B siRNA. Furthermore, we found that high expression of IQGAP3 and PIK3C2B correlated with tumour stage and vessel invasion in human CRC, whereby patients with high expression of both in tumours had a worse prognosis compared with patients with single-positive or double-negative tumours.
Conclusion
The results of our current study and corresponding previous studies provide evidence that IQGAP3 is elevated in CRC and promotes colon cancer growth and metastasis by regulating PIK3C2B activation.
Keywords: IQGAPs, PIK3C2B, colon cancer, invasion, prognosis
Background
Colon cancer, or colorectal cancer (CRC), is a common type of tumour of the digestive system. Previous studies have indicated that the progression of colon epithelial cells from normal adenoma to carcinoma entails multiple steps involving many oncogene or tumour-suppressor gene mutations, such as K-RAS, APC, and T-p53.1,2 Although these mutations are important during CRC development, they are still not able to explain all the mechanisms of malignant behavior in CRC.1,2 Mutation of APC genes in CRC results in constitutive activation of the β-catenin pathway, with transcriptional activation of many downstream genes, such as c-myc and cyclin D1.3,4 Additionally, neoplasm transformation by mutation of Ras and other oncoproteins often contributes to EGFR pathway activation and resistance to B-Raf and tyrosine kinase inhibitor therapies, such as vemurafenib, cetuximab, sorafenib and dabrafenib.5 In fact, approximately 30% of human tumours carry Ras and B-Raf mutations. These mutations are necessary for ERK pathway activation and result in tumour invasion and proliferation promotion.6 Ras mutation and pathway activation are common in CRC. Although mutations in Cdc42 and Rac1 are rare in cancer, many researchers have reported that their abnormal expression of is associated with Ras activation in cancer.7,8 Furthermore, the peroxisome proliferator activated receptor gamma (PPARγ) gene serves as a tumour suppressor in CRC. Abnormal expression of PPARγ leads to an increased risk of carcinogen-induced CRC.9 Therefore, CRC carcinogenesis is a complex process involving epithelial dysplasia and multiple abnormal signaling pathways. Nonetheless, the detailed mechanism of CRC development still needs to be further explored.
Members of the IQ domain‐containing GTPase‐activating protein (IQGAP) family contain an order domain of CH, WW, IQ and GRD from the NH2‐terminus. There are three paralogues of IQGAPs in humans, named IQGAP1, IQGAP2 and IQGAP3, which have similar domain compositions and may have similar or divergent functions, tissue expression and subcellular localization. These proteins participate in several cellular processes, such as cell adhesion, invasion, proliferation, exocytosis, intracellular signaling, cytoskeletal dynamics, and neuronal morphogenesis, and they have been defined as important factors in cytokinesis in yeast.10 IQGAPs are also reported to be involved in mitogen-activated protein kinase (MAPK) pathway regulation.11,12 IQGAP1 is the best characterized IQGAP isoform to date and is overexpressed in many human cancers.13–15 For example, IQGAP1 is associated with increased tumour progression and angiogenesis in breast cancer.16 By interacting with many MAPK pathway components, IQGAP1 inhibits Ras-driven tumorigenesis by targeting the MAPK axis and mediates optimal ERK activation.17,18 In many tumours, such as CRC and pancreatic cancer, activation of Rho GTPases, Cdc42 and Rac1 by Ras and promotion of other oncoproteins occurs. IQGAP1 expression has been reported to enhance activation of Cdc42 and Rac1 by targeting IQGAP1/Cdc42 and IQGAP1/Rac1 complexes in cancer cells, thereby increasing tumourigenesis.19–21 Unlike IQGAP1, which is ubiquitously expressed, IQGAP2 is mainly expressed in the liver and to a lesser extent in other tissues, such as the testis, kidney, platelets, salivary glands, prostate, thyroid and stomach. Despite 62% similarity in the amino acid sequences of IQGAP2 and IQGAP1, IQGAP1 is defined as an oncogene, whereas IQGAP2 appears to act as a tumour-suppressor gene in hepatocellular tumours;22 indeed, they are reciprocally altered in hepatocellular cancer. Schmidt et al23 reported that IQGAP2-knockout mice will automatically develop hepatocellular cancer in an IQGAP1-dependent manner. In addition, lower expression of IQGAP2 has been found in prostate24,25 and gastric carcinomas.26 However, the detailed mechanisms of IQGAP2 in cancer are still unclear. Since it was identified in 2007, IQGAP3 is much less characterized than the other two molecules. Nevertheless, it is reported to be detected in only a few normal tissues, such as the small intestine, lung, brain, colon and testis.27 Recently, some studies revealed have that, similar to IQGAP1, IQGAP3 promotes proliferation and correlates with increased migration and invasion abilities in cancer cells by promoting extracellular signal regulated kinase (ERK) activation.28–30 Although IQGAPs have been suggested to play a core regulatory function in many tumours, their roles in CRC remain mostly unclear.
In our current study, we experimentally investigated expression of IQGAPs in CRC, and the results provide evidence showing that IQGAP3 promotes CRC progression associated with phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta (PIK3C2B or PIK3C2β) regulation and predicts poor prognosis. IQGAP3 overexpression in CRC may therefore play a tumourigenic role in CRC.
Methods
Public Database Sources
ONCOMINE Datasets
ONCOMINE is an online cancer microarray gene expression data site (www.oncomine.org) that we used it to analyze transcriptional expression levels of IQGAPs in CRC. mRNA expression changes of IQGAPs in CRC specimens were compared with normal tissue by using Student’s t-test.
GEPIA Dataset
GEPIA is an interactive web server for analyzing RNA sequencing expression data of tumours and normal samples from the TCGA and GTEx projects. It provides customizable functions according to cancer type, such as differential expression, pathological stage, survival analysis, dimensionality reduction analysis and correlation gene analysis.31
TCGA and cBioPortal Data
A 379-case Colorectal Adenocarcinoma (TCGA, Provisional) dataset was selected for further analyses of IQGAPs using cBioPortal (https://www.cbioportal.org).32,33 The dataset includes gene mutations, copy number alterations (CNA) from GISTIC, mRNA expression Z-scores (RNA Seq V2 RSEM) and protein expression Z-scores (RPPA). A coexpression network was constructed and analyzed according to the online instructions of the cBioPortal website. Then, based on Spearman correlation value (>0.3), a Venn diagram was drawn by using the online Venn diagrams tool (http://bioinformatics.psb.ugent.be/webtools/Venn/).
Samples and Patients
Forty fresh CRC tissue samples were collected from the Department of General Surgery of Colorectal Cancer of Zhejiang Cancer Hospital from 2017 to 2018. The tumour histologic type was diagnosed by three independent pathologists and classified according to the American Joint Committee on Cancer (AJCC) classification (8th and 7th versions) of CRC tumours.34 Matched normal colon epithelial tissues were collected 5 cm away from the tumour site. None of the patients received chemotherapy prior to surgery. This study was approved by the Ethics Committee of Zhejiang Cancer Hospital with written informed consent. Upon admission, an additional 120 CRC FFPE (from 2012–2013) samples were collected for tumour microarray (TMA) preparation. In brief, a core tumour area (tumour occupying >50%) and adjacent normal tissue of every GC patient’s wax block were determined and circled by two pathologists through HE-stained sections. Then, by using a trephine, the core cancer or normal tissues (approximately 2 mm diameter) were taken from individual paraffin-embedded GC blocks and individually arranged into recipient paraffin tissue microarray blocks.
All patients were followed up for 5 years until December 2018, and the survival time was calculated from the date of surgery to the date of death or deadline of the follow-up period. The age of the CRC patients ranged from 17 to 80 years (with a median age of 57.7 years). The clinicopathological characteristics of this cohort are summarized in Table 2.
Table 2.
Association Between IQGAP3, PIK3C2B Expression and Clinicopathological Factors
| Variables | IQGAP3 | PIK3C2B | ||||
|---|---|---|---|---|---|---|
| Negative | Positive | t/χ2, P-value | Negative | Positive | t/χ2, P-value | |
| Age, years | 0.249, 0.618 | 0.013, 0.909 | ||||
| ≤60 | 25(49.0%) | 37(53.6%) | 22(52.4%) | 40(51.3%) | ||
| >60 | 26(51.0%) | 32(46.4%) | 20(47.6%) | 38(48.7%) | ||
| Sex | 0.260, 0.610 | 0.155, 0.694 | ||||
| Male | 34(66.7%) | 49(71.0%) | 30(71.4%) | 53(67.9%) | ||
| Female | 17(33.3%) | 20(29.0%) | 12(28.6%) | 25(32.1%) | ||
| Location | 0.198, 0.906 | 1.767, 0.413 | ||||
| Left | 8(15.7%) | 10(14.5%) | 4(9.5%) | 14(17.9%) | ||
| Right | 17(33.3%) | 21(30.4%) | 13(31.0%) | 25(32.1%) | ||
| Transverse | 26(51.0%) | 38(55.1%) | 25(59.5%) | 39(50.0%) | ||
| Tumor size (cm) | 0.612, 0.434 | 0.006, 0.938 | ||||
| <5 | 31(60.8%) | 37(53.6%) | 24(57.1%) | 44(56.4%) | ||
| ≥5 | 20(39.2%) | 32(46.4%) | 18(42.9%) | 34(43.6%) | ||
| Differentiation | 7.151, 0.028 | 3.070, 0.215 | ||||
| Well | 6(11.8%) | 2(2.9%) | 5(11.9%) | 3(3.8%) | ||
| Moderate | 19(37.3%) | 17(24.6%) | 13(31.0%) | 23(29.5%) | ||
| Poor | 26(51.0%) | 50(72.5%) | 24(57.1%) | 52(66.7%) | ||
| Invasion depth | 4.940, 0.176 | 2.363, 0.501 | ||||
| T1 | 3(5.9%) | 0(0.0%) | 0(0.0%) | 3(3.8%) | ||
| T2 | 11(21.6%) | 12(17.4%) | 8(19.0%) | 15(19.2%) | ||
| T3 | 29(56.9%) | 47(68.1%) | 29(69.0%) | 47(60.3%) | ||
| T4 | 8(15.7%) | 10(14.5%) | 5(11.9%) | 13(16.7%) | ||
| Lymph node metastasis stages | 9.123, 0.010 | 0.725, 0.696 | ||||
| N0 | 15(29.4%) | 6(8.7%) | 9(21.4%) | 12(15.4%) | ||
| N1 | 5(9.8%) | 12(17.4%) | 6(14.3%) | 11(14.1%) | ||
| N2 | 31(60.8%) | 51(73.9%) | 27(64.3%) | 55(70.5%) | ||
| TNM stages | 13.606, 0.003 | 8.749, 0.033 | ||||
| I | 6(11.8%) | 0(0.0%) | 1(2.4%) | 5(6.4%) | ||
| II | 9(17.6%) | 6(8.7%) | 8(19.0%) | 7(9.0%) | ||
| III | 28(54.9%) | 56(81.2%) | 32(76.2%) | 52(66.7%) | ||
| IV | 8(15.7%) | 7(10.1%) | 1(2.4%) | 14(17.9%) | ||
| TNM stages 2 | 8.717, 0.003 | 0.691, 0.406 | ||||
| I+II | 15(29.4%) | 6(8.7%) | 9(21.4%) | 12(15.4%) | ||
| III+IV | 36(70.6%) | 63(91.3%) | 33(78.6%) | 66(84.6%) | ||
| Lymphatic invasion | 3.173, 0.075 | 5.199, 0.023 | ||||
| No | 22(43.1%) | 19(27.5%) | 20(47.6%) | 21(26.9%) | ||
| Yes | 29(56.9%) | 50(72.5%) | 22(52.4%) | 57(73.1%) | ||
| Vessel invasion | 4.125, 0.042 | 9.111, 0.003 | ||||
| No | 24(47.1%) | 20(29.0%) | 23(54.8%) | 21(26.9%) | ||
| Yes | 27(52.9%) | 49(71.0%) | 19(45.2%) | 57(73.1%) | ||
| Distant metastasis | 0.823, 0.364 | 6.049, 0.014 | ||||
| No | 43(84.3%) | 62(89.9%) | 41(97.6%) | 64(82.1%) | ||
| Yes | 8(15.7%) | 7(10.1%) | 1(2.4%) | 14(17.9%) | ||
| PIK3C2B expression | 5.669, 0.017 | |||||
| Negative | 24(47.1%) | 18(26.1%) | ||||
| Positive | 27(52.9%) | 51(73.9%) | ||||
Notes: The pathological staging follows to the WHO’s (2010) pathological classification of CRC; Invasion Depth (T Grade) grade T1a and T1b are classed as T1, T4a and T4b are classed as T4; Lymphatic Metastasis (N Grade) grade N1 includes N1a, N3b and N3c; TNM grade IIa, IIb and IIc are classed as TNM grade II, IIIa, IIIb and IIIc are classed as TNM grade III, includes IVa and IVb are classed as grade IV.
RNA Isolation and Real-Time PCR
Total mRNA was isolated according to the protocol of the RNAsimple Total RNA Kit (DP419, TIANGEN Biotech Co., Ltd., China). cDNA was synthesized by using PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A, TaKaRa Biotechnology Co., Ltd., China). Real-time PCR (qRT-PCR) was performed using a Bio-Rad CFx96 system (Bio-Rad, USA) with SYBR Premix ExTaq Kit (DRR081A; TaKaRa Biotechnology Co., Ltd., China). All experiments were carried out in triplicate. Relative gene expression levels were normalized to the internal standard GAPDH and calculated by using the 2-∆∆ct method. The primers used for candidate genes were selected from PrimerBank (https://pga.mgh.harvard.edu/primerbank/); the sequences are listed in Supplementary file 1, Part 1. The qPCR reaction conditions were as follows: initial denaturation for 4 minutes at 95°C, followed by 35 cycles of denaturation at 95°C for 10 seconds, annealing at 55°C for 10 seconds, and extension at 72°C for 10 seconds.
Cell Culture and Transfection
CRC cell lines (HCT116, SW480, SW620 and DLD1) were purchased from the Cell Bank of Shanghai Institute of Cell Biology and cultured in 10% FBS DMEM culture medium (HyClone, USA) at 37°C and 5% CO2.
The pcDNA3.1-IQGAP3 plasmid and PIK3C2B siRNA were constructed and purchased from Guangzhou RiboBio Co., Ltd. of China. In brief, 4×105 CRC cells per well were seeded into a six-well plate. After the cells were attached, they were transfected with pcDNA3.1-IQGAP3, PIK3C2B siRNA or the corresponding negative controls at a final concentration of 1 µg of plasmid or 10 nM of siRNA, respectively, according to the Lipofectamine 3000 transfection reagent manufacturer’s protocol (Thermo Fisher Scientific, USA). After transfection, the cells were collected for future assays, such as invasion, Western blotting, and qRT-PCR experiments.
Invasion Assays
Invasion assays were performed using invasion chambers precoated with Matrigel (354480; BD, USA). CRC cells pretransfected with or without pcDNA3.1-IQGAP3 plasmid and PIK3C2B siRNA and controls (2×105 for invasion assays) were seeded into the upper chamber with optimal serum-free MEM medium; 30% FBS DMEM medium was added to the lower chamber as a chemoattractant. Twenty-four or forty-eight hours later, the upper chambers were removed, and noninvading cells in the upper chambers were removed by using cotton swabs. The cells remaining were stained with crystal violet staining solution (E607309, Sangon Biotech, China), and the number of migrating or invading cells was calculated in five random fields under a microscope (×200).
Western Blotting
In brief, RIPA lysis buffer (Beyotime Biotech, China) was used to extract total proteins from cells. Thirty micrograms of total protein from each sample was separated by 10% SDS-PAGE and then transferred onto a 0.45-μm PVDF membrane. The PVDF membranes were blocked with 5% BSA in TBST at room temperature for 2 h and then with primary antibodies against IQGAP3 (ab219354, at 1:2000 dilution, Abcam, USA), PIK3C2B (ab231122, at 1:1000 dilution, Abcam, USA), and GAPDH (M1310-2, at 1:3000 dilution; HuaBio Inc, China) overnight at 4°C. The membranes were incubated with the corresponding horseradish peroxidase (HRP)-labelled goat anti-rabbit or anti-mouse IgG antibody for 1h. After washing, the ChemiDoc chemiluminescence system (BIO-RAD, USA) was used to detect the signal of each blot.
CRC TMA Immunohistochemistry and Immune Signal Evaluation
After conventional dewaxing hydration and antigen repair, 4-μm TMA sections were incubated with primary antibodies against IQGAP3 (1:100 dilution, ab219354, Abcam, USA) and PIK3C2B (1:100 dilution, ab231122, Abcam, USA) at 4°C overnight. After the sections were rinsed three times in PBST for 5 min each, they were incubated with HRP-labelled secondary antibodies for 1 h at room temperature. Signal detection was performed using 3,3-diaminobenzidine (DAB) reagent, and the slides were stained with haematoxylin. For each sample, the immunostaining intensity levels were evaluated under a microscope independently by two pathologists in the absence of clinical information, according to the following criteria: the average staining signal intensity (range from 0–3) and the proportion of positively stained tumour cells (0, <5%; 1, 5–25%; 2, 26–50%; 3, 51–75%; and 4, 76–100%). Finally, a composite score was calculated by multiplying the intensity and percentage scores; a score ranging from 0 to 4 was defined as negative, and a score of 5 to 12 was defined as positive.
Statistical Analysis
All statistical analyses were carried out by using Prism software and SPSS version 13.0 (SPSS Inc., USA). Statistical differences in IQGAP and PIK3C2B expression between cancer and normal tissues were calculated by using a 2-tailed paired Student’s t-test. Data from cell invasion, luciferase, WB, and PCR assays are shown as the mean ± SE, and significant differences were determined by independent sample t-tests. The Chi-squared test was applied to analyze the relationship between IQGAP3 and PIK3C2B expression and clinicopathological characteristics. The Kaplan–Meier method and the Log rank test were used to draw survival curves. A multivariate Cox regression model was employed to analyze the significance of survival-related variables in this CRC cohort. A P value smaller than 0.05 (P<0.05) was defined as statistically significant.
Results
IQGAP Expression in Patients with CRC
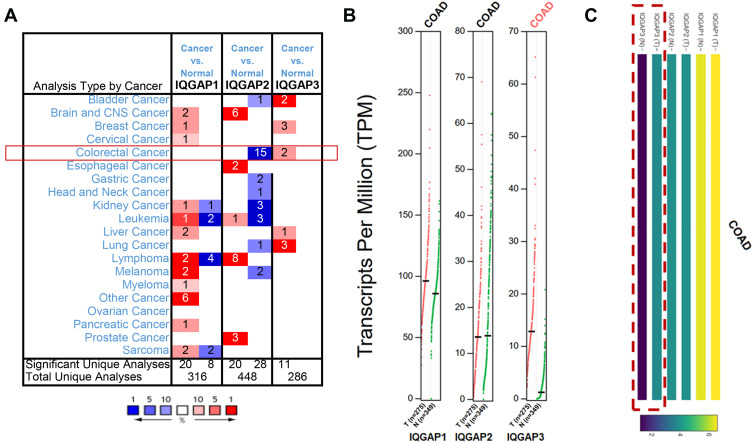
Comparing the transcriptional levels of three IQGAPs from ONCOMINE databases, the mRNA expression levels of IQGAP3 were significantly upregulated in two CRC datasets (Figure 1A).
Figure 1.
Transcriptional levels of IQGAPs in different types of cancer public databases. (A) Transcriptional levels of IQGAPs in CRC in the ONCOMINE public database. (B and C) Transcriptional levels of IQGAPs in CRC in the GEPIA public database. Expression levels of IQGAP3 were higher in CRC tissues than in normal tissues; those of IQGAP1 and IQGAP2 showed no difference.
Abbreviation: COAD, colon adenocarcinoma.
In the TCGA and Hong CRC datasets,35,36 IQGAP3 was overexpressed with fold changes of 2.384 and 2.581 (both P<0.05) compared with normal samples, respectively. However, lower IQGAP2 mRNA expression levels were observed in 15 datasets (Table 1), and there was no dataset for IQGAP1 (Figure 1, Table 1). Then, we used the GEPIA dataset (the Match TCGA normal and GTEx data set) to analyze the difference in IQGAP expression between CRC and normal tissues. With a slight difference from the Oncomine database, the results from GEPIA showed that IQGAP3 levels were higher in CRC tissues, with no difference in IQGAP1 or IQGAP2 (Figure 1B and C).
Table 1.
Significant Changes in IQGAP Expression at the Transcriptional Level Between Different Types of CRC Tissues and Normal Tissues (ONCOMINE Database)
| Gene | Cancer vs Normal | Data Set | Fold Change | P value | t-test | Ref. |
|---|---|---|---|---|---|---|
| IQGAP1 | N/A | N/A | N/A | N/A | N/A | |
| IQGAP2 | Colon Adenocarcinoma vs Normal | Kaiser Colon Statistics | −3.25 | 2.23E-16 | −12.60 | [57] |
| Cecum Adenocarcinoma vs Normal | Kaiser Colon Statistics | −2.656 | 1.59E-7 | −7.826 | ||
| Colon Mucinous Adenocarcinoma vs Normal | Kaiser Colon Statistics | −2.133 | 4.11E-7 | −8.086 | ||
| Rectosigmoid Adenocarcinoma vs Normal | Kaiser Colon Statistics | −2.932 | 2.89E-5 | −6.7210 | ||
| Colon Adenocarcinoma vs Normal | Ki Colon Statistics | −2.829 | 2.11E-5 | −8.995 | [58] | |
| Colorectal Carcinoma vs Normal | Hong Colorectal Statistics | −5.396 | 7.18E-23 | −14.578 | [35] | |
| Rectal Adenocarcinoma vs Normal | Gaedcke Colorectal Statistics | −3.874 | 1.23E-36 | −19.888 | [59] | |
| Colon Adenocarcinoma vs Normal | TCGA Colorectal Statistics | −3.528 | 1.16E-24 | −14.525 | [36] | |
| Rectosigmoid Adenocarcinoma vs Normal | TCGA Colorectal Statistics | −5.547 | 2.49E-9 | −16.820 | ||
| Rectal Adenocarcinoma vs Normal | TCGA Colorectal Statistics | −3.584 | 1.09E-19 | −11.947 | ||
| Colon Adenoma Epithelia vs Normal | Skrzypczak Colorectal 2 Statistics | −4.411 | 1.42E-7 | −22.666 | [60] | |
| Colon Carcinoma vs Normal | Skrzypczak Colorectal 2 Statistics | −2.739 | 5.43E-8 | −12.184 | ||
| Colon Carcinoma Epithelia vs Normal | Skrzypczak Colorectal 2 Statistics | −2.192 | 5.28E-8 | −15.691 | ||
| Colon Adenoma vs Normal | Skrzypczak Colorectal 2 Statistics | −5.899 | 3.74E-6 | −13.834 | ||
| Colorectal Carcinoma vs Normal | Skrzypczak Colorectal Statistics | −2.847 | 1.38E-11 | −8.36 | [60] | |
| IQGAP3 | Rectal Mucinous Adenocarcinoma vs Normal | TCGA Colorectal Statistics | 2.384 | 7.72E-6 | 6.864 | [36] |
| Colorectal Carcinoma vs Normal | Hong Colorectal Statistics | 2.581 | 1.72E-7 | 5.834 | [35] |
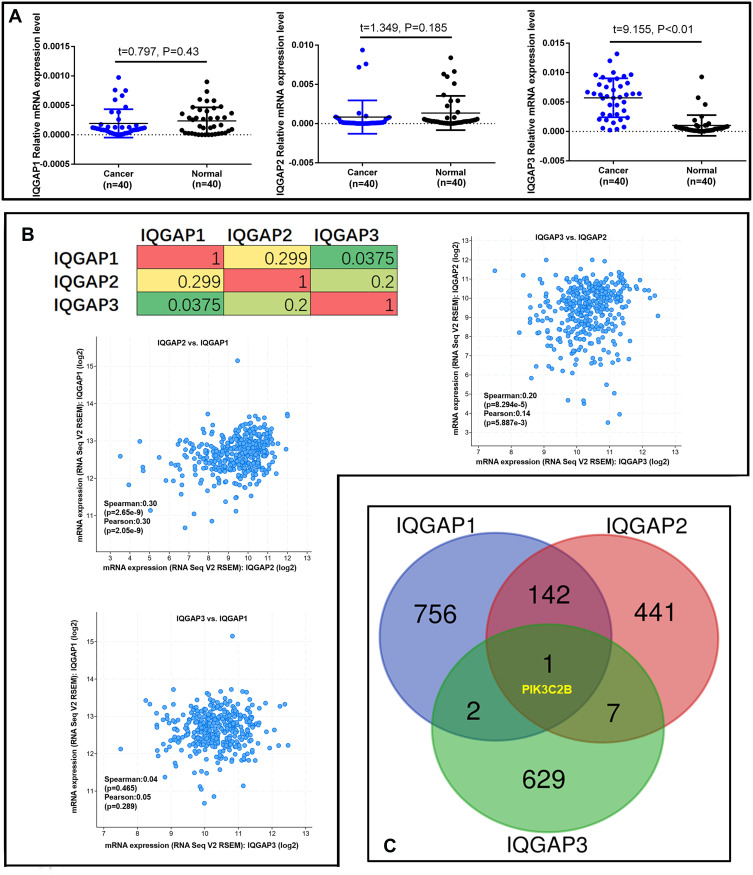
To validate the real expression level of IQGAPs in our clinical CRC tissues, 40 paired CRC and normal colon epidermal tissues were collected for qRT-PCR analysis. Consistent with the GEPIA database, the qRT-PCR results from our clinical samples indicated that IQGAP3 expression in CRC tissues was significantly higher than that in matched normal tissues (Figure 2A). In contrast, IQGAP2 expression was slightly lower in CRC tissue but not significantly different (P>0.05), with no difference in IQGAP1 expression (Figure 2A). Thus, we focused on IQGAP3 and explored the potential mechanisms of CRC progression.
Figure 2.
IQGAP gene expression and frequently altered neighbouring gene analysis in CRC (cBioPortal). (A) Real-time PCR analysis of IQGAP gene expression in 40 paired CRC tissues. P<0.05 was significant. (B) Correlations of IQGAPs with each other by analysing their mRNA expression (RNA Seq V2 RSEM) via the cBioPortal online tool for CRC (TCGA, Provisional). (C) The Venn diagram results show that PIK3C2B is positively associated with IQGAP alterations in CRC.
Abbreviation: t, t value.
Predicted IQGAPs Frequently Alter Neighbouring Genes in CRC Patients
As described above and in a previous study, IQGAP3 may function in a manner similar to that of IQGAP1 in cell proliferation and invasion promotion via extracellular signal regulated kinase (ERK) activation.37,38 We analyzed IQGAP alterations, correlations, and networks by using the Colorectal Adenocarcinoma (TCGA, Provisional) dataset from the cBioPortal database, which contains samples (379 patients/382 samples) with RNA Seq V2 data. Pearson’s correction correlations of IQGAP levels with each other were determined by analyzing their RNA Seq V2 RSEM data in the Colorectal Adenocarcinoma (TCGA, Provisional) dataset via the cBioPortal online tool. The results showed a positive correlation of IQGAP3 with IQGAP2 and of IQGAP1 with IQGAP2 (Figure 2B). We then downloaded coexpressed genes from the Colorectal Adenocarcinoma (TCGA, Provisional) dataset in the cBioPortal database and drew a Venn diagram by setting the cut-off value at Spearman correlation value >0.3 (Supplementary file 1, Parts 3–5). Finally, an interesting result was that only one gene, PIK3C2B, was closely and positively associated with IQGAP alterations in CRC (Figure 2C).
IQGAP3 Promotes CRC Invasiveness Associated with PIK3C2B in vitro
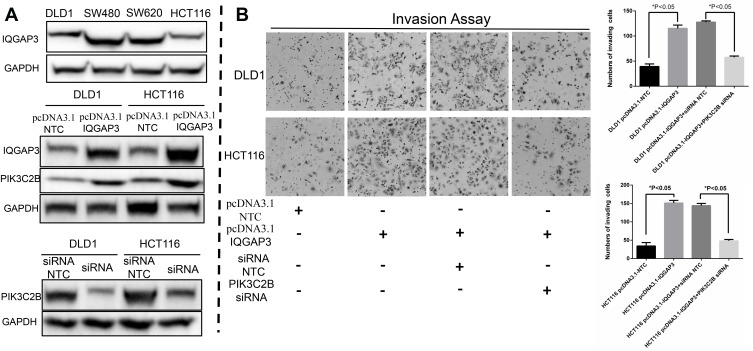
As one of the three isoforms of class II PI3Ks, PIK3C2B has a substrate specificity targeted to PI and PI(4)P, which can subsequently activate pathways of RTKs, such as AKT-mTOR and EGFR signaling pathways. Moreover, PIK3C2B can regulate different cellular functions, such as cell proliferation, invasion and cancer progression.39,40 In our current study, we discovered that IQGAP3 was expressed at relatively lower levels in DLD1 and HCT116 CRC cell lines (Figure 3A) and that cell invasion abilities were significantly increased after exogenous overexpression of IQGAP3 (Figure 3B). At the same time, PIK3C2B expression was increased at both the protein and mRNA transcription levels (Figure 3A and Supplementary file 1, Part 2). Furthermore, these effects of promoting invasion in response to IQGAP3 overexpression in these two CRC cell lines was blocked by pretransfecting them with PIK3C2B siRNA (Figure 3B). These results indicate that PIK3C2B activation is involved in the enhancement of invasiveness during IQGAP3 overexpression.
Figure 3.
IQGAP3 promotes CRC invasiveness associated with PIK3C2B in vitro. (A) WB results of IQGAP3 and PIK3C2B. (B) Results of the invasion assay. CRC cell invasion abilities were significantly increased after exogenous overexpression of IQGAP3, and this invasion-promoting effect was blocked by pretransfecting the cells with PIK3C2B siRNA.
Clinical Significance of IQGAP3 and PIK3C2B in CRC
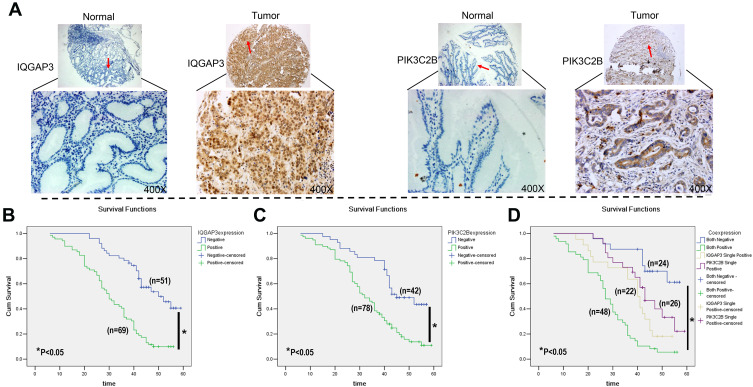
IHC staining for IQGAP3 and PIK3C2B in TMA was used to further analyse correlations of IQGAP3, PIK3C2B, pathological parameters and prognosis (Table 2). In this cohort, the mean age of the patients was 57.70±11.17 years. Elevated levels of IQGAP3 and PIK3C2B were detected in 57.5% (69/120) and 65% (78/120) of cases, respectively (Figure 4A). Further statistical analysis between IQGAP3 and PIK3C2B expression and clinical pathological parameters was performed. Overall, we found that higher levels of IQGAP3-positive staining in tumours correlated with differentiation, lymph node metastasis stage (N stage), TNM stage and vessel invasion (P<0.05, Table 2); we also found that elevated PIK3C2B was associated with TNM stage, lymph node invasion, vessel invasion and distant metastasis (P<0.05, Table 2). Additionally, a positive relationship was found between IQGAP3 and PIK3C2B expression (P<0.05, Table 2). Conversely, there were no associations between age, sex, location, or histological type of CRC with either IQGAP3 or PIK3C2B expression (Table 2).
Figure 4.
IHC staining of IQGAP3 and PIK3C2B in CRC tissue and Kaplan–Meier survival curves. (A) Typical TMA array images of IQGAP3 and PIK3C2B in CRC tissue and normal tissue. Original magnification, ×100 and ×400. (B–D) Kaplan–Meier survival curves of IQGAP3 and PIK3C2B expression in CRC.
In this cohort of CRC patients (n=120), the mean survival time in IQGAP3-positive patients was significantly shorter than that in IQGAP3-negative patients (31.26 ± 1.59 months vs 47.31 ± 1.79 months, P<0.05). Furthermore, the 5-year survival rate in IQGAP3-positive patients was lower than that in IQGAP3-negative patients (10.1% vs 51.0%, P<0.05, Figure 4B, respectively). Patients with positive PIK3C2B expression also had a worse survival time and rate compared to patients without PIK3C2B expression (33.87 ± 1.62 months vs 45.18 ± 2.03 months and 16.7% vs 47.6%, respectively, P<0.05, Figure 4C). Moreover, the patients with positive expression of both IQGAP3 and PIK3C2B had a worse survival rate (6.3%) than did those with positive expression of only IQGAP3 (18.2%) or PIK3C2B (38.5%) or expression of neither (66.7%). Survival time was also shorter in patients positive for both IQGAP3 and PIK3C2B (28.09 ± 1.74 months) than in those positive for IQGAP3 (37.96 ± 2.56 months) or PIK3C2B (43.99 ± 2.46 months) or negative for both (50.07 ± 2.23 months) (P<0.05, Figure 4D). Multivariate Cox regression analysis showed vascular invasion, IQGAP3 expression and PIK3C2B expression to be independent prognostic factors in this cohort of CRC patients (Table 3).
Table 3.
Cox Regression and Multivariates Analysis in 120 CRC Patients
| Clinicopathological Parameters |
95% Confidential Interval | Hazard Ratio | P value | |
|---|---|---|---|---|
| Lower | Upper | |||
| PIK3C2B expression | 1.508 | 4.737 | 11.342 | 0.001 |
| IQGAP3 expression | 2.045 | 6.994 | 17.991 | 0.000 |
| Vascular invasion | 1.251 | 3.668 | 7.707 | 0.006 |
Discussion
In humans, the IQGAP family consists of three proteins, IQGAP1, IQGAP2 and IQGAP3.13 Among them, IQGAP1 is characterized as participating in the activation of several signaling pathways and protein-protein interactions. Although IQGAP2 and IQGAP3 contain all the domains found in IQGAP1, the biological roles of these two molecules remain poorly understood. Many proteins, such as E-cadherin, Cdc42, β-catenin, Rac1 and MAPK kinase components, are reported to bind with IQGAP1 to promote cancer progression.19,27,41 However, the actual role of IQGAPs in CRC is still unknown. Here, we summarize the analysis of IQGAPs in CRC. Although IQGAP1 expression was slightly increased in colon cancer tissue in the GEPIA database, IQGAP2 was found to be widely downregulated in 15 datasets in the ONCOMINE database. In our 40 paired clinical CRC tissues, we did not find a significant change in IQGAP1 and IQGAP2 expression between CRC and adjacent normal tissues. The reason for this difference, especially for IQGAP2, may be that the tumours and normal tissues used in the databases did not originate from matched tissues of the same patient. Interestingly, consistent with the ONCOMINE and GEPIA databases, we confirmed that IQGAP3 was significantly increased in CRC tissues in our cohort. IQGAP family members have similar domain compositions and may have similar functions in cancer progression. IQGAP1 is reported to be upregulated in many human cancers and associated with enhanced malignant behaviours.42–44 In contrast, the characteristics of IQGAP3 are less studied, though it may have a role similar that of to IQGAP1 in tumorigenesis. In our study, we compared IQGAP3 expression in 40 paired CRC tissues, which provided important evidence showing IQGAP3 to be upregulated in CRC tissue. Further analysis in clinical CRC FFPE tissues (TMA) showed that a high level of IQGAP3 was related to tumour differentiation, lymph node metastasis, TNM stage, vascular invasion and poor prognosis, which supports a tumorigenic role for IQGAP3 in CRC. An in vitro transwell assay also indicated that decreased IQGAP3 levels in CRC cells may inhibit cancer cell invasion abilities.
Phosphoinositide 3 kinases (PI3Ks) play an essential role in signal transduction, cell differentiation, proliferation and cancer progression in many tumours.39,40,45 Mammals have eight PI3Ks, and they have been divided into three classes based on substrate sequence similarity. Among them, class I PI3K isoforms are activated by G protein-coupled receptors (GPCRs) or tyrosine kinases and are important for protein kinase B and Akt pathway activation and linked to tumourigenesis.45,46 Based on its core role in intracellular signal transduction, abnormal expression of the PI3K network is a common event in human tumours. In contrast to class I PI3Ks, class II PI3Ks remain poorly characterized. However, class II PI3K can be divided into three isoforms, PIK3C2α, PIK3C2β and PIK3C2γ, serving as downstream targets of tyrosine receptor kinases, such as EGFR, PDGFR, C-KIT and IR. Previous studies have reported that these factors may play an important role during cancer development.46–48 Activation of PIK3C2B is predominately associated with cell motility, differentiation and apoptosis, and it has important roles during tumourigenesis.49 In addition, amplification and somatic mutations of PIK3C2B have been detected in glioblastoma and non-small cell lung cancer,50–52 but the function of these mutations is still unclear. In esophageal squamous cell tumours, expression of PIK3C2B is associated with cell migration and tumour metastasis via Akt signaling pathway activation and cisplatin resistance.53 Furthermore, recent reports have documented that PIK3C2B promotes the invasiveness of cancer cells via Rho family GTPase activation and target inactivation of PIK3C2B, which potentiates insulin signaling and sensitivity.54–56 Thus, PIK3C2B plays a positive role in cancer progression. In our current study, we analyzed coexpressed genes of IQGAPs downloaded from the cBioPortal database and found that PIK3C2B gene expression correlates positively with IQGAP3 gene expression. In addition, in vitro assays showed that the level of PIK3C2B expression was increased after exogenous overexpression of IQGAP3, resulting in a promotion of cell invasion ability. Furthermore, these promotion effects of IQGAP3 overexpression were blocked by pretransfecting the cells with PIK3C2B siRNA. Based on IHC, IQGAP3 was positively associated with high PIK3C2B expression in CRC, and high IQGAP3 expression and high PIK3C2B expression both correlated with TNM stage and vessel invasion in human CRC. In addition, patients with high expression of both IQGAP3 and PIK3C2B in tumours had the worst prognosis compared with patients with single-positive or double-negative tumours.
Considering our previous studies and our current results, we conclude that IQGAP3 may promote CRC invasion and progression by regulating PIK3C2B expression and its associated downstream signaling pathway. Nevertheless, in an effort to suppress CRC progression, additional research is needed to determine the exact mechanisms underlying PIK3C2B-mediated functions in CRC cells, such as the mechanisms involved in targeting the PIK3C2B pathway in CRC treatment.
Conclusion
In summary, we performed an integrative analysis of the IQGAP family in CRC and identified that expression of IQGAP3 is upregulated in CRC. Furthermore, IQGAP3 may promote CRC invasion and progression by regulating PIK3C2B expression. Thus, targeting PIK3C2B or the corresponding PI3K pathway may serve as a new therapeutic strategy for CRC patients with high IQGAP3 expression.
Acknowledgments
The authors thank Wenjuan Xu for the TMA and IHC staining and technical support. We also thank the members of our laboratory for technical support and stimulating discussion.
Funding Statement
The TMA production was funded by the Medicine and Health Research Foundation of Zhejiang (2018256123). All reagents for the in vitro experiments were supported by the China National Natural Science Foundation (81502090). The fees for technical staff and articles regarding English editing were funded by the Natural Science Foundation of Zhejiang Provincial (Y18H160037).
Abbreviations
CRC, Colorectal cancer; IQGAPs, IQ Motif-Containing GTPase-Activating Protein Family; IQGAP1, IQ Motif-Containing GTPase-Activating Protein 1; IQGAP2, IQ Motif-Containing GTPase-Activating Protein 2; IQGAP3, IQ Motif-Containing GTPase-Activating Protein 3; qRT-PCR, Real-time fluorescent quantitative polymerase chain reaction; IHC, Immunohistochemistry; PIK3C2B, Phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 beta; K-RAS, KRAS proto-oncogene; APC, APC regulator of the WNT signaling pathway; T-p53, Tumour protein p53; CDC42, Cell division cycle 42; Rac1, Rac family small GTPase 1; MAPK, Mitogen-activated protein kinase; HRP, Horseradish peroxidase; TMA, Tumour tissue microarray; ERK, Extracellular signal regulated kinase; PI3Ks, Phosphoinositide 3 kinases; EGFR, Epidermal growth factor receptor; PDGFR, Platelet-derived growth factor receptor.
Data Sharing Statement
We declare that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
Ethics Approval and Consent to Participate
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. This article does not contain any studies with animals performed by any of the authors. The tumour tissues and FFPE samples collection in this study were approved by the Ethics Committee of Zhejiang Cancer Hospital with written informed consent. Informed consent was obtained from all individual adult participants included in the study. And the written informed consent was also taken from the parent or legal guardian of any patient who under the age of 18 years old.
Author Contributions
ZL, XH and YZ designed and performed most of the experiments, analysed the data, prepared the figures, including drawings, and wrote the manuscript; JM performed the IHC and data analyses. XL, DL, HJ, YL and YC performed the public database analysis as well as the in vitro and Western blot experiments and data collection. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
There is no conflict of interest in relation to this article for any of the authors.
References
- 1.Hong SN. Genetic and epigenetic alterations of colorectal cancer. Intest Res. 2018;16(3):327–337. doi: 10.5217/ir.2018.16.3.327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jeon CH, Lee HI, Shin IH, Park JW. Genetic alterations of APC, K-ras, p53, MSI, and MAGE in Korean colorectal cancer patients. Int J Colorectal Dis. 2008;23(1):29–35. doi: 10.1007/s00384-007-0373-0 [DOI] [PubMed] [Google Scholar]
- 3.Segditsas S, Tomlinson I. Colorectal cancer and genetic alterations in the Wnt pathway. Oncogene. 2006;25(57):7531–7537. doi: 10.1038/sj.onc.1210059 [DOI] [PubMed] [Google Scholar]
- 4.Novellasdemunt L, Antas P, Li VS. Targeting Wnt signaling in colorectal cancer. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;309(8):C511–521. doi: 10.1152/ajpcell.00117.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Knickelbein K, Zhang L. Mutant KRAS as a critical determinant of the therapeutic response of colorectal cancer. Genes Dis. 2015;2(1):4–12. doi: 10.1016/j.gendis.2014.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nandan MO, Yang VW. An update on the biology of RAS/RAF mutations in colorectal cancer. Curr Colorectal Cancer Rep. 2011;7(2):113–120. doi: 10.1007/s11888-011-0086-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang HW, Shin MG, Lee S, et al. Cooperative activation of PI3K by Ras and Rho family small GTPases. Mol Cell. 2012;47(2):281–290. doi: 10.1016/j.molcel.2012.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li W, Chong H, Guan KL. Function of the Rho family GTPases in Ras-stimulated Raf activation. J Biol Chem. 2001;276(37):34728–34737. doi: 10.1074/jbc.M103496200 [DOI] [PubMed] [Google Scholar]
- 9.Dai Y, Wang WH. Peroxisome proliferator-activated receptor γ and colorectal cancer. World J Gastrointest Oncol. 2010;2(3):159–164. doi: 10.4251/wjgo.v2.i3.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mateer SC, Wang N, Bloom GS. IQGAPs: integrators of the cytoskeleton, cell adhesion machinery, and signaling networks. Cell Motil Cytoskeleton. 2003;55(3):147–155. doi: 10.1002/cm.10118 [DOI] [PubMed] [Google Scholar]
- 11.Adachi M, Kawasaki A, Nojima H, Nishida E, Tsukita S. Involvement of IQGAP family proteins in the regulation of mammalian cell cytokinesis. Genes Cells. 2014;19(11):803–820. doi: 10.1111/gtc.12179 [DOI] [PubMed] [Google Scholar]
- 12.Roy M, Li Z, Sacks DB. IQGAP1 is a scaffold for mitogen-activated protein kinase signaling. Mol Cell Biol. 2005;25(18):7940–7952. doi: 10.1128/MCB.25.18.7940-7952.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hedman AC, Smith JM, Sacks DB. The biology of IQGAP proteins: beyond the cytoskeleton. EMBO Rep. 2015;16(4):427–446. doi: 10.15252/embr.201439834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bardwell AJ, Lagunes L, Zebarjedi R, Bardwell L. The WW domain of the scaffolding protein IQGAP1 is neither necessary nor sufficient for binding to the MAPKs ERK1 and ERK2. J Biol Chem. 2017;292(21):8750–8761. doi: 10.1074/jbc.M116.767087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee BH, Neela PH, Kent MS, Zehnder AM. IQGAP1 is an oncogenic target in canine melanoma. PLoS One. 2017;12(4):e0176370. doi: 10.1371/journal.pone.0176370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jadeski L, Mataraza JM, Jeong HW, Li Z, Sacks DB. IQGAP1 stimulates proliferation and enhances tumorigenesis of human breast epithelial cells. J Biol Chem. 2008;283(2):1008–1017. doi: 10.1074/jbc.M708466200 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y, Liang W, Yang Q, et al. IQGAP1 mediates angiotensin II-induced apoptosis of podocytes via the ERK1/2 MAPK signaling pathway. Am J Nephrol. 2013;38(5):430–444. doi: 10.1159/000355970 [DOI] [PubMed] [Google Scholar]
- 18.Jameson KL, Mazur PK, Zehnder AM, et al. IQGAP1 scaffold-kinase interaction blockade selectively targets RAS-MAP kinase-driven tumors. Nat Med. 2013;19(5):626–630. doi: 10.1038/nm.3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuroda S, Fukata M, Nakagawa M, et al. Role of IQGAP1, a target of the small GTPases Cdc42 and Rac1, in regulation of E-cadherin- mediated cell-cell adhesion. Science. 1998;281(5378):832–835. doi: 10.1126/science.281.5378.832 [DOI] [PubMed] [Google Scholar]
- 20.Carmon KS, Gong X, Yi J, et al. LGR5 receptor promotes cell-cell adhesion in stem cells and colon cancer cells via the IQGAP1-Rac1 pathway. J Biol Chem. 2017;292(36):14989–15001. doi: 10.1074/jbc.M117.786798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hage B, Meinel K, Baum I, Giehl K, Menke A. Rac1 activation inhibits E-cadherin-mediated adherens junctions via binding to IQGAP1 in pancreatic carcinoma cells. Cell Commun Signal. 2009;7:23. doi: 10.1186/1478-811X-7-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.White CD, Khurana H, Gnatenko DV, et al. IQGAP1 and IQGAP2 are reciprocally altered in hepatocellular carcinoma. BMC Gastroenterol. 2010;10:125. doi: 10.1186/1471-230X-10-125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schmidt VA, Chiariello CS, Capilla E, Miller F, Bahou WF. Development of hepatocellular carcinoma in Iqgap2-deficient mice is IQGAP1 dependent. Mol Cell Biol. 2008;28(5):1489–1502. doi: 10.1128/MCB.01090-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y, Yan J, Cutz JC, et al. IQGAP2, a candidate tumour suppressor of prostate tumorigenesis. Biochim Biophys Acta. 2012;1822(6):875–884. doi: 10.1016/j.bbadis.2012.02.019 [DOI] [PubMed] [Google Scholar]
- 25.Xie Y, Zheng L, Tao L. Downregulation of IQGAP2 correlates with prostate cancer recurrence and metastasis. Transl Oncol. 2019;12(2):236–244. doi: 10.1016/j.tranon.2018.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jin SH, Akiyama Y, Fukamachi H, Yanagihara K, Akashi T, Yuasa Y. IQGAP2 inactivation through aberrant promoter methylation and promotion of invasion in gastric cancer cells. Int J Cancer. 2008;122(5):1040–1046. doi: 10.1002/ijc.23181 [DOI] [PubMed] [Google Scholar]
- 27.Wang S, Watanabe T, Noritake J, et al. IQGAP3, a novel effector of Rac1 and Cdc42, regulates neurite outgrowth. J Cell Sci. 2007;120(Pt 4):567–577. [DOI] [PubMed] [Google Scholar]
- 28.Nojima H, Adachi M, Matsui T, Okawa K, Tsukita S, Tsukita S. IQGAP3 regulates cell proliferation through the Ras/ERK signalling cascade. Nat Cell Biol. 2008;10(8):971–978. doi: 10.1038/ncb1757 [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Zhao W, Xu QW, Wang XS, Zhang Y, Zhang J. IQGAP3 promotes EGFR-ERK signaling and the growth and metastasis of lung cancer cells. PLoS One. 2014;9(5):e97578. doi: 10.1371/journal.pone.0097578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu Z, Liu D, Bojdani E, El-Naggar AK, Vasko V, Xing M. IQGAP1 plays an important role in the invasiveness of thyroid cancer. Clin Cancer Res. 2010;16(24):6009–6018. doi: 10.1158/1078-0432.CCR-10-1627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weiser MR. AJCC 8th edition: colorectal cancer. Ann Surg Oncol. 2018;25(6):1454–1455. doi: 10.1245/s10434-018-6462-1 [DOI] [PubMed] [Google Scholar]
- 35.Hong Y, Downey T, Eu KW, Koh PK, Cheah PY. A ‘metastasis-prone’ signature for early-stage mismatch-repair proficient sporadic colorectal cancer patients and its implications for possible therapeutics. Clin Exp Metastasis. 2010;27(2):83–90. doi: 10.1007/s10585-010-9305-4 [DOI] [PubMed] [Google Scholar]
- 36.Cancer Genome Atlas N. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–337. doi: 10.1038/nature11252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roy M, Li Z, Sacks DB. IQGAP1 binds ERK2 and modulates its activity. J Biol Chem. 2004;279(17):17329–17337. doi: 10.1074/jbc.M308405200 [DOI] [PubMed] [Google Scholar]
- 38.Vetterkind S, Lin QQ, Morgan KG. A novel mechanism of ERK1/2 regulation in smooth muscle involving acetylation of the ERK1/2 scaffold IQGAP1. Sci Rep. 2017;7(1):9302. doi: 10.1038/s41598-017-09434-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mavrommati I, Cisse O, Falasca M, Maffucci T. Novel roles for class II Phosphoinositide 3-Kinase C2β in signalling pathways involved in prostate cancer cell invasion. Sci Rep. 2016;6:23277. doi: 10.1038/srep23277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chikh A, Ferro R, Abbott JJ, et al. Class II phosphoinositide 3-kinase C2β regulates a novel signaling pathway involved in breast cancer progression. Oncotarget. 2016;7(14):18325–18345. doi: 10.18632/oncotarget.7761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goto T, Sato A, Shimizu M, et al. IQGAP1 functions as a modulator of dishevelled nuclear localization in Wnt signaling. PLoS One. 2013;8(4):e60865. doi: 10.1371/journal.pone.0060865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu CC, Li H, Xiao Y, et al. Over-expression of IQGAP1 indicates poor prognosis in head and neck squamous cell carcinoma. J Mol Histol. 2018;49(4):389–398. doi: 10.1007/s10735-018-9779-y [DOI] [PubMed] [Google Scholar]
- 43.Li CH, Sun XJ, Niu SS, et al. Overexpression of IQGAP1 promotes the angiogenesis of esophageal squamous cell carcinoma through the AKT and ERKmediated VEGFVEGFR2 signaling pathway. Oncol Rep. 2018;40(3):1795–1802. doi: 10.3892/or.2018.6558 [DOI] [PubMed] [Google Scholar]
- 44.Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ, Pei Y. Overexpression of IQGAP1 in human pancreatic cancer. Hepatobiliary Pancreat Dis Int. 2013;12(5):540–545. doi: 10.1016/S1499-3872(13)60085-5 [DOI] [PubMed] [Google Scholar]
- 45.Liu P, Cheng H, Roberts TM, Zhao JJ. Targeting the phosphoinositide 3-kinase pathway in cancer. Nat Rev Drug Discov. 2009;8(8):627–644. doi: 10.1038/nrd2926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arcaro A, Zvelebil MJ, Wallasch C, Ullrich A, Waterfield MD, Domin J. Class II phosphoinositide 3-kinases are downstream targets of activated polypeptide growth factor receptors. Mol Cell Biol. 2000;20(11):3817–3830. doi: 10.1128/MCB.20.11.3817-3830.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang BH, Liu LZ. PI3K/PTEN signaling in angiogenesis and tumorigenesis. Adv Cancer Res. 2009;102:19–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Maffucci T, Cooke FT, Foster FM, Traer CJ, Fry MJ, Falasca M. Class II phosphoinositide 3-kinase defines a novel signaling pathway in cell migration. J Cell Biol. 2005;169(5):789–799. doi: 10.1083/jcb.200408005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rao SK, Edwards J, Joshi AD, Siu IM, Riggins GJ. A survey of glioblastoma genomic amplifications and deletions. J Neurooncol. 2010;96(2):169–179. doi: 10.1007/s11060-009-9959-4 [DOI] [PubMed] [Google Scholar]
- 51.Kind M, Klukowska-Rotzler J, Berezowska S, Arcaro A, Charles RP. Questioning the role of selected somatic PIK3C2B mutations in squamous non-small cell lung cancer oncogenesis. PLoS One. 2017;12(10):e0187308. doi: 10.1371/journal.pone.0187308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu P, Morrison C, Wang L, et al. Identification of somatic mutations in non-small cell lung carcinomas using whole-exome sequencing. Carcinogenesis. 2012;33(7):1270–1276. doi: 10.1093/carcin/bgs148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Z, Sun C, Zhang Y, Ji Z, Yang G. Phosphatidylinositol 3-kinase-C2β inhibits cisplatin-mediated apoptosis via the Akt pathway in oesophageal squamous cell carcinoma. J Int Med Res. 2011;39(4):1319–1332. doi: 10.1177/147323001103900419 [DOI] [PubMed] [Google Scholar]
- 54.Alliouachene S, Bilanges B, Chicanne G, et al. Inactivation of the class II PI3K-C2β potentiates insulin signaling and sensitivity. Cell Rep. 2015;13(9):1881–1894. doi: 10.1016/j.celrep.2015.10.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katso RM, Pardo OE, Palamidessi A, et al. Phosphoinositide 3-kinase C2β regulates cytoskeletal organization and cell migration via Rac-dependent mechanisms. Mol Biol Cell. 2006;17(9):3729–3744. doi: 10.1091/mbc.e05-11-1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boller D, Doepfner KT, De Laurentiis A, et al. Targeting PI3KC2β impairs proliferation and survival in acute leukemia, brain tumours and neuroendocrine tumours.. Anticancer Res. 2012;32(8):3015–3027. [PubMed] [Google Scholar]
- 57.Kaiser S, Park YK, Franklin JL, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8(7):R131. doi: 10.1186/gb-2007-8-7-r131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ki DH, Jeung HC, Park CH, et al. Whole genome analysis for liver metastasis gene signatures in colorectal cancer. Int J Cancer. 2007;121(9):2005–2012. doi: 10.1002/ijc.22975 [DOI] [PubMed] [Google Scholar]
- 59.Gaedcke J, Grade M, Jung K, et al. Mutated KRAS results in overexpression of DUSP4, a MAP-kinase phosphatase, and SMYD3, a histone methyltransferase, in rectal carcinomas. Genes Chromosomes Cancer. 2010;49(11):1024–1034. doi: 10.1002/gcc.20811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Skrzypczak M, Goryca K, Rubel T, et al. Modeling oncogenic signaling in colon tumors by multidirectional analyses of microarray data directed for maximization of analytical reliability. PLoS One. 2010;5(10):e13091. doi: 10.1371/annotation/8c585739-a354-4fc9-a7d0-d5ae26fa06ca [DOI] [PMC free article] [PubMed] [Google Scholar]