Abstract
Coffin-Siris Syndrome 4 is an autosomal dominant congenital malformation syndrome caused by heterozygous mutations in the SMARCA4 gene with its main features being intellectual disability, developmental delay, behavioral abnormalities, and hypoplastic or absent fifth fingernails and fifth distal phalanges. Here, we report a young woman with developmental delay, moderate intellectual disability, and bilateral sensorineural hearing loss, referred for genetic testing. High-resolution chromosomal microarray analysis identified a 428-kb deletion in chromosome 19 which included the SMARCA4 gene. We conclude that haploinsufficiency of SMARCA4 may be a valid pathophysiological mechanism leading to milder Coffin-Siris syndrome phenotypes.
Keywords: Array CGH, Chromosomal microarray analysis, Cognitive delay, Deletion, Haploinsufficiency, Intellectual disability, Language impairment
Established Facts
Coffin-Siris syndrome 4 is an autosomal dominant congenital malformation syndrome caused by heterozygous mutations in the SMARCA4 gene with its main features being intellectual disability, developmental delay, behavioral abnormalities, and hypoplastic or absent fifth fingernails and fifth distal phalanges.
Reports on focal CNVs affecting SMARCA4 are rare and typically include more genes.
Novel Insights
In this study, we report a young woman with a 428-kb microdeletion including SMARCA4 with a Coffin-Siris syndrome phenotype albeit lacking some of the hallmark features of the syndrome.
Haploinsufficiency of SMARCA4 could be a valid pathophysiological mechanism for the establishment of a milder Coffin-Siris syndrome phenotype.
Coffin-Siris Syndrome 4 (CSS4; OMIM 614609) is an autosomal dominant congenital malformation syndrome caused by heterozygous mutations in the SMARCA4 gene. The main phenotypical features comprise developmental delay, intellectual disability (ID), behavioral abnormalities, hypoplastic or absent fifth fingernails and fifth distal phalanges, feeding difficulties, frequent infections as well as coarse facial features, although these may be less severe than those in patients with typical CSS1 (OMIM 135900), which is caused by mutations in the ARID1B gene [Kosho et al., 2014a].
These core phenotypic features were first described by Coffin and Siris [1970] in 3 unrelated girls with ID lacking the nail and terminal phalanx of the fifth finger. Since then, similar phenotypes have been reported in individuals with mutations in genes encoding subunits of the BRG1-and-BRM-associated (BAF) chromatin remodeling complex (SWI/SNF-like complex) [Ronan et al., 2013], regulating gene expression during development including ARID1A, ARID1B, SMARCB1, SMARCA4, SMARCE1, ARID2, DPF2, SMARCC2, SOX11, and SOX4, designating 10 distinct CSS subtypes (CSS1-CSS10) [Kosho et al., 2014a]. Additionally, mutations in the PHF6 gene have been identified in 2 individuals with CSS phenotypes [Wieczorek et al., 2013].
Other BAF complex genes have been associated with ID syndromes such as Nicolaides-Baraitser syndrome, nonsyndromic ID, autism spectrum disorders (ASD), and schizophrenia [Santen et al., 2012a].
Furthermore, mutations in these genes are associated with sporadic tumors and tumor predisposition syndromes [Santen et al., 2012b]. SMARCA4, which acts as a tumor suppressor gene, has been associated with Rhabdoid tumor predisposition syndrome 2 (OMIM 613325) [Witkowski et al., 2013].
Heterozygous mutations in SMARCA4 have thus far been reported in at least 20 patients with CSS phenotypes [Kosho et al., 2014a; Dsouza et al., 2019; Sekiguchi et al., 2019]. Small in-frame deletions have also been reported [Tsurusaki et al., 2012, 2014; Kosho et al., 2014b]; however, reports on focal copy number variants (CNVs) of 19p13.2 affecting SMARCA4 are rare and typically include additional genes due to the high gene density of the specific chromosomal region [Haberlandt et al., 2012].
Here, we report a young woman with a CSS-like phenotype caused by a 428-kb heterozygous deletion in 19p13.2 including the SMARCA4 gene, which adds to the scientific knowledge of the syndrome.
Case Presentation
The patient is a 30-year-old woman presenting with a history of moderate cognitive and speech delay. She was born at term but was small for dates, with a birth weight of 2.4 kg (2nd centile) and failure to thrive. She had serious feeding difficulties, leading to continuous weight loss and recurrent febrile seizures, which were successfully treated with valproic acid.
Assessment by audiogram at the age of 5 showed moderate to severe bilateral sensorineural hearing loss. Since then, she has been wearing hearing aids in both ears. Assessment with the Wechsler Adult Intelligence Scale Fourth Edition (WAIS-IV) showed moderate ID (IQ 53), with a verbal comprehension score of 65, perceptual reasoning 61, working memory 56, and processing speed index 62. As a child, she had learning difficulties (dysgraphia, dyscalculia). Receptive language was better than production. The patient also presented with mixed anxiety-depressive disorder with aggressive and compulsive manifestations, as well as verbal aggressiveness, especially when confronted with cancellation of planned activities. Serial brain MRI scans did not reveal any brain structure, growth, or myelination abnormalities. Clinical examination revealed scoliosis and sparse scalp hair. Currently she has a height of 158 cm (21st centile), weight of 59 kg (54th centile), and an OFC of 52cm (1st centile), indicative of microcephaly. She was recently diagnosed with Hashimoto hypothyroidism, and receives 75 μg T4 daily. Additionally, she has myopia and is prone to sprains of the lower limbs. No further dysmorphic features or clinical abnormalities were present in our patient, nor reported in her medical record.
Materials and Methods
Genomic DNA was extracted from peripheral blood samples of the patient and her sister using the QiAmp DNA Mini Kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The quality and quantity of the DNA was determined using the NanoDrop 2000c UV-VIS spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Chromosomal microarray analysis was performed using the high-resolution 4 × 180K G3 CGH+SNP microarray platform (G5890A, Design ID 029830, Agilent Technologies, Santa Clara, CA, USA). The specific platform features a total of 110,712 oligonucleotide CGH probes covering the whole genome annotated against NCBI Build 38 (UCSC GRCh38, Dec. 2013), with a median CGH probe spacing of 25.3 kb as well as 59,647 SNP probes for the detection of copy-neutral loss of heterozygosity. The laboratory protocol was carried out according to the manufacturer's instructions (Agilent Oligonucleotide Array-Based CGH for Genomic DNA Analysis) and consisted of enzymatic digestion of each genomic DNA sample as well as a sex-matched reference DNA (Agilent Technologies), followed by differential labeling with Cy5 and Cy3 fluorescent dyes for sample and reference, respectively. The combined labeled DNA samples were applied to the microarray, left to hybridize for 24 h at 67°C, washed and scanned at 3 μm resolution on the SureScan Dx microarray scanner (Agilent Technologies). The tiff images were extracted and analyzed using the Agilent Feature Extraction software and the CytoGenomics v.5.0.1 software suite. The ADM-1 aberration detection algorithm was utilized with the minimum number of probes required for a call set to 4 and the log2 ratio threshold for duplications and deletions set to ±0.25.
Results
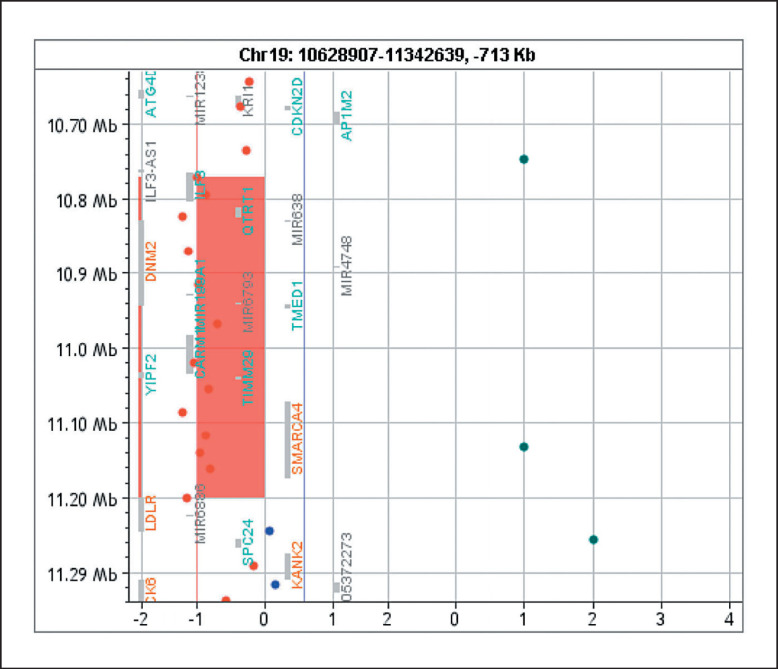
Chromosomal microarray analysis identified a 428-kb microdeletion in the 19p13.2 chromosomal region, arr[GRCh37] 19p13.2(10771654_11199893)×1, which included the SMARCA4 gene (Fig. 1). No parental samples were available for testing.
Fig. 1.
428-kb deletion of the 19p13.2 chromosomal region including the SMARCA4 gene. The deleted region also contains ILF3, QTRT1, DNM2, MIR638, MIR4748, TMED1, C19orf38, CARM1, YIPF2, and TIMM29, many of which are involved in gene expression regulation.
Discussion
Here, we report a 30-year-old female patient whose mild condition may be included in a CSS-related clinical spectrum caused by a heterozygous 428-kb deletion including SMARCA4.
Heterozygous mutations in SMARCA4 have thus far been reported in at least 20 patients with a CSS phenotype [Kosho et al., 2014a; Dsouza et al., 2019; Sekiguchi et al., 2019], and were all non-truncating (either missense or in-frame deletions), leading to gain-of-function or negative-dominant effects. Small in-frame deletions have also been reported [Tsurusaki et al., 2012, 2014; Kosho et al., 2014b]. Clinical features for these patients include developmental delay, ID, mild to moderate growth impairment, feeding difficulties, hypertrichosis, cardiac anomalies, varying facial features, and hypoplastic fifth fingers/fingernails or toes/toenails. Reports on CNVs affecting SMARCA4 are rare. In the most recent study, Sekiguchi et al. [2019] tested a cohort of more than 200 patients with CSS for single nucleotide variants as well as CNVs on all genes so far suspected to contribute to CSS, with no detected CNVs affecting the SMARCA4 gene. Haberlandt et al. [2012] reported on a 4-year-old boy with ID, minor dysmorphisms, and febrile seizures that carried a de novo deletion of 834.2 kb in 19p13.2 encompassing 32 genes, discussing a potential role of SMARCA4 haploinsufficiency. Furthermore, Welham et al. [2015] reported on 10 patients with 19p13.2 microdeletions presenting with behavioral abnormalities of variable degree. Of those, 4 patients had larger deletions in SMARCA4, with only one patient (patient 9) having a partially overlapping deletion of 573 kb which included 7 genes, namely ILF3, DNM2, CARM1, SMARCA4, LDLR, KANK2, and DOCK6 [Welham et al., 2015]. This specific patient was a 16-year- old male reported to be autonomous, with normal vision and hearing. Interestingly, he showed aggressive behavior and property destruction, and had indications of ASD. The DECIPHER database (https://decipher.sanger.ac.uk) holds a record of a larger 1.23-Mb de novo deletion found in a female patient with microphthalmia as well as a record of a patient with a 247-kb microdeletion surrounding SMARCA4 with mild neurodevelopmental and facial anomalies [Errichiello et al., 2017]. Two additional records of patients with much larger deletions that include SMARCA4 exist in the ClinVar database (https://www.ncbi.nlm.nih.gov/clinvar). The main clinical features of our patient were moderate ID, speech delay, feeding difficulties, bilateral sensorineural hearing loss, and mixed anxiety-depressive disorder. However, other hallmark features of CSS, such as fifth finger nail/distal phalanx hypoplasia or aplasia, coarse facial features (thick eyebrows, long eyelashes, wide mouth with thick lips, broad nasal bridge, abnormal ear shape), excess body hair, ophthalmological disorders (e.g., strabismus, cataract, ptosis), or cardiac anomalies (e.g., ventricular septal or atrial septal defects, tetralogy of Fallot) were not present in our patient. Additionally, no autistic traits were observed.
Errichiello et al. [2017] suggested that a haploinsufficiency pathophysiological mechanism for SMARCA4 may explain milder CSS phenotypes, as previously reported for haploinsufficient ARID1B constitutional mutations associated with CSS.
Rhabdoid tumor predisposition syndrome 2 (OMIM 613325) caused by germline mutations in SMARCA4 results in an elevated risk for developing small-cell carcinoma of the ovary-hypercalcemic type, a highly lethal tumor occurring in young women, and thus, pelvic ultrasound every 6 months is advised for ovarian surveillance [Mitchell et al., 2019]. As of now, our patient does not have any manifestations of rhabdoid tumors, but close clinical observation was recommended.
The main causative gene in the deleted region appears to be SMARCA4; however, the region also contains ILF3, QTRT1, DNM2, MIR638, MIR4748, TMED1, C19orf38, CARM1, YIPF2, and TIMM29, many of which are involved in gene expression regulation. Specifically, the ILF3 gene codes for a double-strand RNA-binding protein that regulates gene expression and stabilizes mRNAs, thus participating in a wide range of transcriptional and post-transcriptional processes. QTRT1 encodes the catalytic subunit of tRNA-guanine transglycosylase with an important role in tRNA modification and has been related to ASD and benign epilepsy. DNM2 encodes for dynamin 2, a GTP-binding protein associated with microtubules, implicated in many neuromuscular disorders including Charcot-Marie-Tooth disease type 2M (OMIM 606482), autosomal dominant centronuclear myopathy 1 (OMIM 160150) and lethal congenital contracture syndrome 5 (OMIM 615368), with only single nucleotide variants having been reported to date. Our patient did not exhibit impaired motor skills. MIR638 and MIR4748 are noncoding RNAs involved in post-transcriptional regulation of gene expression. TMED1 encodes for a protein identified by its interaction with interleukin 1 receptor-like 1 (IL1RL1) with a role in innate immunity. CARM1 encodes for a methyltransferase that regulates gene expression by acting specifically on histones and other chromatin-associated proteins. No evidence for haploinsufficiency pathogenicity for any of these genes is available in the ClinGen database; however, the gene regulation activity that many of these genes possess could potentially play a role in shaping the milder clinical phenotype of our patient. The 428-kb deletion found in our patient is one of the smallest that have been reported to date to include SMARCA4, thus making it important for genotype-phenotype correlation.
Our findings suggest that heterozygous SMARCA4 deletions could be related to a CSS phenotypical spectrum, albeit with a milder manifestation, not including all the hallmark features of the syndrome (e.g., fifth finger or toe malformation). We conclude that haploinsufficiency of SMARCA4 is a valid pathophysiological mechanism for the establishment of a CSS-like phenotype.
Statement of Ethics
The present study was conducted based on the guidelines issued by the Hellenic National Bioethics Committee and the Ethics Committee of the National and Kapodistrian University of Athens, in accordance with the Declaration of Helsinki principles. Written informed consent was obtained prior to investigation.
Disclosure Statement
The authors have no conflicts of interest to disclose.
References
- Coffin GS, Siris E. Mental retardation with absent fifth fingernail and terminal phalanx. Am J Dis Child. 1970;119:433–439. doi: 10.1001/archpedi.1970.02100050435009. [DOI] [PubMed] [Google Scholar]
- Dsouza NR, Zimmermann MT, Geddes GC. A case of Coffin-Siris syndrome with severe congenital heart disease and a novel SMARCA4variant. Cold Spring Harb Mol case Stud. 2019;5:a003962. doi: 10.1101/mcs.a003962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errichiello E, Mustafa N, Vetro A, Notarangelo LD, de Jonge H, et al. SMARCA4inactivating mutations cause concomitant Coffin-Siris syndrome, microphthalmia and small-cell carcinoma of the ovary hypercalcaemic type. J Pathol. 2017;243:9–15. doi: 10.1002/path.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberlandt E, Spreiz A, Baumgartner Sigl S, Janetschek C, Röthlisberger B, et al. Microdeletion 19p13.2 in an almost 5-year-old boy. Am J Med Genet A. 2012;158A:1190–1194. doi: 10.1002/ajmg.a.35291. [DOI] [PubMed] [Google Scholar]
- Kosho T, Miyake N, Carey JC. Coffin-Siris syndrome and related disorders involving components of the BAF (mSWI/SNF) complex: historical review and recent advances using next generation sequencing. Am J Med Genet Part C Semin Med Genet. 2014a;166C:241–251. doi: 10.1002/ajmg.c.31415. [DOI] [PubMed] [Google Scholar]
- Kosho T, Okamoto N, Coffin-Siris Syndrome., International Collaborators Genotype-phenotype correlation of coffin-siris syndrome caused by mutations in SMARCB1SMARCA4SMARCE1, and ARID1A. Am J Med Genet Part C Semin Med Genet 2014b. 2014b;166:262–275. doi: 10.1002/ajmg.c.31407. [DOI] [PubMed] [Google Scholar]
- Mitchell SG, Pencheva B, Porter CC. Germline genetics and childhood cancer: emerging cancer predisposition syndromes and psychosocial impacts. Curr Oncol Rep. 2019;21:85. doi: 10.1007/s11912-019-0836-9. [DOI] [PubMed] [Google Scholar]
- Ronan JL, Wu W, Crabtree GR. From neural development to cognition: unexpected roles for chromatin. Nat Rev Genet. 2013;14:347–359. doi: 10.1038/nrg3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santen GWE, Aten E, Sun Y, Almomani R, Gilissen C, et al. Mutations in SWI/SNF chromatin remodeling complex gene ARID1Bcause Coffin-Siris syndrome. Nat Genet. 2012a;44:379–380. doi: 10.1038/ng.2217. [DOI] [PubMed] [Google Scholar]
- Santen GWE, Kriek M, van Attikum H. SWItching from malignancies to intellectual disability. Epigenetics. 2012b;7:1219–1224. doi: 10.4161/epi.22299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi F, Tsurusaki Y, Okamoto N, Teik KW, Mizuno S, et al. Genetic abnormalities in a large cohort of Coffin-Siris syndrome patients. J Hum Genet. 2019;64:1173–1186. doi: 10.1038/s10038-019-0667-4. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Kosho T, Imai Y, et al. Mutations affecting components of the SWI/SNF complex cause Coffin-Siris syndrome. Nat Genet. 2012;44:376–378. doi: 10.1038/ng.2219. [DOI] [PubMed] [Google Scholar]
- Tsurusaki Y, Okamoto N, Ohashi H, Mizuno S, Matsumoto N, et al. Coffin-Siris syndrome is a SWI/SNF complex disorder. Clin Genet. 2014;85:548–554. doi: 10.1111/cge.12225. [DOI] [PubMed] [Google Scholar]
- Welham A, Barth B, Moss J, Penhallow J, Sheth K, et al. Behavioral characteristics associated with 19p13.2 microdeletions. Am J Med Genet Part A. 2015;167:2334–2343. doi: 10.1002/ajmg.a.37180. [DOI] [PubMed] [Google Scholar]
- Wieczorek D, Bögershausen N, Beleggia F, Steiner-Haldenstätt S, Pohl E, et al. A comprehensive molecular study on Coffin-Siris and Nicolaides-Baraitser syndromes identifies a broad molecular and clinical spectrum converging on altered chromatin remodeling. Hum Mol Genet. 2013;22:5121–5135. doi: 10.1093/hmg/ddt366. [DOI] [PubMed] [Google Scholar]
- Witkowski L, Lalonde E, Zhang J, Albrecht S, Hamel N, et al. Familial rhabdoid tumour “avant la letter” - from pathology review to exome sequencing and back again. J Pathol. 2013;231:35–43. doi: 10.1002/path.4225. [DOI] [PubMed] [Google Scholar]