Abstract
The water-soluble vitamin biotin is essential for cellular growth, development, and well-being, but its absorption, distribution, metabolism, and excretion are poorly understood. This paper describes the radiolabeling of biotin with the positron emission tomography (PET) radionuclide carbon-11 ([11C]biotin) to enable the quantitative study of biotin trafficking in vivo. We show that intravenously administered [11C]biotin is quickly distributed to the liver, kidneys, retina, heart, and brain in rodents—consistent with the known expression of the biotin transporter—and there is a surprising accumulation in the brown adipose tissue (BAT). Orally administered [11C]biotin was rapidly absorbed in the small intestine and swiftly distributed to the same organs. Preadministration of nonradioactive biotin inhibited organ uptake and increased excretion. [11C]Biotin PET imaging therefore provides a dynamic in vivo map of transporter-mediated biotin trafficking in healthy rodents. This technique will enable the exploration of biotin trafficking in humans and its use as a research tool for diagnostic imaging of obesity/diabetes, bacterial infection, and cancer.
Introduction
Biotin (vitamin B7) is a cofactor for five carboxylases involved in fatty acid biosynthesis, gluconeogenesis, and catabolism of amino acids and fatty acids.1 Catalysis by mammalian biotin-dependent carboxylases drives the incorporation of a single CO2 carbon unit into molecules such as pyruvate, β-methylcrotonyl-CoA, propionyl-CoA, and acetyl-CoA. Four out of the five carboxylases are located in the mitochondria, and accordingly, biotin deficiencies result in abnormal mitochondrial structures and functions.2 Biotin is taken up by the cells via a sodium-dependent vitamin transporter (SMVT), expressed in the cytoplasm and the mitochondrial membranes. SMVT is present in the gastrointestinal tract, liver, kidneys, retina, heart, brain, and skin.3−11 Cancer cells, including leukemia, ovarian, colon, mastocytoma, lung, renal, and breast cancer cell lines, exhibit elevated SMVT expression and enhanced biotin uptake capability to sustain their rapid cell growth and enhanced proliferation.12
To date, biotin uptake via the SMVT has been studied with tritium (3H)- and carbon-14 (14C)-radiolabeled biotin in cell-based assays and in laboratory animal studies.13−19 However, in vivo investigations using these radiotracers are limited in scope because of the low stability of [3H]biotin in vivo (the tritium atoms at positions 8 and 9 in the valeric acid side chain are removed in vivo via β-oxidation—confounding the tissue distribution analysis).18 For example, about half of the radioactivity associated with the administered dose of [3H]biotin was excreted in 24 h as [3H]acetate/[3H]H2O in the urine of pigs.18 Additional limitations include the need for a postmortem analysis to determine the radiotracer tissue distribution, precluding human translational studies,18,20 and the long radioactive half-life of 3H (12.5 years) and 14C (5700 years)—impeding their use due to the associated risks of long-term radioactivity exposure. In this work, we aimed to overcome these limitations by radiolabeling biotin with a positron-emitting radionuclide that can be used for in vivo imaging, in both animals and humans, by positron emission tomography (PET).
Biotin has already been radiolabeled using PET and single-photon emission computed tomography (SPECT) radionuclides such as 64Cu, 18F, 111In, 125I, 131I, 68Ga, and 99mTc21−28 and conjugated with fluorophores for optical imaging.29−31 However, these “biotin-inspired” imaging probes have been developed via the conjugation of the carboxylic group of biotin to chelators or linkers bearing a radionuclide or a fluorophore. These radiotracers have been used in preclinical and clinical settings to detect Escherichia coli (E. coli) and Staphylococcus aureus (S. aureus) infection, which express biotin transporters (BirB and BioY, respectively)32−36 and tumor antigens by exploiting the strong interaction (Kd ≈ 10–15 M) between radiolabeled biotin and preaccumulated antibody–avidin conjugates (a pretargeting approach).24,28,37,38 The biotin-conjugated fluorescent probes have been used in preclinical studies, and fluorescence images showed the accumulation in tumor tissues of murine xenograft tumor models.29−31 However, these tracers are biotin conjugates with grossly modified structures and their in vivo profile is not consistent with the literature reports of biotin biodistribution and transporter expression.26,27 Indeed, changes to the carboxylic acid function (esterification, amidation, and decarboxylation) or heterocyclic moiety (sulfoxidation and elimination of the ureido carbonyl group) prevent their biorecognition by biotin transporters.39−41
Our aim is to characterize the whole-body pharmacokinetics and tissue-distribution of biotin in vivo. To achieve this, we have developed a method to produce biotin autologously radiolabeled with a short-lived positron-emitting radionuclide carbon-11 ([11C]biotin). Carbon-11 labeling of biotin does not alter its chemical structure or biological properties. Moreover, it allows straightforward translation from preclinical to clinical research, without toxicological assessment, and reduces risks associated with radioactive waste management and radiation exposure to patients due to the short half-life of 11C (half-life = 20.4 min).
Results and Discussion
Radiosynthesis of [11C]biotin
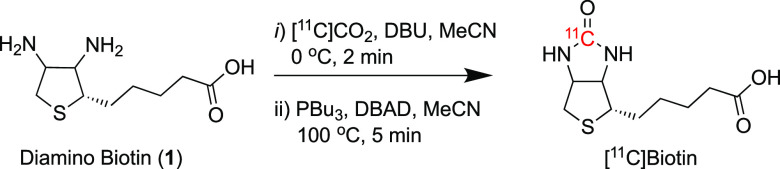
In developing our carbon-11 labeling strategy, we initially considered a previously described [14C]biotin labeling procedure using [14C]phosgene ([14C]COCl2) and diamino biotin via a multistep process to introduce carbon-11 in the urea position.42 Although [11C]COCl2 has previously been obtained from cyclotron-produced carbon-11 carbon dioxide ([11C]CO2), it is impractical, requires a long synthesis time (15–25 min) with a dedicated infrastructure, and is only available in few radiochemistry laboratories worldwide, limiting its usage.43
Inspired by the new 11C-urea labeling method using the primary cyclotron-produced [11C]CO2 synthon,44−46 we conceived a novel route to produce [11C]biotin from diamino biotin. Applying this quick and efficient chemistry to the 11C-labeling of biotin at the urea carbon atom, [11C]biotin was synthesized via a simple two-step, one-pot reaction in a fully automated system (Figure 1 and Figure S1). In the first step, cyclotron-produced [11C]CO2 was bubbled into a reaction vial containing diamino biotin (1) and 1,8-diazabicyclo[5.4.0]undec-7-ene (DBU) dissolved in acetonitrile (MeCN) at 0 °C. Then, a solution of Mitsunobu reagents [di-tert-butyl azodicarboxylate (DBAD) and tributylphosphine (PBu3)] was added to the reaction vial, and the reaction mixture was heated at 100 °C for 5 min (Scheme 1). The reaction was subsequently cooled and quenched with a PBS solution. [11C]Biotin was purified by semipreparative high-performance liquid chromatography (HPLC), and the formulated solution was used for in vitro and in vivo experiments (Figure S2). The total synthesis time including HPLC purification was 32 ± 1 min from the end of [11C]CO2 delivery.
Figure 1.
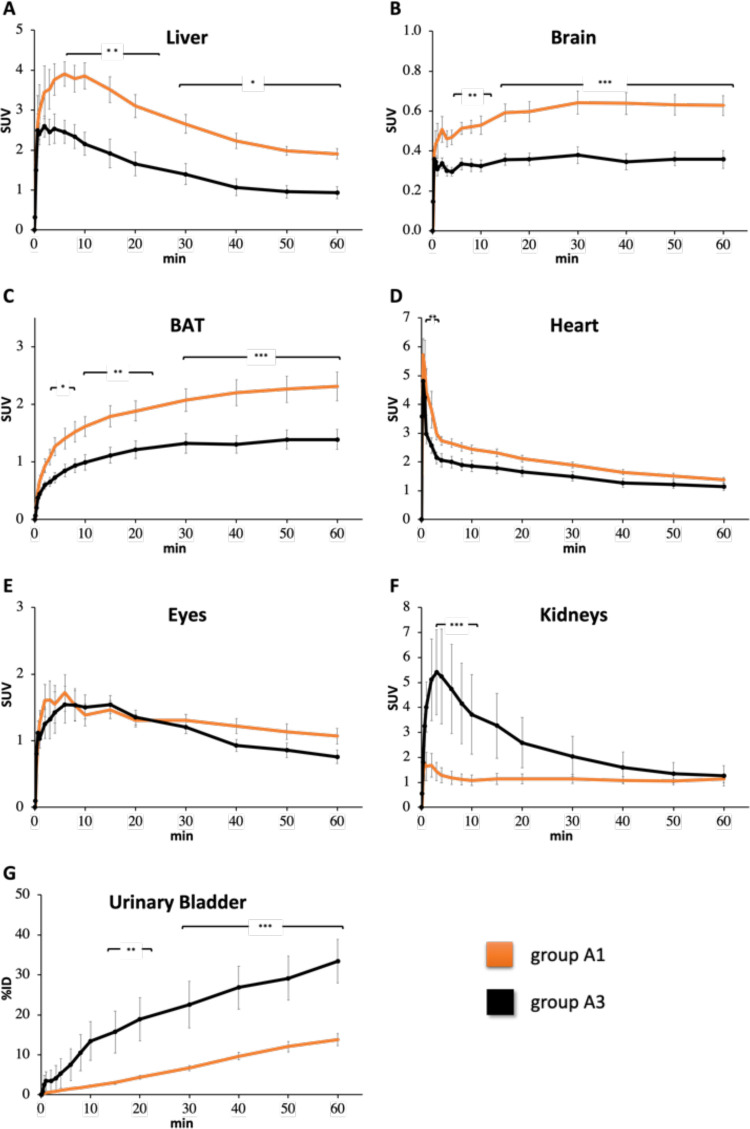
Time–SUV profile (0–60 min) of (A) liver, (B) brain, (C) BAT, (D) heart, (E) eyes, and (F) kidneys in NBA (orange line, group A1) and biotin-challenged (black line, group A3) mice receiving [11C]biotin IV. (G) Radioactivity of urinary bladder is expressed as %ID. Note that the y-scale (SUV) varies between different tissues.
Scheme 1. Radiolabeling of [11C]biotin.
The amount of [11C]biotin obtained was 352 ± 38 MBq in 4–5 mL of phosphate-buffered saline (PBS) with 2.5% ethanol in an injectable solution from 5.9 ± 0.3 GBq of cyclotron-produced [11C]CO2 with an isolated radiochemical yield (RCY) of 19 ± 2%, a radiochemical purity (RCP) of > 99%, and a molar activity (Am) of 7 ± 1 GBq/μmol at the end of the [11C]CO2 delivery (Table S1).
In Vitro Studies
The incubation of [11C]biotin with streptavidin for 5 min at 25 °C showed that 95 ± 1% of the radioactivity is in the form of a [11C]biotin–streptavidin complex (see Supporting Information for assay details and Figure S3).
Preclinical Studies of [11C]biotin
Results of [11C]biotin Injected in Mice
To examine the [11C]biotin trafficking in vivo, [11C]biotin was administered intravenously (IV) in healthy anesthetized mice placed on a high-resolution microPET scanner. Sixteen mice were divided into three groups: group A1 (female, vehicle IV injection 10 min before [11C]biotin IV injection, n = 6, Figure S4), group A2 (male, vehicle IV injection 10 min before [11C]biotin IV injection, n = 5), and group A3 (female, biotin 5 mg/Kg IV injection 10 min before [11C]biotin IV injection, n = 5). The dynamic PET image data were acquired for 60 min.
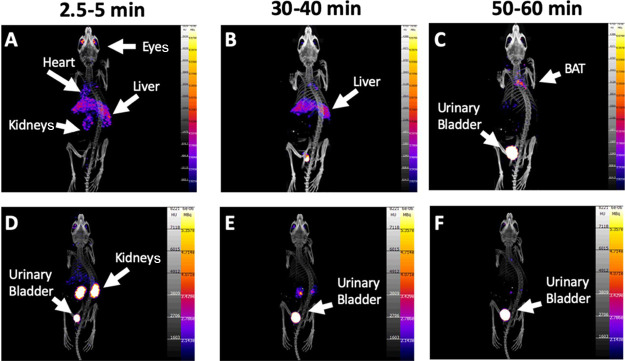
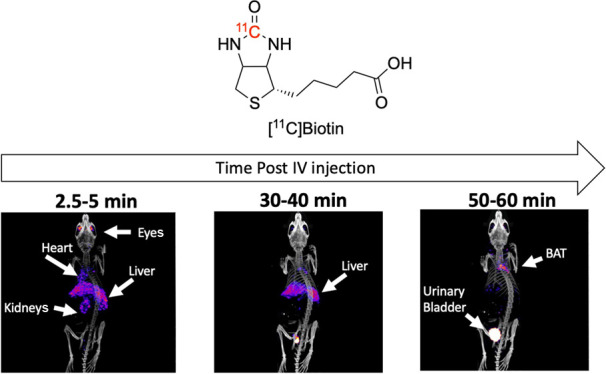
In the no-biotin-added (NBA) group (group A1), PET imaging demonstrated the [11C]biotin uptake in the liver, heart, brain, eyes, kidneys, and interscapular BAT (Figures 1 and 2). The presence of the SMVT in the liver,3,6 heart,7,8 brain,5,10 eyes,4 and kidneys11 provides a rationale for the observed [11C]biotin distribution in vivo. To date, no reports have established the presence of SMVT in the BAT. Further studies (e.g., immunohistochemistry) are needed to confirm the expression of biotin transporters in the BAT. However, the uptake in BAT is consistent with prior knowledge that fatty acid synthesis, glucose metabolism, and lipolysis require biotin-dependent enzymes, and that biotin-deficient rats have decreased adipose biotin-dependent mitochondrial enzyme functions.7,8 Biotin is also known to be essential for the differentiation of preadipocytes into brown adipocytes.47−49 Studies on [18F]fluorodeoxyglucose ([18F]FDG) and fatty acid radiotracers (e.g., [11C]acetate and [123I]beta-methyl-iodophenyl-pentadecanoic acid) have recently revealed that the BAT serves as a metabolic sink for glucose and fatty acids.50−52
Figure 2.
Maximum intensity projections of PET images from NBA (upper row, group A1) and biotin-challenged (lower row, group A3) mice at (A, D) 2.5–5 min, (B, E) 30–40 min, and (C, F) 50–60 min post [11C]biotin IV injection. Tracer uptake is reduced in the biotin-challenged mice in the BAT, liver, heart, and eyes. PET images are displayed according to the intensity scale for the tracer activity, from white (highest) through red (intermediate) to purple (lowest).
To further examine the relation between in vivo [11C]biotin distribution and SMVT expression, male mice (group A2) and female mice challenged with biotin (group A3, Figure S4) were imaged.
To examine the uptake transporter at blood–tissue interfaces, we determined the radioactivity concentrations in the blood of groups A1–A3. An image-derived blood curve was generated by placing the region of interest into the left ventricle of the heart.53 Radioactivity concentrations measured in the last PET time frame (50–60 min postadministration) showed good correlation (r = 0.910, P < 0.0001) with the radioactivity concentration (%ID/g) in the venous blood collected at the end of the PET scan measured using a γ counter (Figure S6). Liver–to–blood, kidney–to–blood, BAT–to–blood, and brain–to–blood area under the curve (AUC) radioactivity ratios (Kp,tissue, AUC) were calculated (eq 1). The “integration plot” analysis53−56 was used to calculate the liver uptake (CLuptake,liver), kidney uptake (CLuptake,kidney), BAT uptake (CLuptake,BAT), brain uptake (CLuptake,brain), and intrinsic urinary excretion (CLint,urine) clearances, which represent the uptake in the liver/kidneys/BAT/brain and intrinsic efflux activity across the corticomedullary region of the kidneys, respectively.
Blood radioactivity concentrations were lower (p < 0.05) for the groups A2 and A3 than that for the control group. The AUCliver 0–60 min was higher in group A2 and lower in group A3 than that in the control mice group (group A1). The liver–to–blood AUC ratio (Kp,liver,AUC) and CLuptake,liver value were higher in group A2 than those in the control mice group. No statistically significant difference was observed for Kp,liver,AUC and CLuptake,liver of group A3 versus the control mice group. The Kp,kidney,AUC value was not different between groups A1 and A2; however, the CLuptake,kidney value in group A2 was significantly higher than that in the control mice group (Table 1). Both Kp,kidney,AUC and CLuptake,kidney (Table 1) values were significantly higher in group A3 than those in the control mice group. Although AUCBAT, 0–60 min and AUCbrain, 0–60 min values were lower in groups A2 and A3 than that in the control mice group, the Kp,BAT,AUC, CLuptake,BAT, Kp,brain,AUC, and CLuptake,brain values of group A1 versus group A2 or A3 were not statistically different (Table 1).
Table 1. Pharmacokinetic Parameters of [11C]biotin in Groups A1–A3.
| group A1 control group | group A2 | group A3 | |
|---|---|---|---|
| body weighta | 17 ± 1 | 24 ± 1* | 17 ± 1 |
| AUCblood, 0–60 min (SUV·min)a | 135.8 ± 10.1 | 96.0 ± 8.8* | 97.2 ± 9.4* |
| AUCliver, 0–60 min (SUV·min)a | 160.5 ± 12.1 | 220.8 ± 6.6** | 86.1 ± 11.0** |
| AUCBAT, 0–60 min (SUV·min)a | 116.2 ± 10.1 | 63.1 ± 5.1** | 69.5 ± 6.9** |
| AUCkidneys, 0–60 min (SUV·min) | 77.2 ± 9.0 | 98.9 ± 13.8 | 172.3 ± 41.1 |
| AUCbrain, 0–60 min (SUV·min)a | 35.9 ± 2.6 | 26.3 ± 2.1** | 20.5 ± 1.4*** |
| Kp,liver, AUC, 60 min (mL/g of tissue)a | 1.22 ± 0.17 | 2.35 ± 0.15*** | 0.88 ± 0.06 |
| Kp,kidneys, AUC, 60 min (mL/g of tissue)a | 0.59 ± 0.08 | 1.02 ± 0.05 | 1.74 ± 0.39* |
| Kp,BAT, AUC, 60 min (mL/g of tissue) | 0.87 ± 0.08 | 0.66 ± 0.03 | 0.73 ± 0.08 |
| Kp,brain,AUC, 60 min (mL/g of tissue) | 0.27 ± 0.03 | 0.28 ± 0.02 | 0.21 ± 0.01 |
| VT,livera mL/g of tissue | 1.27 ± 0.16 | 2.54 ± 0.18*** | 0.91 ± 0.06 |
| VT,kidneysa mL/g of tissue | 0.58 ± 0.08 | 1.00 ± 0.04** | 1.99 ± 0.49* |
| Ki, BAT | 0.021 ± 0.004 | 0.051 ± 0.012 | 0.027 ± 0.007 |
| Ki,brain | 0.005 ± 0.001 | 0.015 ± 0.004 | 0.004 ± 0.001 |
| CLuptake,liver (mL/min/g tissue)a | 0.37 ± 0.07 | 1.35 ± 0.34** | 0.37 ± 0.12 |
| CLuptake,kidneysa (mL/min/g tissue) | 0.28 ± 0.09 | 2.02 ± 0.54* | 1.37 ± 0.28* |
| CLuptake,BAT (mL/min/g tissue) | 0.13 ± 0.03 | 0.24 ± 0.07 | 0.12 ± 0.03 |
| CLuptake,brain (mL/min/g tissue) | 0.05 ± 0.01 | 0.18 ± 0.07 | 0.05 ± 0.01 |
| Xurine,0–60 min (% dose)a | 13.8 ± 1.5 | 30.4 ± 3.3** | 34.7 ± 5.5* |
| Clrenal-blood (ml/min/Kg) | 1.7 ± 0.4 | 16.6 ± 5.6 | 7.2 ± 2.2 |
| CLint,urine† ml/min/Kg | 1.9 ± 0.2 | 3.8 ± 0.14*** | 2.8 ± 1.0 |
One-way ANOVA, p < 0.05. Body weight, AUC, Kp, X, and Ki were analyzed using the one-way ANOVA. For this analysis, p < 0.05 was considered statistically significant, and posthoc t-tests were used to explore the differences between the pairs of groups. The Levene’s test was used to test for homogeneity of variances between the three groups and where this was significant (p < 0.05), the Welch’s test for equality of means was used in place of ANOVA and t-tests assuming that unequal variances were used to explore the group differences where appropriate. The groups A2 and A3 were compared to group A1 and were indicated with (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001.
Next, we used graphical methods57−59 to establish the binding kinetics of [11C]biotin in the liver, kidneys, brain, and BAT. We estimated the volume of distribution (VT, Table 1) for reversible binding and the net uptake rate constant (Ki) for irreversible binding using the Logan and Patlak graphical methods, respectively.57−59 Logan plots for liver and kidneys are shown in Figure S7, and the plot linearity indicates a reversible tracer behavior for these tissues in all groups.
Logan plots for the BAT and brain (Figure S7) were nonlinear indicating a nonreversible tracer behavior in these tissues. This is confirmed by the Patlak analysis, which shows good fits for the BAT and brain indicating that [11C]biotin is irreversibly trapped in these tissues for the duration of the scan (Figure S8). Notably, the Ki values are similar (Table 1); however, the flux of [11C]biotin from blood–to–BAT and blood–to–brain is the product of Ki and the concentration of free biotin in the blood. Although we did not directly measure the concentration of biotin in the blood, it is reasonable to assume that group A3 had a much higher biotin concentration in the blood compared to group A1 and therefore, with equal Ki, the flux of biotin trapped in the BAT and brain was proportionally higher. As with many PET quantitative techniques, graphical methods can be sensitive to ROI placement. Further studies are required to elucidate the biological mechanisms which determine biotin uptake.
A significant increase of [11C]biotin excretion was observed in both the male (group A2) and the biotin-challenged female groups (group A3) compared to the NBA group (group A1) (Figure 1). A remarkable increase in [11C]biotin uptake was observed in the kidneys of groups A2 and A3 mice versus the control group (group A1) in the first 10 min (Figure 1). From 10 to 60 min, the radioactivity in the kidneys in both groups decreased with a concomitant increase in the urinary bladder. The urinary excretion was 2.2 and 2.5 times higher in the A2 and A3 groups, respectively, than that in the control group. Gender has a significant impact in renal clearance with increased CLrenal,blood and CLint,urine higher in male versus female mice (Table 1). Regarding the increase of [11C]biotin in urinary excretion in biotin-challenged mice, we postulate that this may be a consequence of saturation of SMVT expressed in the proximal tubular epithelial cells.11,40,60,61 Saturation would preclude biotin reabsorption and facilitate the clearance of [11C]biotin, consistent with our observations of radioactivity excretion through the kidneys and urinary bladder (Figure 2D–F).
These studies demonstrated that the fraction of [11C]biotin excreted is dose-dependent. This implies that the homeostasis of vitamins is fine-tuned by the body, and the administration of high concentrations of biotin increases its excretion (total biotin administered in the biotin-challenged mice was ∼ 40 times higher than that in the NBA mice). In line with these results, high excretion rates have been observed in biotin-challenged rats after intraperitoneal (IP) injection of [14C]biotin at doses of 0.005, 0.04, and 10 mg/Kg where 47, 83, and > 90% of the administered radioactivity, respectively, was found in the urine within 12 h.17
Ex Vivo Biodistribution Post-[11C]biotin Injection
The ex vivo biodistribution analysis of groups A1–A3 at 70 min post-[11C]biotin injection showed a radioligand uptake of > 10% ID/g in the liver, kidneys, and heart (Figure S9). The liver/blood ratio uptake (Table S3) was higher in male than in female mice. This ex vivo biodistribution result was in agreement with the in vivo uptake parameter (Kp,liver,AUC) obtained from PET imaging at 60 min post-[11C]biotin administration. The biodistribution analysis of biotin-challenged mice (group A3) showed a lower liver/blood ratio compared to the control group (group A1, Table S3).
Radiometabolite Analysis Post-[11C]biotin Injection
The radiometabolite analysis by TLC-autoradiography was performed by following the protocol developed by McCormick D. B. et al., which has been validated to separate the main [14C]biotin radiometabolites in the rat urine.17 In our experiments (see Supporting Information for assay details), only intact [11C]biotin was detected in the urine of NBA and biotin-challenged groups, consistent with the previous studies of [14C]biotin metabolomics in biotin-challenged rats (10 mg/kg, 12 h post-IP injection) and NBA pigs (24 ng/kg, 24 h post-IV injection).17,18 These studies also reported that the administration of low doses of [14C]biotin (10 ng/kg, IP injection) in rats showed two radiometabolites: [14C]bisnorbiotin (29%) and [14C]biotin sulfoxide (10%) at a later time point of 3 h postadministration.19 It is important to note that bisnorbiotin and biotin sulfoxide are not substrates for SMVT.40 The absence of [11C]biotin radiometabolites in our experiments might be due to species differences (mice vs rats), concentration of biotin in the formulation (2 μg/kg vs 10 ng/kg), the early urine sampling time point (70 min vs 3 h), or the radiotracer route of administration (IV vs IP injection). A limitation of our preclinical rodent study is that we were unable to perform the plasma metabolite analysis during the PET image acquisition.
Oral Administration of [11C]biotin in Mice
The intake of biotin in humans and other mammals is via gastrointestinal absorption.9 Thus, to examine the gastrointestinal absorption of biotin and its body circulation in vivo, [11C]biotin was orally administered in mice. Ten anesthetized female mice were divided into two groups: group B1 (orogastric (OG) administration of vehicle 10 min before [11C]biotin OG administration, n = 5, Figure S4) and group B2 (biotin OG administration, 5 mg/Kg, 10 min before [11C]biotin OG administration, n = 5).
To establish the relative contribution of intestinal SMVT to biotin OG absorption, small-animal PET imaging studies were conducted in the isoflurane-anesthetized mice for 120 min postadministration of [11C]biotin. Demonstration of differential oral absorption between these two groups is highly valuable as it provides new insights into the role of SMVT expressed in the intestinal epithelium for the absorption of dietary biotin and delivery for circulation. Thus, we investigated the gastric emptying, intestinal absorption, and tissue distribution of OG-delivered [11C]biotin (Figure 3A–C).
Figure 3.
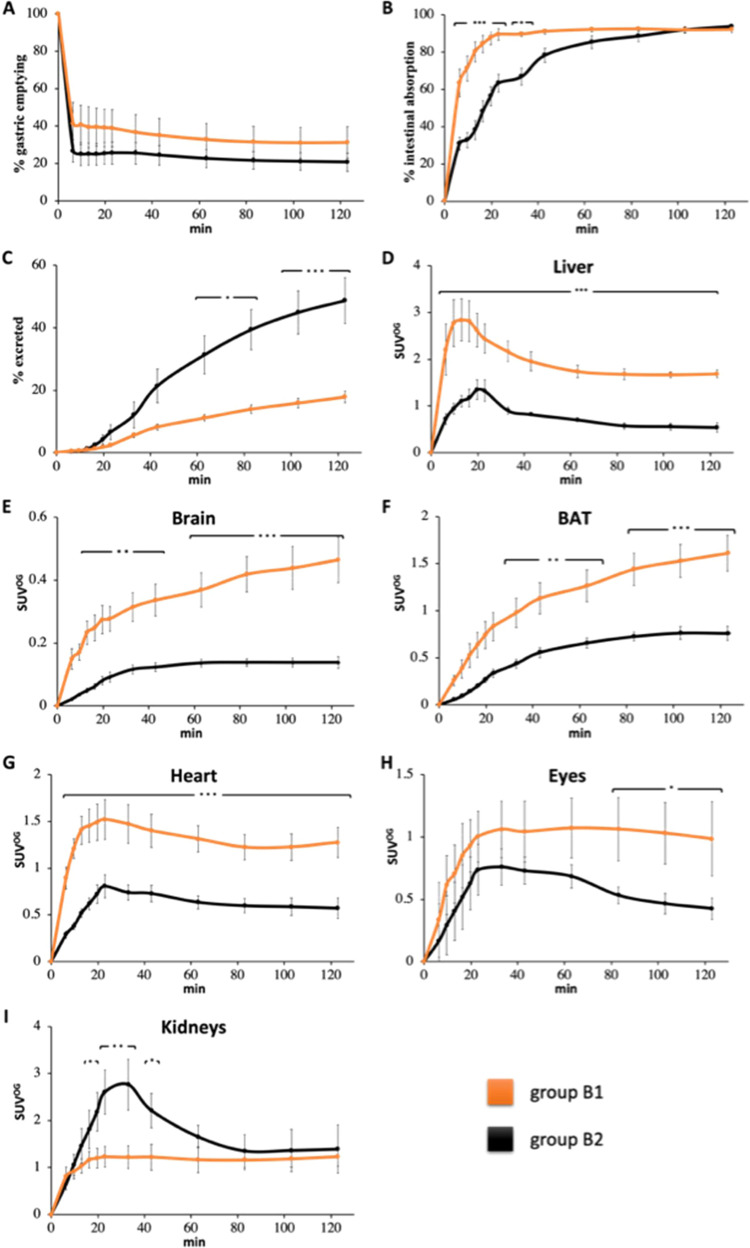
(A) Time (0–120 min)–% gastric emptying profile, (B) time–% intestinal absorption profile, (C) time–% excretion profile, and time–SUVOG profile for (D) liver, (E) brain, (F) BAT, (G) heart, (H) eyes, and (I) kidneys in NBA (orange line, group B1) and biotin-challenged (black line, group B2) mice receiving [11C]biotin OG. Data are the mean ± SEM. Note that the y-scale (SUVOG) varies between different tissues.
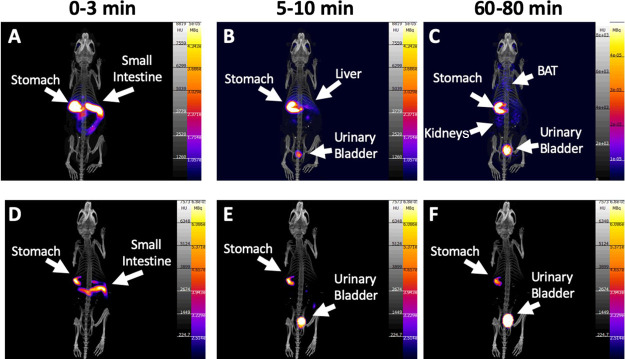
Gastric emptying by 2 h was ∼75% (Figure 3A and Figure S10A) with no differences between groups. Complete gastric emptying was not observed as the anesthesia reduces the gastrointestinal peristaltic movement.62 Indeed, dynamic PET studies showed that gastric emptying was a major factor in limiting complete [11C]biotin absorption because once delivered into the intestine, [11C]biotin was rapidly absorbed in the duodenum (no radioactivity was observed in the large intestine, Figure 4 and Figure S10B). Almost all [11C]biotin delivered to the small intestine (92%) was absorbed after 20 min in the NBA group (group B1), in contrast with a much lower [11C]biotin absorption in the biotin-challenged mice (56% in group B2) (Figures 3B and 4). The biotin-challenged mice absorbed almost all [11C]biotin delivered to the small intestine (80%) at 80 min post radiotracer administration.
Figure 4.
Maximum intensity projections of PET images from NBA (upper row, group B1) and biotin-challenged (lower row, group B1) mice receiving [11C]biotin OG at (A, D) 0–3 min, (B, E) 5–10 min, and (C, F) 60–80 min after the start of the PET imaging study. PET images are displayed according to the intensity scale for tracer activity, from white (highest) through red (intermediate) to purple (lowest). Stomach, intestine, urinary bladder, liver, kidneys and BAT are indicated where visible.
The [11C]biotin absorbed by the intestine entered the systemic circulation and was distributed throughout the body (Figure 4A–C) in the liver, heart, eyes, brain, and interscapular BAT. The organ distribution of [11C]biotin administered orally matches with that observed after IV administration. Compared with the NBA group (group B1), the biotin-challenged mice (group B2) showed a 2- to 4-fold decrease in [11C]biotin uptake in all SMVT-expressing organs (liver, brain, BAT, heart, and eyes) from 10 to 120 min postadministration and more than 2-fold higher concentration in the kidneys at 30 min (Figure 4D–F). The NBA and biotin-challenged mice excreted 18 and 49% of the total administered radioactivity within 120 min, respectively (Figure 3C), suggesting the saturation effect of the administered biotin, which results in faster elimination of bulk biotin through the kidneys to the urinary bladder.
Potential Use of [11C]biotin as a Research Tool for Diagnostic Imaging of Obesity/Diabetes, Bacterial Infection, and Cancer
Based on these results, [11C]biotin might be a valuable tool for imaging the function of BAT in humans, particularly in light of clinical studies suggesting an association between the decreased BAT function and obesity and diabetes.63,64 In translational antiobesity and antidiabetic research, pharmacological activation of BAT thermogenesis is a potential strategy for increasing energy expenditure. The gold-standard PET radiotracer for the in vivo visualization of BAT is [18F]FDG, but glucose uptake might not reflect BAT thermogenesis as fatty acids obtained from intracellular triglyceride lipolysis, not glucose, are the primary substrate for activated BAT.65,66 Therefore, the involvement of biotin-dependent enzymes in the fatty acid metabolism suggests that BAT uptake of [11C]biotin might reflect the BAT thermogenesis activity. The validation of [11C]biotin as a tool to monitor the BAT thermogenesis activity will require further studies in rodents under cold exposure or pharmacological interventions (e.g., norepinephrine or propranolol).
Other potential applications of [11C]biotin include the diagnosis and localization of bacterial infection and cancer via a direct-labeling or pretargeting strategy. First, the uptake of biotin by E. coli is highly efficient, but the modification of the carboxylic acid moiety of biotin reduces the affinity to bacterial transporters by 50-fold.67 [11C]Biotin, possessing a free carboxylic acid, might have similar or better prospects for imaging bacterial infection compared with the “biotin-inspired” radiotracers in which the carboxylic group is modified. Second, various aggressive cancer cell lines such as ovarian, leukemia, mastocytoma, colon, breast renal, and lung cancer cell lines overexpress SMVT.12 In anticancer drug development, the biotin–drug conjugation enhances tumor delivery of drugs and anticancer effects.68 The use of [11C]biotin in cancer might be useful for cancer diagnosis/management and for understanding the role of SMVT in tumorigenesis.
Besides the potential of [11C]biotin as an imaging biomarker in pathological conditions, its clinical translation will allow the study of whole-body biotin trafficking in healthy humans receiving an adequate intake level (30 mg/day) supplemented (10 mg/day) with daily doses of biotin to form the basis of a rational dosage regimen for this essential micronutrient. High biotin intake (300 mg/day) has shown positive effects in reducing disability progression in multiple sclerosis patients by activating myelin synthesis and enhancing energy production in demyelinated nerves.69,70 A multicenter randomized double-blind placebo-controlled study evaluating 642 multiple sclerosis patients administered with 300 mg/day of biotin is underway (clinical trial: NCT02936037). In our work, [11C]biotin administered by OG mimics the main route of intake of biotin in humans and other mammals. Intestinal absorption of [11C]biotin in mouse is rapid and virtually completes within minutes. Our OG studies indicate that [11C]biotin can be used to further investigate factors such as age, the effect of vitamin supplements, drugs, food, and pathological conditions of the gastrointestinal tract that may affect biotin absorption and trafficking. Gastrointestinal absorption in humans might be difficult to predict from the results of these preclinical animal studies due to species differences and the administration of anesthesia, which lowers the gastrointestinal peristaltic movements. However, this preclinical PET imaging study provides a fundamental methodology to investigate gastrointestinal absorption and whole-body biotin trafficking in humans.
Conclusions
In this report, we have demonstrated for the first time the feasibility of using a short-lived radioactive carbon-11 isotopologue of biotin to noninvasively study biotin trafficking in vivo using microPET imaging. This was made possible by the radiosynthesis of [11C]biotin, achieved using cyclotron-produced [11C]CO2 to radiolabel a commercially available precursor using a novel radiochemical reaction with a fully automated radiolabeling procedure. Here, we advance our understanding of the in vivo biodistribution and kinetics of [11C]biotin and the molecular mechanisms of biotin absorption in mice by studying the different routes of administration (IV vs OG), gender differences (male vs female), and the effect of biotin administration (NBA vs BC). The IV or OG administration of [11C]biotin to female mice revealed accumulation in SMVT-expressing organs such as the heart, eyes, brain, liver, and BAT.
Interestingly, the in vivo distribution of “biotin-inspired” imaging agents is not consistent with the known expression of SMVT, confirming that the use of [11C]biotin is crucial to elucidate the true behavior of biotin-dependent mechanisms in vitro and in vivo. In conclusion, the current work lays the foundation for mapping SMVT in disease models, the understanding of biotin’s role in BAT thermogenesis, and the prospect of a new translational tool to study biotin absorption and trafficking in healthy and diseased humans.
Experimental Section
Production of [11C]biotin
The automated radiosynthesis of [11C]biotin was performed on an Eckert & Ziegler (E&Z) Modular Lab system with the configuration as shown in Figure S1. Prior to production, an automated “flow test” sequence in Eckert & Ziegler (E&Z) software was performed by applying helium pressure into the system to check that the flow of gases was not obstructed, and the system was gas-tight. The preparation for each run also included an automated “conditioning” sequence. [11C]Biotin was prepared according to the following stepwise procedure.
[11C]CO2 was produced in a cyclotron using the 14N(p,α)11C nuclear reaction in a carbon-11 gas target filled with N2 containing 0.5% O2. The standard parameter for production was 15 μA for 8 min, with an estimated yield at the EOB of approximately 7–8 GBq of [11C]CO2. Cyclotron-produced [11C]CO2 was bubbled in a stream of helium gas directly into a solution (Vial A) containing diamino biotin (5 mg, 22.9 μmol, 1 equiv.) and DBU (4.5 equiv.) in MeCN (300 μL) at 0 °C for 1.75 min. At the end of the [11C]CO2 delivery, DBAD (6 equiv.) and PBu3 (6 equiv.) dissolved in 200 μL of MeCN (Vial C) were transferred to the reaction vial under a positive pressure of helium. The vial was heated at 100 °C for 5 min (Figure S1) with a helium flow rate of 60 mL/min, and the distilled solvent in Vial B was kept at −20 °C. Then, the Vial A was cooled at 25 °C, and the PBS solution from Vial D was transferred with helium (100 mL/min) to Vial A. The crude mixture was transferred to an HPLC injection loop using a vent filter. The reaction mixture was purified by semipreparative HPLC using a mobile phase composed of 2.5% ethanol in 10 mM PBS buffer solution (pH = 7.4). The mixture was transferred to an HPLC loop (2 mL) for subsequent semipreparative HPLC purification using a reversed-phase HPLC column (Discovery C18 HPLC Column, 5 μm particle size, 25 cm length, 10 mm internal diameter) equipped with a radioactivity detector (Radio-RP-HPLC) and eluted with a mobile phase composed of 2.5% ethanol in PBS at pH = 7.4 and at a flow rate of 4 mL/min (retention time: 12.6 min). The [11C]biotin peak was collected in a vented sterile vial through a 0.22 μm filter. An analytical HPLC analysis for quality control (QC) of the final tracer product was carried out on an HPLC analytical C18 column (Discovery C18 HPLC Column, 5 μm particle size, 15 cm length, 4.6 mm internal diameter, Figure S2A–C).
Quality Control (QC) of [11C]biotin
The analytical HPLC analysis for the QC of the final tracer product was carried out on an Agilent 1200 HPLC system equipped with a UV detector (λ = 210 nm) and a β + −flow detector coupled in series. The samples were injected onto an analytical Discovery C18 column (Discovery C18 HPLC Column, 5 μm particle size, 15 cm length, 4.6 mm internal diameter), which was eluted with a mobile phase of 2.5% ethanol in 10 mM PBS. The column flow rate is 1 mL/min and was kept at 25 °C (Figure S2A). The typical retention time of biotin is 4:08 min for UV absorbance (the delay of the radioactivity detector is 15 seconds at 1 mL/min further downstream from the UV detector, Figure S2B), and the radiochemical purity (RCP) was > 99%. A linear regression is determined for the UV absorbance peak areas of the standards. This constitutes the calibration curve. Then the UV peak area of the [11C]biotin formulation is fit on the calibration curve to determine the biotin concentration in the formulation and Am.
Biodistribution and PET Imaging Studies with [11C]biotin In Vivo
In vivo studies were carried out in male and female mice (Balb/C, Charles River UK Ltd). All animal studies were carried out in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986. Experiments complied with the UK Research Councils’ and the Medical Research Charities guidelines on responsibility in the use of animals in bioscience research, under UK Home Office project and personal licenses. The reporting of this study complied with the Animal Research: Reporting in vivo experiments (ARRIVE) guidelines (https://www.nc3rs.org.uk/arrive-guidelines). In order to assess and compare the in vivo distribution of [11C]biotin, the mice were divided into five groups. Each group of mice was allocated to: (1) the NBA IV female group, (2) the NBA IV male group, (3) the biotin-challenged IV female group, (4) the NBA OG female group, and (5) the biotin-challenged OG female. Mice of age 56–66 days were selected because they have an ideal size to perform total body imaging in our small-animal PET. PET/CT scans were taken once on each mouse using one administration route (Figure S4). A limitation of the study in this work is the challenge to carry out dynamic microPET experiments on fully conscious rodents because of ethical and practical concerns, so the mice were under anesthesia for the entire protocol study.
PET/CT Imaging in Mice
Dynamic PET scans (1:5 coincidence mode; 5 ns coincidence time window) were performed using a nanoScan PET/CT 8 W scanner (Mediso Ltd., Budapest, Hungary) over 60 (groups A1–A3) or 120 min (groups B1–B2) followed by CT scans (180 projections, 55 kVp X-ray source, 600 ms exposure time, 1:4 binning and semicircular acquisition) using proprietary acquisition software (Nucline 1.07). Ten minutes before the in vivo protocol study was scheduled to start, mice were anesthetized in a heated induction box by inhalation of 2% isoflurane in 100% oxygen.
For the IV injection protocol study, 30 μL of vehicle composed of 2.5% ethanol in 10 mM phosphate-buffered solution (PBS, pH 7.4, NBA group) or biotin (3.3 mg/mL, 2.5% ethanol in PBS, biotin-challenged group) was administered IV. After 10 min, [11C]biotin (146 ± 8 μL, 4.6 ± 0.8 MBq, Table S2) was administered IV (Figure S4A). The IV administrations were given via the tail vein cannula in mice positioned on a heated PET-CT small animal holder. There was no lag between the injection of [11C]biotin and the start of the PET acquisition. The syringe radioactivity was measured before and after administration to calculate the injected dose (ID).
For the OG administration protocol study, 30 μL of vehicle (2.5% ethanol in PBS, NBA group) or biotin (3.3 mg/mL, 2.5% ethanol in PBS, biotin-challenged group) was administered by OG. After 10 min, [11C]biotin (175 ± 6 μL, 6.1 ± 1.1 MBq) was administered by OG (Figure S4B). For dynamic microPET studies, anesthetized animals were placed on the PET-CT holder immediately after OG administration of [11C]biotin and a 2 h PET scan started; a delay of approximately 2.5–3 min was observed between the tracer delivery due to the time taken to administer [11C]biotin by OG and position the animal in the scanner. PET scan initiation has been taken when representing the data in Figures 3 and 4. The syringe radioactivity was measured before and after administration to calculate the administered dose.
After completion of the PET data acquisition, computed tomography (CT) scans were performed to provide anatomical information. CT images were acquired over 7 min. After the PET/CT scans, animals were culled (70 and 130 min post radiotracer IV injection and OG administration, respectively).
Analysis of PET Data
Whole-body Tera-Tomo (Mediso) 3-dimensional reconstruction was performed (400–600-keV energy window, 1–3 coincidence mode, 4 iterations, and 6 subsets) using an isotropic voxel size of 0.4 mm3. Images were corrected for attenuation, scatter, and decay. The acquired data were binned into 16 image frames (2 × 10, 2 × 20, 3 × 60, 3 × 120, 2 × 300, and 4 × 600 s) for Figures 1 and 2 and 12 image frames (6 × 200 and 2 × 600 and 4 × 1200 s) for Figures 3 and 4. VivoQuant software (Version 3.5, InviCRO Inc., http://www.vivoquant.com/) was used for image display and volume-of-interest (VOI) analysis. In each experiment, VOI for the whole mouse, liver, kidneys, brain, heart, BAT, and eyes were drawn manually. For animals receiving [11C]biotin IV, SUV = [decay-corrected tissue radioactivity concentration (Bq/ml)/(injected dose (Bq)] × body weight (g).
The apparent tissue–to–blood area under the curve (AUC) ratio (Kp,liver,AUC, Kp,kidney,AUC, Kp,BAT,AUC, and Kp,brain,AUC) was calculated using the following equation:
| 1 |
where AUCtissue,0–60 min represents the area under the tissue radioactivity concentration–time curve for the time period 0–60 min, and AUCblood,0–60 min represents the area under the blood radioactivity concentration–time curve for the time period 0–60 min.
The renal clearance (CLrenal,blood,0–60 min) with respect to the blood radioactivity concentration was calculated using the following equation:
| 2 |
where Xurine, 0–60 min represents the amount of radioactivity secreted into the urine between 0 and 60 min.
For animals receiving [11C]biotin by OG, SUVOG = [decay-corrected tissue radioactivity concentration (Bq/ml)/ (administered dose minus radioactivity remaining in the stomach (Bq)] × body weight (g). In each experiment, VOI for the whole mouse, stomach, intestine, liver, kidneys, brain, heart, BAT, eyes, and urinary bladder was drawn manually, and the radioactivity (Bq) in each VOI was estimated using VivoQuant software.
Gastric emptying and intestinal absorption in animals71 receiving [11C]biotin by OG were estimated by determining the radioactivity amount in the stomach and small intestine, respectively, as a function of time. Gastric emptying was estimated from the ratio of [11C]biotin in the stomach and the amount in the whole body. Intestinal absorption was estimated from the ratio of the amount of [11C]biotin in the small intestine versus the amount in the whole body minus the amount remaining in the stomach.
Statistical Analysis
Quantitative data were expressed as mean ± SEM. For Figures 1 and 3, and Figure S5, organ SUV at various time points of [11C]biotin was compared between the groups using a repeated measure mixed-effect model with an auto-regressive covariance structure. Posthoc analyses were corrected for multiple comparisons (IBM SPSS Statistics, Version 24.0). Differences at the 95% confidence level (P < 0.05) were considered significant. Data were indicated with (*) for p < 0.05, (**) for p < 0.01, and (***) for p < 0.001.
Acknowledgments
This work was supported by the Medical Research Council [MRC, MR/K022733/1], the European Commission FP7-PEOPLE-2012-ITN [316882, RADIOMI] and the Wellcome Trust [WT 084052/Z/07/Z] and the Wellcome/EPSRC Centre for Medical Engineering [WT 203148/Z/16/Z]. The research was supported by the National Institute for Health Research (NIHR) Biomedical Research Centre based at Guy’s and St Thomas’ NHS Foundation Trust and King’s College London. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, or the Department of Health.
Glossary
Abbreviations
- Am
molar activity
- AUC
area under the curve
- BAT
brown adipose tissue
- 11C
carbon-11
- 14C
carbon-14
- CLint,urine
intrinsic urinary excretion clearance, CO2, carbon dioxide
- DBU
8-diazabicyclo[5.4.0]undec-7-ene
- DBAD
di-tert-butyl azodicarboxylate
- [18F]FDG
[18F]Fluorodeoxyglucose
- ID
injected dose
- 3H
tritium
- HPLC
high-performance liquid chromatography
- IV
intravenous
- IP
intraperitoneal
- Kp,tissue, AUC
tissue–to–blood area under the curve (AUC) radioactivity ratio
- MeCN
acetonitrile
- NBA
no-biotin-added
- OG
orogastric
- PBS
phosphate-buffered saline
- PET
positron emission tomography
- PBu3
tributylphosphine
- QC
quality control
- RCP
radiochemical purity
- SMVT
sodium-dependent vitamin transporter
- SPECT
single-photon emission computed tomography
- SUV
standardized uptake value
- TLC
thin-layer chromatography.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acs.jmedchem.0c00494.
Preparation of the synthesis module, schematic diagram of the automated synthesis of [11C]biotin, in vitro [11C]biotin–streptavidin binding, radio-HPLC radiochromatogram of formulated [11C]biotin on analytical HPLC, schematic illustration of the PET/CT image acquisition of intravenous (IV) and orogastric (OG) administration, biodistribution and metabolite studies in mice, time–activity curves of male and female mice receiving [11C]biotin IV, correlation of blood–radioactivity concentration from in vivo PET images versus ex vivo biodistribution, Logan plots of kidneys, liver, BAT, and brain of groups A1–A3, and Patlak plots of BAT and brain of groups A1–A3 (PDF)
Molecular formula strings (CSV)
Author Contributions
S.B. and A.G. conceived the study. S.B. performed the radiosynthesis. K.S. and S.B. designed the preclinical protocol. J.B. and T.S. and S.B. performed the preclinical experiments. T.S. and S.B. performed the ex vivo biodistribution and metabolite analysis. S.B. and J.D. analysed the data. S.B., J.D., P. B., and A.G. interpreted the data and wrote the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Said H. M. Biotin: biochemical, physiological and clinical aspects. Subcell. Biochem. 2012, 56, 1–19. 10.1007/978-94-007-2199-9_1. [DOI] [PubMed] [Google Scholar]
- Ochoa-Ruiz E.; Diaz-Ruiz R.; Hernandez-Vazquez Ade J.; Ibarra-Gonzalez I.; Ortiz-Plata A.; Rembao D.; Ortega-Cuellar D.; Viollet B.; Uribe-Carvajal S.; Corella J. A.; Velázquez-Arellano A. Biotin deprivation impairs mitochondrial structure and function and has implications for inherited metabolic disorders. Mol. Genet. Metab. 2015, 116, 204–214. 10.1016/j.ymgme.2015.08.009. [DOI] [PubMed] [Google Scholar]
- Balamurugan K.; Ortiz A.; Said H. M. Biotin uptake by human intestinal and liver epithelial cells: role of the SMVT system. Am. J. Physiol. Gastrointest. Liver Physiol. 2003, 285, G73–G77. 10.1152/ajpgi.00059.2003. [DOI] [PubMed] [Google Scholar]
- Ohkura Y.; Akanuma S.; Tachikawa M.; Hosoya K. Blood-to-retina transport of biotin via Na+–dependent multivitamin transporter (SMVT) at the inner blood-retinal barrier. Exp. Eye Res. 2010, 91, 387–392. 10.1016/j.exer.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Uchida Y.; Ito K.; Ohtsuki S.; Kubo Y.; Suzuki T.; Terasaki T. Major involvement of Na(+)-dependent multivitamin transporter (SLC5A6/SMVT) in uptake of biotin and pantothenic acid by human brain capillary endothelial cells. J. Neurochem. 2015, 134, 97–112. 10.1111/jnc.13092. [DOI] [PubMed] [Google Scholar]
- Said H. M.; McAlister-Henn L.; Mohammadkhani R.; Horne D. W. Uptake of biotin by isolated rat liver mitochondria. Am. J. Physiol. 1992, 263, G81–G86. 10.1152/ajpgi.1992.263.1.G81. [DOI] [PubMed] [Google Scholar]
- Mallet R. T.; Sun J. Mitochondrial metabolism of pyruvate is required for its enhancement of cardiac function and energetics. Cardiovasc. Res. 1999, 42, 149–161. 10.1016/S0008-6363(98)00300-9. [DOI] [PubMed] [Google Scholar]
- Velázquez-Arellano A.; Hernandez-Esquivel Mde L.; Sanchez R. M.; Ortega-Cuellar D.; Rodriguez-Fuentes N.; Cano S.; Leon-Del-Rio A.; Carvajal K. Functional and metabolic implications of biotin deficiency for the rat heart. Mol. Genet. Metab. 2008, 95, 213–219. 10.1016/j.ymgme.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Chatterjee N. S.; Kumar C. K.; Ortiz A.; Rubin S. A.; Said H. M. Molecular mechanism of the intestinal biotin transport process. Am. J. Physiol. 1999, 277, C605–C613. 10.1152/ajpcell.1999.277.4.C605. [DOI] [PubMed] [Google Scholar]
- Park S.; Sinko P. J. The blood-brain barrier sodium-dependent multivitamin transporter: a molecular functional in vitro-in situ correlation. Drug Metab. Dispos. 2005, 33, 1547–1554. 10.1124/dmd.105.005231. [DOI] [PubMed] [Google Scholar]
- Podevin R. A.; Barbarat B. Biotin uptake mechanisms in brush-border and basolateral membrane vesicles isolated from rabbit kidney cortex. Biochim. Biophys. Acta 1986, 856, 471–481. 10.1016/0005-2736(86)90138-0. [DOI] [PubMed] [Google Scholar]
- Ren W. X.; Han J.; Uhm S.; Jang Y. J.; Kang C.; Kim J. H.; Kim J. S. Recent development of biotin conjugation in biological imaging, sensing, and target delivery. Chem. Commun. 2015, 51, 10403–10418. 10.1039/C5CC03075G. [DOI] [PubMed] [Google Scholar]
- Dakshinamurti K.; Mistry S. P. Tissue and intracellular distribution of biotin-C-1400H in rats and chicks. J. Biol. Chem. 1963, 238, 294–296. [PubMed] [Google Scholar]
- Kang Y. S.; Saito Y.; Pardridge W. M. Pharmacokinetics of [3H]biotin bound to different avidin analogues. J. Drug Target. 1995, 3, 159–165. 10.3109/10611869509059215. [DOI] [PubMed] [Google Scholar]
- Robinson B. H.; Oei J.; Saunders M.; Gravel R. [3H]Biotin-labeled proteins in cultured human skin fibroblasts from patients with pyruvate carboxylase deficiency. J. Biol. Chem. 1983, 258, 6660–6664. [PubMed] [Google Scholar]
- Wang K. S.; Kearns G. L.; Mock D. M. The clearance and metabolism of biotin administered intravenously to pigs in tracer and physiologic amounts is much more rapid than previously appreciated. J. Nutr. 2001, 131, 1271–1278. 10.1093/jn/131.4.1271. [DOI] [PubMed] [Google Scholar]
- Lee H. M.; Wright L. D.; McCormick D. B. Metabolism of carbonyl-labeled 14 C-biotin in the rat. J. Nutr. 1972, 102, 1453–1463. 10.1093/jn/102.11.1453. [DOI] [PubMed] [Google Scholar]
- Mock D. M.; Wang K. S.; Kearns G. L. The pig is an appropriate model for human biotin catabolism as judged by the urinary metabolite profile of radioisotope-labeled biotin. J. Nutr. 1997, 127, 365–369. 10.1093/jn/127.2.365. [DOI] [PubMed] [Google Scholar]
- Wang K. S.; Patel A.; Mock D. M. The metabolite profile of radioisotope-labeled biotin in rats indicates that rat biotin metabolism is similar to that in humans. J. Nutr. 1996, 126, 1852–1857. 10.1093/jn/126.7.1852. [DOI] [PubMed] [Google Scholar]
- Fraenkel-Conrat J.; Fraenkel-Conrat H. Metabolic fate of biotin and of avidin-biotin complex upon parenteral administration. Biochim. Biophys. Acta 1952, 8, 66–70. 10.1016/0006-3002(52)90008-5. [DOI] [PubMed] [Google Scholar]
- Rusckowski M.; Fogarasi M.; Virzi F.; Hnatowich D. J. Influence of endogenous biotin on the biodistribution of labelled biotin derivatives in mice. Nucl. Med. Commun. 1995, 16, 38–46. 10.1097/00006231-199501000-00008. [DOI] [PubMed] [Google Scholar]
- Claesener M.; Breyholz H. J.; Hermann S.; Faust A.; Wagner S.; Schober O.; Schafers M.; Kopka K. Efficient synthesis of a fluorine-18 labeled biotin derivative. Nucl. Med. Biol. 2012, 39, 1189–1194. 10.1016/j.nucmedbio.2012.08.001. [DOI] [PubMed] [Google Scholar]
- Shoup T. M.; Fischman A. J.; Jaywook S.; Babich J. W.; Strauss H. W.; Elmaleh D. R. Synthesis of fluorine-18-labeled biotin derivatives: biodistribution and infection localization. J. Nucl. Med. 1994, 35, 1685–1690. [PubMed] [Google Scholar]
- Kudo T.; Ueda M.; Konishi H.; Kawashima H.; Kuge Y.; Mukai T.; Miyano A.; Tanaka S.; Kizaka-Kondoh S.; Hiraoka M.; Saji H. PET imaging of hypoxia-inducible factor-1-active tumor cells with pretargeted oxygen-dependent degradable streptavidin and a novel 18F-labeled biotin derivative. Mol. Imaging Biol. 2011, 13, 1003–1010. 10.1007/s11307-010-0418-6. [DOI] [PubMed] [Google Scholar]
- Lewis M. R.; Wang M.; Axworthy D. B.; Theodore L. J.; Mallet R. W.; Fritzberg A. R.; Welch M. J.; Anderson C. J. In vivo evaluation of pretargeted 64Cu for tumor imaging and therapy. J. Nucl. Med. 2003, 44, 1284–1292. [PubMed] [Google Scholar]
- Hnatowich D. J.; Virzi F.; Rusckowski M. Investigations of avidin and biotin for imaging applications. J. Nucl. Med. 1987, 28, 1294–1302. [PubMed] [Google Scholar]
- Foulon C. F.; Alston K. L.; Zalutsky M. R. Synthesis and preliminary biological evaluation of (3-iodobenzoyl)norbiotinamide and ((5-iodo-3-pyridinyl)carbonyl)norbiotinamide: two radioiodinated biotin conjugates with improved stability. Bioconjugate Chem. 1997, 8, 179–186. 10.1021/bc970006m. [DOI] [PubMed] [Google Scholar]
- Prakash S.; Hazari P. P.; Meena V. K.; Jaswal A.; Khurana H.; Kukreti S.; Mishra A. K. Biotinidase resistant 68gallium-radioligand based on biotin/avidin interaction for pretargeting: synthesis and preclinical evaluation. Bioconjug. Chem. 2016, 27, 2780–2790. 10.1021/acs.bioconjchem.6b00576. [DOI] [PubMed] [Google Scholar]
- Li K.; Dong W.; Liu Q.; Lv G.; Xie M.; Sun X.; Qiu L.; Lin J. A biotin receptor-targeted silicon(IV) phthalocyanine for in vivo tumor imaging and photodynamic therapy. J. Photochem. Photobiol. B 2019, 190, 1–7. 10.1016/j.jphotobiol.2018.09.001. [DOI] [PubMed] [Google Scholar]
- Guo R.; Huang F.; Zhang B.; Yan Y.; Che J.; Jin Y.; Zhuang Y.; Dong R.; Li Y.; Tan B.; Song R.; Hu Y.; Dong X.; Li X.; Lin N. GSH activated niotin-tagged near-infrared probe for efficient cancer imaging. Theranostics 2019, 9, 3515–3525. 10.7150/thno.32742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhuniya S.; Maiti S.; Kim E. J.; Lee H.; Sessler J. L.; Hong K. S.; Kim J. S. An activatable theranostic for targeted cancer therapy and imaging. Angew. Chem. Int. Ed. Engl. 2014, 53, 4469–4474. 10.1002/anie.201311133. [DOI] [PubMed] [Google Scholar]
- Erba P. A.; Cataldi A. G.; Tascini C.; Leonildi A.; Manfredi C.; Mariani G.; Lazzeri E. 111In-DTPA-Biotin uptake by Staphylococcus aureus. Nucl. Med. Commun. 2010, 31, 994–997. 10.1097/MNM.0b013e32833ce32c. [DOI] [PubMed] [Google Scholar]
- Lazzeri E.; Erba P.; Perri M.; Tascini C.; Doria R.; Giorgetti J.; Mariani G. Scintigraphic imaging of vertebral osteomyelitis with 111In-biotin. Spine 2008, 33, E198–E204. 10.1097/BRS.0b013e31816960c9. [DOI] [PubMed] [Google Scholar]
- Rusckowski M.; Paganelli G.; Hnatowich D. J.; Magnani P.; Virzi F.; Fogarasi M.; DiLeo C.; Sudati F.; Fazio F. Imaging osteomyelitis with streptavidin and indium-111-labeled biotin. J. Nucl. Med. 1996, 37, 1655–1662. [PubMed] [Google Scholar]
- Lazzeri E.; Manca M.; Molea N.; Marchetti S.; Consoli V.; Bodei L.; Bianchi R.; Chinol M.; Paganelli G.; Mariani G. Clinical validation of the avidin/indium-111 biotin approach for imaging infection/inflammation in orthopaedic patients. Eur. J. Nucl. Med. 1999, 26, 606–614. 10.1007/s002590050428. [DOI] [PubMed] [Google Scholar]
- Lazzeri E.; Pauwels E. K.; Erba P. A.; Volterrani D.; Manca M.; Bodei L.; Trippi D.; Bottoni A.; Cristofani R.; Consoli V.; Palestro C. J.; Mariani G. Clinical feasibility of two-step streptavidin/111In-biotin scintigraphy in patients with suspected vertebral osteomyelitis. Eur. J. Nucl. Med. Mol. Imaging 2004, 31, 1505–1511. 10.1007/s00259-004-1581-2. [DOI] [PubMed] [Google Scholar]
- Paganelli G.; Magnani P.; Zito F.; Villa E.; Sudati F.; Lopalco L.; Rossetti C.; Malcovati M.; Chiolerio F.; Seccamani E.; Siccardi A. G.; Fazio F. Three-step monoclonal antibody tumor targeting in carcinoembryonic antigen-positive patients. Cancer Res. 1991, 51, 5960–5966. [PubMed] [Google Scholar]
- Kalofonos H. P.; Rusckowski M.; Siebecker D. A.; Sivolapenko G. B.; Snook D.; Lavender J. P.; Epenetos A. A.; Hnatowich D. J. Imaging of tumor in patients with indium-111-labeled biotin and streptavidin-conjugated antibodies: preliminary communication. J. Nucl. Med. 1990, 31, 1791–1796. [PubMed] [Google Scholar]
- Spencer R. P.; Brody K. R. Biotin transport by small intestine of rat, hamster, and other species. Am. J. Physiol. 1964, 206, 653–657. 10.1152/ajplegacy.1964.206.3.653. [DOI] [PubMed] [Google Scholar]
- Baur B.; Wick H.; Baumgartner E. R. Na(+)-dependent biotin transport into brush-border membrane vesicles from rat kidney. Am. J. Physiol. 1990, 258, F840–F847. 10.1152/ajprenal.1990.258.4.F840. [DOI] [PubMed] [Google Scholar]
- Ma T. Y.; Dyer D. L.; Said H. M. Human intestinal cell line Caco-2: a useful model for studying cellular and molecular regulation of biotin uptake. Biochim. Biophys. Acta 1994, 1189, 81–88. 10.1016/0005-2736(94)90283-6. [DOI] [PubMed] [Google Scholar]
- Melville D. B.; Pierce J. G.; Partridge C. W. The preparation of C14-labeled biotin and a study of its stability during carbon dioxide fixation. J. Biol. Chem. 1949, 180, 299–305. [PubMed] [Google Scholar]
- Rotstein B. H.; Liang S. H.; Placzek M. S.; Hooker J. M.; Gee A. D.; Dolle F.; Wilson A. A.; Vasdev N. 11C=O bonds made easily for positron emission tomography radiopharmaceuticals. Chem. Soc. Rev. 2016, 45, 4708–4726. 10.1039/C6CS00310A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dheere A. K. H.; Bongarzone S.; Taddei C.; Yan R.; Gee A. D. Synthesis of C-11-labelled symmetrical ureas via the rapid incorporation of [C-11]CO2 into aliphatic and aromatic amines. Synlett 2015, 26, 2257–2260. 10.1055/s-0034-1381055. [DOI] [Google Scholar]
- Dheere A. K. H.; Yusuf N.; Gee A. Rapid and efficient synthesis of [C-11]ureas via the incorporation of [C-11]CO2 into aliphatic and aromatic amines. Chem. Commun. 2013, 49, 8193–8195. 10.1039/c3cc44046j. [DOI] [PubMed] [Google Scholar]
- Bongarzone S.; Ferocino A.; Gee A. Expanding the scope of carbon-11 labelled ureas: a universal method to access short-lived click reagents for in vivo PET imaging. J. Labelled Compd. Radiopharm. 2019, 62, S244–S245. [Google Scholar]
- Lee P.; Swarbrick M. M.; Zhao J. T.; Ho K. K. Y. Inducible brown adipogenesis of supraclavicular fat in adult humans. Endocrinology 2011, 152, 3597–3602. 10.1210/en.2011-1349. [DOI] [PubMed] [Google Scholar]
- Lee M. J.; Fried S. K. Optimal protocol for the differentiation and metabolic analysis of human adipose stromal cells. Methods Enzymol. 2014, 538, 49–65. 10.1016/B978-0-12-800280-3.00004-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuri-Harcuch W.; Wise L. S.; Green H. Interruption of the adipose conversion of 3T3 cells by biotin deficiency: differentiation without triglyceride accumulation. Cell 1978, 14, 53–59. 10.1016/0092-8674(78)90300-8. [DOI] [PubMed] [Google Scholar]
- Saito M.; Okamatsu-Ogura Y.; Matsushita M.; Watanabe K.; Yoneshiro T.; Nio-Kobayashi J.; Iwanaga T.; Miyagawa M.; Kameya T.; Nakada K.; Kawai Y.; Tsujisaki M. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 2009, 58, 1526–1531. 10.2337/db09-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet V.; Labbe S. M.; Blondin D. P.; Phoenix S.; Guerin B.; Haman F.; Turcotte E. E.; Richard D.; Carpentier A. C. Brown adipose tissue oxidative metabolism contributes to energy expenditure during acute cold exposure in humans. J. Clin. Invest. 2012, 122, 545–552. 10.1172/JCI60433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F.; Hao G.; Shao M.; Nham K.; An Y.; Wang Q.; Zhu Y.; Kusminski C. M.; Hassan G.; Gupta R. K.; Zhai Q.; Sun X.; Scherer P. E.; Oz O. K. An adipose tissue atlas: an image-guided identification of human-like BAT and beige depots in rodents. Cell Metab. 2018, 27, 252–262.e3. e253 10.1016/j.cmet.2017.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxl A.; Wanek T.; Mairinger S.; Stanek J.; Filip T.; Sauberer M.; Muller M.; Kuntner C.; Langer O. Breast cancer resistance protein and p-glycoprotein influence in vivo disposition of 11C-erlotinib. J. Nucl. Med. 2015, 56, 1930–1936. 10.2967/jnumed.115.161273. [DOI] [PubMed] [Google Scholar]
- Shingaki T.; Hume W. E.; Takashima T.; Katayama Y.; Okauchi T.; Hayashinaka E.; Wada Y.; Cui Y.; Kusuhara H.; Sugiyama Y.; Watanabe Y. Quantitative evaluation of mMate1 function based on minimally invasive measurement of tissue concentration using PET with [(11)C]metformin in mouse. Pharm. Res. 2015, 32, 2538–2547. 10.1007/s11095-015-1642-1. [DOI] [PubMed] [Google Scholar]
- Takano A.; Kusuhara H.; Suhara T.; Ieiri I.; Morimoto T.; Lee Y. J.; Maeda J.; Ikoma Y.; Ito H.; Suzuki K.; Sugiyama Y. Evaluation of in vivo p-glycoprotein function at the blood-brain barrier among MDR1 gene polymorphisms by using 11C-verapamil. J. Nucl. Med. 2006, 47, 1427–1433. [PubMed] [Google Scholar]
- Takashima T.; Wu C.; Takashima-Hirano M.; Katayama Y.; Wada Y.; Suzuki M.; Kusuhara H.; Sugiyama Y.; Watanabe Y. Evaluation of breast cancer resistance protein function in hepatobiliary and renal excretion using PET with 11C-SC-62807. J. Nucl. Med. 2013, 54, 267–276. 10.2967/jnumed.112.110254. [DOI] [PubMed] [Google Scholar]
- Bertoldo A.; Rizzo G.; Veronese M. Deriving physiological information from PET images: from SUV to compartmental modelling. Clin. Transl. Imaging 2014, 2, 239–251. 10.1007/s40336-014-0067-x. [DOI] [Google Scholar]
- Logan J. Graphical analysis of PET data applied to reversible and irreversible tracers. Nucl. Med. Biol. 2000, 27, 661–670. 10.1016/S0969-8051(00)00137-2. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Ye W.; Brasic J. R.; Wong D. F. Multi-graphical analysis of dynamic PET. Neuroimage 2010, 49, 2947–2957. 10.1016/j.neuroimage.2009.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan K.; Vaziri N. D.; Said H. M. Biotin uptake by human proximal tubular epithelial cells: cellular and molecular aspects. Am. J. Physiol. Renal. Physiol. 2005, 288, F823–F831. 10.1152/ajprenal.00375.2004. [DOI] [PubMed] [Google Scholar]
- Spencer P. D.; Roth K. S. On the uptake of biotin by the rat renal tubule. Biochem. Med. Metab. Biol. 1988, 40, 95–100. 10.1016/0885-4505(88)90109-0. [DOI] [PubMed] [Google Scholar]
- Yamashita S.; Takashima T.; Kataoka M.; Oh H.; Sakuma S.; Takahashi M.; Suzuki N.; Hayashinaka E.; Wada Y.; Cui Y.; Watanabe Y. PET imaging of the gastrointestinal absorption of orally administered drugs in conscious and anesthetized rats. J. Nucl. Med. 2011, 52, 249–256. 10.2967/jnumed.110.081539. [DOI] [PubMed] [Google Scholar]
- van Marken Lichtenbelt W. D.; Vanhommerig J. W.; Smulders N. M.; Drossaerts J. M.; Kemerink G. J.; Bouvy N. D.; Schrauwen P.; Jaap Teule G. J. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 2009, 360, 1500–1508. 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- Nedergaard J.; Bengtsson T.; Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am. J. Physiol. Endocrinol. Metab. 2007, 293, E444–E452. 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- Sampath S. C.; Sampath S. C.; Bredella M. A.; Cypess A. M.; Torriani M. Imaging of brown adipose tissue: state of the art. Radiology 2016, 280, 4–19. 10.1148/radiol.2016150390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong F. J.; Ahmed B. A.; Oreskovich S. M.; Blondin D. P.; Haq T.; Konyer N. B.; Noseworthy M. D.; Haman F.; Carpentier A. C.; Morrison K. M.; Steinberg G. R. Recent advances in the detection of brown adipose tissue in adult humans: a review. Clin. Sci. (Lond) 2018, 132, 1039–1054. 10.1042/CS20170276. [DOI] [PubMed] [Google Scholar]
- Piffeteau A.; Zamboni M.; Gaudry M. Biotin transport by a biotin-deficient strain of Escherichia coli. Biochim. Biophys. Acta 1982, 688, 29–36. 10.1016/0005-2736(82)90575-2. [DOI] [PubMed] [Google Scholar]
- Maiti S.; Paira P. Biotin conjugated organic molecules and proteins for cancer therapy: a review. Eur. J. Med. Chem. 2018, 145, 206–223. 10.1016/j.ejmech.2018.01.001. [DOI] [PubMed] [Google Scholar]
- Tourbah A.; Lebrun-Frenay C.; Edan G.; Clanet M.; Papeix C.; Vukusic S.; De Seze J.; Debouverie M.; Gout O.; Clavelou P.; Defer G.; Laplaud D. A.; Moreau T.; Labauge P.; Brochet B.; Sedel F.; Pelletier J.; MD1003 (high-dose biotin) for the treatment of progressive multiple sclerosis: a randomised, double-blind, placebo-controlled study. Mult. Scler. 2016, 22, 1719–1731. 10.1177/1352458516667568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedel F.; Bernard D.; Mock D. M.; Tourbah A. Targeting demyelination and virtual hypoxia with high-dose biotin as a treatment for progressive multiple sclerosis. Neuropharmacology 2016, 110, 644–653. 10.1016/j.neuropharm.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Sala-Rabanal M.; Ghezzi C.; Hirayama B. A.; Kepe V.; Liu J.; Barrio J. R.; Wright E. M. Intestinal absorption of glucose in mice as determined by positron emission tomography. J. Physiol. 2018, 596, 2473–2489. 10.1113/JP275934. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.