Abstract
Gamma aminobutyric acid (GABA) is the principal inhibitory neurotransmitter playing a key role in anxiety and depression disorders in mammals. Recent studies revealed that members of the gut microbiota are able to produce GABA modulating the gut–brain axis response. Among members of the human gut microbiota, bifidobacteria are well known to establish many metabolic and physiologic interactions with the host. In this study, we performed genome analyses of more than 1,000 bifidobacterial strains publicly available revealing that Bifidobacterium adolescentis taxon might represent a model GABA producer in human gastrointestinal tract. Moreover, the in silico screening of human/animal metagenomic datasets showed an intriguing association/correlation between B. adolescentis load and mental disorders such as depression and anxiety. Interestingly, in vitro screening of 82 B. adolescentis strains allowed identifying two high GABA producers, i.e. B. adolescentis PRL2019 and B. adolescentis HD17T2H, which were employed in an in vivo trial in rats. Feeding Groningen rats with a supplementation of B. adolescentis strains, confirmed the ability of these microorganisms to stimulate the in vivo production of GABA highlighting their potential implication in gut–brain axis interactions.
Subject terms: Microbiology, Microbial genetics
Introduction
Gamma-Aminobutyric acid (GABA) is a non-protein amino acid that is widely distributed in plants, animals and microorganisms1,2. GABA is synthetized by a pyridocal-5′-phosphate (PLP)—dependent glutamate decarboxylase (GAD) enzyme by irreversible α-decarboxylation of l-glutamate and consummation of one cytoplasmic proton1,2. GABA has several well-known physiological and psychological functions. Different studies highlighted that it is predominantly present in the brain where it acts as a major inhibitory neurotransmitter in the mammalian central nervous system (CNS)1,2. Specifically, dysfunctions in GABA metabolism are involved in anxiety and depression3–5. Furthermore, it is involved in the regulation of blood pressure and heart rate and plays a role in the perception of pain and anxiety5,6. Other potential health benefits of GABA are control of growth hormone secretion, protection against glycerol-induced acute renal failure in rats and anti-proliferative activity7.
Recently, the term “psychobiotic” has been introduced to designate live bacterial strains, including lactobacilli and bifidobacteria, which are able to influence the CNS function8. There are several compounds produced by these bacteria, such as proteins, peptides and components of cell wall that are potential mediators between bacteria and their hosts. Neurotransmitters, such as GABA, represent an example of neuroactive molecules produced by psychobiotics and members of the human gut microbiota that have been found to modulate neural signals which affect neurological and psychiatric parameters, as well as sleep, appetite, mood and cognition8. Genetically, it has been found the presence of gad genes, predicted to encode for glutamate decarboxylase or glutamic acid decarboxylase, in the genomes of Lactic Acid Bacteria (LAB) and bifidobacteria that are supposed to be responsible of the GABA production5,9–12. Recent studies revealed that the increased level of GABA in the human gut could be derived by the ability of the intestinal microbiota or ingested probiotic, such as bifidobacteria and lactobacilli, to metabolize dietary monosodium glutamate (MSG)5,9–12. Nevertheless, the ability to produce GABA by gut-derived bifidobacteria strains remains poorly studied. Until now, only three bifidobacterial species, such as Bifidobacterium dentium, Bifidobacterium longum subsp. infantis and Bifidobacterium adolescentis were shown to produce GABA by means of in vitro studies5.
The aim of this study is to understand if the production of GABA in bifidobacteria is a strain-specific feature, analyzing the genomic sequence of 1,022 bifidobacterial strains belonging to the currently known 77 Bifidobacterium taxa, representing 70 species and seven subspecies, coupling the in silico information with an in vitro measurements of GABA levels generated by these bacteria. Notably, the production of GABA by those B. adolescentis strains displaying the highest in vitro GABA-synthesis performance was further evaluated through an in vivo trial involving rats. In addition, the screening of metagenomic datasets of clinical population and rat models of depression and anxiety revealed an intriguing association/correlation between B. adolescentis load and these mental disorders.
Results and discussion
Distribution of GABA genes among the Bifidobacterium genus
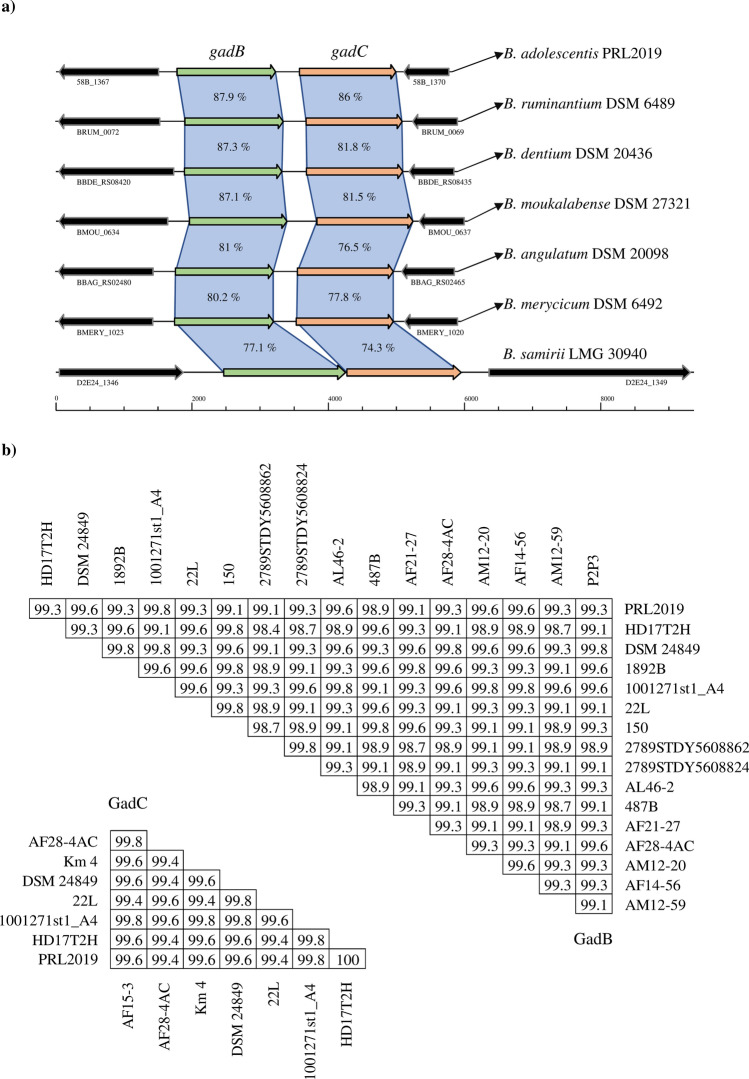
The ability to produce GABA by few gut-derived bifidobacterial taxa have been previously described13. Thus, a comprehensive screening of GABA production by bifidobacteria for each of the currently recognized species belonging to the genus Bifidobacterium was warranted. In order to fulfill this gap of knowledge a genetic survey involving 1,022 genomes from 81 (sub)species of the genus Bifidobacterium14–16, including taxa isolated from the gut of humans and animals, was performed to shed light into which taxa possess the appropriate genetic makeup for the synthesis of GABA. The dissected proteome of 1,022 bifidobacterial strains retrieved from the genomic NCBI database as well as our bifidobacterial genome database (Table S1), revealed that 81 strains encode for both GadB and GadC, encompassing seven different species, i.e., B. adolescentis, Bifidobacterium angulatum, B. dentium, Bifidobacterium merycicum, Bifidobacterium moukalabense, Bifidobacterium ruminantium and Bifidobacterium samirii (Table S3). Interestingly, four of the identified species that share the GAD/GABA antiporter locus belongs to members of the B. adolescentis phylogenetic group14, including 75 out of 81 analyzed genomes. Based on the sequence identity values obtained between the identified protein sequences, we observed a higher conservation among members of the B. adolescentis phylogenetic group, while lower values of identity were found in B. angulatum, B. merycicum and B. samirii taxa, which reflect their belonging to other bifidobacterial phylogenetic groups14,15 (Fig. 1a). Overall, among the identified bifidobacterial species sharing the GAD/GABA antiporter locus, B. adolescentis, B. angulatum and B. dentium are of human origins, while the other five taxa are usually associated with the gut of other mammals, such as monkeys and bovines17–19. Between the above listed taxa of human origins, members of the B. adolescentis species are the most scrutinized for both genomic and proved production of GABA20–22. Intriguingly, the high level of prevalence of GAD/GABA antiporter locus within the 50 B. adolescentis genomes analyzed (94%) (Table S3), coupled with the fact that such bifidobacterial species are occurring in the human gut20,23, suggests that this bifidobacterial taxon might represent a model GABA producer.
Figure 1.
Bifidobacterium genetic map of GAD/GABA antiporter locus. Panel (a) displays genetic maps belonging to different Bifidobacterium species in which the locus has been identified. The gadB and gadC genes are highlighted with the relative color. Each arrow indicates an open reading frames (ORF), whereas the length of the arrow is proportional to the length of the predicted ORF. Panel (b) depicts the amino acid sequence identity values of GadB and GadC between the analyzed B. adolescentis genomes. Duplicates of both genes were removed to highlight non-redundant values between strains.
Gut microbiota composition in depression and anxiety
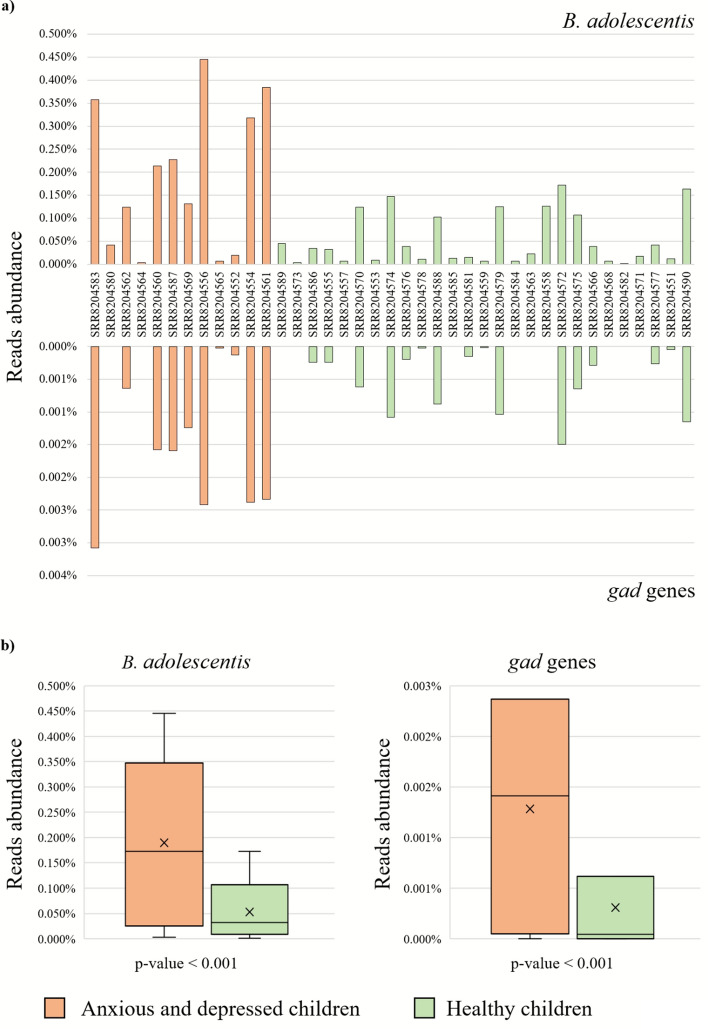
Since GABA, which is the primary inhibitory neurotransmitter known to counterbalance the action of the excitatory neurotransmitter glutamate, plays an important role in the treatment of anxiety and depressive disorders24,25, we decided to investigate the presence of B. adolescentis genomes and associated gad gene sequences in two public human gut microbiome datasets related to these illnesses (PRJNA496479 and PRJNA474710). Thus, metagenomic samples collected from children (PRJNA496479) were screened for reads corresponding to gad genes and B. adolescentis chromosome sequences, unveiling dissimilar profiles between samples (Fig. 2). The number of metagenomic reads belonging to B. adolescentis ranged from 76,127 to none, with higher values especially in samples belonging to anxious and depressed children (t test p value < 0.001, df = 37, Cohen's d = 0.97 and effect-size r = 0.43) (sample size estimation of 12 between groups, based on B. adolescentis abundance) (Fig. 2). Accordingly, metagenomic reads belonging to gad genes were found to be statistically higher in the samples displaying higher abundance of B. adolescentis (t test p value < 0.001, df = 37, Cohen's d = 1.02 and effect-size r = 0.45) (Fig. 2). Therefore, these data highlighted a clear correlation between the higher relative abundance of B. adolescentis sequences, together with related gad genes, and children with subclinical symptoms of anxiety and depression. In contrast, metagenomic samples from rats (PRJNA474710) displayed the complete absence of any trace of sequences related to B. adolescentis chromosome and gad genes. Such finding could be explained by the fact that B. adolescentis are not naturally occurring in the ceca of rats23. Based on these results, B. adolescentis was found to be an excellent model organism to investigate its ability to produce GABA in the gut environment.
Figure 2.
Relative abundance of B. adolescentis and gad genes within analyzed children gut microbiomes. Panel (a) shows the overall abundance of B. adolescentis- and gad genes-associated reads within the filtered children gut microbiome samples (PRJNA496479). The y-axis represents the percentage of reads identified, whereas the x-axis reports the sample numbers. Values associated to gad genes are reported in reverse order. The anxious and depressed children samples are represented as orange-colored bars, whereas healthy subjects in green. Panel (b) exhibits two Whisker plots based on relative abundances of B. adolescentis and gad genes in the gut microbiota data, which results in both chases with a p value of < 0.001 between depressed and healthy children (Student’s t test). The y axis shows the percentage of reads identified. Boxes represent 50% of the data set, distributed between the 1st and 3rd quartiles. The median divides the boxes into the interquartile range, while the X represents the mean. The lines extending vertically outside the boxes show the outlier range.
Production of GABA by B. adolescentis strains
In order to investigate the production of GABA in B. adolescentis species, a collection of 82 bifidobacterial strains was scrutinized for this feature employing an in vitro approach. The investigated strains were mainly isolated from fecal samples or colon biopsy of healthy humans (Table 1). In accordance to the in silico data previously described, in vitro GABA production was revealed as a frequent trait of B. adolescentis taxon, since 79% of the tested B. adolescentis strains displayed the ability to transform the precursor monosodium glutamate (GMS) to GABA. Specifically, 23% of all the tested B. adolescentis strains were classified as high GABA producers, as they were capable to efficiently convert more than 65% of the precursor to GABA (Fig. 3). In view of these results, two representative strains classified as high GABA producers, i.e. B. adolescentis PRL2019 and B. adolescentis HD17T2H, were chosen as model bifidobacterial strains to further investigate this intriguing metabolic feature in an in vivo model.
Table 1.
GABA production levels determined in overnight cultures from the 82 Bifidobacterium strains included in this work.
| Species | Strain | Strain origin | [GABA] mM | % GMS conversion to GABA | ||
|---|---|---|---|---|---|---|
| Average | SD | Average | SD | |||
| B. adolescentis | 14B | Intestine of adult | 8.77 | 0.434 | 80.755 | 3.999 |
| B. adolescentis | 153B | Intestine of adult | 1.72 | 0.534 | 15.877 | 4.921 |
| B. adolescentis | 1BCM1 | Colon biopsy | 6.04 | 2.272 | 55.587 | 20.918 |
| B. adolescentis | 1CCM5 | Colon content | 5.38 | 2.126 | 49.489 | 19.573 |
| B. adolescentis | 22L | Human milk | 1.92 | 0.367 | 17.707 | 3.375 |
| B. adolescentis | 235B | Intestine of adult | 6.78 | 0.726 | 62.397 | 6.687 |
| B. adolescentis | 236B | Intestine of adult | 9.16 | 1.914 | 84.355 | 17.622 |
| B. adolescentis | 2BCM1 | Colon biopsy | 3.73 | 1.858 | 34.358 | 17.106 |
| B. adolescentis | 2BCM2 | Colon biopsy | 7.37 | 0.816 | 67.855 | 7.511 |
| B. adolescentis | 2CCM6 | Colon content | 5.67 | 0.862 | 52.196 | 7.940 |
| B. adolescentis | 2CCM7 | Colon content | 5.62 | 1.064 | 51.787 | 9.798 |
| B. adolescentis | 42B | Human faeces | 4.60 | 0.328 | 42.314 | 3.023 |
| B. adolescentis | 487B | Human faeces | 4.45 | 2.076 | 40.928 | 19.111 |
| B. adolescentis | 4CCM2 | Colon content | 2.82 | 0.887 | 25.944 | 8.170 |
| B. adolescentis | 50B | Intestine of adult | 4.30 | 0.628 | 39.619 | 5.780 |
| B. adolescentis | 53B | Intestine of adult | 8.62 | 0.614 | 79.410 | 5.656 |
| B. adolescentis | 55B | Intestine of adult | 5.15 | 0.493 | 47.407 | 4.536 |
| B. adolescentis | 56B | Intestine of adult | 5.19 | 0.193 | 47.798 | 1.774 |
| B. adolescentis | 57B | Intestine of adult | 5.98 | 0.537 | 55.014 | 4.944 |
| B. adolescentis | PRL2019 | Intestine of adult | 7.06 | 0.213 | 64.965 | 1.963 |
| B. adolescentis | 61B | Intestine of adult | 4.50 | 0.254 | 41.447 | 2.342 |
| B. adolescentis | 62B | Intestine of adult | 4.26 | 0.286 | 39.204 | 2.636 |
| B. adolescentis | 6BCM1 | Colon biopsy | 7.80 | 0.366 | 43.102 | 39.418 |
| B. adolescentis | 6CCM3 | Colon content | 7.13 | 0.803 | 65.673 | 7.390 |
| B. adolescentis | 703B | Human faeces | 0.59 | 0.008 | 5.454 | 0.070 |
| B. adolescentis | 70B | Human faeces | 3.59 | 0.115 | 33.065 | 1.059 |
| B. adolescentis | 712B | Human faeces | 0.91 | 0.232 | 8.369 | 2.135 |
| B. adolescentis | 713B | Intestine of adult | 4.53 | 0.676 | 41.696 | 6.224 |
| B. adolescentis | 714B | Intestine of adult | 0.86 | 0.079 | 7.881 | 0.731 |
| B. adolescentis | 740B | Intestine of adult | 0.71 | 0.059 | 6.496 | 0.545 |
| B. adolescentis | 74B | Intestine of adult | 8.31 | 0.939 | 76.539 | 8.649 |
| B. adolescentis | 75B | Intestine of adult | 5.31 | 0.839 | 29.338 | 27.333 |
| B. adolescentis | 76B | Intestine of adult | 6.31 | 1.565 | 58.064 | 14.410 |
| B. adolescentis | 77B | Intestine of adult | 3.33 | 0.356 | 30.679 | 3.280 |
| B. adolescentis | 780B | Intestine of adult | 0.73 | 0.012 | 6.712 | 0.108 |
| B. adolescentis | 796B | Intestine of adult | 4.35 | 0.419 | 40.034 | 3.857 |
| B. adolescentis | 79B | Intestine of adult | 2.05 | 0.088 | 12.611 | 10.937 |
| B. adolescentis | 809B | Intestine of adult | 7.73 | 0.542 | 71.159 | 4.990 |
| B. adolescentis | 856B | Intestine of adult | 0.73 | 0.022 | 6.690 | 0.201 |
| B. adolescentis | 859B | Intestine of adult | 0.64 | 0.017 | 5.875 | 0.161 |
| B. adolescentis | 951B | Intestine of adult | 2.11 | 0.364 | 19.421 | 3.353 |
| B. adolescentis | 952B | Intestine of adult | 1.11 | 0.385 | 10.219 | 3.549 |
| B. adolescentis | 954B | Intestine of adult | 3.02 | 0.333 | 27.779 | 3.063 |
| B. adolescentis | 971B | Intestine of adult | 1.82 | 0.095 | 16.745 | 0.871 |
| B. adolescentis | AD2-8 | Human faeces | 5.71 | 1.839 | 52.601 | 16.936 |
| B. adolescentis | AL12-4 | Human faeces | 0.64 | 0.072 | 5.889 | 0.659 |
| B. adolescentis | HD17T1d | Human faeces | 5.75 | 0.902 | 52.918 | 8.301 |
| B. adolescentis | HD17T1h | Human faeces | 0.87 | 0.036 | 8.024 | 0.332 |
| B. adolescentis | HD17T2h | Human faeces | 9.43 | 1.492 | 86.802 | 13.741 |
| B. adolescentis | HD17T3h | Human faeces | 0.97 | 0.027 | 8.959 | 0.247 |
| B. adolescentis | HD17T9h | Human faeces | 6.54 | 0.506 | 60.201 | 4.655 |
| B. adolescentis | HD19T1h | Human faeces | 4.29 | 0.692 | 39.526 | 6.367 |
| B. adolescentis | HD19T2d | Human faeces | 8.47 | 1.033 | 77.998 | 9.507 |
| B. adolescentis | HD19T3h | Human faeces | 2.85 | 0.209 | 26.263 | 1.921 |
| B. adolescentis | HD23T1h | Human faeces | 8.01 | 1.371 | 73.779 | 12.621 |
| B. adolescentis | HD23T3d | Human faeces | 3.42 | 0.819 | 31.461 | 7.539 |
| B. adolescentis | HD23T4d | Human faeces | 3.87 | 0.202 | 35.595 | 1.862 |
| B. adolescentis | HD23T4h | Human faeces | 5.03 | 0.140 | 46.349 | 1.289 |
| B. adolescentis | HD23T6h | Human faeces | 6.18 | 1.348 | 67.461 | 3.516 |
| B. adolescentis | HD23T8h | Human faeces | 5.25 | 0.290 | 48.347 | 2.669 |
| B. adolescentis | HD24T1h | Human faeces | 3.81 | 0.168 | 35.061 | 1.549 |
| B. adolescentis | HD24T5h | Human faeces | 9.32 | 0.367 | 85.788 | 3.379 |
| B. adolescentis | HD24T7h | Human faeces | 8.44 | 0.233 | 77.694 | 2.142 |
| B. adolescentis | HD28T1d | Human faeces | 7.45 | 1.133 | 68.605 | 10.431 |
| B. adolescentis | HD28T2d | Human faeces | 0.81 | 0.077 | 7.481 | 0.710 |
| B. adolescentis | HD28T7h | Human faeces | 0.66 | 0.079 | 6.049 | 0.729 |
| B. adolescentis | HD35T1h | Human faeces | 5.24 | 0.156 | 48.250 | 1.439 |
| B. adolescentis | HD35T1h | Human faeces | 7.96 | 1.541 | 85.557 | 0.072 |
| B. adolescentis | HD35T2d | Human faeces | 5.66 | 0.677 | 52.066 | 6.237 |
| B. adolescentis | HD35T4d | Human faeces | 5.82 | 0.708 | 53.553 | 6.517 |
| B. adolescentis | HD35T5h | Human faeces | 6.49 | 1.448 | 59.745 | 13.331 |
| B. adolescentis | HD36T1h | Human faeces | 0.87 | 0.066 | 8.052 | 0.605 |
| B. adolescentis | HD36T2d | Human faeces | 1.14 | 0.001 | 10.528 | 0.007 |
| B. adolescentis | HD36T4h | Human faeces | 0.94 | 0.082 | 8.609 | 0.755 |
| B. adolescentis | HD36T6h | Human faeces | 1.02 | 0.059 | 9.391 | 0.547 |
| B. adolescentis | HD36T8h | Human faeces | 0.91 | 0.008 | 8.369 | 0.078 |
| B. adolescentis | HD4T2h | Human faeces | 8.73 | 0.953 | 80.332 | 8.774 |
| B. adolescentis | LMG10502 | Culture collection, adult intestine | 0.66 | 0.044 | 6.031 | 0.401 |
| B. adolescentis | LMG10733 | Culture collection, adult intestine | 0.66 | 0.018 | 4.034 | 3.495 |
| B. adolescentis | LMG10734 | Culture collection, adult intestine | 2.82 | 0.864 | 25.942 | 7.959 |
| B. adolescentis | LMG11579 | Culture collection, bovine rumen | 1.35 | 0.275 | 12.403 | 2.529 |
| B. adolescentis | LMG18897 | Culture collection, human feces | 5.94 | 0.171 | 54.670 | 1.574 |
| B. moukalabense | DSM27231 | Faeces of a wild lowland gorilla (Gorilla gorilla) | 7.41 | 0.272 | 70.058 | 3.174 |
| B. stercoris | JCM15918 | Culture collection, human faeces | 1.62 | 0.100 | 14.9381 | 0.9226 |
| B. angulatum | LMG11039 | Culture collection, human feaces | 2.78 | 0.297 | 25.5759 | 2.7344 |
| B. dentium | LMG11045 | Human dental caries | 5.57 | 0.056 | 51.327 | 0.517 |
| B. merycicum | LMG11341 | Culture collection, bovine rumen | 0.62 | 0.014 | 5.747 | 0.133 |
| B. ruminatium | LMG21811 | Culture collection, bovine rumen | 0.64 | 0.017 | 5.902 | 0.156 |
Figure 3.
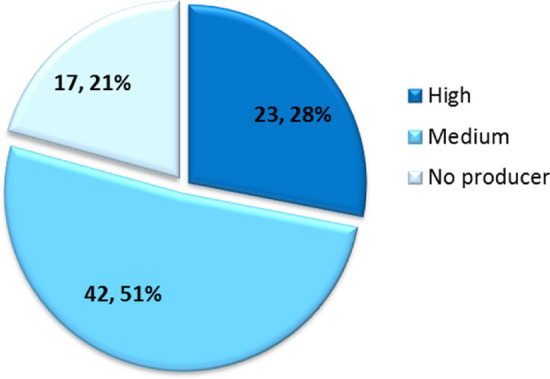
B. adolescentis distribution according to the production of GABA quantified by means of HPLC.
Genetic features of B. adolescentis PRL2019 and B. adolescentis HD17T2H
The genome sequence length of selected representative strains classified as high GABA producers, namely B. adolescentis PRL2019 and B. adolescentis HD17T2H, consist of 2,212,477 and 2,163,875 bp with an average G + C content of 59.17% and 59.23%, respectively, which are similar to those of other sequenced bifidobacterial genomes, being consistent with the range of G + C mol % values previously described for Actinobacteria26 (Table 2). Furthermore, the genome of PRL2019 and HD17T2H possess 54 and 55 tRNA genes, respectively, and both genomes encompass four rRNA gene operons. Identification of protein-coding sequences revealed 1,796 open reading frames (ORFs) in PRL2019 strain and 1,753 ORFs in HD17T2H strain. Chromosome sequences of both strains were scrutinized allowing identifying genes encoding glutamate decarboxylase (gadB) and glutamate/GABA antiporter (gadC). The resulting amino acid sequences were compared to those of GadB and GadC belonging to 47 B. adolescentis strains possessing the GAD/GABA antiporter locus (Table S3). Sequence alignments highlighted GadB as a conserved protein among the B. adolescentis species, with sequence identities ranging from 98.4% to 100% (Fig. 1b). Moreover, GadC was identified as an even more highly conserved protein, sharing an amino acid identity sequence ranging from 99.4 to 100% between the analyzed B. adolescentis predicted proteomes (Fig. 1b). Additionally, based on search for homologous genes, we also identified in both genomes of B. adolescentis PRL2019 and HD17T2H the gene pdxST involved in vitamin B6 metabolism in bifidobacteria. In particular, pyridoxal 5′-phosphate (PLP), the metabolically active form of vitamin B6, represents an important cofactor in the biosynthesis of several neurotransmitters, including GABA27,28.
Table 2.
General genetic features.
| B. adolescentis PRL2019 | B. adolescentis HD17T2H | |
|---|---|---|
| Biological origin | Human gut | Human feces |
| Average coverage | 279 | 91 |
| Number of assembled contigs | 1 | 12 |
| Genome length (pb) | 2,212,477 | 2,163,875 |
| Average GC percentage | 59.17 | 59.23 |
| Number of predicted ORFs | 1,796 | 1,753 |
| tRNA | 54 | 55 |
| rRNA | 4 | 4* |
| Accession number | PRJNA628852 | PRJNA628660 |
*Predicted number of rRNA loci.
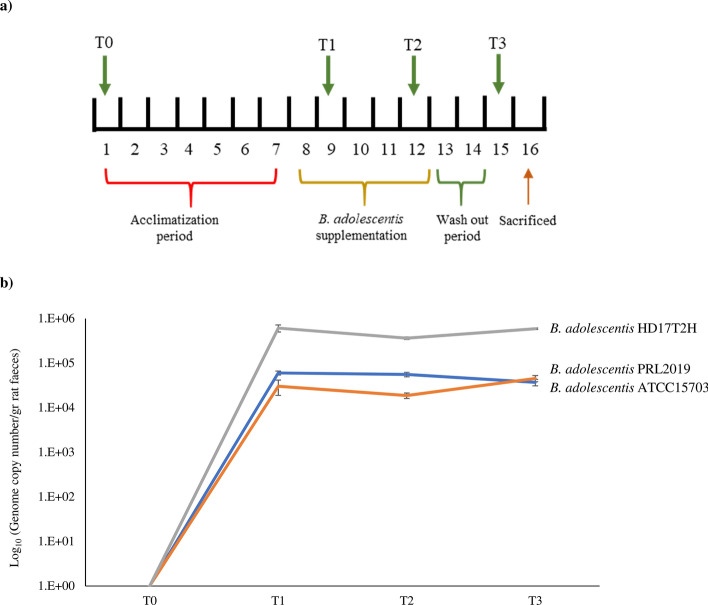
GABA production of B. adolescentis strains in a rat model
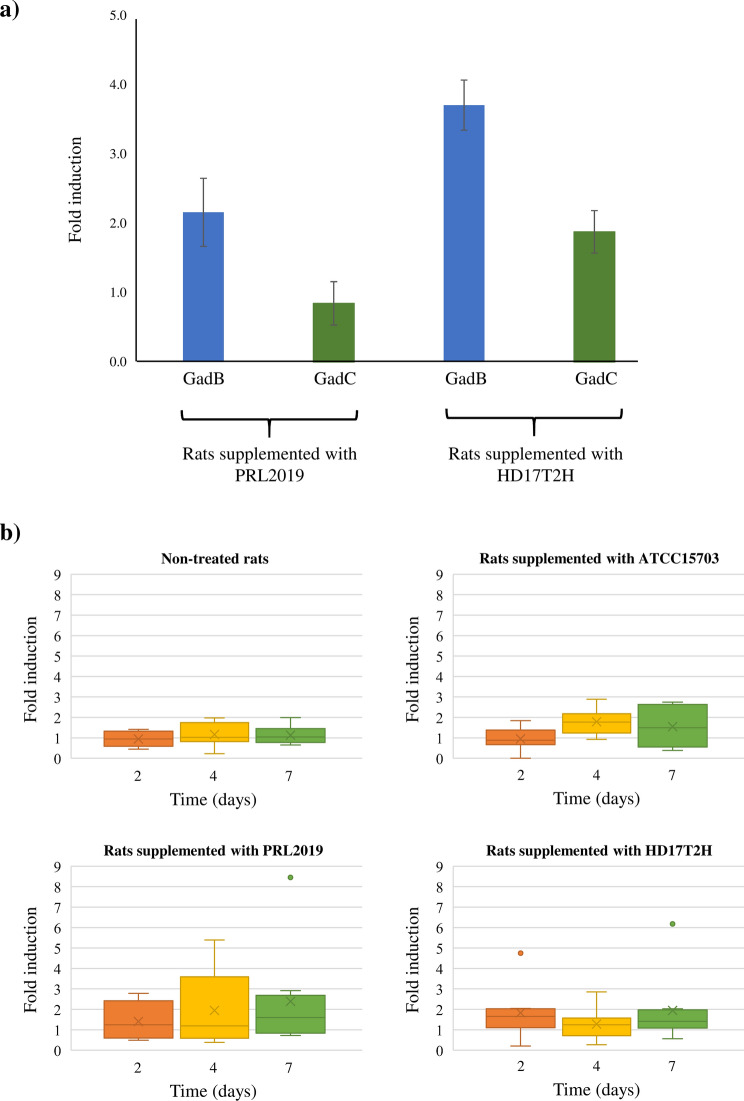
Three groups of rats (Rattus norvegicus) were supplemented for 5 days with a single daily dose of 109 colony forming unit (CFU) of B. adolescentis strains isolated from the human gut, i.e. B. adolescentis ATCC15703, B. adolescentis PRL2019 and B. adolescentis HD17T2H (Fig. 4a). Notably, as above described, the genome of B. adolescentis ATCC15703 lacks gadB and gadC genes (Table S2). Furthermore, a fourth group of rats, representing the control group, was supplemented with a sucrose solution without any bifidobacterial strains. Subsequently, the abundance of B. adolescentis ATCC15703, B. adolescentis PRL2019 and B. adolescentis HD17T2H was monitored during the experiment using a qPCR approach based on strain-specific primers. Interestingly, data collected from the qPCR analysis revealed an estimated abundance of all supplemented B. adolescentis strains ranging from 104 to 105 CFU/gr (Fig. 4b). These data highlighted a stable bifidobacterial abundance between samples collected from T1 to T3 that correspond with the bacterial supplementation (see materials and methods) (Fig. 4b). Furthermore, in order to investigate the expression level of genes involved in the GABA metabolism of PRL2019 and HD17T2H, we performed transcription profiling of gadB and gadC genes using a qRT-PCR approach on rats’ feces collected at T3. Interestingly, the transcription level of PRL2019 and HD17T2H gad genes revealed that gadB expression was significantly enhanced, ranging from 1.5- to sevenfold induction, while the expression of gadC ranged from 0.1 to fourfold induction (Fig. 5a). The enhanced expression of genes belonging to the GAD/GABA antiporter locus, revealed that B. adolescentis PRL2019 and B. adolescentis HD17T2H are able to stimulate the GABA production in rat model.
Figure 4.
Schematic representation of in vivo trials. Panel (a) displays the schedule of the experimental procedures. Panel (b) shows the average of DNA presence of the B. adolescentis strains in faecal samples observed during the bifidobacterial administration. Each point represents the average of the log-population size ± standard deviation for eight rats.
Figure 5.
GadB and gadC gene expressions and GABA levels in rat feces. Panel (a) highlight the expression of gadB and gadC genes under in vivo conditions. Data are expressed as means ± standard deviation. Each experiment was performed in triplicate. The y axis represents the level of expression as normalized expression (ΔΔCt) in respect to the housekeeping rpoB and atpB genes. Panel (b) shows the fold induction of GABA in faeces of rats non-treated and treated for 5 days with B. adolescentis ATCC15703, B. adolescentis PRL2019 or B. adolescentis HD17T2H in respect to the GABA basal level in the corresponding T0. Box-plot represents the median (bold line), interquartile range (box), mean (X) and minimum and maximum values.
In order to evaluate the GABA level in rats involved in these experiments, we performed an ELISA assay among fecal samples collected at different time points, i.e. T0, T1, T2 and T3. Interestingly, the concentration of GABA (μg/g) seemed to increase in rats treated with B. adolescentis PRL2019 and B. adolescentis HD17T2H, but no statistical differences were found with respect to rats treated with no-GABA producer strain B. adolescentis ATCC15703 and with respect to rats not supplemented by B. adolescentis strains (control group) (Fig. S1). The normalized concentration of GABA, normalized respect to the T0 data, revealed higher GABA levels in rats treated with GABA-producer B. adolescentis strains, but also in the non-producer ATCC15703 strain when compared with the control group (Fig. 5b). In particular, rats treated with B. adolescentis PRL2019 revealed a twofold increase of GABA level after 4 days of treatment, while rats treated with B. adolescentis HD17T2H highlighted an enhancement of 1.4-fold after the first 2 days of treatment. Despite the higher abundance of B. adolescentis HD17T2H (Fig. 4b) and the higher gad genes expression fold induction in respect to PRL2019 (Fig. 5a), the GABA concentration at T3 was lower (Fig. 5b), suggesting that the amount of in vivo produced GABA was not proportional between strains. Furthermore, the increased concentration of GABA even in rats fed with B. adolescentis ATCC15703 that does not harbor gad genes, could suggest that the administration of this species of Bifidobacterium could modulate the intestinal microbiota of rats favoring those endogenous populations able to synthesize this neurotransmitter.
Conclusions
In the current study, we performed a comprehensive in silico survey of 1,022 bifidobacterial genomes highlighting the genetic arsenal requested for the synthesis of GABA in seven different bifidobacterial species, i.e. B. adolescentis, B. angulatum, B. dentium, B. merycicum, B. moukalabense, B. ruminantium and B. samirii. Intriguingly, B. adolescentis strains showed the highest level of prevalence of gad genes in their genomes, suggesting this bifidobacterial taxon as a model GABA producer within the Bifidobacterium genus. Furthermore, metagenomics-based analyses involving datasets collected from children with subclinical symptoms of depression and anxiety revealed an intriguing association/correlation with reads belonging to B. adolescentis as well as B. adolescentis gad genes.
The in vitro screening of 82 B. adolescentis strains isolated from the human gut allowed to highlight those exhibiting the highest performances in the synthesis of GABA. Among B. adolescentis isolates, strains PRL2019 and HD17T2H were employed in an in vivo trial, highlighting an enhanced expression of GABA level in rats following the treatment with these bacteria. However, in vivo trials with animal models of anxiety/depression disorders will need to be performed in order to further support these findings and validate the role of B. adolescentis in the modulation of gut–brain axis signaling. Nonetheless, the achieved results contribute the expanding of the current knowledge about a possible role of B. adolescentis in the modulation of the gut microbiota-brain axis, since PRL2019 and HD17T2H strains represent intriguing GABA-producing gut microbes isolated from humans.
Materials and methods
Bifidobacterium adolescentis strains and growth conditions
All strains used in this study were cultivated in an anaerobic atmosphere (10% H2, 10% CO2 and 80% N2) in an anaerobic MG500 chamber (Don Whitley Scientific, West Yorkshire, United Kingdom) on De Man-Rogosa-Sharp (MRS) broth (BD-Difco Biosciences, San Diego, CA) supplemented with 0.25% (w/v) l-cysteine hydrochloride (Sigma-Aldrich) and incubated at 37 °C for variable times (Table 1).
Measurement of GABA production
To determine GABA production, strains were subcultured in MRS supplemented with 2 mM monosodium glutamate (GMS, Sigma-Aldrich) and grown for 48 h anaerobically at 37ºC. GABA production was evaluated by HPLC on cell-free supernatants following diethyl ethoxymethylenemalonate (DEEM, Sigma-Aldrich) derivatization according to the following indications29. After centrifugation (18,000 g for 10 min), supernatants were filtered through a syringe filters (13 mm diameter, 0.22 µm pore size, PTFE membrane, VWR International, Radnor, PA, USA). Aliquots of 100 µl were thoroughly mixed by vortexing with 175 µl of borate buffer (1 M boric acid, pH 9.0), 75 µl methanol, 3 µl DEEM and 2 µl of 2-l-amino adipic acid (stock solution at 2 mgml−1) (Sigma-Aldrich), as an internal standard. Mixtures were held in an ultrasound water bath at 30º C for three 15 min cycles. Then samples were maintained at 70ºC in a water bath for 2 h to remove DEEM excess. Finally, samples were centrifuged for 5 min at 11,000 g and supernatants were further filtered through 0.22 µm membranes.
GABA was determined by reverse-phase (RP)-HPLC in the Ascentis C18 (250 × 4.6 mm, 5 μm) column coupled with a pre-column Supelguard Ascentis C18 (20 × 4.0,0 mm) (Supelco, Sigma-Aldrich, St. Louis, MO), using a chromatographic system composed of the Alliance 2,695 separation module, the UV–visible PDA 2,996 detector and the acquisition/analysis software Empower (Waters, Milford, MA, USA). Separation was carried out at 35ºC with a gradient of the mobile phase: 25 mM acetate buffer pH 6.7 plus 0.02% sodium azide (eluent A), acetonitrile (eluent B) and methanol (eluent C)30. Samples (5 l) were injected, separated at 1 ml min−1 flow rate (total rum 100 min) and the GABA was detected at 280 nm. Quantification was performed using external calibration pattern using known concentrations of GABA standard (Sigma), submitted to the same derivatization procedure, to obtain the corresponding linear regression equation (R2 > 0.99). All determinations were performed, at least, in two independent biological replicates.
Genome sequencing and assemblies
Based on the results achieved from the production of GABA between 82 B. adolescentis strains, two representative strains classified as high GABA producers namely B. adolescentis PRL2019 and B. adolescentis HD17T2H, were submitted to shotgun genome sequencing. DNA extracted from B. adolescentis PRL2019 and B. adolescentis HD17T2H cultures was subjected to whole-genome sequencing using MiSeq (Illumina, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Illumina, UK). Moreover, in order to improve the genome quality of B. adolescentis PRL2019, its DNA was extracted and submitted to whole-genome sequencing using a MinION approach (Oxford Nanopore, UK) at GenProbio srl (Parma, Italy) according to the supplier’s protocol (Oxford Nanopore, UK). Fastq files of the paired-end reads obtained from targeted genome sequencing of isolated strains were utilized as input for genome assemblies through the MEGAnnotator pipeline31. SPAdes software was used for de novo assembly of each Bifidobacterium adolescentis genome sequence32,33, while open reading frames (ORFs) were predicted using Prodigal34. The coverage depth of these newly isolated B. adolescentis chromosomes ranged from 91- to 279-fold, which upon assembly generated 12 contigs and a complete chromosome sequence, respectively.
GAD/GABA antiporter locus identification
We retrieved the proteome of 1,022 Bifidobacterium strains from the National Center for Biotechnology Information (NCBI) public database (Table S1). Accordingly, we assessed which bifidobacterial species encode the genes required for GABA production by means of local alignment search against the NCBI bifidobacterial reference glutamate decarboxylase (GadB) and glutamate/GABA antiporter (GadC) amino acid sequences (Accession: ADB10338.1 and VEG24324.1). Putative GadB and GadC proteins of the 1,022 Bifidobacterium strains were identified by means of BLASTP (cutoff E value, 1 × 10−30 and 50% identity over at least 80% of both protein sequences).
Shotgun metagenomic screening of B. adolescentis and gad gene sequences
In order to investigate the presence of B. adolescentis and to explore the occurrence of gad genes into the microbiota of individuals exhibiting depression and anxiety behaviors, we analyzed two public metagenomic datasets related to these illnesses (PRJNA496479 and PRJNA474710). In this context, we collected the metagenomic data of a cohort of early school-aged children with a combination of subclinical mental health symptoms of depression and anxiety (PRJNA496479) and those of a well-characterized model of stress vulnerable Sprague Dawley rats showing depressive- and anxiety-like behaviors due to social defeat (PRJNA474710). Each data set was filtered to obtain only high quality reads (minimum mean quality score 20; window size 5; quality threshold 25; minimum length 80) using the fastq-mcf script (https://expressionanalysis.github.io/ea-utils/). The resulting reads were aligned against the Homo sapiens and Rattus norvegicus genomes using the Burrows-Wheeler Aligner program35 (BWA-MEM algorithm with trigger reseeding, 1.5; minimum seed length, 19; matching score, 1; mismatch penalty, 4; gap open penalty, 6; and gap extension penalty, 1) and further processed with the SAMtools software package36 in order to remove human and rats reads. Finally, the filtered reads were used to identify B. adolescentis-associated reads within the data set for each sample by means of Bowtie237 through multiple-hit mapping and a “very sensitive” policy. The mapping was performed using a minimum score threshold function (–score-min C, -13,0) in order to limit reads of arbitrary length to two mismatches and retain those matches with at least 98% full-length identity. The software employed to calculate read counts corresponding to bifidobacterial genes was HTSeq38, running in union mode.
Experimental design of the in vivo trials
Experiments involved 5-month-old male wild-type Groningen rats (R. norvegicus). This rat strain, originally derived from the University of Groningen (The Netherlands), was bred in the animal facility of the University of Parma under standard conditions. From the initiation of the experiments, rats were housed individually in polymethyl methacrylate (Plexiglas) cages (39 cm × 23 cm × 15 cm). Rats were kept in rooms with controlled temperature (22 ± 2 °C) and humidity (60 ± 10%) and maintained in a 12/12 light/dark cycle (light on from 19:00 to 7:00 h), with food and water ad libitum. The first week represented an acclimatization period, during which rats continued to consume a standard chow diet supplemented with an oral administration of 500 µl of sucrose solution (2%) in order to adapt to drink from a syringe. For the following 5 days, rats (n = 32) were randomized to 4 groups and orally supplemented using a syringe with: (1) B. adolescentis ATCC15703; (2) B. adolescentis PRL2019; (3) B. adolescentis HD17T2H; (4) sucrose solution only (i.e., negative control) (Table S2). The treatment with B. adolescentis strains was daily administered at 109 CFU per rat by syringe. Before the treatment, microbial cultures were cultivated as previously described, and fecal samples of rats were analyzed to ensure the absence of B. adolescentis strains by means of specific primers. Subsequently, bacterial cultures were harvested by centrifugation (3,000 rpm for 8 min), washed and resuspended in 500 µL of 2% (w/v) sucrose solution. The viable count of each inoculum was determined by retrospective plating on MRS. In order to evaluate bifidobacterial colonization fecal samples were collected at four different time points. The first sample collection was performed before the oral administration of bifidobacteria (T0), in order to access the baseline concentration of GABA in each rat. Then, we collected fecal samples at 2, 4 and 7 days (T1, T2 and T3) to cover with multiple sampling the days the oral bifidobacterial supplementation (Fig. 4a). Faeces were stored at − 80 °C until use.
DNA extraction and qPCR
Bacterial DNA extraction from rat’s fecal samples was performed following the manufacturer’s protocol of the QIAamp Fast DNA stool Mini Kit (Qiagen Ltd, Strasse, Germany). Bifidobacterial DNA presence was evaluated in rat’s fecal samples. Quantitative PCR (qPCR) was performed as described previously39. Strain specific primers were designed for the identification of different B. adolescentis strains in fecal samples. Primers Bado_PRL2019_fw (5′-GAGCAGGCAAGGACACTTTC-3′) and Bado_PRL2019_rev (5′-CTGAAGAGGCAAGCTTGAGG-3′) were used for B. adolescentis PRL2019; primers Bado_HD17T2M_fw (5′-CGGCTACAGGTTCGCTTATC-3′) and Bado_ HD17T2H_rev (5′-TTCCGCAGTAATTCGAGCTT-3′) were used for B. adolescentis HD17T2H; and Bado_ATCC15703_fw (5′-GGTGATTACGCAGCATCCTT-3′) and Bado_ATCC15703_rev (5′-CTTCCTCACAAACGTCAGCA-3′) were used for B. adolescentis ATCC15703. PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle of 95 °C for 2 min, followed by 42 cycles of 95 °C for 15 s and Tm 62 °C for B. adolescentis PRL2019, 64 °C B. adolescentis HD17T2H and 60 °C B. adolescentis ATCC15703, for 30 s. The melting curve was 65 °C to 95 °C with increments of 0.5 °C/s. In each run, a negative control (no DNA) for each primer set was included.
RNA extraction and qRT-PCR
In order to evaluate the expression of genes involved in GABA production, we have extracted the total RNA from faecal samples of rats. 0.4 g of stool sample were mixed to 1 mL of QIAzoL Lysis Reagent (Qiagen, UK) and were transferred in a sterile tube containing glass beads (Merck, Germany). The cells were lysed using Precellys 24 homogenizer (Bertin instruments, France). The protocol provides 2 min of stirring the mix alternating with 2 min of static cooling; this step was repeated three times. The cells were centrifuged at 12,000 rpm for 15 min and the upper phase was recovered. The RNA samples were purified using the RNAesy Mini Kit (Qiagen, UK) following the manufacturer’s protocol. RNA concentration and purity were evaluated by a Picodrop microliter spectrophotometer (Picodrop, UK). cDNA was synthesized and purified using the iScript cDNAsynthesis kit (Bio-Rad, CA, USA) according to the supplier’s instructions. Primers used for the normalization of the data were designed on housekeeping genes, i.e. rpoB and atpB, as described previously40, while for gadB gene were used primers GadB_fw (5′-CACATGCTCGCCGATCTATG-3′) and GadB_rev (5′-TCGACCGGCTCATACATACC-3′), whereas for gadC gene were used primers GadC_fw (5′-GTCTCGCTTCCATTCTGCTG-3′) and GadC_rev (5′-CGAACACATACGACAGGCTG-3′). qRT-PCR was performed using the CFX96 system (Bio-Rad, CA, USA). PCR products were detected with SYBR green fluorescent dye and amplified according to the following protocol: one cycle of 95 °C for 2 min, followed by 42 cycles of 95 °C for 15 s and 60 °C for 30 s. The melting curve was 65 °C to 95 °C with increments of 0.5 °C/s. In each run, a negative control (no cDNA) for each primer set was included. The expression ratio of the selected genes was calculated and analyzed using CFX Manager Expression software (Bio-Rad, CA, USA).
GABA measurement in rat faeces
Faeces of each rat at different time points were diluted 1:10 (w/v) in milli-Q water in order to obtain faecal waters. Each sample was mixing until disaggregation of faeces and centrifuging at 5,000 rpm for 5 min and keeping the supernatant fraction. This aqueous fraction was used for quantification of GABA using the GABA ELISA kit (LDN Diagnostics, Germany) following manufacturer instructions. Dilution factor was taken into account for GABA calculation.
Statistical analyses
SPSS software v. 25 (IBM, Italy) was used to perform statistical analysis between shogun metagenomic data of anxious and depressed children, and healthy subjects (BioProjects PRJNA496479) by Student’s t test. The sample size between groups was evaluated by means of Statulator (https://statulator.com/SampleSize/ss2M.html).
Ethical statement
All experimental procedures and protocols involving animals were approved by the Italian Ministry of Health and the Veterinarian Animal Care and Use Committee of Parma University (Authorization Number 370/2018) and conducted in accordance with the European Community Council Directives dated 22 September 2010 (2010/63/UE).
Supplementary information
Acknowledgements
This work was funded by the EU Joint Programming Initiative—A Healthy Diet for a Healthy Life (JPI HDHL, https://www.healthydietforhealthylife.eu/) to MV (in conjunction with MIUR, Italy). IPLA was funded by the projects RTI2018-096339-B-I00 (MCIU/AEI/FEDER, UE), RTI2018-095021-J-I00 (MCIU/AEI/FEDER, UE) and IDI_2018_000236 (PCTI Govierno Principado de Asturias / FEDER); H. Tamés acknowledges the “Severo Ochoa” Research grant from the Government of “Principado de Asturias”. We thank GenProbio srl for financial support of the Laboratory of Probiogenomics. Part of this research is conducted using the High Performance Computing (HPC) facility of the University of Parma. Thanks are given to Isabel Cuesta, from the Technical Services of IPLA-CSIC, and Diana Luaces for their excellent technical assistance.
Author contributions
S.D, L.R. and G.A.L designed the experiments and wrote the manuscript. S.D., G.A.L., W.M., L.R. and H.T. performed the experiments. G.L. performed libraries preparation and illumina sequencing. G.A.L., C.M. and L.M. performed the bioinformatics analyses. L.C. and W.M. performed the in vivo experiments. M.V., A.M and A.S. participated in the design and supervised the study. F.T. and P.R.M. conceived the study, participated in its design and coordination, and contributed to the manuscript preparation. All authors reviewed the manuscript. All authors read and approved the final manuscript.
Data availability
Newly isolated B. adolescentis genomes were sequenced and deposited at DDBJ/ENA/GenBank under the accession numbers reported in Table 2 (BioProject No. PRJNA628660 and PRJNA628852).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Sabrina Duranti, Lorena Ruiz and Gabriele Andrea Lugli.
Contributor Information
Patricia Ruas-Madiedo, Email: ruas-madiedo@ipla.csic.es.
Francesca Turroni, Email: francesca.turroni@unipr.it.
Supplementary information
is available for this paper at 10.1038/s41598-020-70986-z.
References
- 1.Dhakal R, Bajpai VK, Baek KH. Production of gaba (gamma—aminobutyric acid) by microorganisms: a review. Braz. J. Microbiol. 2012;43:1230–1241. doi: 10.1590/S1517-83822012000400001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sarasa SB, et al. A brief review on the non-protein amino acid, gamma-amino butyric acid (GABA): its production and role in microbes. Curr. Microbiol. 2020;77:534–544. doi: 10.1007/s00284-019-01839-w. [DOI] [PubMed] [Google Scholar]
- 3.Cryan JF, Kaupmann K. Don't worry 'B' happy!: a role for GABA(B) receptors in anxiety and depression. Trends Pharmacol. Sci. 2005;26:36–43. doi: 10.1016/j.tips.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 4.Schousboe A, Bak LK, Sickmann HM, Sonnewald U, Waagepetersen HS. Energy substrates to support glutamatergic and GABAergic synaptic function: role of glycogen, glucose and lactate. Neurotoxicol. Res. 2007;12:263–268. doi: 10.1007/bf03033909. [DOI] [PubMed] [Google Scholar]
- 5.Barrett E, Ross RP, O'Toole PW, Fitzgerald GF, Stanton C. gamma-aminobutyric acid production by culturable bacteria from the human intestine. J. Appl. Microbiol. 2012;113:411–417. doi: 10.1111/j.1365-2672.2012.05344.x. [DOI] [PubMed] [Google Scholar]
- 6.Mody I, De Koninck Y, Otis TS, Soltesz I. Bridging the cleft at GABA synapses in the brain. Trends Neurosci. 1994;17:517–525. doi: 10.1016/0166-2236(94)90155-4. [DOI] [PubMed] [Google Scholar]
- 7.Kim HY, Yokozawa T, Nakagawa T, Sasaki S. Protective effect of gamma-aminobutyric acid against glycerol-induced acute renal failure in rats. Food Chem. Toxicol. 2004;42:2009–2014. doi: 10.1016/j.fct.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 8.Kali A. Psychobiotics: an emerging probiotic in psychiatric practice. Biomed. J. 2016;39:223–224. doi: 10.1016/j.bj.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siragusa S, et al. Synthesis of gamma-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl. Environ. Microbiol. 2007;73:7283–7290. doi: 10.1128/AEM.01064-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ueno Y, Hayakawa K, Takahashi S, Oda K. Purification and characterization of glutamate decarboxylase from Lactobacillus brevis IFO 12005. Biosci. Biotechnol. Biochem. 1997;61:1168–1171. doi: 10.1271/bbb.61.1168. [DOI] [PubMed] [Google Scholar]
- 11.Nomura M, Kimoto H, Someya Y, Furukawa S, Suzuki I. Production of gamma-aminobutyric acid by cheese starters during cheese ripening. J. Dairy Sci. 1998;81:1486–1491. doi: 10.3168/jds.S0022-0302(98)75714-5. [DOI] [PubMed] [Google Scholar]
- 12.Nomura M, Kimoto H, Someya Y, Suzuki I. Novel characteristic for distinguishing Lactococcus lactis subsp. lactis from subsp. cremoris. Int. J. Syst. Bacteriol. 1999;49(Pt 1):163–166. doi: 10.1099/00207713-49-1-163. [DOI] [PubMed] [Google Scholar]
- 13.Yunes RA, et al. GABA production and structure of gadB/gadC genes in Lactobacillus and Bifidobacterium strains from human microbiota. Anaerobe. 2016;42:197–204. doi: 10.1016/j.anaerobe.2016.10.011. [DOI] [PubMed] [Google Scholar]
- 14.Lugli GA, et al. Tracking the taxonomy of the genus bifidobacterium based on a phylogenomic approach. Appl. Environ. Microbiol. 2018 doi: 10.1128/AEM.02249-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lugli GA, et al. Isolation of novel gut bifidobacteria using a combination of metagenomic and cultivation approaches. Genome Biol. 2019;20:96. doi: 10.1186/s13059-019-1711-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Duranti S, et al. Characterization of the phylogenetic diversity of two novel species belonging to the genus Bifidobacterium: Bifidobacterium cebidarum sp. nov. and Bifidobacterium leontopitheci sp. nov. Int. J. Syst. Evol. Microbiol. 2020 doi: 10.1099/ijsem.0.004032. [DOI] [PubMed] [Google Scholar]
- 17.Biavati B, Mattarelli P. Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int. J. Syst. Bacteriol. 1991;41:163–168. doi: 10.1099/00207713-41-1-163. [DOI] [PubMed] [Google Scholar]
- 18.Lugli GA, et al. The genome sequence of bifidobacterium moukalabense DSM 27321 highlights the close phylogenetic relatedness with the Bifidobacterium dentium Taxon. Genome Announc. 2014 doi: 10.1128/genomeA.00048-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duranti S, et al. Characterization of the phylogenetic diversity of five novel species belonging to the genus Bifidobacterium: Bifidobacterium castoris sp. nov., Bifidobacterium callimiconis sp. nov., Bifidobacterium goeldii sp. nov., Bifidobacterium samirii sp. nov. and Bifidobacterium dolichotidis sp. nov. Int. J. Syst. Evol. Microbiol. 2019;69:1288–1298. doi: 10.1099/ijsem.0.003306. [DOI] [PubMed] [Google Scholar]
- 20.Duranti S, et al. Evaluation of genetic diversity among strains of the human gut commensal Bifidobacterium adolescentis. Sci. Rep. 2016;6:23971. doi: 10.1038/srep23971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jang HM, Jang SE, Han MJ, Kim DH. Anxiolytic-like effect of Bifidobacterium adolescentis IM38 in mice with or without immobilisation stress. Benef. Microbes. 2018;9:123–132. doi: 10.3920/BM2016.0226. [DOI] [PubMed] [Google Scholar]
- 22.Yunes RA, et al. A multi-strain potential probiotic formulation of GABA-producing Lactobacillus plantarum 90sk and bifidobacterium adolescentis 150 with antidepressant effects. Probiot. Antimicrob. Proteins. 2019 doi: 10.1007/s12602-019-09601-1. [DOI] [PubMed] [Google Scholar]
- 23.Milani C, et al. Unveiling bifidobacterial biogeography across the mammalian branch of the tree of life. ISME J. 2017;11:2834–2847. doi: 10.1038/ismej.2017.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kalueff AV, Nutt DJ. Role of GABA in anxiety and depression. Depress. Anxiety. 2007;24:495–517. doi: 10.1002/da.20262. [DOI] [PubMed] [Google Scholar]
- 25.Lydiard RB. The role of GABA in anxiety disorders. J. Clin. Psychiatry. 2003;64(Suppl 3):21–27. [PubMed] [Google Scholar]
- 26.Milani C, et al. The first microbial colonizers of the human gut: composition, activities, and health implications of the infant gut microbiota. Microbiol. Mol. Biol. Rev. 2017 doi: 10.1128/MMBR.00036-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El Qaidi S, Yang J, Zhang JR, Metzger DW, Bai G. The vitamin B(6) biosynthesis pathway in Streptococcus pneumoniae is controlled by pyridoxal 5'-phosphate and the transcription factor PdxR and has an impact on ear infection. J. Bacteriol. 2013;195:2187–2196. doi: 10.1128/JB.00041-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parra M, Stahl S, Hellmann H. Vitamin B(6) and its role in cell metabolism and physiology. Cells. 2018 doi: 10.3390/cells7070084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gomez-Alonso S, Hermosin-Gutierrez I, Garcia-Romero E. Simultaneous HPLC analysis of biogenic amines, amino acids, and ammonium ion as aminoenone derivatives in wine and beer samples. J. Agric. Food Chem. 2007;55:608–613. doi: 10.1021/jf062820m. [DOI] [PubMed] [Google Scholar]
- 30.Redruello B, et al. A fast, reliable, ultra high performance liquid chromatography method for the simultaneous determination of amino acids, biogenic amines and ammonium ions in cheese, using diethyl ethoxymethylenemalonate as a derivatising agent. Food Chem. 2013;139:1029–1035. doi: 10.1016/j.foodchem.2013.01.071. [DOI] [PubMed] [Google Scholar]
- 31.Lugli GA, Milani C, Mancabelli L, van Sinderen D, Ventura M. MEGAnnotator: a user-friendly pipeline for microbial genomes assembly and annotation. FEMS Microbiol. Lett. 2016 doi: 10.1093/femsle/fnw049. [DOI] [PubMed] [Google Scholar]
- 32.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nurk S, et al. Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. J. Comput. Biol. 2013;20:714–737. doi: 10.1089/cmb.2013.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hyatt D, et al. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Anders S, Pyl PT, Huber W. HTSeq–a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Turroni F, et al. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front. Microbiol. 2015;6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milani C, et al. The Sortase-dependent fimbriome of the genus bifidobacterium: extracellular structures with potential to modulate microbe-host dialogue. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.01295-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Newly isolated B. adolescentis genomes were sequenced and deposited at DDBJ/ENA/GenBank under the accession numbers reported in Table 2 (BioProject No. PRJNA628660 and PRJNA628852).