Abstract
Cell-free protein synthesis (CFPS) systems enable the production of protein without the use of living, intact cells. An emerging area of interest is to use CFPS systems to characterize individual elements for genetic programs [e.g. promoters, ribosome binding sites (RBS)]. To enable this research area, robust CFPS systems must be developed from new chassis organisms. One such chassis is the Gram-negative Pseudomonas bacteria, which have been studied extensively for their diverse metabolism with promises in the field of bioremediation and biosynthesis. Here, we report the development and optimization of a high-yielding (198 ± 5.9 µg/ml) batch CFPS system from Pseudomonas putida ATCC 12633. Importantly, both circular and linear DNA templates can be applied directly to the CFPS reaction to program protein synthesis. Therefore, it is possible to prepare hundreds or even thousands of DNA templates without time-consuming cloning work. This opens the possibility to rapidly assess and validate genetic part performance in vitro before performing experiments in cells. To validate the P. putida CFPS system as a platform for prototyping genetic parts, we designed and constructed a library consisting of 15 different RBSs upstream of the reporter protein sfGFP, which covered an order of magnitude range in expression. Looking forward, our P. putida CFPS platform will not only expand the protein synthesis toolkit for synthetic biology but also serve as a platform in expediting the screening and prototyping of gene regulatory elements.
Keywords: synthetic biology, Pseudomonas putida, cell-free protein synthesis, prototyping, gene regulatory elements, TX-TL
Introduction
Initially used to elucidate the genetic code, crude extract based cell-free protein synthesis (CFPS) systems have emerged as a research tool to accelerate both fundamental and applied biology, with a range of applications in high-throughput protein expression, protein evolution and synthetic biology (1–19). CFPS systems complement in vivo production approaches, while offering some advantages. For example, the open nature of the reaction allows the user to directly influence the biochemical systems of interest. As a result, new components can be added or synthesized, and these can be maintained at precise concentrations. In addition, the reduced cell viability constraints offered by cell-free systems enable the production of complex proteins at titers that might otherwise be toxic in living cells. Furthermore, processes that take days or weeks to design, prepare and execute in vivo can be done more rapidly in a cell-free system, leading to high-throughput production campaigns on a whole-proteome scale (20) with the ability to automate (21). Finally, CFPS reactions can be freeze-dried for storage at ambient temperature for up to 1 year (18, 19, 22).
The aforementioned advantages have led to a wave of new interest in the development and use of CFPS systems. However, the vast majority of previous efforts have been focused on a select few model systems: Escherichia coli, Saccharomyces cerevisiae and Chinese Hamster Ovary cells (23–28). Unfortunately, these well-developed systems may fall short at times (29), especially in efforts to mimic the native host for prototyping applications. In this context, several new CFPS systems have been developed, including some from Streptomyces species and Bacillus subtilis (30–33).
Pseudomonas putida is a Gram-negative bacterium that has emerged as a reliable and robust chassis microorganism for laboratory research and industrial production (34). For example, P. putida as a microbial cell factory has been applied to produce recombinant antibody fragments, biofuels and natural products (35–39). As a model organism, P. putida is well understood at the biochemical level, its genome has been completely sequenced, and versatile genetic tools are readily available for engineering the host strain (34, 40). In addition, P. putida can be easily cultivated and grown quickly under precise conditions in either shake flasks or bioreactors (41, 42). Despite these features, there has yet to be, to our knowledge, a high-yielding CFPS developed from P. putida that could permit design-build-test-learn iterations without the need to re-engineer organisms. Previous efforts have explored the possibilities of using other Pseudomonas organisms for CFPS, such as P. fluorescens. However, its complicated cell extract preparation method (periplasmic fraction removal before cell lysis) and relatively low protein yield <100 µg/ml leaves room for improvement (43).
In this study, we aim to establish a robust and high-yielding P. putida-based CFPS system. After establishing the baseline ability to synthesize super-folder green fluorescent protein (sfGFP) as a reporter, we set out to increase protein synthesis yields to greater than 100 µg/ml by systematically optimizing process parameters. Specifically, we assessed the impact of temperature, lysate content, plasmid concentration and magnesium ion concentration on protein synthesis yields. We observed a ∼10-fold increase in sfGFP yields relative to the unoptimized case, resulting in a final titer of approximately 200 µg/ml in 4-h batch reaction. We went on to demonstrate that CFPS reactions could be programed with linear DNA templates. By use of overlap extension polymerase chain reaction (PCR), we show that transcription and translation elements (i.e. T7 promoter) can be rapidly added to the coding region of gene, which efficiently directs a combined transcription and translation process in vitro. Finally, we applied our P. putida-based CFPS system to the expression of a library of genetic parts, specifically ribosome binding sites (RBSs), which covered a 10-fold range of expression values. Looking forward, we anticipate that our P. putida-based CFPS system will provide broad utility for functional annotation, metabolic engineering and synthetic biology.
Materials and methods
Bacterial strains, culture medium and plasmids
The P. putida strain, ATCC 12633, used in this work was a generous gift from Prof. Keith Tyo (Northwestern University, Evanston, IL, USA), and it was grown in LB medium (10 g/l tryptone, 5 g/l yeast extract and 5 g/l sodium chloride). The reporter protein construct used is pJL1-sfGFP, and the sequence of the pJL1 vector can be obtained from Addgene #69496.
To construct the RBS library, the pJL1-sfGFP vector was first linearized by PCR amplification with the pJL1-NdeI-FP and pJL1-XbaI-RP primer pair (Supplementary Table S1), and then ligated with oligonucleotide pairs to replace the original RBS. The RBS library is shown in Supplementary Table S2. DNA plasmids used in CFPS were obtained from cultures of E. coli DH5α strain (Invitrogen, Thermo Fisher Scientific) using Qiagen Plasmid Maxi Prep kits. All plasmids used in this experiment were sequence verified.
PCR templates
All PCR templates were amplified with Phusion high fidelity DNA polymerase (New England BioLabs). Primers used for amplification were purchased from Integrated DNA Technologies with no modifications, or with 5′biotinylated and phosphorothioated modifications (Supplementary Table S3). The amplified PCR products were purified with QIAquick PCR Purification Kit (Qiagen) or QIAquick Gel Extraction Kit (Qiagen).
Cell extract preparation
Pseudomonas putida ATCC 12633 was first grown from a glycerol stock in a standard glass culture tube with 5 ml of LB medium at 26°C, and the cells were allowed to recover overnight. Next, 1 ml of the overnight culture was used to inoculate 50 ml fresh LB medium in a 250 ml baffled culture flask overnight. Then, 20 ml of the overnight culture was used to inoculate 1 l fresh LB medium in a 2.5 l full baffled Tunair flask (IBI Scientific, Peosta, IA, USA), and cells were allowed to grow to an OD600 of ∼2.5 before harvesting. The cells were harvested at 5000 g and 4°C for 15 min. Wet cell pellets were washed three times with S30 extract buffer (10 mM tris-acetate at pH 8.2, 14 mM magnesium acetate, 60 mM potassium acetate and 1 mM of dithiothreitol). After the final wash, the wet cell pellet was resuspended with 1 ml of the same S30 extract buffer for each gram of the wet cell pellet. After a complete resuspension of the cell pellet, the cell mixture was set in an ice-water bath to cool. Cell lysis was carried out with a Q125 Sonicator (Qsonica, Newtown, CT, USA) at 50% amplitude with a 45-s on and 59-s off lysis cycle for a total of 1500 J (44). This procedure follows a protocol previously optimized for E. coli, but that seems to work equally well here. The homogeneous cell lysate was clarified for two rounds at 16 000 g and 4°C for 30 min each. Then, the supernatant was collected as the final cell extract and flash frozen in liquid nitrogen immediately. The final cell extract was stored at −80°C until further use.
Cell-free protein synthesis
A standard CFPS reaction was carried out in a 200 μl PCR tube in a thermocycler with the following components to a final volume of 15 μl: 16 mM magnesium acetate, 80 mM ammonium acetate, 230 mM potassium glutamate, 57.2 mM HEPES-KOH (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid-potassium hydroxide) pH 7.5, 1.2 mM ATP (adenosine triphosphate), 0.86 mM GTP/UTP/CTP (guanosine triphosphate/uridine triphosphate/cytidine triphosphate), 34.0 μg/ml folinic acid, 170.6 μg/ml E.coli tRNA mixture, 2.0 mM each of the 20 amino acids, 33.3 mM phosphoenolpyruvate, 20 μg/ml T7 RNA polymerase, 13.3 ng/μl DNA plasmid template, 20% v/v cell extract and nuclease-free water. Note the composition of the standard CFPS reaction was adjusted during the optimization process as described in the text. The CFPS reaction was incubated at 23°C for 4 h unless otherwise indicated. Quantification of sfGFP product was accomplished using a previously established method (30), and a standard curve that converts fluorescence readings to protein yields was developed to expedite protein yield quantification process (Supplementary Figure S1).
Results and discussion
Development of P. putida CFPS platform
With the goal of trying to build a robust, high-yielding P. putida CFPS system in mind, we started by trying to adopt the protocol used for E. coli CFPS systems (11, 23), which routinely yield g/l quantities of model proteins. After preparing extracts from cells harvested at late exponential phase (OD600 = 5.0), we carried out CFPS of sfGFP in a 15 µl batch reaction for 4 h at 26°C, the optimum growth temperature for P. putida. Unfortunately, this approach only achieved sfGFP yields of <20 µg/ml.
Since the composition of the cellular machinery at the time of harvest directly affects the CFPS potential of the crude extract, we hypothesized that the harvest conditions needed to be adjusted. Accordingly, we grew P. putida cells to different OD600 values: 5, as initially done, but also ∼3.8 and ∼2.5, which spanned a range of mid to late exponential growth in our batch growth conditions (see Supplementary Figure S2 for a growth curve). Then, we prepared batches of crude extract from each of these cultivations. After preparing crude extracts from each batch of the cells, we compared their potential for CFPS. The results indicated that cell extracts prepared from OD600 of ∼2.5 synthesized the highest yield of sfGFP at 79.5 ± 3.6 µg/ml (Figure 1). The most active extracts of P. putida were obtained from the mid-exponential growth phase, which is similar as the E. coli CFPS system (41). Taken together, our data highlight the importance of cell harvest as a critical factor for development and optimization of CFPS systems.
Figure 1.
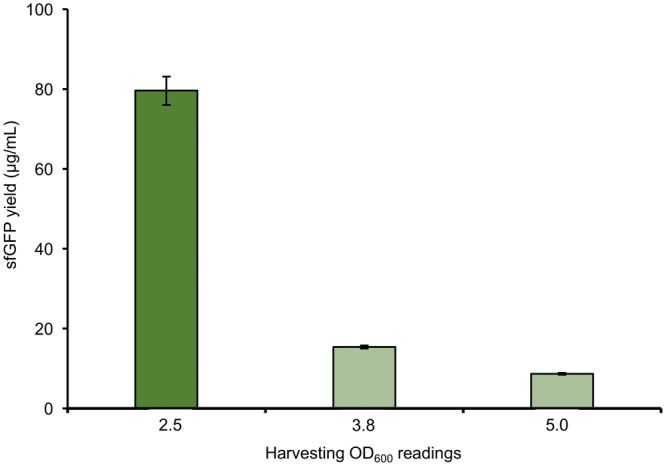
Comparison of sfGFP yield with P. putida biomass harvested at different optical densities (OD600). Reaction conditions: 33% (v/v) lysate content, 20 ng/µl plasmid template, 12 mM magnesium concentration and 26°C. Average sfGFP yield with error bars representing standard deviations of three independent experiments is shown.
Optimization of the P. putida-based CFPS platform
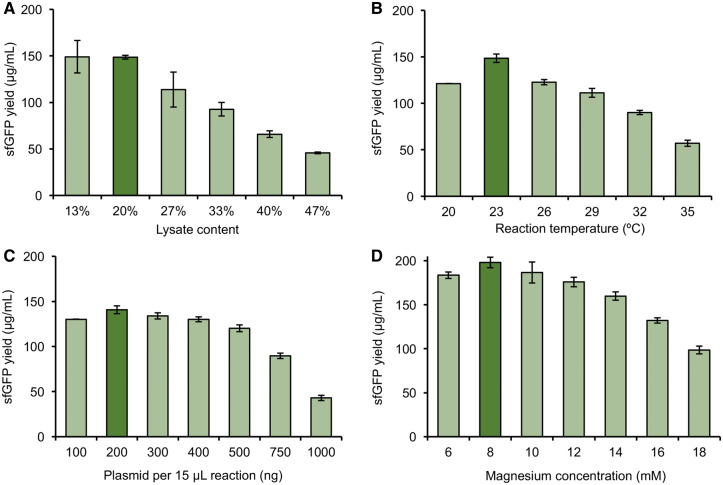
We subsequently carried out a series of optimization experiments to try to increase the sfGFP yield with our P. putida CFPS system. We explored the effects of reaction temperature, cell extract per reaction, plasmid concentration and Mg2+ concentration. We first examined the impact of lysate content on sfGFP yields. To synthesize proteins of interest, crude extract based CFPS systems harness an ensemble of catalytic components (e.g. RNA polymerases, ribosomes, aminoacyl-tRNA synthetases, translation initiation and elongation factors etc.) that are essential for protein synthesis from cell lysates. For this reason, we hypothesized that increasing cell extract concentration in the CFPS reaction might result in an increase in reporter protein yields, as we previously showed for Streptomyces (31). We tested a range of extract concentrations by adjusting the volume of extract in the reaction from 13.3 to 46.7% (v/v), meanwhile, the concentrations of all other components were kept constant in CFPS during the optimization process. We found that 13.3% and 20.0% (v/v) lysate content gave the highest sfGFP yields of 149.1 ± 17.5 and 148.6 ± 2.0 µg/ml, respectively (Figure 2A). Notably, when we increased lysate content further to 46.7% (v/v), a drastic drop in sfGFP yield was observed, which we hypothesize is a result of the physiochemical environment not being optimized as observed in other CFPS systems (31).
Figure 2.
Cell-free protein synthesis optimization for P. putida enhances sfGFP expression yields. The CFPS reaction was optimized by surveying a range of (A) lysate content; (B) reaction temperatures; (C) plasmid concentrations; (D) magnesium concentrations. Initial reaction conditions before the optimization: 33% (v/v) lysate content, 20 ng/µl plasmid template, 12 mM magnesium concentration and 26°C. Average sfGFP yield with error bars representing standard deviations of three independent experiments is shown.
Next, we surveyed a range of reaction temperatures surrounding the preferred growth temperature of P. putida, which is 26°C. Our results showed that at slightly decreased temperature at 23°C, we were able to achieve a 20% increase in sfGFP yield when compared to reactions carried out at 26°C (Figure 2B). This is similar to another CFPS system that we recently developed from Streptomyces, where the optimal CFPS reaction temperature (23°C) is lower than the preferred cell growth temperature (30°C) (30).
We next sought to investigate the influence of plasmid content over an order of magnitude range of increase (100–1000 ng of plasmid template per 15 μl reaction) on the performance of our P. putida CFPS system. Our data indicated that the sfGFP expression peaked when 200 ng of plasmid DNA was supplied as template (Figure 2C).
Finally, we explored the impact of magnesium concentration of CFPS yields, which has been previously shown to be perhaps the most critical component of CFPS reactions (23, 24, 30, 45). Magnesium is used to balance the charge present from nucleic acid phosphate groups and other anionic species. Therefore, optimization of magnesium is essential for many protein–nucleic acid interactions, ribosome assembly, and the proper function of biological processes of the cell, including protein synthesis. CFPS reactions were carried out at a range of magnesium ion concentrations from 6 to 18 mM. We observed that 8 mM gave the highest yield of 198 ± 5.9 µg/ml (Figure 2D). Overall, our newly designed CFPS system, which includes lowered temperature, increased total extract concentration, increased plasmid concentration and optimized magnesium concentration, resulted in a more than 10-fold improvement of CFPS yield relative to the non-optimized reaction conditions shown in Figure 1, in which we observed expression for the first time.
Using PCR amplicon as CFPS template
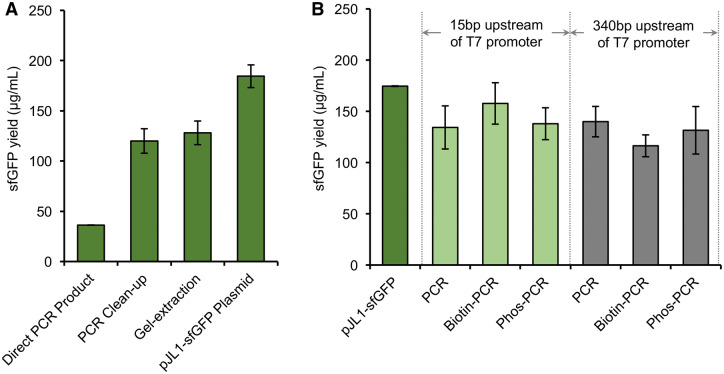
Upon demonstration of robust and high yielding protein expression from our P. putida combined transcription and translation system, we then set out to demonstrate the potential for using linear DNA templates for P. putida-based CFPS. High-throughput protein expression has become a key technology in systems biology and synthetic biology. Using linear DNA molecules, i.e. PCR product, expedites the process since there are no laborious cloning steps. To this end, we substituted direct PCR amplicons, together with purified PCR products (QIAquick PCR Purification Kit or QIAquick Gel Extraction Kit), for plasmid DNA in the CFPS reactions. For the purified PCR products, we could achieve ∼70% of the sfGFP yield when plasmid DNA was used as a template (Figure 3A). Representative time courses of sfGFP synthesis using plasmid and linear DNA templates with online fluorescence measurement are shown in Supplementary Figure S3. This is important because it enables a high-throughput platform where one can go from DNA sequence to protein in under 6 h.
Figure 3.
sfGFP yield with various PCR templates and purification schemes compared to that of the pJL1-sfGFP plasmid. (A) sfGFP yield with different post-PCR purification. The purified PCR template used per reaction was normalized to the same molar concentration of the plasmid pJL1-sfGFP. (B) 15/340 bp upstream: space upstream of T7 promoter; all templates were purified by PCR clean-up. Average sfGFP yield with error bars representing standard deviations of three independent experiments is shown.
One of the existing challenges with using PCR templates is their potential instability in CFPS. Thus, we next asked whether or not the sfGFP synthesis yields could be improved from linear template DNA by modifying the expression template in two ways. First, and in light of some previous studies (46, 47), we wondered if the addition of extra non-coding DNA sequence upstream of the T7 promoter might aid in expression yields. Unfortunately, we did not observe significant difference between PCR amplicons with 15 and 340 base-pairs extra upstream sequences in sfGFP yield (Figure 3B). Second, we tested the use of 5′biotinylated or phosphorothioated primers during PCR amplification to produce amplicons with less susceptibility to degradation, as has been done before in CFPS (48, 49). We observed that the sfGFP yield was the same with or without the use of protected primer ends (Figure 3B). Taken together, our results indicated that linear PCR amplicons were stable in the P. putida-based CFPS system without the need to add the purified protein GamS for the protection of PCR templates, which was found to be necessary in the E. coli-based CFPS reaction (16). While our efforts to modify the PCR amplicon template did not improve CFPS yields in the P. putida cell-free system, we chose to use linear templates moving forward, as the template can be used directly and prepared by two-step PCR from a small amount of sample.
RBS library
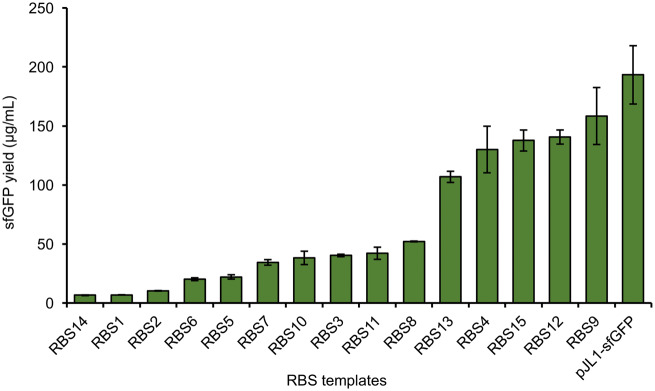
Our new P. putida-based CFPS opens the possibility to rapidly assess and validate genetic part performance by preparing hundreds or even thousands of DNA templates without time-consuming cloning work. As a proof of concept demonstration for such screening capabilities, we wanted to examine the ability of a small RBS library to tune sfGFP expression in our P. putida CFPS system. This is important because one of the key ideas driving this research is that our system can provide an extremely rapid way to screen and test libraries. To carry out this test, we designed a total of 15 different RBSs replacing the original RBS sequence between the T7 promoter sequence and the start of the sfGFP gene. These 15 sequences were designed based on the Salis Ribosome Binding Site Calculator with a wide spectrum of predicted expression levels of sfGFP from minimal (RBS sequence 1) to maximal (RBS sequence 15) (RBS sequences are listed in Supplementary Table S2) (50, 51). Following library construction, PCR amplicons of the RBS library were used to initiate 15 μl batch CFPS reactions for 4 h at 23°C. We observed a 20-fold range of sfGFP expression yields (Figure 4). Thus, as expected the RBS library was able to tune sfGFP expression. Interestingly, the expression pattern did not follow the designed trend where, in theory, RBS1 should have the lowest expression whereas RBS15 should have the highest. For example, RBS14 demonstrated the weakest expression. As a result, our results here join an emerging wave of reports that highlight how CFPS screening platforms can be used to assess if genetic designs function as expected.
Figure 4.
Different levels of sfGFP expression with the RBS library highlights the ability of P. putida-based CFPS system for genetic part characterization. Rapid turn-over time for screening and characterization of DNA regulatory elements such as RBSs can be achieved with our P. putida CFPS system. Average sfGFP yield with error bars representing standard deviations of three independent experiments is shown.
Conclusions
In this study, we described the development and optimization of a P. putida-based CFPS system, and we were able to achieve sfGFP yields of ∼200 µg/ml in 4 h batch reactions. For comparison, although the current yield is lower than that of the E. coli-based CFPS system (>1000 µg/ml), which has been developed over the past 20 years (52), it is ten times higher than the recently developed S. cerevisiae-based CFPS system (<20 µg/ml) (45). The high yielding CFPS capacity, together with the heterologous protein expressing capability of P. putida, makes it a valuable addition to the current existing CFPS platforms. Just as several bacterial CFPS systems (e.g. E. coli and B. subtilis) have been developed to mimic the native host for prototyping applications (33, 53), we believe the P. putida platform will be useful for rapid prototyping of native genetic parts before evaluating a smaller design set in cells. Since P. putida has a relatively high guanine-cytosine (GC)-content genome (>60% GC) (37), our new platform may also be able to efficiently express GC-rich genes with codon usage bias. Although recently Streptomyces-based CFPS systems were established to express high GC-content genes (30, 31), the process for the preparation of Streptomyces cell extract was slower and more laborious than that of P. putida due to the growth of Streptomyces microorganisms. Moreover, CFPS yields were lower than those observed in this study.
In addition to showing high yielding CFPS, we demonstrated that the P. putida-based CFPS system is compatible with PCR amplicon as expression templates with minimal purification required. We also showed our system’s capability as a gene regulatory screening platform by monitoring sfGFP expression from a library of RBSs. We were able to characterize the expression efficiencies of 15 RBS sequences in less than a day.
Looking forward, we believe this work not only expands protein synthesis toolkit for synthetic biology, but also sets the stage for future application of a P. putida-based CFPS screening platform for gene regulatory elements. The next step moving forward would be closing the gap between in vitro and in vivo prediction, and we anticipate that future efforts will further advance its relevance in characterizing and discovering more DNA parts for synthetic biology by expediting screening and prototyping efforts.
SUPPLEMENTARY DATA
Supplementary Data are available at SYNBIO Online, including [reference citation(s)].
Supplementary Material
Acknowledgments
M.C.J. thanks the David and Lucile Packard Foundation and the Camille-Dreyfus Teacher-Scholar Program for their generous support.
Funding
DARPA 1KM program [HR0011-15-C-0084].
Conflict of interest statement. None declared.
References
- 1. Kanter G., Yang J., Voloshin A., Levy S., Swartz J.R., Levy R. (2007) Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood, 109, 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Yang J., Kanter G., Voloshin A., Michel-Reydellet N., Velkeen H., Levy R., Swartz J.R. (2005) Rapid expression of vaccine proteins for B-cell lymphoma in a cell-free system. Biotechnol. Bioeng., 89, 503–511. [DOI] [PubMed] [Google Scholar]
- 3. Min S.E., Lee K.-H., Park S.-W., Yoo T.H., Oh C.H., Park J.-H., Yang S.Y., Kim Y.-S., Kim D.-M. (2016) Cell-free production and streamlined assay of cytosol-penetrating antibodies. Biotechnol. Bioeng., 113, 2107–2112. [DOI] [PubMed] [Google Scholar]
- 4. Stech M., Kubick S. (2015) Cell-free synthesis meets antibody production: a review. Antibodies, 4, 12–33. [Google Scholar]
- 5. Bundy B.C., Franciszkowicz M.J., Swartz J.R. (2008) Escherichia coli-based cell-free synthesis of virus-like particles. Biotechnol. Bioeng., 100, 28–37. [DOI] [PubMed] [Google Scholar]
- 6. Lu Y., Chan W., Ko B.Y., VanLang C.C., Swartz J.R. (2015) Assessing sequence plasticity of a virus-like nanoparticle by evolution toward a versatile scaffold for vaccines and drug delivery. Proc. Natl. Acad. Sci. USA, 112, 12360–12365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Henrich E., Hein C., Dötsch V., Bernhard F. (2015) Membrane protein production in Escherichia coli cell-free lysates. FEBS Lett., 589, 1713–1722. [DOI] [PubMed] [Google Scholar]
- 8. Sachse R., Dondapati S.K., Fenz S.F., Schmidt T., Kubick S. (2014) Membrane protein synthesis in cell-free systems: from bio-mimetic systems to bio-membranes. FEBS Lett., 588, 2774–2781. [DOI] [PubMed] [Google Scholar]
- 9. Boyer M.E., Stapleton J.A., Kuchenreuther J.M., Wang C.W., Swartz J.R. (2008) Cell-free synthesis and maturation of [FeFe] hydrogenases. Biotechnol. Bioeng., 99, 59–67. [DOI] [PubMed] [Google Scholar]
- 10. Kwon Y.-C., Oh I.-S., Lee N., Lee K.-H., Yoon Y.J., Lee E.Y., Kim B.-G., Kim D.-M. (2013) Integrating cell-free biosyntheses of heme prosthetic group and apoenzyme for the synthesis of functional P450 monooxygenase. Biotechnol. Bioeng., 110, 1193–1200. [DOI] [PubMed] [Google Scholar]
- 11. Li J., Lawton T.J., Kostecki J.S., Nisthal A., Fang J., Mayo S.L., Rosenzweig A.C., Jewett M.C. (2016) Cell-free protein synthesis enables high yielding synthesis of an active multicopper oxidase. Biotechnol. J., 11, 212–218. [DOI] [PubMed] [Google Scholar]
- 12. Hong S.H., Kwon Y.-C., Martin R.W., Des Soye B.J., de Paz A.M., Swonger K.N., Ntai I., Kelleher N.L., Jewett M.C. (2015) Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. ChemBioChem, 16, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hong S.H., Ntai I., Haimovich A.D., Kelleher N.L., Isaacs F.J., Jewett M.C. (2014) Cell-free protein synthesis from a release factor 1 deficient Escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth. Biol., 3, 398–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Garamella J., Marshall R., Rustad M., Noireaux V. (2016) The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol., 5, 344–355. [DOI] [PubMed] [Google Scholar]
- 15. Karim A.S., Jewett M.C. (2016) A cell-free framework for rapid biosynthetic pathway prototyping and enzyme discovery. Metab. Eng., 36, 116–126. [DOI] [PubMed] [Google Scholar]
- 16. Sun Z.Z., Yeung E., Hayes C.A., Noireaux V., Murray R.M. (2014) Linear DNA for rapid prototyping of synthetic biological circuits in an Escherichia coli based TX-TL cell-free system. ACS Synth. Biol., 3, 387–397. [DOI] [PubMed] [Google Scholar]
- 17. Takahashi M.K., Chappell J., Hayes C.A., Sun Z.Z., Kim J., Singhal V., Spring K.J., Al-Khabouri S., Fall C.P., Noireaux V.. et al. (2015) Rapidly characterizing the fast dynamics of RNA genetic circuitry with cell-free transcription–translation (TX-TL) systems. ACS Synth. Biol., 4, 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pardee K., Green A.A., Ferrante T., Cameron D.E., DaleyKeyser A., Yin P., Collins J.J. (2014) Paper-based synthetic gene networks. Cell, 159, 940–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Pardee K., Green A.A., Takahashi M.K., Braff D., Lambert G., Lee J.W., Ferrante T., Ma D., Donghia N., Fan M.. et al. (2016) Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell, 165, 1255–1266. [DOI] [PubMed] [Google Scholar]
- 20. Goshima N., Kawamura Y., Fukumoto A., Miura A., Honma R., Satoh R., Wakamatsu A., Yamamoto J., Kimura K., Nishikawa T.. et al. (2008) Human protein factory for converting the transcriptome into an in vitro–expressed proteome. Nat. Methods, 5, 1011–1017. [DOI] [PubMed] [Google Scholar]
- 21. Matsuoka K., Komori H., Nose M., Endo Y., Sawasaki T. (2010) Simple screening method for autoantigen proteins using the N-terminal biotinylated protein library produced by wheat cell-free synthesis. J. Proteome Res., 9, 4264–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Salehi A.S.M., Smith M.T., Bennett A.M., Williams J.B., Pitt W.G., Bundy B.C. (2016) Cell-free protein synthesis of a cytotoxic cancer therapeutic: onconase production and a just-add-water cell-free system. Biotechnol. J., 11, 274–281. [DOI] [PubMed] [Google Scholar]
- 23. Jewett M.C., Swartz J.R. (2004) Mimicking the Escherichia coli cytoplasmic environment activates long-lived and efficient cell-free protein synthesis. Biotechnol. Bioeng., 86, 19–26. [DOI] [PubMed] [Google Scholar]
- 24. Martin R.W., Majewska N., Chen C.X., Albanetti T.E., Jimenez R.B.C., Schmelzer A.E., Jewett M.C., Roy V. (2017) Development of a CHO-based cell-free platform for synthesis of active monoclonal antibodies. ACS Synth. Biol., 6, 1370–1379. [DOI] [PubMed] [Google Scholar]
- 25. Hodgman C.E., Jewett M.C. (2013) Optimized extract preparation methods and reaction conditions for improved yeast cell-free protein synthesis. Biotechnol. Bioeng., 110, 2643–2654. [DOI] [PubMed] [Google Scholar]
- 26. Schoborg J.A., Hodgman C.E., Anderson M.J., Jewett M.C. (2014) Substrate replenishment and byproduct removal improve yeast cell-free protein synthesis. Biotechnol. J., 9, 630–460. [DOI] [PubMed] [Google Scholar]
- 27. Gan R., Jewett M.C. (2014) A combined cell-free transcription-translation system from Saccharomyces cerevisiae for rapid and robust protein synthesis. Biotechnol. J., 9, 641–651. [DOI] [PubMed] [Google Scholar]
- 28. Anderson M.J., Stark J.C., Hodgman C.E., Jewett M.C. (2015) Energizing eukaryotic cell-free protein synthesis with glucose metabolism. FEBS Lett., 589, 1723–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zemella A., Thoring L., Hoffmeister C., Kubick S. (2015) Cell-free protein synthesis: pros and cons of prokaryotic and eukaryotic systems. ChemBioChem, 16, 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li J., Wang H., Kwon Y.C., Jewett M.C. (2017) Establishing a high yielding Streptomyces-based cell-free protein synthesis system. Biotechnol. Bioeng., 114, 1343–1353. [DOI] [PubMed] [Google Scholar]
- 31. Li J., Wang H., Jewett M.C. (2018) Expanding the palette of Streptomyces-based cell-free protein synthesis systems with enhanced yields. Biochem. Eng. J., 130, 29–33. [Google Scholar]
- 32. Moore S.J., Lai H.E., Needham H., Polizzi K.M., Freemont P.S. (2017) Streptomyces venezuelae TX-TL—a next generation cell-free synthetic biology tool. Biotechnol. J., 12, 1600678. [DOI] [PubMed] [Google Scholar]
- 33. Kelwick R., Webb A.J., MacDonald J.T., Freemont P.S. (2016) Development of a Bacillus subtilis cell-free transcription-translation system for prototyping regulatory elements. Metab. Eng., 38, 370–381. [DOI] [PubMed] [Google Scholar]
- 34. Nikel P.I., Martinez-Garcia E., de Lorenzo V. (2014) Biotechnological domestication of pseudomonads using synthetic biology. Nat. Rev. Microbiol., 12, 368–379. [DOI] [PubMed] [Google Scholar]
- 35. Dammeyer T., Steinwand M., Krüger S.-C., Dübel S., Hust M., Timmis K.N. (2011) Efficient production of soluble recombinant single chain Fv fragments by a Pseudomonas putida strain KT2440 cell factory. Microb. Cell Fact., 10, 11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nikel P.I., de Lorenzo V. (2014) Robustness of Pseudomonas putida KT2440 as a host for ethanol biosynthesis. N. Biotechnol., 31, 562–571. [DOI] [PubMed] [Google Scholar]
- 37. Loeschcke A., Thies S. (2015) Pseudomonas putida-a versatile host for the production of natural products. Appl. Microbiol. Biotechnol., 99, 6197–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Martinez-Garcia E., Nikel P.I., Aparicio T., de Lorenzo V. (2014) Pseudomonas 2.0: genetic upgrading of P. putida KT2440 as an enhanced host for heterologous gene expression. Microb. Cell Fact., 13, 159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zobel S., Benedetti I., Eisenbach L., de Lorenzo V., Wierckx N., Blank L.M. (2015) Tn7-based device for calibrated heterologous gene expression in Pseudomonas putida. ACS Synth. Biol., 4, 1341–1351. [DOI] [PubMed] [Google Scholar]
- 40. Nelson K.E., Weinel C., Paulsen I.T., Dodson R.J., Hilbert H., Martins dos Santos V.A.P., Fouts D.E., Gill S.R., Pop M., Holmes M.. et al. (2002) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ. Microbiol., 4, 799–808. [DOI] [PubMed] [Google Scholar]
- 41. Pecoraro V., Zerulla K., Lange C., Soppa J. (2011) Quantification of ploidy in proteobacteria revealed the existence of monoploid, (mero-)oligoploid and polyploid species. PLoS One, 6, e16392.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fonseca P., Moreno R., Rojo F. (2011) Growth of Pseudomonas putida at low temperature: global transcriptomic and proteomic analyses. Environ. Microbiol. Rep., 3, 329–339. [DOI] [PubMed] [Google Scholar]
- 43. Nakashima N., Tamura T. (2004) Cell-free protein synthesis using cell extract of Pseudomonas fluorescens and CspA promoter. Biochem. Biophys. Res. Commun., 319, 671–676. [DOI] [PubMed] [Google Scholar]
- 44. Kwon Y.C., Jewett M.C. (2015) High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci. Rep., 5, 8663.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schoborg J.A., Clark L.G., Choudhury A., Hodgman C.E., Jewett M.C. (2016) Yeast knockout library allows for efficient testing of genomic mutations for cell-free protein synthesis. Synth. Syst. Biotechnol., 1, 2–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sawasaki T., Ogasawara T., Morishita R., Endo Y. (2002) A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA, 99, 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jun S.Y., Kang S.H., Lee K.H. (2008) Continuous-exchange cell-free protein synthesis using PCR-generated DNA and an RNase E-deficient extract. BioTechniques, 44, 387–391. [DOI] [PubMed] [Google Scholar]
- 48. Vosberg H.P., Eckstein F. (1982) Effect of deoxynucleoside phosphorothioates incorporated in DNA on cleavage by restriction enzymes. J. Biol. Chem., 257, 6595–6599. [PubMed] [Google Scholar]
- 49. Yabuki T., Motoda Y., Hanada K., Nunokawa E., Saito M., Seki E., Inoue M., Kigawa T., Yokoyama S. (2007) A robust two-step PCR method of template DNA production for high-throughput cell-free protein synthesis. J. Struct. Funct. Genomics, 8, 173–191. [DOI] [PubMed] [Google Scholar]
- 50. Salis H.M., Mirsky E.A., Voigt C.A. (2009) Automated design of synthetic ribosome binding sites to control protein expression. Nat. Biotechnol., 27, 946–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Espah Borujeni A., Channarasappa A.S., Salis H.M. (2014) Translation rate is controlled by coupled trade-offs between site accessibility, selective RNA unfolding and sliding at upstream standby sites. Nucleic Acids Res., 42, 2646–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. (2012) Cell-free protein synthesis: applications come of age. Biotechnol. Adv., 30, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chappell J., Jensen K., Freemont P.S. (2013) Validation of an entirely in vitro approach for rapid prototyping of DNA regulatory elements for synthetic biology. Nucleic Acids Res., 41, 3471–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.