Abstract
Synthetic biology requires students and scientists to draw upon knowledge and expertise from many disciplines. While this diversity is one of the field’s primary strengths, it also makes it challenging for newcomers to acquire the background knowledge necessary to thrive. To address this gap, we developed a course that provides a structured approach to learning the biological principles and theoretical underpinnings of synthetic biology. Our course, Principles of Synthetic Biology (PoSB), was released on the massively open online course platform edX in 2016. PoSB seeks to teach synthetic biology through five key fundamentals: (i) parts and layers of abstraction, (ii) biomolecular modeling, (iii) digital logic abstraction, (iv) circuit design principles and (v) extended circuit modalities. In this article, we describe the five fundamentals, our formulation of the course, and impact and metrics data from two runs of the course through the edX platform.
Keywords: synthetic biology, massive open online course (MOOC), edX, education, curriculum building
1. Introduction
Over the past two decades, the field of synthetic biology has grown to encompass a powerful set of integrated molecular and cellular engineering technologies, linked by an increasingly coherent set of standards for design and characterization (1, 2). The field has had successful applications in health, agriculture and chemical/pharmaceutical production (1, 3–5). However, the practice of synthetic biology draws on skills spanning cell and molecular biology, biophysics, chemical engineering, computer science, control theory and statistics. Due to this diversity, it is difficult to generate comprehensive educational and training resources for students interested in entering this field.
In recent years, several approaches have arisen to expand synthetic biology education. At the collegiate and high school level, the most well-known method of getting involved with synthetic biology is the annual International Genetically Engineered Machine (iGEM) competition (6, 7). In iGEM, students can gain research experience through the design, construction and sharing of novel genetic circuits. While iGEM teams do learn relevant molecular biology and modeling techniques, a structured framework for understanding synthetic biology is not present. Another growing high school-level program is BioBuilder (8), through which students are introduced to synthetic biology via a variety of hands-on labs and activities (9). While BioBuilder provides an excellent introductory experience for students to learn basic synthetic biology theory and molecular biology skills, college-level students can benefit from a deeper theoretical foundation.
To accomplish the goal of providing a strong, unified framework for synthetic biology, we adapted Principles of Synthetic Biology (PoSB)—an upper-division course taught jointly at MIT and UC Berkeley by Professors Ron Weiss and Adam Arkin—for the massively online open course (MOOC) platform, edX (https://www.edx.org/course/principles-synthetic-biology-mitx-20-305x-0). MOOCs are a modern educational development that makes college-level education material accessible to anyone with an internet connection. MOOCs can push beyond the traditional educational capacity of textbooks via a combination of videos, lecture notes, problems and user forums, which have been shown to increase information retention rates and positively influence motivation (10, 11). Importantly, MOOCs are uniquely suited for emerging disciplines like synthetic biology because MOOCs can be easily updated every year as new literature is published. Finally, MOOCs can reach a large audience; edX is one of the most successful MOOC platforms with over 2.4 million unique users accessing one or more of its 290 courses between 2012 and 2016 (12).
In this article, we lay out the course content of PoSB and the core concepts used to guide its creation. Then, data from the first two runs of the course are discussed to assess the impact of the course and suggest alterations. We envision this article being of particular interest to educators and prospective students. Educators interested in integrating synthetic biology into their own curricula should find the Course Fundamentals and Course Outline sections particularly useful. These sections provide pedagogical principles and specific references to the course content that should be helpful in adapting portions of our course into new curricula. Prospective students should focus on the Course Outline and Course Formulation sections. These sections can be used to identify and target specific knowledge gaps as well as provide an understanding into the process of taking PoSB through the edX platform.
We expect that students with a background in biology will particularly benefit from the course’s insights into model design, digital logic and design-build-test approaches. Those with engineering backgrounds can learn how to model gene expression, how biological ‘machines’ can implement familiar functions, and how biological systems have fundamental limitations. PoSB is an upper-division course and, as such, we recommend prerequisite courses covering: molecular biology, math through integral calculus, basic programming and at least one organic chemistry course. Recommended subjects include: differential equations, electronic circuits, control theory and biochemistry.
2. Course fundamentals
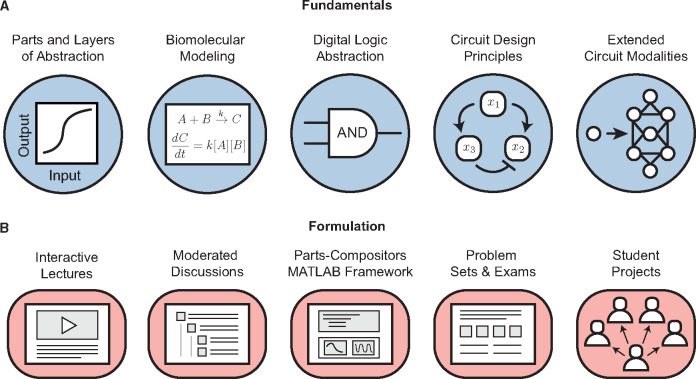
PoSB is structured around five fundamentals: (i) parts and layers of abstraction; (ii) biomolecular modeling; (iii) digital abstractions; (iv) circuit design principles; and (v) extended circuit modalities (Figure 1A). This next section details these specific fundamentals and some of their pedagogical choices.
Figure 1.
Course fundamentals and formulation. (A) The fundamental methodologies and abstractions used to design the course curriculum. (B) The many different modes that students use to engage with the course.
2.1 Parts and layers of abstraction
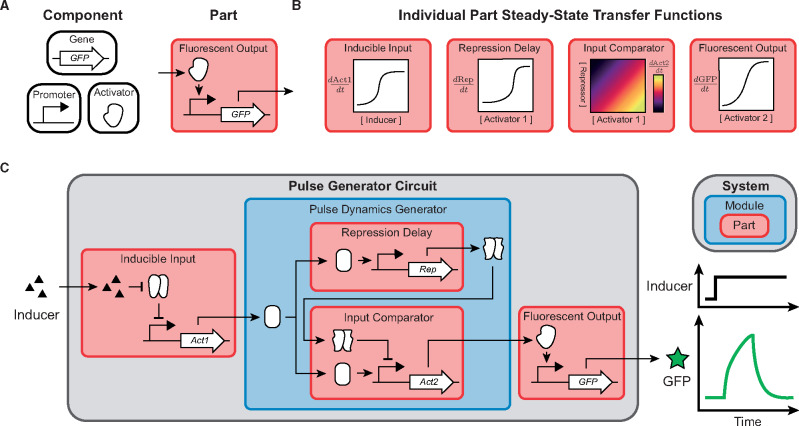
One of the first things we seek to teach in this course is our definition of a part in synthetic biology. From an engineering perspective, a ‘part’ is a useful abstraction for understanding and building larger systems (13). For synthetic biologists, these systems are often genetic regulatory networks. A biologist—caring about the mechanistic underpinnings of the system—may define their ‘part’ as a protein or a piece of DNA; a synthetic biologist—concerned with the capabilities of the system—will define their ‘part’ differently. For synthetic biology, we choose to define a part as a process that modifies the state of the cell. While the cell state modifications are enacted by a set of physical components within the cell, we seek to abstract ourselves from those components and define the part in terms of how it changes our system of interest (Figure 2A). In PoSB, we define the input of a part to be the current state of the cell (i.e. the concentrations of molecular species) and the output as the rate of change of those species (i.e. a differential equation for each of the species). We note that this definition of part is not yet a universally accepted definition within synthetic biology. Different approaches can benefit from other part definitions; however, this abstraction is well-suited for the analytical approaches used in PoSB and we believe this definition also has relevance for the general community. In the following paragraphs, we demonstrate the general utility of a PoSB part by using it to construct a hypothetical pulse generator circuit via hierarchical composition (Figure 2).
Figure 2.
Parts and layers of abstractions in the context of a pulse generator circuit. (A) The difference between physical components and functional parts in the example of a fluorescent reporter. (B) The individually profiled steady-state transfer functions of each pulse generator circuit part in isolation. The Input Comparator part has two inputs and its output is represented as a heatmap color (B) A full pulse-generator genetic circuit system (gray) made by combining parts (red) and modules (blue). A hypothetical time trace is shown for GFP fluorescence upon a step input of inducer.
Transcription factor (TF) activated GFP production is an example of a part that is presented early in the course. The input into this part is the concentration of an activator protein and the output is the production rate of GFP (Figure 2A, Part). We note that this functional definition of a part is distinct from the physical components that make up the part: an activator, a promoter and the GFP gene (Figure 2A, components).
A key aspect of parts is that they can be characterized in isolation and then predictably composed into larger systems. Higher-order functionalities can then be designed when individual parts are combined through well-defined input/output responses and interfaces. This compositional approach can be used in a hierarchical manner to design modules from individual parts and then systems from individual modules. In the case of the Pulse Generator Circuit system, the key functionality originates from the Pulse Dynamics Generator module. This module is created from two parts that take the activating Act1 input through two branches (Figure 2B). In the Input Comparator part, Act1 activates the output of the module, Act2. Soon after, delayed production of a repressor from Repressor Delay part overrides the activation in the Input Comparator part. As a whole, this results in only a brief spike of Act2 production. Once the Pulse Dynamics Generator module has been created, we can interface it with parts that have experimentally relevant inputs and outputs. The Inducible Input part allows for the initiation of a pulse and the Fluorescent Output part facilitates the readout of circuit state (Figure 2B). The final result is the Pulse Generator Circuit system (Figure 2C).
Layers of abstraction are a key engineering principle that allows the separation of high-level functional design from physical implementation. Our Pulse Generator Circuit is an example of how layers of abstraction can be leveraged to build complex systems. The first layer of abstraction is the definition of a functional part from physical components (Figure 2A). This abstracts the designer from the physical components that act on the system. The next layer of abstraction is the creation of a Pulse Dynamics Generator module from the Repression Delay and Input Comparator parts (Figure 2C). When working with the module, the designer only needs to know that providing an appropriate amount of Act1 input will result in a spike of Act2 production. They do not need to concern themselves with the specifics of two parts within the module, nor the numerous physical components that make up each part. The highest layer of abstraction is the whole Pulse Generator Circuit system. This system is built by combining the Pulse Dynamics Generator module with input- and output-interfacing parts. While a strong understanding of biology is required to design and connect these modular components, if the system is properly designed, much of the complex biology can be abstracted away. The designer is then free to focus on understanding higher-order functionalities of the system, such as the effect of inducer addition and removal on pulse shape and timing.
While the Pulse Generator Circuit system is the highest level of abstraction presented here, one can imagine how this system could, itself, become a part of an even larger system using the same abstraction methodologies.
2.2 Biomolecular modeling
A central premise of engineering disciplines is that model-driven design and diagnostics are critical for scaling devices, improving their reliability and learning from failures. As such, mathematical modeling has been an important aspect of synthetic biology since its inception (14, 15). It has helped guide design, understand failure modes and propose new mechanisms for genetic circuits (16–18). Models can vary widely in complexity and ease of manipulation. As most of the analyses in PoSB focus on small genetic circuits, we chose to use relatively simple, state-space models built on mass-action kinetics principles (19). We first teach how to apply these models to very simple systems such as enzymatic reactions (Michaelis–Menten kinetics). Then, using the notion of parts, these models are progressively built up to eventually encapsulate entire genetic circuits. These models allow students to investigate the effect of parameters on circuit functionality in silico. To complement these models, we also include discussion on how and when our deterministic models may fail to capture real-world genetic circuits functionality due to phenomena such as stochasticity, small molecular counts and noise propagation. We mention that for these scenarios, probabilistic approaches such as Gillespie algorithms or chemical master equations are more informative. Building and manipulation of all models is completed within the edX system through an integrated MATLAB toolbox (Section 4.3).
2.3 Digital logic abstraction
After the first few sections of the course, students have learned how to compose and simulate the basic components of genetic circuits. Once the input–output functions for a part are defined, we demonstrate how in some parameter regimes, certain parts can exhibit ultrasensitive response functions to yield digital-like behavior. We then use these parts as building blocks to develop higher-order genetic circuit functionality like computing digital logic functions. We focus on digital abstractions because they are a useful tool for simplifying the high-level design and analysis of genetic circuits.
Students are taught digital circuitry from an electrical engineering perspective to motivate and guide the construction of circuits with complex capabilities. This requires students to first learn how to define problems and solutions in terms of combinational logic problems (e.g. identify a cell as a cancer cell if certain miRNAs are present and/or absent). Once combinational circuits are outlined and motivated, we apply some basic computer science tools for reducing and manipulating digital logic circuits: Boolean cubes and Karnaugh maps. After students have learned to form basic digital circuits, we present the biological components that can be used to enact digital logic.
2.4 Circuit design principles
After the basic components of genetic circuits have been presented, we dive in to some of the higher-order properties that emerge when larger circuits are built. First, we discuss simple cascades in both steady-state and dynamical situations. We then discuss common circuit topologies such as feedforward and feedback and highlight their unique dynamical properties. From there, we study the design features of toggle switches and oscillators to demonstrate how they can be used to create advanced circuits for coordinated timing and control of cellular state. Finally, problems of scalability and modularity are discussed to illustrate current engineering challenges in the field.
Throughout these discussions, we emphasize using modeling methods to identify and design around possible failure modes for circuits. Collectively, these topics teach students about fundamental biological circuit motifs, their composition into larger circuits, and how to reduce trial-and-error when experimentally implementing a circuit. This helps students understand how modeling can be used to avoid problems such as timing hazards and mismatched transfer functions.
2.5 Extended circuit modalities
In the final section of the course, we introduce a variety of genetic circuit modalities that exhibit expanded input/output types, dynamics, safety and robustness. Since many of the early course concepts are demonstrated with TF-based circuitry, we start with a survey of scalable TF-based circuit components including TetR homologs, zinc-finger nucleases (ZFNs), TALEs and CRISPRi. We then present RNA-based components in both bacterial and mammalian systems. For the RNA components, we highlight the unique sensing capabilities of miRNA circuits and aptazymes, as well as the improved safety of RNA-only circuits, which do not require DNA encoding. Entirely protein-based circuits are then discussed and their fast dynamics are highlighted through the load driver application (20). Lastly, we present recombinase circuits and highlight their improved robustness for genetically encoded memory. Together, these diverse modalities expose students to the breadth of engineering capabilities that synthetic biologists use to control cellular processes.
3. Course outline
The fundamental goal of the PoSB curriculum is to bring together engineering principles with biological implementations. In Table 1, we outline the primary course contents of the most recent PoSB iteration (as of this writing, Spring 2017) and organize them with respect to each course fundamental. A brief description accompanies each topic along with pointers to the relevant course material (L indicating lectures and PS indicating problem sets). As synthetic biology fundamentally necessitates and builds upon biology, relevant biological background material is provided in the ungraded Problem Set 0 (PS0). Within PS0, we also provide links to free online introductory biology courses through MIT OpenCourseWare. As shown in Table 1, PS0 also contains background materials for differential equations, digital logic and measurement methods.
Table 1.
Primary course content
| Fundamental | Specific content | Description | Pointer |
|---|---|---|---|
| Parts and layers of abstraction | Top-down design | Methods for designing composable and modular biological circuits | L2/3 |
| Parts and composition | Defining parts as processes in biological systems and simple modeling introduction | L5/6 | |
| Biomolecular modeling | ODE background | Introduction to mass-action kinetics and using ODEs to describe cellular processes | PS0 |
| Michaelis–Menten | Modeling and engineering enzymatic reactions with Michaelis–Menten kinetics | L6 | |
| Gene expression | Capturing simple transcription and translation with mathematical equations | L7 | |
| Digital logic abstractions | Digital background | Introduction to basic digital logic ideas including gates, circuits and truth tables | PS0 |
| Digital biology | Digital logic abstractions in biology with transcriptional repressors as NOT/NOR gates | L9 | |
| Logic minimization | Designing and simplifying arbitrary digital circuits with uniting theorem and Karnaugh maps | L16 | |
| Designing with cello | Description of how Cello genetic circuit design software works (21) | L15 | |
| Circuit design principles | Cascades | Layering transcriptional components and predicting function and timing hazards | L10/11 |
| Feedback loops | Negative and positive gene autoregulation and 1D graphical analysis of ODEs | L8 | |
| Feedforward loops | Feedforward loop network motifs, their dynamical properties and biological implications | L12 | |
| Toggle Switches | Using feedback to build memory devices and 2D graphical ODE analysis | L13 | |
| Oscillators | Using feedback and feedforward to build oscillating biological circuits | L14 | |
| Extended circuit modalities | Transcription factors | Survey of scalable transcriptional regulators: TetR homologs, TALEs, ZFNs and CRISPRi/a | L17 |
| Prokaryotic RNAs | Survey of RNA control in prokaryotes: RNA-IN/OUT and aptamers | L18 | |
| Eukaryotic RNAs | Survey of RNA control in eukaryotes: RNAi and replicons | L19 | |
| Protein circuits | Survey of protein-based circuits: phosphorelays, allostery, scaffolding and the load driver | L20 | |
| Recombinases | Survey of recombinase-based circuits used for memory and digital logic | L21 |
In addition to the content centered around the five course fundamentals, we also included material on methods and applications in synthetic biology, as depicted in Table 2.
Table 2.
Additional course content
| Topic | Specific content | Description | Pointer |
|---|---|---|---|
| Methods | Measurement | How biological measurements are made: reporters, flow cytometry, microscopy | PS0 |
| DNA assembly | Survey of DNA assembly: restriction enzymes, Gibson, Golden Gate, Gateway | L4 | |
| Applications | Tissue homeostasis | Design of a synthetic circuit to maintain β cell populations in type I diabetes patients | L3 |
| Cancer classifier | Design of a circuit to kill cancer cells through miRNA detection and digital logic | L19 | |
| Morphogenesis | Designing cellular patterning and organoids with cell-to-cell communication | L22 |
4. Course formulation
The PoSB online content interleaves lecture videos, written notes, practice problems and discussion boards. The course is designed as an instructor-paced course running the length of a semester. Each week, two complete lectures are released. There are five problem sets, a midterm, a final exam and a final course project. In this section, we will describe the formulation of the course, including how we developed lectures, the types of problems we created and how we managed the students’ final projects (Figure 1B). While the course content was initially designed to fit into a semester timeline, we believe it could be adapted to a two-quarter course, with the first quarter focusing on the core theory and the second quarter expanding upon this theory to more complex circuits and applications. Following this strategy, our recommendation for the first quarter would be to teach the first four fundamentals: (i) parts and layers of abstraction, (ii) biomolecular modeling, (iii) digital logic abstraction and (iv) circuit design principles. The second quarter could then cover the final fundamental, Extended Circuit Modalities, as well as the Methods and Applications sections.
4.1 Interactive lectures
Most of the PoSB course content is delivered via interactive lectures, which include: video lectures delivered by professors Weiss and Arkin, detailed lecture notes, practice problems and discussion boards tied to specific questions raised during the lectures. As a residential course, PoSB is cotaught via a videoconference set up between MIT and Berkeley. In the edX adaptation of these lectures, we chose to include some in-class discussions from the residential course. EdX students are given a chance to share their thoughts on these in-class discussion topics with their peers via embedded discussion boards. Each video segment is also accompanied by detailed notes that reiterate the major points, provide additional context and supply links to referenced papers.
To help reinforce student learning, we added occasional problems in line with the lecture notes. Some of these problems are designed to give students practice with technical topics, while other problems encourage student exploration of model systems. For example, there are many live MATLAB coding boxes which can be used to modify and simulate the models discussed in lecture. We also built a genetic circuit interpreter that students can use to evaluate the logic of TF and miRNA-based regulation. When combined with the discussion elements, these types of problems allow for deeper student engagement beyond simple multiple-choice problems.
4.2 Moderated discussions
Discussion boards are placed alongside lecture content and can also be accessed through a central forum. These discussion boards end up serving many different purposes. Discussion boards within interactive lectures have thought-provoking questions such as, ‘is a cell more like a computer or a burrito?’. Discussion boards in problem sets allow students to work together to solve the problems as if they are in a study group. For the final project, discussion boards help students develop and share their ideas.
The discussion boards are pivotal to scaling the course to thousands of students. Although our resources are limited, students can help to answer each other’s questions through the boards. Additionally, the discussion boards allow us to easily solicit course improvements and identify bugs. For many students, getting to work together and interact with other students provides a unique opportunity to engage with synthetic biology in a way that may have otherwise been inaccessible.
4.3 Parts-Compositors MATLAB framework
To put into practice our concept of a part, we built a custom MATLAB-based numerical simulation framework that students can use to quickly build and simulate genetic circuits (https://github.com/Weiss-Lab/Parts-Compositors). The framework is designed to be used in an object-oriented manner, with each part of the biological system being an object. Our framework allows students to simulate models without having to directly code ordinary differential equation (ODE) simulations themselves. MATLAB code is run directly in the browser using interactive coding boxes placed within lectures, problem sets and exams. The coding boxes were developed by MathWorks, the creators of MATLAB.
4.4 Problem sets and exams
Problem set and exam questions in the course can be divided into four categories: multiple choice, drag-and-drop, free response and MATLAB-based. Many of our drag-and-drop problems involve placing labels for genetic components onto circuit diagrams. These test the students’ ability to process a description of a model system, reason through the logic of how components interact, and then place those components into a valid arrangement that satisfies a given specification. Free-response problems require students to enter quantitatively correct information. For example, some questions request students to analyze logic gates, input reaction rates and compute steady-state solutions to a given model. The MATLAB-based questions have students use the Parts-Compositors framework to gain insights into circuit function through various questions integrated with the MATLAB code boxes.
Before being released on edX, the problem sets and exams were prototyped with residential classes at UC Berkeley and MIT. We used feedback from residential students and members of the Weiss and Arkin labs to improve problem design, adjust problem sets/exam composition and debug complicated problems in advance of the wider edX release.
4.5 Student projects
A final project is included to encourage students to design and analyze a synthetic biology system by building upon what they learned in the course. For this project, students identify a biological problem of interest, design a synthetic biology-based solution and model their synthetic system to identify key parameters and design constraints. Through the discussion boards, students can discuss their ideas, solicit feedback and eventually share their final projects for all to see. To facilitate the grading of projects, we require students submitting final projects to peer-evaluate each other’s work. This allows for the large-scale evaluation of student projects. While peer review can be inaccurate (22), review accuracy may be improved by moving from numerical to ranked peer-scoring in future iterations of PoSB (23). Ultimately, peer evaluation provides one final opportunity for students to engage with the diverse subject matter of synthetic biology before finishing the course.
5. Impact and metrics
The edX version of PoSB has had two runs: Spring 2016 (v1) and Spring 2017 (v2). As of November 2018, there were 28 623 signups for v1 and v2. However, only 17 229 students viewed any course material (a 60% follow-through rate compared with an average edX follow-through rate of 53%) (12). We further stratify students who viewed the course material into three types: (i) 11 768 on-time students (signed up before the second problem set was due); (ii) 1595 late students (signed up after the second problem set was due, but before the course archive date); and (iii) 3866 archival students (signed up after the course was archived). All further analyses only consider students who signed up on time and therefore experienced the desired course pacing (N = 11 768). In this next section, we analyze these students’ demographics and course activity in the context other edX STEM courses.
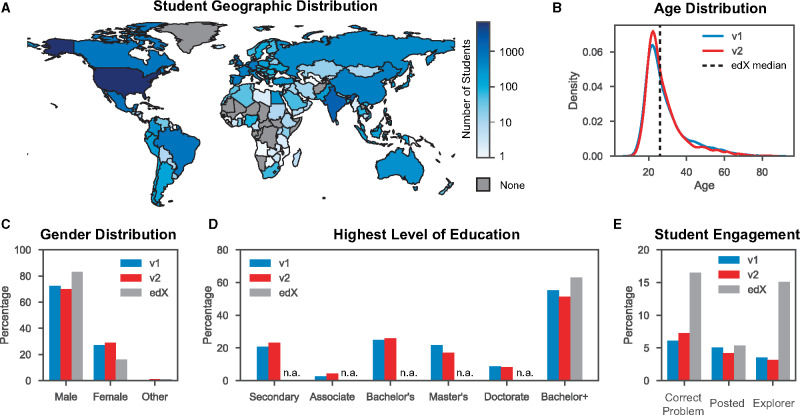
The broad reach of our course is apparent when looking at the global distribution of students. Across both runs of the course, students hailed from 150 different countries (Figure 3A). Students most frequently came from the USA (28%). This is similar to the median domestic percentage of all edX STEM courses (25%) (12). The next most frequent countries were India (7%) and Mexico (5%). Additionally, the median PoSB student age was 26, identical to that of all edX STEM courses (Figure 3B) (12). Interestingly, our student demographics deviated from edX median values in two ways: we had a higher proportion of female-declared participants (Figure 3C) and a lower percentage of bachelor-degree-holding students (Figure 3D). The reasons for these deviations were not clear.
Figure 3.
Course demographics and engagement metrics. (A) Counts of the total number of participating students from each country shown on a logarithmic scale. Countries with no students are grey. (B) Age distribution for both runs of the course. The median age for edX STEM courses is indicated with a dashed line. (C) The self-identified gender distributions of v1 and v2 of the course alongside edX STEM median gender values. (D) The highest level of education indicated by students. Cumulative edX data were only available for the Bachelor+ category (Bachelor, Masters and/or PhD). (E) Percentages of students that correctly answered at least one problem (‘Correct Problem’), made at least one post in the discussion board (‘Posted’), and viewed more than 50% of the course content (‘Explorer’). Median values for edX STEM course are displayed alongside.
When looking at student engagement in the course, we found some fairly large differences from the edX STEM median values (Figure 3E). Students in PoSB were significantly less likely to correctly answer at least one problem in the course than the edX median (6.5% compared with the edX median of 16%). This may be the result of higher problem difficulty or less general interest in completing problems. Students were also less likely to become ‘explorers’ (a term for students who viewed at least half of the course). This indicated a higher than usual course attrition rate. As shown by student posting rates, the discussion board activity was healthy and similar to standard edX STEM courses (Figure 3E). We initially considered certification rate as an engagement metric, but it is an unreliable method of comparing courses due to the fact that edX courses vary greatly in their requirements for certification (grade value, payment required, problem difficulty, etc.) (12). Other MOOCs have had their effectiveness evaluated by comparing baseline knowledge tests given before and after students take the course (24). While we did not administer these tests, we plan to do this in future iterations of the course.
Between the two runs of the course, the main difference was the enrollment number. The first run of the course attracted 9282 participating students, while the second run had only 2459 (26% of the original). We note that it is normal to observe a drop in enrollment for the second run of a course (edX median is 75% of the original course enrollment rate) (12). While our drop in enrollment was a bit more than the edX median, the demographic and engagement profiles were similar for both runs of our course. This indicates that the drop is likely due to differences in advertising and/or course demand rather than specific differences in course content/management.
Overall, the two runs of PoSB have managed to reach a broad audience that is generally similar to the student demographics of a typical edX STEM course. While we do see a healthy and active discussion board, we acknowledge that our course does have a higher-than-average attrition rate. We plan to address this issue in future iterations of the course by streamlining content to improve approachability and by adding easier and more frequent in-lecture problems to improve engagement.
6. Conclusions
Our goal with the PoSB edX course was to create a broadly accessible, structured introduction to synthetic biology. From the first two runs of the course, we believe that we were successful in this goal. Over two years, PoSB has managed to reach over 10 000 students from more than 150 countries. This is a scale of education that is only possible through a MOOC platform like edX.
Synthetic biology is a rapidly progressing field and PoSB represents our effort to deliver the latest research insights while simultaneously grounding students in the engineering and biological principles needed to understand the field. With this article, we aimed to break down the course structure, content and ideas so that students and educators interested in learning about and teaching synthetic biology can begin to engage with and use our content. For the students and educators who are interested in a more detailed understanding of the course, all PoSB material is accessed through the edX platform. We note that for the first two iterations of the course, all PoSB content was freely available—in perpetuity—to all students. A recent policy change by edX requires students to pay a small fee for access to exam/project materials and archived versions of the course. However, the core aspects of PoSB—lecture videos/notes, practice problems and problem sets—will still be freely available for the duration of course runs.
SUPPLEMENTARY DATA
Course demographics and engagement data are not publicly available for reasons of student privacy.
Acknowledgements
We would like to thank everyone who helped make PoSB on edX possible. Former TAs that helped develop class content: Jeremy Gam, Eerik Kaseniit, Jacob Becraft, Mohammad Soheilypour, Tracy Washington, Sarvesh Varma, Sebastian Palacios, Guillaume Kugener and Sepideh Dolatshahi. Current TAs working on PoSB v3: Yuge Ji, Raphaël Gayet and Ching Fang. MITx Staff: Brad Goodman, Kyle Boots, Colleen Cressman, Lana Scott, Lisa Eichel, Garrett Beazley and Edwin Cabrera. MATLAB support: Michael Reardon. Administrative support: Olga Parkin. The Synthetic Biology Institute at UC Berkeley and Synthetic Biology Center at MIT. The Departments of Biological Engineering at MIT and Bioengineering at UC Berkeley. And all the MIT, UC Berkeley and edX students who gave feedback on the problem sets and exams.
Funding
Development of PoSB on edX was supported by funding from the MIT Department of Biological Engineering, MIT Department of Electrical Engineering and Computer Science, UC Berkeley Department of Bioengineering, and a generous donation from the Wertheimer Foundation. Additional funding was provided by the National Science Foundation [1521925].
Conflict of interest statement. None declared.
References
- 1. Cameron D.E., Bashor C.J., Collins J.J. (2014) A brief history of synthetic biology. Nat. Rev. Microbiol., 12, 381–390. [DOI] [PubMed] [Google Scholar]
- 2. Bartley B.A., Rodriguez C.A., Misirli G., Hillson N.J., Galdzicki M., Endy D., Grünberg R., Hallinan J., Roehner N., Stan G.-B.. et al. (2014) The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat. Biotechnol., 32, 545–550. [DOI] [PubMed] [Google Scholar]
- 3. Khalil A.S., Collins J.J. (2010) Synthetic biology: applications come of age. Nat. Rev. Genet., 11, 367–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Weber W., Fussenegger M. (2012) Emerging biomedical applications of synthetic biology. Nat. Rev. Genet., 13, 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shih P.M., Liang Y., Loqué D. (2016) Biotechnology and synthetic biology approaches for metabolic engineering of bioenergy crops. Plant J., 87, 103–117. [DOI] [PubMed] [Google Scholar]
- 6. Mitchell R., Dori Y.J., Kuldell N.H. (2011) Experiential engineering through iGEM—an undergraduate summer competition in synthetic biology. J. Sci. Educ. Technol., 20, 156–160. [Google Scholar]
- 7. Dubé S., Orr D., Dempsey B., Wieden H.J. (2017) A synthetic biology approach to integrative high school STEM training. Nat. Biotechnol., 35, 591–595. [DOI] [PubMed] [Google Scholar]
- 8. Dixon J., Kuldell N. (2011) Chapter twelve - BioBuilding: Using Banana-Scented Bacteria to Teach Synthetic Biology In: Voigt,C. (ed). Synthetic Biology, Part A. Vol. 497 Academic Press, pp. 255–271. [DOI] [PubMed] [Google Scholar]
- 9. Kuldell N, Bernstein R., Ingram K., Hart K.M. (2015) Chapter 5: Introduction to the BioBuilder Labs. In: Loukides M., MacDonald B. (eds). BioBuilder | Synthetic Biology in the Lab First. O’Reilly Media, Sebastopol, CA, pp. 91–94. [Google Scholar]
- 10. Epstein M.L., Lazarus A.D., Calvano T.B., Matthews K.A., Hendel R.A., Epstein B.B., Brosvic G.M. (2002) Immediate feedback assessment technique promotes learning and corrects inaccurate first responses. Psychol. Rec., 52, 187–201. [Google Scholar]
- 11. Barak M., Watted A., Haick H. (2016) Motivation to learn in massive open online courses: examining aspects of language and social engagement. Comput. Educ., 94, 49–60. [Google Scholar]
- 12. Chuang I., Ho A. (2016) HarvardX and MITx: four years of open online courses—fall 2012-summer 2016. Available at SSRN: https://ssrn.com/abstract=2889436 or 10.2139/ssrn.2889436. [DOI]
- 13. Endy D. (2005) Foundations for engineering biology. Nature, 438, 449–453. [DOI] [PubMed] [Google Scholar]
- 14. Elowitz M.B., Leibler S. (2000) A synthetic oscillatory network of transcriptional regulators. Nature, 403, 335–338. [DOI] [PubMed] [Google Scholar]
- 15. Gardner T.S., Cantor C.R., Collins J.J. (2000) Construction of a genetic toggle switch in Escherichia coli. Nature, 403, 339–342. [DOI] [PubMed] [Google Scholar]
- 16. Endler L., Rodriguez N., Juty N., Chelliah V., Laibe C., Li C., Novere N.L. (2009) Designing and encoding models for synthetic biology. J. R. Soc. Interface, 6, S405–S417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chandran D., Copeland W.B., Sleight S.C., Sauro H.M. (2008) Mathematical modeling and synthetic biology. Drug Discov. Today Dis. Model, 5, 299–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hucka M., Finney A., Sauro H.M., Bolouri H., Doyle J.C., Kitano H., Arkin A.P., Bornstein B.J., Bray D., Cornish-Bowden A.. et al. (2003) The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics, 19, 524–531. [DOI] [PubMed] [Google Scholar]
- 19. Domitilla D.V.,, Murray R.M. (2014) Biomolecular Feedback Systems. Princeton University Press, Princeton and Oxford. ISBN: 9780691161532. [Google Scholar]
- 20. Mishra D., Rivera P.M., Lin A., Del Vecchio D., Weiss R. (2014) A load driver device for engineering modularity in biological networks. Nat. Biotechnol., 32, 1268–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nielsen A.A.K., Der B.S., Shin J., Vaidyanathan P., Paralanov V., Strychalski E.A., Ross D., Densmore D., Voigt C.A. (2016) Genetic circuit design automation. Science, 352, aac7341. [DOI] [PubMed] [Google Scholar]
- 22. Suen H.K. (2014) Peer assessment for Massive Open Online Courses (MOOCs. ). Int. Rev. Res. Open Distrib. Learn., 15, 312–327. [Google Scholar]
- 23. Shah N.B., Bradley J.K., Parekh A., Wainwright M., Ramchandran K. (2013) A case for ordinal peer-evaluation in MOOCs. NIPS Workshop on Data Driven Education, pp. 1–8. [Google Scholar]
- 24. Colvin K.F., Champaign J., Liu A., Zhou Q., Fredericks C., Pritchard D.E. (2014) Learning in an introductory physics MOOC: all cohorts learn equally, including an on-campus class. Int. Rev. Res. Open Distrib. Learn., 15, 263–283. [Google Scholar]