Abstract
The incorporation of cell-free transcription and translation systems into high-throughput screening applications enables the in situ and on-demand expression of peptides and proteins. Coupled with modern microfluidic technology, the cell-free methods allow the screening, directed evolution and selection of desired biomolecules in minimal volumes within a short timescale. Cell-free high-throughput screening applications are classified broadly into in vitro display and on-chip technologies. In this review, we outline the development of cell-free high-throughput screening methods. We further discuss operating principles and representative applications of each screening method. The cell-free high-throughput screening methods may be advanced by the future development of new cell-free systems, miniaturization approaches, and automation technologies.
Keywords: synthetic biology, high-throughput screening, cell-free systems, directed evolution
Introduction
High-throughput screening methods analyze the interactions between a large number of chemical compounds or biomolecules and specific targets in a robust, time efficient and highly reproducible format. This screening technology was initially driven by the increase in the number of compounds and molecules available for testing, and the necessity to accelerate the research and development of new drugs and therapies (1–4). For the last two decades, high-throughput screening assays have advanced significantly based on the development of miniaturized systems (5, 6), more sensitive detection methods (7–9), better data analysis (10) and automation of screening procedures (4, 11). Indeed, high-throughput screening has been used successfully in a wide range of applications such as drug discovery (12), evolution of proteins (13), enzyme engineering (14), as well as screening and discovery of chemical probes (15), small molecules (16, 17) and lipopeptides (18).
Despite the considerable increase of sample processing capacity and quality of the screening assays, the field has relied primarily on the use of in vivo (or cell-based) approaches, and the use of highly purified or chemically synthesized target proteins. In vivo approaches exploit the change of cellular phenotype to screen for the presence of specific molecules (Table 1). The in vivo approaches are useful when the presence or the activity of the biomolecule of interest can be determined using fluorescence, luminescence, or altered cellular physiology. However, they are not optimal for the screening of binding between two molecules (e.g. identification of small molecule targets of an antibody), or if the assayed molecule is toxic to the host cells. The use of purified or chemically synthesized proteins in high-throughput screening assays reduces interference with other molecules and false positives. Both approaches exhibit certain disadvantages; chemical synthesis is only capable of generating small peptides, and the use of purified proteins requires purification steps that increase the cost and time of the screening procedure.
Table 1.
Advantages and disadvantages of in vitro and in vivo methods for the high-throughput evolution, screening and selection of biomolecules
| In vitro (cell-free) approaches | In vivo (cell-based) approaches | |
|---|---|---|
| Principle | The approaches use cell-free transcription–translation systems for the synthesis of biomolecules encoded in genetic materials. The biomolecules are then tested in reconstituted environments | The approaches use live cells for the synthesis and subsequent screening of biomolecules. The biomolecules are screened in natural cellular environments. |
| Advantages |
|
|
| Disadvantages |
|
|
These limitations have been addressed through the development of high-throughput screening assays that use cell-free transcription and translation systems (Table 1). The cell-free systems synthesize biomolecules encoded on DNA libraries, which are then tested in defined media for an increased enzymatic activity through affinity chromatography methods or detection of fluorescence reporters. The use of cell-free systems adds an additional layer of complexity due to the high diversity of molecules present in the analysis solution, the increased possibility of non-specific interactions with cellular components and the myriad ways of controlling protein expression. Despite this complexity, cell-free protein expression systems bring forth several advantages over conventional cell-based approaches, including the capability to express toxic or insoluble proteins (19), the incorporation of unnatural or isotope-labeled amino acids into the peptide chain (20, 21), reduced processing time (22) and reaction volumes (23), as well as the lack of gene-cloning steps (24). These advantages make cell-free systems ideal for high-throughput applications (25, 26).
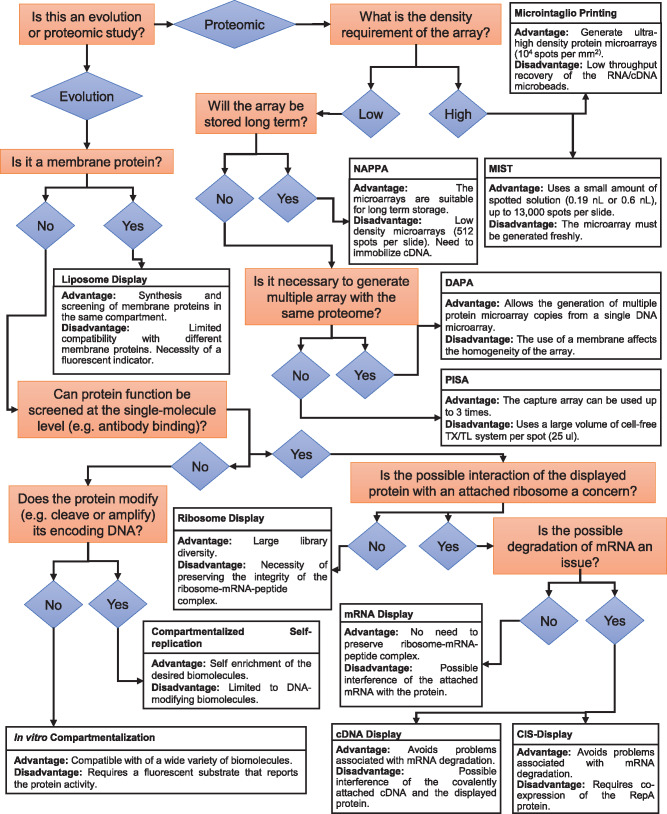
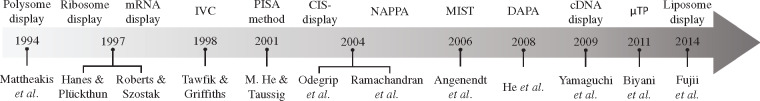
The techniques that use in vitro cell-free transcription/translation systems for the screening, selection and evolution of biomolecules in a high-throughput format can be classified as in vitro display and on-chip technologies. Here, we define in vitro display technologies as methods that link genotype and phenotype by covalent linkage between proteins, ribosomes, DNA and RNA. After expression, these complexes are directly screened and tested for activity. The linked-genetic sequence is then used in subsequent rounds of enrichment. On the other hand, we define on-chip or microarray technologies as methods that immobilize expressed proteins into treated surfaces without the source RNA or DNA. Several reviews have summarized and analyzed different aspects of these two classes of cell-free high-throughput screening techniques (8, 27–29). In contrast, we aim to underline the development of the central techniques that broaden the adaptation of cell-free expression systems for the high-throughput screening, selection and evolution of biomolecules. We focus our efforts on explaining how each one of the technologies was developed in a connected timeline and modified to tackle problems in the preceding techniques. We also review the techniques with increasing complexity of compartmentalization: (i) without physical compartmentalization; (ii) 2D microarrays in which the biomolecules are attached to a treated surface; (iii) 3D micrometer scale compartments. Furthermore, we summarize all the techniques regarding their main advantages and disadvantages (Figure 1). We also emphasize the chronological development of these technologies (Figure 2) to show how the advancement of in vitro display and on-chip technologies has been enabled by discoveries in other scientific areas.
Figure 1.
A summary of the techniques that incorporate cell-free protein expression systems for the high-throughput screening, selection and evolution of biomolecules. This flowchart outlines the choice of a technique based on the experimental requirements as well as provide a brief description of the advantage and disadvantage of each technique.
Figure 2.
Chronology of the techniques used for the high-throughput screening of biomolecules using cell-free gene expression systems. The timeline shows the authors and their invented techniques that use cell-free transcription/translation systems for the high-throughput screening of biomolecules.
Screening of biomolecules in a single pot without physical compartmentalization
Polysome and ribosome display
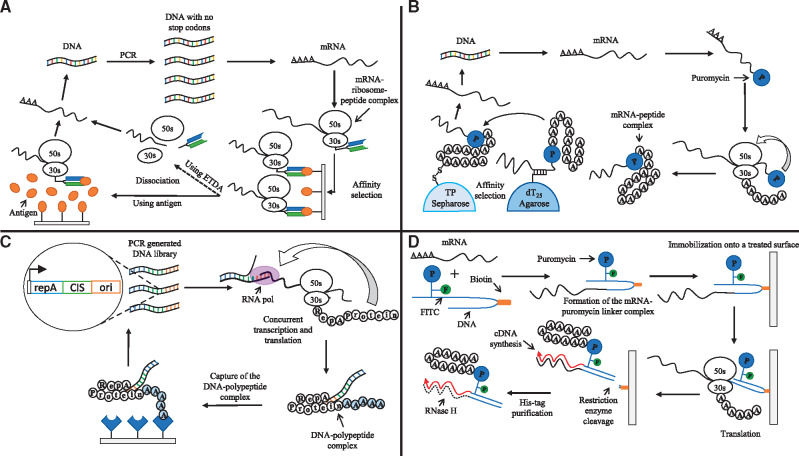
Polysome and ribosome display are the first fully in vitro approaches developed for the selection and evolution of small peptides and native folded proteins respectively (30, 34) (Figure 3A). Ribosome display adapted the pioneering method used by in vitro polysome display for the screening of a large library of decapeptides displayed on polysomes using affinity selection towards an immobilized antibody (34). The method intentionally omits the stop codon so that a peptide expressed using an E. coli S30 system remains attached to the ribosome while ensuring that the peptide folds properly. In this study, the authors synthesized single-chain fragments of an antibody with disulfide bonds and screened the antibody fragments using an antigen. Through this proof of concept, the authors showed that it was possible to carry out phenotypic selection (i.e. ligand binding) using completely native proteins that were synthesized in vitro. The method has two main advantages over the cell-based selection and screening technologies. First, the diversity of the library is not limited by the transformation efficiency of host cells. Instead, the quality of the library is controlled only by the number of ribosomes and mRNA. Second, a new round of diversification can be introduced to the library after the end of each selection round, hence overcoming the need to transform cells after each diversification step in the cell-based screening methods (35). The ribosome display method has been applied to select peptides with increased activity towards prostate-specific antigen (36), streptavidin (STA) binding peptides that confer an increased affinity to bovine heart fatty acid-binding protein (FABP) (37) and picloram-specific variable fragments of heavy chain antibodies (38). Polysome and ribosome display are the first methods that demonstrate the in vitro linkage between genotype (mRNA) and phenotype (small peptides and proteins respectively). However, their main disadvantage is the difficulty in displaying certain peptides due to their unpredictable interactions with the attached ribosome (39). A new display technology that addresses this issue was developed swiftly.
Figure 3.
Techniques for the screening of biomolecules without physical compartmentalization. (A) Graphic representation of in vitro ribosome display, adapted from (30). This technique uses polymerase chain reaction (PCR) to amplify DNA without stop codons, allowing the synthesized mRNA to stay attached to the ribosome, while the generated peptide folds properly. Then, the mRNA-ribosome-peptide complexes are affinity selected for subsequent reverse transcription into cDNA. (B) Graphic representation of in vitro selection (mRNA display), adapted from (31). mRNA display uses mRNA molecules linked to puromycin (P) to generate a mRNA-peptide complex. The resulting molecule is then selected by affinity chromatography and reverse transcribed to generate DNA. (C) Graphic representation of the CIS-display technology, adapted from (32). This method uses a property of the RepA protein that transiently interacts with the CIS element. A peptide is co-expressed with the RepA protein. When the RepA protein interacts with the CIS element found in the DNA, the interaction produces a linkage between the DNA and the polypeptide. The resulting molecule can be captured using an immobilized target and PCR amplified for the next round of selection. (D) Graphic representation of the cDNA display method using a puromycin linker DNA, adapted from (33). mRNA and the puromycin linker are ligated using T4 ligase. The product of this reaction is then immobilized onto a treated surface using a biotin adapter attached to the DNA linker. The immobilized mRNA is then translated using a cell-free expression system. The resulting protein stays attached to the ribosome, and it fuses to the linker through the puromycin molecule. The mRNA is reverse transcribed, and the immobilized complex is released using a restriction enzyme. After affinity chromatography purification, the mRNA is digested with RNase to confirm the presence of the cDNA-peptide fusion.
mRNA display
Shortly after the development of the ribosome display technology, the mRNA display method is developed for the in vitro selection and directed evolution of proteins (31) (Figure 3B). This methodology uses synthetic mRNAs that carry puromycin at their 3ʹ end. This approach avoids the necessity of preserving the integrity of the ribosome-mRNA-peptide complex required in the ribosome display methodology. Instead, it uses a mRNA molecule that serves as both a messenger and an adapter. Puromycin is attached to the 3ʹ end of the synthetic mRNA, and it is an antibiotic that mimics the aminoacyl end of tRNA. Using this characteristic, the protein of interest is expressed using a reticulocyte lysate. The expressed protein then inhibits the translation by entering the ribosomal A site yielding a peptidyl-puromycin fusion molecule as a result of the peptidyl transferase activity of the ribosome. Taking advantage of this reaction, this method generates a stable mRNA-peptide fusion molecule that can be then purified using affinity chromatography followed by immunoprecipitation. The DNA sequence can also be recovered by reverse transcribing the mRNA.
To demonstrate the utility of this technique, a fusion of the long myc template (LP154) and its synthetic mRNA was enriched from a pool of random sequence templates (LP160). Comparing the change of the ratio between pool/myc, the authors determined that the myc sequence was enriched 20- to 40-fold. The mRNA display has been used in the evolution of antibody mimics (40) and single-chain antibodies (41), the selection of high-affinity aptamers (42) and the identification of drug receptors (43). We point the readers to a few comprehensive reviews about this technique (44–46).
CIS-display
The CIS-display method (32) takes advantage of the cis-activity inherent of the RepA protein (a DNA replication initiator protein) that binds to the template DNA from which it has been expressed (Figure 3C). The template DNA contains a CIS element that composes of a Rho-dependent transcriptional terminator, which stalls the RNA polymerase. The stalling of RNA polymerase allows the newly synthesized RepA protein to bind to the CIS element that switches RepA to the adjacent ori site. To create the DNA library, DNA fragments of random DNA sequence are ligated with a DNA fragment that encodes RepA and carries the CIS element and an ori site. These DNA fragments are used as templates in the E. coli S30 expression system that generates protein-DNA complexes due to the stable association of each protein with the DNA that encodes.
CIS-display technology is intended to overcome the size limitations of in vitro selection systems based on emulsion encapsulation that generates DNA mutant libraries of 109 to 1010 per ml. The authors demonstrated the utility of the CIS-display method by creating a library encoding >1012 random 18-mer peptides and showed >1000-fold enrichment of peptides in each round of affinity selection against a relevant target. The selection of peptide ligands was performed using two well-characterized antibodies (anti-P53 DO1 and anti-FLAG M2) and lysozyme. This technique has also been used select a high-affinity binder to the extracellular region of human vascular endothelial growth factor receptor isoform 2 (VEGFR-2) (47) and has been coupled to next-generation sequencing and bioinformatics to facilitate the design of peptides with a potential therapeutic target (48).
cDNA display
The cDNA display method (Figure 3D) is an improvement of the mRNA display technology, originally developed as an approach for the screening of disulfide-rich peptides (33). The method uses a cell-free translation extract, a puromycin-linker DNA containing a ligation site, a biotin site that also serves as the reverse transcription primer site and a restriction enzyme recognition site. These features allow several crucial processes to happen: (i) the rapid ligation of mRNA and the linker, (ii) a biotin/STA-based purification step and (iii) cDNA synthesis by reverse transcription. Taken together, these characteristics prevent the degradation of mRNA, reduce the time employed in the procedure and allow the conversion of mRNA display (mRNA-protein fusion) to cDNA display (cDNA-protein fusion).
As a proof of concept, the authors screened several test proteins (i.e. immunoglobulin G, POU-specific DNA-binding domain of Oct-1 and anti-FLAG antibody) for specific disulfide patterns. This technology has been successfully applied to the screening of Growth Hormone Secretagogue Receptor-binding peptide (49). Since its introduction, several studies have improved the method by making it more robust, practical and convenient through the design of new puromycin-linkers (50, 51), a pull-down method that uses biotinylated bait protein (52) and a puromycin-linker containing 3-cyanovinylcarbazole nucleoside (cnvK) (53).
Screening of proteins on 2D microarrays
Cell-free systems can be used to synthesize proteins using genomic sequences for high-throughput proteomic studies (54). One major class of methods synthesize either each protein one-by-one (19, 55) or the entire proteome in a single reaction (56). Another class of methods synthesizes defined proteome on microarrays for repeated high-throughput assays. In this review, we focus on the later microarray-based methods.
Protein in situ array method
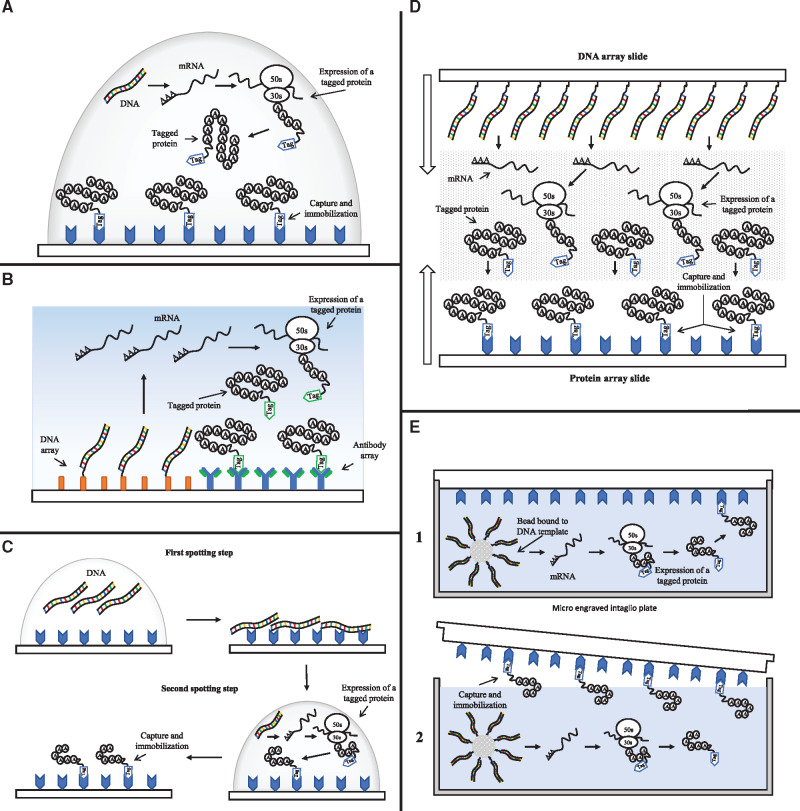
A protein microarray or protein chip is a solid surface (that typically consists of chemically treated glass) on which different proteins are immobilized in a high-density format (57). Protein in situ array (PISA) method is the first protein microarray technology incorporating a cell-free expression system for protein synthesis based on DNA as a template (58) (Figure 4A). This method is renamed DiscernArrayTM in a subsequent study in which the authors used polymerase chain reaction (PCR) to generate genes or gene fragments as templates to produce histidine-tagged proteins using a rabbit reticulocyte transcription/translation system (62). Irrespective of the expression systems, the synthesized proteins are attached to a Ni-Nitrilotriacetic acid coated surface shortly after their expression to generate the protein array. In the first description of PISA method, it was used to produce arrays of single-chain antibodies and luciferase in reaction volumes of 25 μl. In a subsequent study, the volume of cell-free protein expression was decreased to 100 nl using piezoelectric dispensers and by incorporating a commercially available Escherichia coli in vitro transcription translation system. The authors of this study demonstrated high-throughput screening of the enzyme β-galactosidase in nanowells and generated proteins in sufficient quantity for detection by confocal laser scanning (63).
Figure 4.
Techniques for the generation of 2D protein microarrays. (A) Graphic representation of ‘Protein in situ array’ (PISA), adapted from (59). A droplet of cell-free transcription/translation system and DNA is spotted over a surface modified with a tag-capture agent. After the expression of a tagged protein, it is captured and immobilized on the surface. (B) Graphic representation of ‘Nucleic acid programmable protein array’ (NAPPA), adapted from (59). In this technique, DNA and an antibody that is used as a capture agent are first arrayed onto a surface and then incubated with a transcription/translation system. The proteins expressed using the arrayed DNA as a template are captured promptly by the arrayed antibodies. (C) Graphic representation of ‘Multiple spotting technique’ (MIST), adapted from (29). MIST technique uses two sequential spotting steps for the deposition of a DNA template and a cell-free transcription/translation system. After the second spotting step, the protein is expressed and then captured onto the protein array. (D) Graphic representation of ‘DNA array to protein array’ (DAPA), adapted from (60). A slide with a protein array is brought in contact with another slide with a DNA array and a permeable membrane carrying a cell-free expression system. This configuration allows the protein produced from the DNA array to permeate and bind to the slide with the protein array. (E) Graphic representation of microintaglio printing (μP) technique, adapted from (61). This technique uses micro-wells generated by photolithography and a bead bound by a single-copy amplified DNA template. The micro-well is filled with a cell-free transcription/translation system and then sealed with a treated glass surface. After protein expression, the protein is captured and immobilized onto the glass surface, generating a protein array.
Nucleic acid programmable protein array
The next advancement of the protein array technology is described in a study that introduces nucleic acid programmable protein array (NAPPA) as a high-throughput methodology to study protein function through the expression of cDNA clone collections using mammalian reticulocyte lysate (64) (Figure 4B). High-density peptide/protein chips are fabricated by capturing nascent polypeptides during translation onto a solid surface (65). This peptide-capture mechanism takes advantage of the delay in the release of a ribosome that encounters a double-stranded RNA or RNA–DNA hybrid region at the end of the mRNA. This phenomenon provides both the time and the required physical conditions for a puromycin, grafted on an oligo, to enter the A site of the to-be-released ribosome, resulting in synthesized peptides or proteins that are immobilized on a solid glass surface. The authors demonstrated the utility of NAPPA by mapping the interactions among 29 proteins involved in the human DNA replication complex. In a further improvement of the original NAPPA method, the authors demonstrated the expression of over a thousand different human proteins including membrane proteins and proteins over 100 kDa on a microarray surface (66). NAPPA has also been used in the detection of antibodies that bind to tumor antigens in breast cancer (67), the profiling of serological autoantibodies of type 1 diabetes (68) and the profiling of circulating and synovial antibodies of juvenile arthritis patients (69). We refer the reader to other review papers that discuss the applications of the technique in details (70, 71).
Multiple spotting technique
A couple of years after the development of the NAPPA method, a technique called multiple spotting technique (MIST) emerged (Figure 4C). The authors applied a multiple spotting technique (72) for the detection of the fluorescent signal and protein expression in nanoliter volume (63). Later, they used this methodology to develop in situ synthesized protein arrays (73). MIST uses two consecutive spotting steps on an activated glass slide: the first one is used for the deposition of a DNA template and the second one transfers a rapid translation system (RTS) 100 from E. coli on the top of the first spot. After the second spotting step, the slide is incubated in a humid hybridization chamber for rehydration. The rehydration creates separated reaction entities that allow the start of protein expression. When this process has finished, the proteins are detected using specific antibodies. The authors used wild-type green fluorescent proteins (GFP) to demonstrate the creation of microarrays composed of up to 13 000 spots per slide. In addition, they demonstrated that either plasmids or unpurified PCR products could be used as a template. Finally, they synthesized protein arrays using a library of 384 cDNA from human fetal brain cells (73). This technique has also been used in the analysis and characterization of soluble monoclonal antibody fragments (74).
DNA array to protein array
The DNA array to protein array or DAPA is developed for the generation of multiple arrays from an immobilized DNA template (60) (Figure 4D). This approach allows the printing of at least 20 protein arrays from a single DNA template array. The DAPA method uses an array of DNA fragments covalently immobilized to a slide that acts as a template for the synthesis of proteins. This template slide is placed face-to-face with a second slide functionalized with a tag-capturing reagent. The protein expression is carried out using E. coli lysate as a cell-free transcription/translation system that is incorporated in a permeable membrane that is sandwiched between the slides that have the DNA array and the tag-capturing array. The proteins once expressed become rapidly immobilized on the capture slide, creating a protein array that can be duplicated several times. DAPA has been extended to print arrays of 116 different proteins (75). It has also been optimized using different array support coatings, which increase its sensitivity towards the printed proteins (76).
Improvement of protein arrays
Some methods have focused on the improvement of the density achieved in protein arrays. Microintaglio printing (μTP, Figure 4E), allows the parallel spotter-free printing of in situ synthesized proteins for the creation of high-density protein microarrays directly from mRNAs (77) and later from DNA microbeads (61). The μTP approach uses a micro-engraved intaglio plate fabricated by photolithography and replica molding that generates an array of uniformly 5-μm diameter microchambers at a density of 104 per mm2. This micromold plate can be filled with rabbit reticulocyte lysate used to synthesize tagged proteins, which thereafter are captured by a treated glass surface. This approach was further improved using poly(dimethylsiloxane) (PDMS) for the creation of a soft lithography-based microintaglio printing method to generate high-density protein arrays (78). A year later, temperature controlled microintaglio printing (TC-μTP) was developed and used to generate RNA microarrays using DNA-immobilized magnetic beads (79). The second and very similar method that aimed to improve the density of protein arrays uses on-chip micro compartmentalization of protein synthesis in arrayed micrometer scale chambers from confined DNA template molecules (80). The authors compartmentalized single genes and using E. coli lysate they expressed different fluorescent proteins in microchambers of 7 μm in diameter and 5 μm in depth with a volume close to 200 fl. Similarly, they achieved a density of 104 spots per mm2.
Protein arrays have been improved using light-directed localization of genes and protein traps (antibodies specific to a peptide tag) through a novel chimera molecule that serves as a surface coating agent (Nω-Nvoc-amine-Nα-[3-(triethoxysilyl)propyl]-carboxamide-polyethylene glycol or ‘Daisy’) (81). The chimera molecule allows the immobilization of dsDNA molecules with a length of thousands of base pairs into a ‘Daisy-coated’ silicon dioxide surface. The DNA is then transcribed using T7 or sp6 RNA polymerases and translated using a commercial wheat germ-based protein expression system. Using the DNA array, they demonstrated a cell-free gene circuit with a two-stage cascade. This technique was further used to carry out local gene activation using a dual-DNA brush digestion–ligation cascade (82). This digestion-cascade leads to the swapping and rearrangement of DNA resulting in a DNA brush that encodes the complete sequence of GFP under a promoter suitable for expression using an E. coli transcription/translation system. The same coating technique was also used in the development of a new method for the parallel synthesis, assembly and imaging of a protein at the nanoscale (83). This approach was used to demonstrate efficient self-assembly of protein nanotubes using GFP, gp 18 (the tail sheath forming a protein of phage T4), STA-conjugated DNA and RecA proteins.
Screening of biomolecules in 3D micrometer scale compartments
In vitro compartmentalization
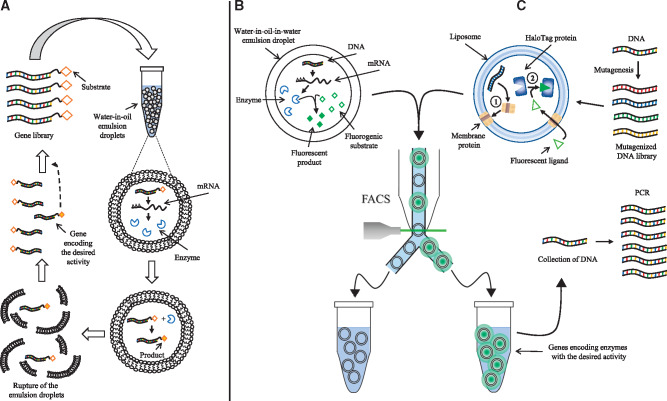
In vitro compartmentalization (IVC) uses an aqueous compartment containing a transcription/translation or a transcription-only system to enclose and link both the phenotype (protein or peptide of interest) and the genotype (coding DNA). ‘Genescis’, or gene selection in a compartmentalized in vitro system is the first technique to demonstrate the use of compartmentalization for high-throughput screening of biomolecules (84). They compartmentalized an E. coli S30 transcription/translation system inside water-in-oil emulsions and used it to express catalytic proteins that were selected based on product formation (Figure 5A). The authors screened for DNA-methylation activity by enriching a methyltransferase (HaeIII) from a 107-fold excess of genes encoding different variants of the enzyme. This essay took advantage of the resistance of the product (methylated DNA) to restriction digestion, allowing only the enrichment of the genes that encode HaeIII methyltransferase.
Figure 5.
Techniques for the screening of biomolecules in 3D micrometer scale compartments. (A) Graphic representation of ‘Gene selection by compartmentalization’, the first reported technique involving IVC, adapted from (84). A gene library is compartmentalized inside droplets of water-in-oil emulsions. The compartmentalized genes are then transcribed and translated. The synthesized enzymes modify a product linked to the gene inside its compartment. After stopping the reaction, the emulsions are broken, and the genes linked to the product are selectively enriched and can be used for a new round of selection. (B) Graphic representation of ‘Selection by IVC in water-in-oil-in-water emulsions’, adapted from (85). Single genes are compartmentalized in droplets of water-in-oil emulsions and translated in the presence of a fluorogenic substrate. The expressed enzyme converts the substrate into a fluorescent product. The droplets are subsequently isolated using FACS. (C) Graphic representation of the liposome display method, adapted from (86). A mutagenized gene library is compartmentalized inside liposomes. Membrane proteins are expressed based on the gene library using a PURE transcription/translation system (see 1 in the figure). The activity of the membrane protein is detected using a fluorescent ligand that is incorporated inside the liposome (see 2 in the figure), and subsequently selected by FACS.
A year later, ‘STABLE’ or STA-biotin linkage in emulsions is introduced. This method is the first to apply in vitro protein selection based on physical association (87). The method is intended for the evolution of proteins and for the selection of cDNA libraries. STABLE uses a water-in-oil compartmentalized E. coli S30 transcription/translation system to synthesize STA-fused polypeptides that are attached to their coding DNAs through a biotin label. The resulting protein-DNA fusion molecules are then recovered from the emulsions and subjected to affinity selection with Ni-agarose resin. To test this approach, the authors assessed the efficiency of enrichment of STA-His fusion molecules synthesized from a mixture of STA-His DNA and STA-Random DNA as a model experiment. The results showed a 10-fold enrichment of STA-His relative to STA-random after a single round of selection. However, the efficacy of the protein–DNA fusion formation was estimated at 1% of the total DNA.
A further study presents a variant of the STABLE system called DNA display (88). The main differences of this method to the original STABLE are the use of a wheat germ transcription/translation system (instead of the E. coli S30) to express cDNAs and the incorporation of an antibody as immobilized bait for the recovery of the DNA-peptide conjugates. This approach was used to generate a library of 109–1010 decapeptides from which 21 were selected using a monoclonal antibody anti-FLAG M2. The efficacy of the formation of protein–DNA fusion increased to >95% due to the use of the wheat germ transcription/translation system.
A method that uses only a polymerase that replicates its own genome, called compartmentalized self-replication (CSR), is developed for the directed evolution of enzymes or auxiliary factors involved in DNA replication (89). Here, the authors use bacterial cells (E. coli) as delivery vehicles for a thermostable polymerase and its coding DNA. The resulting product is then cloned again in E. coli cells, and the process is repeated, yielding polymerase variants with increased thermostability. As a proof of principle, the authors used three rounds of CSR to evolve a polymerase with 11-fold higher thermostability and 130-times more resistance to the inhibitor heparin.
Multiple variations of IVC have been developed. For instance, the microbead display method (90) uses microbeads to link proteins to their coding DNA, which can be selected using fluorescence-activated cell sorting (FACS). The authors demonstrated a 106-fold enrichment of the gene encoding hemagglutinin after two rounds of selection. This same approach has also been used for the selection of specific ligase ribozymes that act in trans (91). A subsequent study describes the use of FACS for the selection and screening of double emulsion water-in-oil droplets containing a fluorescent marker or a fluorogenic substrate (Figure 5B). This approach is used for the enrichment of genes that are trapped together with a fluorescent marker (FITC-BSA) (85). A different variant of IVC called ‘Selective gene amplification’ (SGA) uses product-specific antibodies for the selection of enzymes and ribozymes (92). SNAP-display is an alternative method that covalently links proteins to their coding DNAs using a SNAP-tag (93).
IVC has been used for the directed evolution, selection and screening of a number of biomolecules. In Table 2, we summarize the applications of IVC found in the literature.
Table 2.
Biomolecules evolved, selected and screened using IVC
| Biomolecule | References | |
|---|---|---|
| Directed evolution | Ebg β-galactosidases | (94) |
| p53 variants | (95) | |
| β-Glucosidases | (96) | |
| Cellulases | (97) | |
| Hexose oxidases | (98) | |
| Glucose oxidases | (99) | |
| [FeFe] hydrogenases | (100) | |
| Proteases | (101) | |
| Hydrolases | (102) | |
| Cutinases | (103) | |
| Arylsulfatases | (104) | |
| Anti-HER2 DARPins | (105) | |
| (106) | ||
| Ligase ribozymes | (107) | |
| Meganucleases | (108) | |
| Ligases | (109) | |
| Screening and selection | Zinc finger proteins | (110) |
| DNA-nuclease inhibitors | (111) | |
| Ribozymes | (112) | |
| RNA polymerases | (113) | |
| Restriction endonucleases | (114) | |
| RNA-binding proteins | (115) | |
| Streptavidin variants | (116) | |
| Bacteriophage λ integrases | (117) | |
| Hydrogen peroxide generating enzymes | (118) | |
| T7 promoter variants | (119) | |
| Esterases | (120) |
Liposome display
The previous in vitro display methodologies are useful for the evolution, screening and selection of soluble proteins and peptides. However, these techniques are not applicable to proteins that require integration in lipid bilayer membranes. To address the challenge, liposome display is designed for the evolution and selection of membrane proteins (86) (Figure 5C). In this approach, membrane proteins of interest are displayed on the surface of liposome membranes after being translated from single DNA molecules using encapsulated PURE cell-free translation system (121). The PURE system is selected for this approach due to its lack of membrane proteins that may interfere with the activity of the membrane protein of interest. After expression and display, the membrane proteins embedded in the liposomes fold into their functional forms, and the activity of these proteins can then be quantified with a fluorescent indicator. This methodology uses FACS to select the liposomes that display highly functional proteins correlated with a higher fluorescence emission. The sorted liposomes are then lysed to obtain the DNA fragments encoding the highly functional membrane proteins. The authors used this method for the evolution of pore-forming activity of the protein α-hemolysin from Staphylococcus aureus. After 20 rounds of selection using liposome display, they obtained a mutant version with 30-fold higher pore-forming activity than the wild-type. A recent improvement of this methodology incorporates the Sec translocon into the liposome display technique to increase the membrane integration efficiency and to expand the range of membrane proteins that can be inserted using this technology (122). Liposome display has been used for the in vitro evolution of a mutant pyrrolysyl-tRNA synthetase with improved N-benzyloxycarbonyl-l-lysine incorporation activity (123), and for the evolution of alpha-hemolysin with increased nano-pore activity (124).
Conclusions and visions
We review the high-throughput screening, selection and evolution of biomolecules that use cell-free transcription/translation systems, with special emphasis on how these techniques have been developed, improved and applied. The most recent in vitro display and microarray approaches have focused on increasing the range of applications and the screening of more complex biomolecules, such as membrane proteins and proteins that require multiple disulfide bonds. High-throughput screening of biomolecules using cell-free transcription and translation systems may be improved based on advances in several foundational technologies, including the development of better data analysis tools, the creation of new and improved cell-free protein expression systems (125), the advancement in automation of the screening processes (23) and further miniaturization to single molecule level that can still yield a detectable signal.
The vast majority of high-throughput cell-free screening methods strongly relies on the use of FACS or confocal optical instruments for the detection of the biomolecules of interest. However, the use of such equipment in high-throughput screening assays represents a critical bottleneck because they select a few positive hits among a large number of negative results. The use of specialized equipment also slows down the widespread adaptation of these technologies in laboratories with no or very limited access to the equipment. The challenges, if overcome, will speed up the screening procedure and allow for the exploration of larger candidate library in a shorter time than current screening technology. We anticipate the surge of innovative approaches in the near future that will surpass the restriction imposed by screening on 2D surfaces. These new approaches will perform ultra-high-throughput screening assays that exhibit novel characteristics such as autonomous selection, self-amplification of the DNA encoding the biomolecule of interest and a throughput limited only by the size of the library instead of the choice of screening assay.
Acknowledgments
We appreciate the discussion of the manuscript with members of Tan Lab.
Funding
Society-in-Science: Branco-Weiss Fellowship and Human Frontier Science Program (RGY0080/2015 to C.T.); UC MEXUS-CONACYT doctoral fellowship to L.E.C.-L.
References
- 1. Sundberg S.A. (2000) High-throughput and ultra-high-throughput screening: solution- and cell-based approaches. Curr. Opin. Biotechnol., 11, 47–53. [DOI] [PubMed] [Google Scholar]
- 2. Pereira D.A., Williams J.A. (2007) Origin and evolution of high throughput screening. Br. J. Pharmacol., 152, 53–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mayr L.M., Fuerst P. (2008) The future of high-throughput screening. J. Biomol. Screen., 13, 443–448. [DOI] [PubMed] [Google Scholar]
- 4. Mayr L.M., Bojanic D. (2009) Novel trends in high-throughput screening. Curr. Opin. Pharmacol., 9, 580–588. [DOI] [PubMed] [Google Scholar]
- 5. Battersby B.J., Trau M. (2002) Novel miniaturized systems in high-throughput screening. Trends Biotechnol., 20, 167–173. [DOI] [PubMed] [Google Scholar]
- 6. He N., Liu T., Liu B. (2016) Technologies and applications in micro-volume liquid handling. J. Nanosci. Nanotechnol., 16, 58–66. [DOI] [PubMed] [Google Scholar]
- 7. Zhu Y., Fang Q. (2013) Analytical detection techniques for droplet microfluidics—a review. Anal. Chim. Acta, 787, 24–35. [DOI] [PubMed] [Google Scholar]
- 8. Xiao H., Bao Z., Zhao H. (2015) High throughput screening and selection methods for directed enzyme evolution. Ind. Eng. Chem. Res., 54, 4011–4020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lin J.L., Wagner J.M., Alper H.S. (2017) Enabling tools for high-throughput detection of metabolites: metabolic engineering and directed evolution applications. Biotechnol. Adv., 35, 950–970. [DOI] [PubMed] [Google Scholar]
- 10. Gubler H. (2016). High-throughput screening data analysis In: Zhang L. (ed). Nonclinical Statistics for Pharmaceutical and Biotechnology Industries. Springer International Publishing, Cham, pp. 83–139. [Google Scholar]
- 11. Wildey M.J., Haunso A., Tudor M., Webb M., Connick J.H. (2017) High-throughput screening. In: Goodnow R (ed.) Platform Technologies in Drug Discovery Validation, First edn, Vol. 50, Academic Press, Cambridge, Massachusetts, 2017, pp. 149–195.
- 12. Prevel C., Kurzawa L., Van T.N., Morris M.C. (2014) Fluorescent biosensors for drug discovery new tools for old targets—screening for inhibitors of cyclin-dependent kinases. Eur. J. Med. Chem., 88, 74–88. [DOI] [PubMed] [Google Scholar]
- 13. Packer M.S., Liu D.R. (2015) Methods for the directed evolution of proteins. Nat. Rev. Genet., 16, 379–394. [DOI] [PubMed] [Google Scholar]
- 14. Longwell C.K., Labanieh L., Cochran J.R. (2017) High-throughput screening technologies for enzyme engineering. Curr. Opin. Biotechnol., 48, 196–202. [DOI] [PubMed] [Google Scholar]
- 15. Inglese J., Johnson R.L., Simeonov A., Xia M., Zheng W., Austin C.P., Auld D.S. (2007) High-throughput screening assays for the identification of chemical probes. Nat. Chem. Biol., 3, 466–479. [DOI] [PubMed] [Google Scholar]
- 16. Janzen W.P. (2014) Screening technologies for small molecule discovery: the state of the art. Chem. Biol., 21, 1162–1170. [DOI] [PubMed] [Google Scholar]
- 17. Doyle S.K., Pop M.S., Evans H.L., Koehler A.N. (2016) Advances in discovering small molecules to probe protein function in a systems context. Curr. Opin. Chem. Biol., 30, 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Biniarz P., Łukaszewicz M., Janek T. (2017) Screening concepts, characterization and structural analysis of microbial-derived bioactive lipopeptides: a review. Crit. Rev. Biotechnol., 37, 393–410. [DOI] [PubMed] [Google Scholar]
- 19. Sawasaki T., Ogasawara T., Morishita R., Endo Y. (2002) A cell-free protein synthesis system for high-throughput proteomics. Proc. Natl. Acad. Sci. USA, 99, 14652–14657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rosenblum G., Cooperman B.S. (2014) Engine out of the chassis: cell-free protein synthesis and its uses. FEBS Lett., 588, 261–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zemella A., Thoring L., Hoffmeister C., Kubick S. (2015) Cell-free protein synthesis: pros and cons of prokaryotic and eukaryotic systems. Chembiochem, 16, 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kanter G., Yang J., Voloshin A., Levy S., Swartz J.R., Levy R. (2007) Cell-free production of scFv fusion proteins: an efficient approach for personalized lymphoma vaccines. Blood, 109, 3393–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fan J., Villarreal F., Weyers B., Ding Y., Tseng K.H., Li J., Li B., Tan C., Pan T. (2017) Multi-dimensional studies of synthetic genetic promoters enabled by microfluidic impact printing. Lab Chip, 17, 2198–2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kigawa T., Yabuki T., Matsuda N., Matsuda T., Nakajima R., Tanaka A., Yokoyama S. (2004) Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J. Struct. Funct. Genomics, 5, 63–68. [DOI] [PubMed] [Google Scholar]
- 25. Katzen F., Chang G., Kudlicki W. (2005) The past, present and future of cell-free protein synthesis. Trends Biotechnol., 23, 150–156. [DOI] [PubMed] [Google Scholar]
- 26. Carlson E.D., Gan R., Hodgman C.E., Jewett M.C. (2012) Cell-free protein synthesis: applications come of age. Biotechnol. Adv., 30, 1185–1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dower W.J., Mattheakis L.C. (2002) In vitro selection as a powerful tool for the applied evolution of proteins and peptides. Curr. Opin. Chem. Biol., 6, 390–398. [DOI] [PubMed] [Google Scholar]
- 28. Chandra H., Srivastava S. (2010) Cell-free synthesis-based protein microarrays and their applications. Proteomics, 10, 717–730. [DOI] [PubMed] [Google Scholar]
- 29. Kilb N., Burger J., Roth G. (2014) Protein microarray generation by in situ protein expression from template DNA. Eng. Life Sci., 14, 352–364. [Google Scholar]
- 30. Hanes J., Plückthun A. (1997) In vitro selection and evolution of functional proteins by using ribosome display. Proc. Natl. Acad. Sci. USA, 94, 4937–4942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roberts R.W., Szostak J.W. (1997) RNA-peptide fusions for the in vitro selection of peptides and proteins. Proc. Natl. Acad. Sci. USA, 94, 12297–12302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Odegrip R., Coomber D., Eldridge B., Hederer R., Kuhlman P.A., Ullman C., FitzGerald K., McGregor D. (2004) CIS display: in vitro selection of peptides from libraries of protein-DNA complexes. Proc. Natl. Acad. Sci. USA, 101, 2806–2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yamaguchi J., Naimuddin M., Biyani M., Sasaki T., Machida M., Kubo T., Funatsu T., Husimi Y., Nemoto N. (2009) cDNA display: a novel screening method for functional disulfide-rich peptides by solid-phase synthesis and stabilization of mRNA-protein fusions. Nucleic Acids Res., 37, e108.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mattheakis L.C., Bhatt R.R., Dower W.J. (1994) An in vitro polysome display system for identifying ligands from very large peptide libraries. Proc. Natl. Acad. Sci. USA, 91, 9022–9026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zahnd C., Amstutz P., Pluckthun A. (2007) Ribosome display: selecting and evolving proteins in vitro that specifically bind to a target. Nat. Methods, 4, 269–279. [DOI] [PubMed] [Google Scholar]
- 36. Gersuk G.M., Corey M.J., Corey E., Stray J.E., Kawasaki G.H., Vessella R.L. (1997) High-affinity peptide ligands to prostate-specific antigen identified by polysome selection. Biochem. Biophys. Res. Commun., 232, 578–582. [DOI] [PubMed] [Google Scholar]
- 37. Lamla T., Erdmann V.A. (2003) Searching sequence space for high-affinity binding peptides using ribosome display. J. Mol. Biol., 329, 381–388. [DOI] [PubMed] [Google Scholar]
- 38. Yau K.Y.F., Groves M.A.T., Li S., Sheedy C., Lee H., Tanha J., MacKenzie C.R., Jermutus L., Hall J.C. (2003) Selection of hapten-specific single-domain antibodies from a non-immunized llama ribosome display library. J. Immunol. Methods, 281, 161–175. [DOI] [PubMed] [Google Scholar]
- 39. Gold L. (2001) mRNA display: diversity matters during in vitro selection. Proc. Natl. Acad. Sci. USA, 98, 4825–4826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Xu L., Aha P., Gu K., Kuimelis R.G., Kurz M., Lam T., Lim A.C., Liu H., Lohse P.A., Sun L., Weng S., Wagner R.W., Lipovsek D. (2002) Directed evolution of high-affinity antibody mimics using mRNA display. Chem. Biol., 9, 933–942. [DOI] [PubMed] [Google Scholar]
- 41. Fukuda I., Kojoh K., Tabata N., Doi N., Takashima H., Miyamoto-Sato E., Yanagawa H. (2006) In vitro evolution of single-chain antibodies using mRNA display. Nucleic Acids Res., 34, e127.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wilson D.S., Keefe A.D., Szostak J.W. (2001) The use of mRNA display to select high-affinity protein-binding peptides. Proc. Natl. Acad. Sci. USA, 98, 3750–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. McPherson M., Yang Y., Hammond P.W., Kreider B.L. (2002) Drug receptor identification from multiple tissues using cellular-derived mRNA display libraries. Chem. Biol., 9, 691–698. [DOI] [PubMed] [Google Scholar]
- 44. Takahashi T.T., Austin R.J., Roberts R.W. (2003) mRNA display: ligand discovery, interaction analysis and beyond. Trends Biochem. Sci., 28, 159–165. [DOI] [PubMed] [Google Scholar]
- 45. Lipovsek D., Pluckthun A. (2004) In-vitro protein evolution by ribosome display and mRNA display. J. Immunol. Methods, 290, 51–67. [DOI] [PubMed] [Google Scholar]
- 46. Wang H., Liu R. (2011) Advantages of mRNA display selections over other selection techniques for investigation of protein-protein interactions. Expert Rev. Proteomics, 8, 335–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Patel S., Mathonet P., Jaulent A.M., Ullman C.G. (2013) Selection of a high-affinity WW domain against the extracellular region of VEGF receptor isoform-2 from a combinatorial library using CIS display. Protein Eng. Des. Sel., 26, 307–315. [DOI] [PubMed] [Google Scholar]
- 48. Mathonet P., Ioannou A., Betley J., Ullman C. (2011) CIS display, a DNA-based in vitro selection technology for therapeutic peptides. Chim. Oggi-Chem. Today, 29, 10–12. [Google Scholar]
- 49. Ueno S., Yoshida S., Mondal A., Nishina K., Koyama M., Sakata I., Miura K., Hayashi Y., Nemoto N., Nishigaki K., Sakai T. (2012) In vitro selection of a peptide antagonist of growth hormone secretagogue receptor using cDNA display. Proc. Natl. Acad. Sci. USA, 109, 11121–11126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mochizuki Y., Biyani M., Tsuji-Ueno S., Suzuki M., Nishigaki K., Husimi Y., Nemoto N. (2011) One-pot preparation of mRNA/cDNA display by a novel and versatile puromycin-linker DNA. ACS Comb. Sci., 13, 478–485. [DOI] [PubMed] [Google Scholar]
- 51. Ueno S., Kimura S., Ichiki T., Nemoto N. (2012) Improvement of a puromycin-linker to extend the selection target varieties in cDNA display method. J. Biotechnol., 162, 299–302. [DOI] [PubMed] [Google Scholar]
- 52. Mochizuki Y., Kohno F., Nishigaki K., Nemoto N. (2013) A pull-down method with a biotinylated bait protein prepared by cell-free translation using a puromycin linker. Anal. Biochem., 434, 93–95. [DOI] [PubMed] [Google Scholar]
- 53. Mochizuki Y., Suzuki T., Fujimoto K., Nemoto N. (2015) A versatile puromycin-linker using cnvK for high-throughput in vitro selection by cDNA display. J. Biotechnol., 212, 174–180. [DOI] [PubMed] [Google Scholar]
- 54. Yokoyama S., Hirota H., Kigawa T., Yabuki T., Shirouzu M., Terada T., Ito Y., Matsuo Y., Kuroda Y., Nishimura Y., Kyogoku Y., Miki K., Masui R., Kuramitsu S. (2000) Structural genomics projects in Japan. Nat. Struct. Biol., 7(Suppl), 943–945. [DOI] [PubMed] [Google Scholar]
- 55. Shih Y.P., Kung W.M., Chen J.C., Yeh C.H., Wang A.H., Wang T.F. (2002) High-throughput screening of soluble recombinant proteins. Protein Sci., 11, 1714–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shin J., Jardine P., Noireaux V. (2012) Genome replication, synthesis, and assembly of the bacteriophage T7 in a single cell-free reaction. ACS Synth. Biol., 1, 408–413. [DOI] [PubMed] [Google Scholar]
- 57. Hu S., Xie Z., Qian J., Blackshaw S., Zhu H. (2011) Functional protein microarray technology. Wiley Interdiscip. Rev. Syst. Biol. Med., 3, 255–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. He M., Taussig M.J. (2001) Single step generation of protein arrays from DNA by cell-free expression and in situ immobilisation (PISA method). Nucleic Acids Res., 29, 73e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ueno S., Biyani M., Sato S., Raj Kumal S., Kuramochi H., Akagi T., Ichiki T. (2015) Present status and trends in development of high density microarray. Bunseki Kagaku, 64, 421–429. [Google Scholar]
- 60. He M., Stoevesandt O., Palmer E.A., Khan F., Ericsson O., Taussig M.J. (2008) Printing protein arrays from DNA arrays. Nat. Methods, 5, 175–177. [DOI] [PubMed] [Google Scholar]
- 61. Biyani M., Moriyasu J., Tanaka Y., Sato S., Ueno S., Ichiki T. (2013) Microintaglio printing of in situ synthesized proteins enables rapid printing of high-density protein microarrays directly from DNA microarrays. Appl. Phys. Express, 6, 087001. [Google Scholar]
- 62. He M., Taussig M.J. (2003) DiscernArray™ technology: a cell-free method for the generation of protein arrays from PCR DNA. J. Immunol. Methods, 274, 265–270. [DOI] [PubMed] [Google Scholar]
- 63. Angenendt P., Nyarsik L., Szaflarski W., Glökler J., Nierhaus K.H., Lehrach H., Cahill D.J., Lueking A. (2004) Cell-free protein expression and functional assay in nanowell chip format. Anal. Chem., 76, 1844–1849. [DOI] [PubMed] [Google Scholar]
- 64. Ramachandran N., Hainsworth E., Bhullar B., Eisenstein S., Rosen B., Lau A.Y., Walter J.C., LaBaer J. (2004) Self-assembling protein microarrays. Science, 305, 86–90. [DOI] [PubMed] [Google Scholar]
- 65. Tao S.-C., Zhu H. (2006) Protein chip fabrication by capture of nascent polypeptides. Nat. Biotechnol., 24, 1253–1254. [DOI] [PubMed] [Google Scholar]
- 66. Ramachandran N., Raphael J.V., Hainsworth E., Demirkan G., Fuentes M.G., Rolfs A., Hu Y., LaBaer J. (2008) Next-generation high-density self-assembling functional protein arrays. Nat. Methods, 5, 535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Anderson K.S., Ramachandran N., Wong J., Raphael J.V., Hainsworth E., Demirkan G., Cramer D., Aronzon D., Hodi F.S., Harris L., Logvinenko T., LaBaer J. (2008) Application of protein microarrays for multiplexed detection of antibodies to tumor antigens in breast cancer. J. Proteome Res., 7, 1490–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Miersch S., Bian X., Wallstrom G., Sibani S., Logvinenko T., Wasserfall C.H., Schatz D., Atkinson M., Qiu J., LaBaer J. (2013) Serological autoantibody profiling of type 1 diabetes by protein arrays. J. Proteomics, 94, 486–496. [DOI] [PubMed] [Google Scholar]
- 69. Gibson D.S., Qiu J., Mendoza E.A., Barker K., Rooney M.E., LaBaer J. (2012) Circulating and synovial antibody profiling of juvenile arthritis patients by nucleic acid programmable protein arrays. Arthritis Res. Ther., 14, R77.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ramachandran N., Srivastava S., Labaer J. (2008) Applications of protein microarrays for biomarker discovery. Proteomics Clin. Appl., 2, 1444–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Diez P., Gonzalez-Gonzalez M., Lourido L., Degano R.M., Ibarrola N., Casado-Vela J., LaBaer J., Fuentes M. (2015) NAPPA as a real new method for protein microarray generation. Microarrays (Basel), 4, 214–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Angenendt P., Glökler J., Konthur Z., Lehrach H., Cahill D.J. (2003) 3D protein microarrays: performing multiplex immunoassays on a single chip. Anal. Chem., 75, 4368–4372. [DOI] [PubMed] [Google Scholar]
- 73. Angenendt P., Kreutzberger J., Glokler J., Hoheisel J.D. (2006) Generation of high density protein microarrays by cell-free in situ expression of unpurified PCR products. Mol. Cell Proteomics, 5, 1658–1666. [DOI] [PubMed] [Google Scholar]
- 74. Konthur Z., Wilde J. (2010). Evaluation of recombinant antibodies on protein microarrays applying the multiple spotting technique In: Kontermann R, Dübel S (eds). Antibody Engineering. Springer, Berlin. [Google Scholar]
- 75. Stoevesandt O., Vetter M., Kastelic D., Palmer E.A., He M., Taussig M.J. (2011) Cell free expression put on the spot: advances in repeatable protein arraying from DNA (DAPA). N. Biotechnol., 28, 282–290. [DOI] [PubMed] [Google Scholar]
- 76. Schmidt R., Cook E.A., Kastelic D., Taussig M.J., Stoevesandt O. (2013) Optimised ′on demand′ protein arraying from DNA by cell free expression with the ′DNA to Protein Array′ (DAPA) technology. J. Proteomics, 88, 141–148. [DOI] [PubMed] [Google Scholar]
- 77. Biyani M., Osawa T., Nemoto N., Ichiki T. (2011) Microintaglio printing of biomolecules and its application to in situ production of messenger ribonucleic acid display microarray. Appl. Phys. Express, 4, 047001. [Google Scholar]
- 78. Biyani M., Tanaka Y., Sato S., Ueno S., Ichiki T. (2014) Evaluation of poly(dimethylsiloxane) microreactors for pattern size miniaturization of microintaglio-printing-based protein microarray. Jpn. J. Appl. Phys., 53, 06JL04. [Google Scholar]
- 79. Kobayashi R., Biyani M., Ueno S., Kumal S.R., Kuramochi H., Ichiki T. (2015) Temperature-controlled microintaglio printing for high-resolution micropatterning of RNA molecules. Biosens. Bioelectron., 67, 115–120. [DOI] [PubMed] [Google Scholar]
- 80. Kim S.H., Yoshizawa S., Takeuchi S., Fujii T., Fourmy D. (2013) Ultra-high density protein spots achieved by on chip digitalized protein synthesis. Analyst, 138, 4663–4669. [DOI] [PubMed] [Google Scholar]
- 81. Buxboim A., Bar-Dagan M., Frydman V., Zbaida D., Morpurgo M., Bar-Ziv R. (2007) A single-step photolithographic interface for cell-free gene expression and active biochips. Small, 3, 500–510. [DOI] [PubMed] [Google Scholar]
- 82. Bar M., Bar-Ziv R.H. (2009) Spatially resolved DNA brushes on a chip: gene activation by enzymatic cascade. Nano Lett., 9, 4462–4466. [DOI] [PubMed] [Google Scholar]
- 83. Heyman Y., Buxboim A., Wolf S.G., Daube S.S., Bar-Ziv R.H. (2012) Cell-free protein synthesis and assembly on a biochip. Nat. Nanotechnol., 7, 374–378. [DOI] [PubMed] [Google Scholar]
- 84. Tawfik D.S., Griffiths A.D. (1998) Man-made cell-like compartments for molecular evolution. Nat. Biotechnol., 16, 652–656. [DOI] [PubMed] [Google Scholar]
- 85. Bernath K., Hai M., Mastrobattista E., Griffiths A.D., Magdassi S., Tawfik D.S. (2004) in vitro compartmentalization by double emulsions: sorting and gene enrichment by fluorescence activated cell sorting. Anal. Biochem., 325, 151–157. [DOI] [PubMed] [Google Scholar]
- 86. Fujii S., Matsuura T., Sunami T., Nishikawa T., Kazuta Y., Yomo T. (2014) Liposome display for in vitro selection and evolution of membrane proteins. Nat. Protoc., 9, 1578–1591. [DOI] [PubMed] [Google Scholar]
- 87. Doi N., Yanagawa H. (1999) STABLE: protein-DNA fusion system for screening of combinatorial protein libraries in vitro. FEBS Lett., 457, 227–230. [DOI] [PubMed] [Google Scholar]
- 88. Yonezawa M. (2003) DNA display for in vitro selection of diverse peptide libraries. Nucleic Acids Res., 31, 118e–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Ghadessy F.J., Ong J.L., Holliger P. (2001) Directed evolution of polymerase function by compartmentalized self-replication. Proc. Natl. Acad. Sci. USA, 98, 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Sepp A., Tawfik D.S., Griffiths A.D. (2002) Microbead display by in vitro compartmentalisation: selection for binding using flow cytometry. FEBS Lett., 532, 455–458. [DOI] [PubMed] [Google Scholar]
- 91. Levy M., Griswold K.E., Ellington A.D. (2005) Direct selection of trans-acting ligase ribozymes by in vitro compartmentalization. RNA, 11, 1555–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kelly B.T., Griffiths A.D. (2007) Selective gene amplification. Protein Eng. Des. Sel., 20, 577–581. [DOI] [PubMed] [Google Scholar]
- 93. Stein V., Sielaff I., Johnsson K., Hollfelder F. (2007) A covalent chemical genotype-phenotype linkage for in vitro protein evolution. Chembiochem, 8, 2191–2194. [DOI] [PubMed] [Google Scholar]
- 94. Mastrobattista E., Taly V., Chanudet E., Treacy P., Kelly B.T., Griffiths A.D. (2005) High-throughput screening of enzyme libraries: in vitro evolution of a β-galactosidase by fluorescence-activated sorting of double emulsions. Chem. Biol., 12, 1291–1300. [DOI] [PubMed] [Google Scholar]
- 95. Fen C.X., Coomber D.W., Lane D.P., Ghadessy F.J. (2007) Directed evolution of p53 variants with altered DNA-binding specificities by in vitro compartmentalization. J. Mol. Biol., 371, 1238–1248. [DOI] [PubMed] [Google Scholar]
- 96. Hardiman E., Gibbs M., Reeves R., Bergquist P. (2010) Directed evolution of a thermophilic beta-glucosidase for cellulosic bioethanol production. Appl. Biochem. Biotechnol., 161, 301–312. [DOI] [PubMed] [Google Scholar]
- 97. Korfer G., Pitzler C., Vojcic L., Martinez R., Schwaneberg U. (2016) In vitro flow cytometry-based screening platform for cellulase engineering. Sci. Rep., 6, 26128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Ostafe R., Prodanovic R., Commandeur U., Fischer R. (2013) Flow cytometry-based ultra-high-throughput screening assay for cellulase activity. Anal. Biochem., 435, 93–98. [DOI] [PubMed] [Google Scholar]
- 99. Ostafe R., Prodanovic R., Nazor J., Fischer R. (2014) Ultra-high-throughput screening method for the directed evolution of glucose oxidase. Chem. Biol., 21, 414–421. [DOI] [PubMed] [Google Scholar]
- 100. Stapleton J.A., Swartz J.R. (2010) Development of an in vitro compartmentalization screen for high-throughput directed evolution of [FeFe] hydrogenases. PLoS One, 5, e15275.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Tu R., Martinez R., Prodanovic R., Klein M., Schwaneberg U. (2011) A flow cytometry-based screening system for directed evolution of proteases. J. Biomol. Screen., 16, 285–294. [DOI] [PubMed] [Google Scholar]
- 102. Gupta R.D., Goldsmith M., Ashani Y., Simo Y., Mullokandov G., Bar H., Ben-David M., Leader H., Margalit R., Silman I., Sussman J.L., Tawfik D.S. (2011) Directed evolution of hydrolases for prevention of G-type nerve agent intoxication. Nat. Chem. Biol., 7, 120–125. [DOI] [PubMed] [Google Scholar]
- 103. Hwang B.-Y. (2012) Directed evolution of cutinase using in vitro compartmentalization. Biotechnol. Bioprocess Eng., 17, 500–505. [Google Scholar]
- 104. Kintses B., Hein C., Mohamed M.F., Fischlechner M., Courtois F., Laine C., Hollfelder F. (2012) Picoliter cell lysate assays in microfluidic droplet compartments for directed enzyme evolution. Chem. Biol., 19, 1001–1009. [DOI] [PubMed] [Google Scholar]
- 105. Houlihan G., Gatti-Lafranconi P., Kaltenbach M., Lowe D., Hollfelder F. (2014) An experimental framework for improved selection of binding proteins using SNAP display. J. Immunol. Methods, 405, 47–56. [DOI] [PubMed] [Google Scholar]
- 106. Houlihan G., Gatti-Lafranconi P., Lowe D., Hollfelder F. (2015) Directed evolution of anti-HER2 DARPins by SNAP display reveals stability/function trade-offs in the selection process. Protein Eng. Des. Sel., 28, 269–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Petrie K.L., Joyce G.F. (2014) Limits of neutral drift: lessons from the in vitro evolution of two ribozymes. J. Mol. Evol., 79, 75–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Takeuchi R., Choi M., Stoddard B.L. (2014) Redesign of extensive protein-DNA interfaces of meganucleases using iterative cycles of in vitro compartmentalization. Proc. Natl. Acad. Sci. USA, 111, 4061–4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Gianella P., Snapp E.L., Levy M. (2016) An in vitro compartmentalization-based method for the selection of bond-forming enzymes from large libraries. Biotechnol. Bioeng., 113, 1647–1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Sepp A., Choo Y. (2005) Cell-free selection of zinc finger DNA-binding proteins using in vitro compartmentalization. J. Mol. Biol., 354, 212–219. [DOI] [PubMed] [Google Scholar]
- 111. Bernath K., Magdassi S., Tawfik D.S. (2005) Directed evolution of protein inhibitors of DNA-nucleases by in vitro compartmentalization (IVC) and nano-droplet delivery. J. Mol. Biol., 345, 1015–1026. [DOI] [PubMed] [Google Scholar]
- 112. Agresti J.J., Kelly B.T., Jäschke A., Griffiths A.D. (2005) Selection of ribozymes that catalyse multiple-turnover Diels–Alder cycloadditions by using in vitro compartmentalization. Proc. Natl. Acad. Sci. USA, 102, 16170–16175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Zaher H.S., Unrau P.J. (2007) Selection of an improved RNA polymerase ribozyme with superior extension and fidelity. RNA, 13, 1017–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Zheng Y., Roberts R.J. (2007) Selection of restriction endonucleases using artificial cells. Nucleic Acids Res., 35, e83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Chen Y., Mandic J., Varani G. (2008) Cell-free selection of RNA-binding proteins using in vitro compartmentalization. Nucleic Acids Res., 36, e128.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Levy M., Ellington A.D. (2008) Directed evolution of streptavidin variants using in vitro compartmentalization. Chem. Biol., 15, 979–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tay Y., Ho C., Drőge P., Ghadessy F.J. (2010) Selection of bacteriophage λ integrases with altered recombination specificity by in vitro compartmentalization. Nucleic Acids Res., 38, e25.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Prodanovic R., Ostafe R., Blanusa M., Schwaneberg U. (2012) Vanadium bromoperoxidase-coupled fluorescent assay for flow cytometry sorting of glucose oxidase gene libraries in double emulsions. Anal. Bioanal. Chem., 404, 1439–1447. [DOI] [PubMed] [Google Scholar]
- 119. Paul S., Stang A., Lennartz K., Tenbusch M., Uberla K. (2013) Selection of a T7 promoter mutant with enhanced in vitro activity by a novel multi-copy bead display approach for in vitro evolution. Nucleic Acids Res., 41, e29.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Ma F., Xie Y., Huang C., Feng Y., Yang G. (2014) An improved single cell ultrahigh throughput screening method based on in vitro compartmentalization. PLoS One, 9, e89785.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Shimizu Y., Inoue a., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K., Ueda T. (2001) Cell-free translation reconstituted with purified components. Nat. Biotechnol., 19, 751–755. [DOI] [PubMed] [Google Scholar]
- 122. Ohta N., Kato Y., Watanabe H., Mori H., Matsuura T. (2016) In vitro membrane protein synthesis inside Sec translocon-reconstituted cell-sized liposomes. Sci. Rep., 6, 36466.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Uyeda A., Watanabe T., Kato Y., Watanabe H., Yomo T., Hohsaka T., Matsuura T. (2015) Liposome-based in vitro evolution of aminoacyl-tRNA synthetase for enhanced pyrrolysine derivative incorporation. Chembiochem, 16, 1797–1802. [DOI] [PubMed] [Google Scholar]
- 124. Fujii S., Matsuura T., Yomo T. (2015) In vitro directed evolution of alpha-hemolysin by liposome display. Biophysics (Nagoya-Shi), 11, 67–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Villarreal F., Contreras-Llano L.E., Chavez M., Ding Y., Fan J., Pan T., Tan C. (2018) Synthetic microbial consortia enable assembly of pure translation machinery. Nat. Chem. Biol., 14, 29–35. [DOI] [PubMed] [Google Scholar]