Abstract
Within the last 6 years, CRISPR-Cas systems have transitioned from adaptive defense systems in bacteria and archaea to revolutionary genome-editing tools. The resulting CRISPR technologies have driven innovations for treating genetic diseases and eradicating human pests while raising societal questions about gene editing in human germline cells as well as crop plants. Bringing CRISPR into the classroom therefore offers a means to expose students to cutting edge technologies and to promote discussions about ethical questions at the intersection of science and society. However, working with these technologies in a classroom setting has been difficult because typical experiments rely on cellular systems such as bacteria or mammalian cells. We recently reported the use of an E. coli cell-free transcription-translation (TXTL) system that simplifies the demonstration and testing of CRISPR technologies with shorter experiments and limited equipment. Here, we describe three educational modules intended to expose undergraduate students to CRISPR technologies using TXTL. The three sequential modules comprise (i) designing the RNAs that guide DNA targeting, (ii) measuring DNA cleavage activity in TXTL and (iii) testing how mutations to the targeting sequence or RNA backbone impact DNA binding and cleavage. The modules include detailed protocols, questions for group discussions or individual evaluation, and lecture slides to introduce CRISPR and TXTL. We expect these modules to allow students to experience the power and promise of CRISPR technologies in the classroom and to engage with their instructor and peers about the opportunities and potential risks for society.
Keywords: CRISPR, Cas9, education modules, synthetic biology, TXTL
1. Introduction
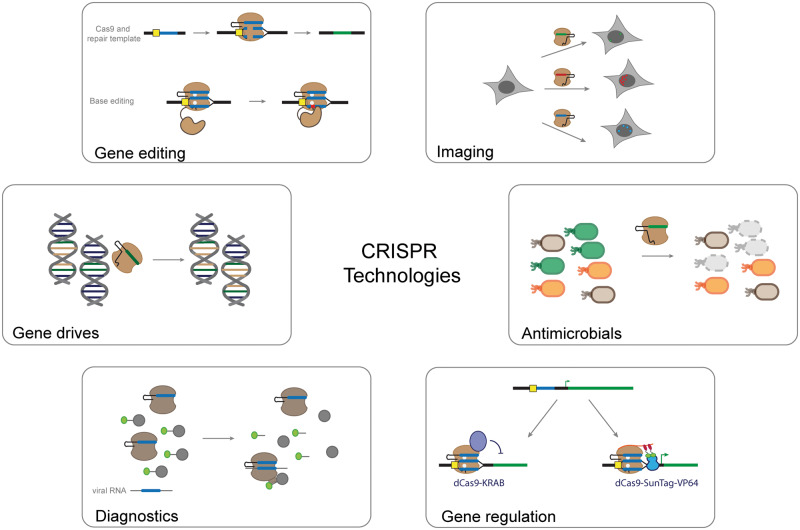
In the past few years, CRISPR-Cas prokaryotic adaptive immune systems have formed the basis of revolutionary technologies in the field of synthetic biology.1 CRISPR-Cas systems have been repurposed for numerous applications, including genome editing, transcriptional regulation, molecular diagnostic tools, epigenetics regulators, antimicrobials and gene drives (Figure 1).
Figure 1.
Overview of CRISPR technologies.
Central components of CRISPR-Cas systems are RNA-guided nucleases that target and cleave a specific DNA or RNA template. There are two targeting rules for any given DNA target: (i) complementarity to the RNA guide and (ii) the presence of a short DNA sequence called the protospacer adjacent motif (PAM) on the target DNA strands. Several classes of CRISPR systems have been discovered that can be used for a growing number of applications based on this molecular mechanism.1 In addition to cleaving DNA, catalytically dead versions of CRISPR enzymes, such as dCas9, have been engineered to be used as a synthetic re-programmable transcription factor in a mechanism called CRISPR interference (CRISPRi), achieving repression of more than two orders-of-magnitude.2 dCas9 binds tightly to a target DNA and inhibits transcription initiation or elongation. Recently, dCas9 was devised to activate transcription,3 thus providing a CRISPR activation (CRISPRa) mechanism counterpart to CRISPRi.
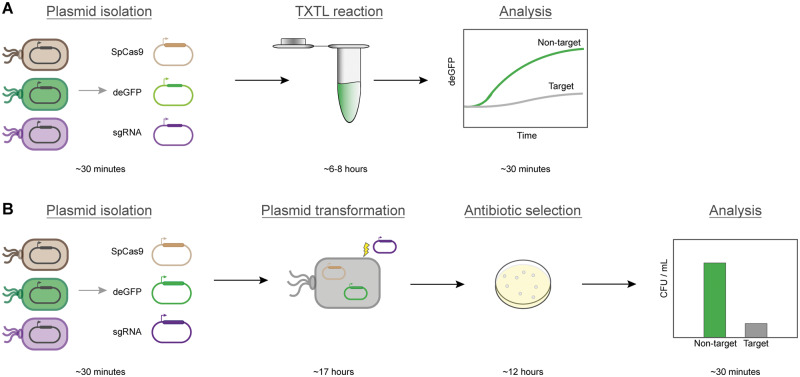
With the ever-growing number of CRISPR-based technologies, it is becoming important for biology-based multidisciplinary researchers, especially at the early stages of their education and careers, to gain a practical and fundamental understanding of CRISPR. However, the educational material on CRISPR technologies has lagged behind research and innovation. Undergraduate students are not typically exposed to the latest developments and research areas in biotechnology. Exposure to the field of synthetic biology through an undergraduate class is an opportunity to reach more students than traditionally possible with summer research programs that have a limited number of spots. Creating educational modules to develop skills on a technology like CRISPR is challenging and even elementary CRISPR experiments performed in vivo are time consuming and require significant laboratory infrastructures. Traditional CRISPR experiments in vivo require laboratory experience and familiarity with advanced techniques such as mammalian cell culture and transfection. Recently, we demonstrated that cell-free transcription-translation (TXTL) can be used to rapidly prototype and characterize CRISPR technologies.4,5 Less than 1 hour of hands-on time is required to add the plasmid components to TXTL reactions. Therefore, the students would be able to setup the TXTL reactions during one class period and receive the results the following day. Because of its fast experiment turnover, TXTL can also be a powerful platform to discover CRISPR both in research and educational capacities. This method is ideal for undergraduate students with little to no experience with complicated laboratory techniques because the students only need to pipet plasmid DNA into the commercially available mastermix. Other cell-free systems, namely the BioBits Explorer6 and BioBits Bright,7 also have been created to make synthetic biology experiments more accessible to educators. TXTL offers a way to carry out simple experiments and get quantitative results in a couple of days, without any large, sophisticated or expensive equipment (Figure 2).
Figure 2.
Schedule of TXTL and in vivo experiments. (A) TXTL schedule totaling approximately one laboratory day. (B) In vivo schedule totaling approximately two laboratory days.
Here, we present three laboratory modules suitable for college-level introductory molecular biology courses, whose participants do not require prior knowledge of CRISPR and TXTL or any wet bench experience. Along with the modules, we provide lecture material to be presented before or at the time of the laboratory modules and a list of the DNA parts that can be easily ordered through Addgene. The modules are designed such that they can be either integrated into an existing course or used for a short, stand-alone course. The modules focus on testing both Cas9 and dCas9 from Streptococcus pyogenes (SpyCas9 and Spy-dCas9, respectively) in TXTL. These modules are the basic sets of experiments that will teach students: the fundamental components of DNA targeting by CRISPR-Cas systems; how to design guide RNAs, perform an off-target analysis, and determine the critical aspects of guide RNA structure; the impact of targeting various locations on a gene; how to use dCas9 for transcriptional repression; and a TXTL system. These modules can be adapted and built upon for more advanced courses to encompass other CRISPR nucleases. Variations of these modules have been successfully implemented in the Synthetic Biology summer course at Cold Spring Harbor Laboratories and in the Biotechnology program at North Carolina State University.
2. Results and discussion
2.1 Previous implementation of TXTL CRISPR modules
Variations of the modules presented here have been successfully implemented in two different contexts. At Cold Spring Harbor Laboratories, the modules to test SpyCas9 and Spy-dCas9 targeting of degfp in TXTL have been incorporated into the 2-week summer course ‘Synthetic Biology’ that enrolled graduate students, postdoctoral researchers, professors and industry scientists. At North Carolina State University, the module to test SpyCas9 targeting degfp in TXTL was incorporated into the ‘Genome Engineering: CRISPR Technologies’ course that enrolled undergraduate and graduate students in the Biotechnology program.
2.2 TXTL CRISPR module description
The material in each module builds on the previous module and should be brought into a laboratory course in sequential order. It is recommended that the introductory lecture material (Introduction I and II) be presented before the modules to familiarize students with the basics of CRISPR-Cas systems and TXTL. The full length protocols, instructor’s manual and associated lecture material can be found in the Supplementary data.
Module 1 provides students the opportunity to design guide RNAs based on the two targeting rules for CRISPR-Cas systems. Within the same module, the students evaluate potential off-target sites for the guides they have designed. Module 2 allows students to explore the necessary components of Cas9 cleavage and the sensitivity to mismatches in the spacer sequence and truncations of the guide RNA handle. It consists of wet laboratory work using TXTL to express deGFP (enhanced green fluorescent protein), SpyCas9 and guide RNAs. Module 3 demonstrates how catalytically dead SpyCas9 (Spy-dCas9) can be used to silence gene expression without cleaving DNA. As a part of this, students express deGFP, Spy-dCas9 and guide RNAs that target various locations on degfp in TXTL. Brief summaries of the modules and protocols are included below while the full protocols can be found in the Supplementary data.
2.3 Module 1: guide RNA design and off-target analysis
Overview. The purpose of this module is to design guide RNAs that can direct Cas proteins to bind and cleave specific DNA sequences. This module introduces students to the fundamental aspects of CRISPR biology, such as its discovery as a prokaryotic adaptive immune system7 and the use of Cas9 as a DNA cutting tool in gene editing.8,9 Other CRISPR-Cas systems are introduced, such as Cas12a/Cpf1 (which will be herein referred to as Cpf1), as they relate to gene editing as well.10 Students are taught the targeting rules for each of the CRISPR nucleases and practice how to design guide RNAs for each. With the designed guides, students learn to evaluate off-target effects.11,12 The ubiquitous S. pyogenes Cas9 is the focus of these modules but students also learn how to apply these design rules to diverse CRISPR-Cas systems.
Learning objectives:
Define common terms, such as CRISPR, guide, guide RNA, target and PAM.
Identify potential target locations for different CRISPR nucleases.
Design guide RNAs for different CRISPR nucleases when given a genetic sequence to target.
Determine potential off-target sites for a given guide sequence.
Materials needed:
Computer with installed web browser, Microsoft word and a PDF viewer.
Introduction I and Module I lecture material.
Brief summary of protocol (full protocol in Supplementary data):
Access segments of genomic DNA from online NCBI tools.
Using the targeting rules (discussed in the lecture material and full protocols), identify the canonical PAMs within the sequence of DNA that could be targeted by the CRISPR nucleases discussed within the protocol.
Using the targeting rules (discussed in the lecture material and full protocols), design guides for each of the Cas effector proteins listed in the protocol.
Identify potential off-target sites for each guide using the off-target analysis tools within the Benchling software.
Reflect on guide RNA design using the Discussion Questions outlined in the full protocol.
2.4 Module 2: targeting degfp with SpyCas9 in TXTL
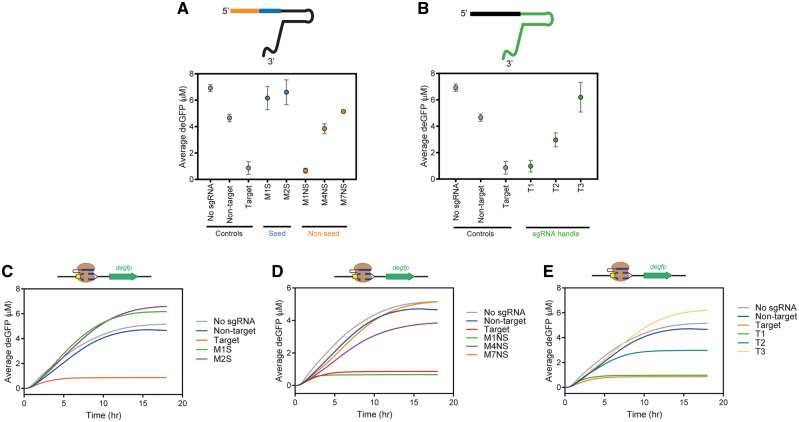
Overview. The purpose of this laboratory is to demonstrate the requirements of the spacer’s complementarity to the target DNA sequence, sgRNA handle and effector protein (SpyCas9) for cleavage of a target DNA sequence.9 This experiment is based on measuring the fluorescence of deGFP in a plate reader over time. In TXTL,13,14 students produce deGFP from a plasmid encoding degfp. Plasmids encoding SpyCas9 and sgRNA target the degfp plasmid.4 Upon cleavage of the target, the linear DNA is degraded and thus deGFP is no longer produced. Students quantify the reduction in deGFP, as compared with negative controls, using fluorescent data from the plate reader. As part of this module, students test the sensitivity of mismatches in the seed and nonseed region of the spacer sequence and corresponding impact on deGFP fluorescence. Furthermore, students test the stability of the sgRNA: Cas9 complex by targeting the degfp gene with sgRNAs that have various truncations in the handle portion of the guide RNA. Expected results and sample data for this module can be seen in Figure 3. Supplementary Table S1 lists options for temperature controlled fluorescence plate readers. If a temperature controlled fluorescence plate reader is unavailable, endpoint TXTL reactions can be qualitatively analyzed by eye; however, because it is difficult to determine small fluorescence differences by eye, a simple ON/OFF may be all that can be concluded. Alternatively, a Nanodrop can be used.
Figure 3.
Sample data for Module 2 where degfp is cleaved by SpyCas9. (A) End point deGFP concentration for mismatches within the spacer sequence. Mismatches in the seed region (shown in blue) increase sequentially from the 3ʹ end of the spacer. Mismatches in the nonseed region (shown in orange) increase sequentially from the 5ʹ end of the spacer. Error bars represent SEM of three independent replicates. (B) End point deGFP concentration for truncations of the sgRNA handle (shown in green). Error bars represent SEM of three independent replicates. The truncations of the handle increase sequentially from the 3ʹ end of the handle. (C) Time course data of the average deGFP concentration (n = 3) for mismatches within the seed region of the spacer. (D) Time course data of the average deGFP concentration (n = 3) for mismatches outside of the seed region of the spacer. (E) Time course data of the average deGFP concentration (n = 3) for truncations within the sgRNA handle. Explanation of trends: Production of deGFP is possible from the expression of the degfp gene off of the intact plasmid. Therefore, if the plasmid is targeted and cleaved by SpyCas9: sgRNA then deGFP will not be produced and accumulate. (A, C, and D) The ‘Target’ sgRNA guided the SpyCas9 to target and cleave the degfp gene resulting in minimal production of deGFP (shown by the low levels of deGFP fluorescence read by the plate reader). The two sgRNAs containing mismatches in the seed region resulting in poor targeting and cleavage by SpyCas9 and therefore deGFP was produced at high levels. Meanwhile, in the nonseed region, two or three mismatches were required for a reduction in SpyCas9’s ability to target and cleave the degfp plasmid. (B and E) Truncation 1, ‘T1’, was still able to guide SpyCas9 to target degfp therefore deGFP was not produced. Increasing the sequence truncated from the 3ʹ end of the sgRNA handle, demonstrated by T2 and T3, resulted in a reduction of SpyCas9’s ability to target and cleave the degfp plasmid.
Learning objectives:
Describe the necessary components for DNA cleavage by the Cas9 nuclease.
Interpret fluorescence data from a plate reader and draw conclusions from a sample set of experimental results.
Briefly describe how SpyCas9 blocks deGFP accumulation in TXTL.
Determine the impact of having mutations within the seed and nonseed regions of the spacer sequence as well as the guide RNA backbone.
Materials needed:
Plate reader capable of temperature control and measuring deGFP fluorescence.
myTXTL Sigma 70 Master Mix (Arbor Biosciences, Cat. #: 507024).
-
Plasmids encoding a sgRNA:
pColE1_sgRNA_NT_AmpR
pColE1_sgRNA_pos6_AmpR
pColE1_sgRNA_M1S_AmpR
pColE1_sgRNA_M2S_AmpR
pColE1_sgRNA_M1NS_AmpR
pColE1_sgRNA_M4NS_AmpR
pColE1_sgRNA_M7NS_AmpR
pColE1_sgRNA_T1_AmpR
pColE1_sgRNA_T2_AmpR
pColE1_sgRNA_T3_AmpR
-
Plasmid encoding SpyCas9:
p15A_SpyCas9_CmR
-
Plasmid encoding deGFP
pColE1_70a_deGFP_AmpR
96 well plate, clear, V-bottom (CLS3357), with caps (CLS3080) or sealing tape (Thermo 232701)
Vortex mixer (VWR, Cat. #: 10153-838; or equivalent)
Centrifuge (Thermo Fisher, Cat. #: 75004241; or equivalent)
Pipets (P10, P20, and P200) and associated pipet tips
ZymoPURE-Express Midi-prep kit (Zymo Research, SKU: D4213; or equivalent)
DNA clean and concentrator kit (Zymo Research, SKU: D4013; or equivalent)
Introduction I, II and Module 2 lecture material
Brief summary of protocol (full protocol in Supplementary data):
Calculate the amount of each plasmid that is needed for each experimental reaction using the TXTL Excel file (included as Supplementary data).
Add deGFP, SpyCas9 and the sgRNA plasmids to the TXTL mastermix to express the components.
Measure the fluorescence output of each reaction over time in the plate reader.
Analyze the data.
2.5 Module 3: targeting different locations in degfp with Spy-dCas9 in TXTL
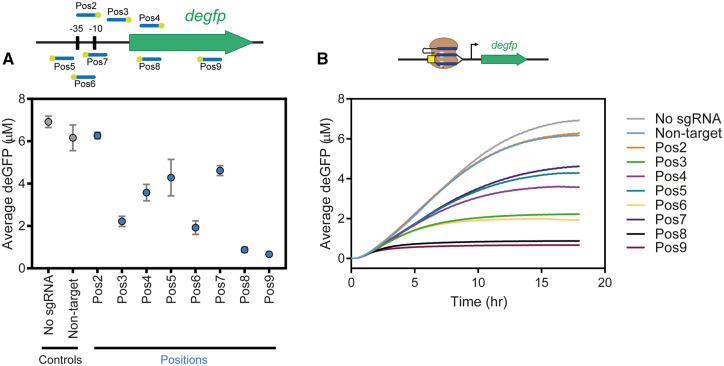
Overview. The purpose of this module is to show that we can achieve gene repression by using the catalytically dead version of SpyCas9 (Spy-dCas9), which tightly binds to, but does not cleave the DNA.2 The bound dCas9 can prevent transcription initiation or elongation, giving a reduction of deGFP production, which we can measure. Sample data for this module can be seen in Figure 4. The experiment also investigates the strength of repression depending on the targeted sequence by testing in vitro targeting in the protomer, 5’ untranslated region, and degfp gene, as well as both the template and nontemplate strands.
Figure 4.
Sample data for Module 3 where the promoter, RBS, and degfp gene are targeted by Spy-dCas9. (A) End point deGFP concentration for various positions being targeted on the degfp plasmid (indicated by blue lines with PAM orientation demonstrated by yellow dots). Error bars represent SEM of three independent replicates. (B) Time course data of the average deGFP concentration (n = 3) for various positions being targeted on the degfp plasmid. Explanation of trends: Production of deGFP is possible from the expression of the degfp gene encoded in the plasmid. Spy-dCas9: sgRNA will target sequences complementary to the guide and remain at the target location without cleaving. Therefore, if the target is near the promoter, RBS or ORF of a gene, then targeting by Spy-dCas9 will block RNA polymerase machinery from expressing the gene. As a result, targeting by Spy-dCas9 will result in reduced deGFP production and accumulation. (A and B) The sgRNAs labeled ‘Pos#’ target in various locations on the degfp plasmid. Pos8 and Pos9 sgRNAs are able to guide Spy-dCas9 to target the degfp plasmid well and result in minimal production of deGFP. Spy-dCas9 targeting by Pos2 sgRNA, however, is weak and there was still an abundant accumulation of deGFP in that sample.
Learning objectives:
Describe mechanistically how Spy-dCas9 blocks deGFP accumulation in TXTL.
Predict the strength of repression for targeting various locations on a sample DNA sequence with annotated gene components (promoter, RBS, UTR, ORF and terminator).
Predict the strength of repression for targeting the template strand versus the nontemplate strand for a sample DNA sequence.
Materials needed:
Plate reader capable of temperature control and measuring deGFP fluorescence.
MyTXTL Sigma 70 Master Mix (Arbor Biosciences, Cat. #: 507024).
-
Plasmids encoding sgRNAs:
pColE1_sgRNA_NT_AmpR
pColE1_sgRNA_pos2_AmpR
pColE1_sgRNA_pos3_AmpR
pColE1_sgRNA_pos4_AmpR
pColE1_sgRNA_pos5_AmpR
pColE1_sgRNA_pos6_AmpR
pColE1_sgRNA_pos7_AmpR
pColE1_sgRNA_pos8_AmpR
pColE1_sgRNA_pos9_AmpR
-
Plasmid encoding Spy-dCas9:
p15A_J23108_SpydCas9_CmR
-
Plasmid encoding deGFP:
pColE1_70a_deGFP_AmpR
96 well plate, clear, V-bottom (CLS3357), with caps (CLS3080) or sealing tape (Thermo 232701)
Vortex mixer (VWR, Cat. #: 10153-838; or equivalent)
Centrifuge (Thermo Fisher, Cat. #: 75004241; or equivalent)
Pipets (P10, P20, and P200) and associated pipet tips
ZymoPURE-Express Midi-prep kit (Zymo Research, SKU: D4213; or equivalent)
DNA clean and concentrator kit (Zymo Research, SKU: D4013; or equivalent)
Introduction I, II, and Module 3 lecture materials
Brief Summary of Protocol (full protocol in Supplementary data):
Calculate the amount of each plasmid that is needed for each experimental reaction using the TXTL Excel file (included as Supplementary data).
Add deGFP, Spy-dCas9 and the sgRNA plasmids to the TXTL mastermix to express the components.
Measure the fluorescence output of each reaction over time in the plate reader.
Analyze the data.
Further exploration. TXTL is a versatile cell-free platform that offers the opportunity for rapid characterization. With the ability to get quantitative results in less than a day, there are many more potential experiments that can be done to investigate CRISPR elements in an educational laboratory setting. Here, we provide a short list of ideas for further experiments if time allows. This list is not limiting, and students should feel encouraged to test their own ideas if they have them.
Varying DNA concentrations: Do we see different repression ratios if we vary the concentrations of Cas9 DNA, sgRNA DNA and reporter DNA? At what concentrations of each component do we saturate cleavage or gene repression?
Testing combinations of sgRNA: Do we achieve greater repression by targeting multiple locations? Is the repression additive? This will depend on the relative concentrations of the sgRNA DNA template.
Preincubating DNA templates: The (d)Cas9 must be expressed from a plasmid before it can cleave or repress. What if we incubate a TXTL reaction with the (d)Cas9 DNA template and the sgRNA DNA template for 2 h before we add the reporter target plasmid? TXTL allows the user to spike in DNAs at any time, and many different experiments can take advantage of this.
3. Perspectives and conclusions
The in vivo educational modules on CRISPR technologies are typically difficult to implement in teaching contexts because of the time required to grow and manipulate cells. Here, we describe modules in a cell-free context to enable instructors to teach students the basics of CRISPR-Cas systems and their use as tools in biotechnology. TXTL is the ideal platform for educational purposes because results are produced quickly and easily. Students will benefit from being able to study more aspects of CRISPR technologies in a shorter amount of time with modules that are suitable for laboratory courses.
Author contributions
D.C., R.M., S.P.C., C.L.B. and V.N. designed the modules. R.M. collected the data presented in the manuscript. D.C. generated the figures. D.C., R.M., C.L.B. and V.N. wrote the manuscript. All authors read and approved the manuscript.
Supplementary Material
Funding
This work was supported by the Defense Advanced Research Projects Agency [Contract HR0011-16-C-01-34 to V.N. and C.L.B.] and the German Research Foundation [405891106 to C.L.B.]. This work was supported and implemented originally at Cold Spring Harbor Laboratory and at the Biotechnology program at North Carolina State University.
Conflict of interest statement. The Noireaux laboratory receives research funds from Arbor Biosciences, a distributor of the myTXTL cell-free protein synthesis kit.
References
- 1. Waller M.C., Bober J.R., Nair N.U., Beisel C.L. (2017) Toward a genetic tool development pipeline for host-associated bacteria. Curr. Opin. Microbiol., 38, 156–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013) Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell, 152, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dong C., Fontana J., Patel A., Carothers J.M., Zalatan J.G. (2018) Synthetic CRISPR-Cas gene activators for transcriptional reprogramming in bacteria. Nat. Commun., 9, 2489.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Marshall R., Maxwell C.S., Collins S.P., Jacobsen T., Luo M.L., Begemann M.B., Gray B.N., January E., Singer A., He Y.. et al. (2018) Rapid and scalable characterization of CRISPR technologies using an E. coli cell-free transcription-translation system. Mol. Cell, 69, 146–157.e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maxwell C.S., Jacobsen T., Marshall R., Noireaux V., Beisel C.L. (2018) A detailed cell-free transcription-translation-based assay to decipher CRISPR protospacer-adjacent motifs. Methods, 143, 48–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang A., Nguyen P.Q., Stark J.C., Takahashi M.K., Donghia N., Ferrante T., Dy A.J., Hsu K.J., Dubner R.S., Pardee K.. et al. (2018) BioBits explorer: a modular synthetic biology education kit. Sci. Adv., 4, eaat5105.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Stark J.C., Huang A., Nguyen P.Q., Dubner R.S., Hsu K.J., Ferrante T.C., Anderson M., Kanapskyte A., Mucha Q., Packett J.S.. et al. (2018) BioBits bright: a fluorescent synthetic biology education kit. Sci. Adv., 4, eaat5107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hsu P.D., Lander E.S., Zhang F. (2014) Development and applications of CRISPR-Cas9 for genome engineering. Cell, 157, 1262–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jinek M., Chylinski K., Fonfara I., Hauer M., Doudna J.A., Charpentier E. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science, 337, 816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zetsche B., Gootenberg J.S., Abudayyeh O.O., Slaymaker I.M., Makarova K.S., Essletzbichler P., Volz S.E., Joung J., van der Oost J., Regev A.. et al. (2015) Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell, 163, 759–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Doench J.G., Fusi N., Sullender M., Hegde M., Vaimberg E.W., Donovan K.F., Smith I., Tothova Z., Wilen C., Orchard R.. et al. (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol., 34, 184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hsu P.D., Scott D.A., Weinstein J.A., Ran F.A., Konermann S., Agarwala V., Li Y., Fine E.J., Wu X., Shalem O.. et al. (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol., 31, 827–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Shin J., Noireaux V. (2012) An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth. Biol., 1, 29–41. [DOI] [PubMed] [Google Scholar]
- 14. Garamella J., Marshall R., Rustad M., Noireaux V. (2016) The all E. coli TX-TL toolbox 2.0: a platform for cell-free synthetic biology. ACS Synth. Biol., 5, 344–355. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.