Abstract
Cyanobacteria are promising ‘low-cost’ cell factories since they have minimal nutritional requirements, high metabolic plasticity and can use sunlight and CO2 as energy and carbon sources. The unicellular Synechocystis sp. PCC 6803, already considered the ‘green’ Escherichia coli, is the best studied cyanobacterium but to be used as an efficient and robust photoautotrophic chassis it requires a customized and well-characterized toolbox. In this context, we evaluated the possibility of using three self-replicative vectors from the Standard European Vector Architecture (SEVA) repository to transform Synechocystis. Our results demonstrated that the presence of the plasmid does not lead to an evident phenotype or hindered Synechocystis growth, being the vast majority of the cells able to retain the replicative plasmid even in the absence of selective pressure. In addition, a set of heterologous and redesigned promoters were characterized exhibiting a wide range of activities compared to the reference PrnpB, three of which could be efficiently repressed. As a proof-of-concept, from the expanded toolbox, one promoter was selected and assembled with the ggpS gene [encoding one of the proteins involved in the synthesis of the native compatible solute glucosylglycerol (GG)] and the synthetic device was introduced into Synechocystis using one of the SEVA plasmids. The presence of this device restored the production of the GG in a ggpS deficient mutant validating the functionality of the tools/device developed in this study.
Keywords: cyanobacteria, promoters, pSEVA plasmids, Synechocystis, synthetic toolbox
1. Introduction
Cyanobacteria are photoautotrophic organisms with simple nutritional requirements due to their ability to use sunlight and CO2 as energy and carbon sources (1). The unicellular Synechocystis sp. PCC 6803 (hereafter Synechocystis) was the first photosynthetic organism to have its genome sequenced (2) and rapidly became the model strain for cyanobacteria. Moreover, the large amount of data generated over the last decades allowed the construction of genome-scale metabolic models (3, 4) leading to this organism being recognized as the ‘green’ Escherichia coli (5). Nevertheless, the vast majority of the available synthetic biology tools have been developed for heterotrophic chassis like E. coli or Saccharomyces cerevisiae (6, 7), and most of the regulatory elements characterized in E. coli function rather poorly or not at all in cyanobacteria (8, 9). Consequently, a considerable effort has been placed on the design and construction of efficient, predictable and easy-to-use molecular tools for cyanobacteria (8, 10–14). Within this context, native promoters such as PpsbA2, PnrsB or the ‘super strong’ PcpcG560 have been previously used to drive gene expression for specialty chemicals production (11, 15, 16). These promoters have the obvious advantage of responding to environmental signals that cells are used to integrate and modulate (17, 18). Nevertheless, the predictability of the system is compromised since these promoters are not insulated from the organism’s regulatory networks (often involving multiple poorly understood control mechanisms), which represents a clear disadvantage for the design of genetic circuits. Chemical-induced promoters have an additional drawback due to the difficulty of removing the inducer from the culture, resulting in a continuous gene control. In metal-induced promoters, the metal can be actively pumped out of the cells, being a restraint when a strong inducible system is required (11). In contrast, light-inducible promoters do not require the addition of a specific molecule but its regulation is confined to specific growth conditions e.g. wavelength and light intensity (12, 19). In this context, the rational design of heterologous promoters has been used as a strategy to increase insulation and obtain more efficient promoters, with several attempts to engineer the well-described systems regulated by TetR or LacI e.g. replacing bases between the -10 box and the transcription start site or changing the distance between -10 and -35 boxes (10, 20). Although some improvements have been achieved there are still limitations for the available systems and room for expanding the cyanobacterial toolbox. The implementation of synthetic devices/circuits into the chassis also presents its challenges. While replicative vectors allow a faster generation/assessment of the mutants, integrative ones ensure long-term stability (21). The availability of replicative vectors for cyanobacterial synthetic biology applications is still scarce (8, 21), while a set of integrative plasmids targeting Synechocystis chromosomal neutral sites were recently developed and validated by us and others (13, 22). Cyanobacteria are promising cell factories that have already been engineered to be used as sustainable platforms to produce a variety of value-added compounds such as ethylene, sugars and biofuels (23–25). Therefore, the development and validation of a cyanobacterial synthetic biology toolbox is of the utmost importance to render processes more efficient and economically viable. With this purpose, we validated the use of three Standard European Vector Architecture (SEVA) replicative plasmids in Synechocystis, developed a novel set of heterologous/redesigned promoters and, as a proof-of-concept, used some of these tools to bypass the native control of the compatible solute glucosylglycerol (GG) synthesis.
2. Materials and methods
2.1 Chemicals and reagents
The chemicals anhydrotetracycline hydrochloride (aTc, Sigma-Aldrich), Isopropyl β-D-1-thiogalactopyranoside (IPTG, lactose analog, BioVectra Inc.), lactose (Merck Millipore), N-(β-ketocaproyl)-L-homoserine lactone (AHL, 3OC6HSL, Santa Cruz Biotech.), and L-arabinose (Sigma-Aldrich) were used. All other chemicals and reagents were purchased from ThermoFisher Scientific or Sigma-Aldrich.
2.2 Organisms and culture conditions
Wild-type and mutants of the cyanobacterium Synechocystis sp. PCC 6803 substrain Kazusa (26, 27) (obtained from the Pasteur Culture Collection, Paris, France) were maintained in Erlenmeyer flasks batch cultures with BG11 medium (28) at 30°C with rotary shaking (100 rpm) under a 12 h light/12 h dark regimen. Light intensity was 20 μE/m2/s in all experiments and cosine-corrected irradiance was measured using a Dual Solar/Electric Quantum Meter (Spectrum Technologies, Inc.). For solid BG11, the medium was supplemented with 1.5% (wt/vol) noble agar (Difco), 0.3% (wt/vol) sodium thiosulfate and 10 mM TES-KOH buffer, pH 8.2. For the selection and maintenance of mutants, BG11 medium was supplemented with kanamycin (Km, 10–500 µg/ml), chloramphenicol (Cm, 10–20 µg/ml) or streptomycin and spectinomycin (Sp and Sm, 1.5 µg/ml each).
For cloning purposes, E. coli strains DH5α (Stratagene) or TOP10 (ThermoFisher Scientific) were used. Cells were grown at 37°C in LB medium (29), supplemented with Amp (100 µg/ml), Km (50 µg/ml), Cm (34 µg/ml) or Sm (50 µg/ml).
2.3 Synthetic promoters design
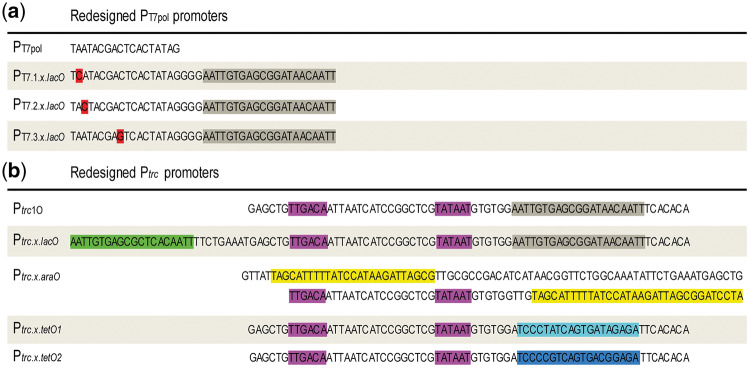
The synthetic T7 promoter variants used in this study (PT7.1.x.lacO, PT7.2.x.lacO and PT7.3.x.lacO) were based on three existing variants of the T7 RNA polymerase promoter sequence containing single nucleotide substitutions—PT7.1, PT7.2 and PT7.3 (30, 31) and on Novagen® pET expression vectors. The existing variants were redesigned including the lacO operator sequence originating the PT7.1.x.lacO, PT7.2.x.lacO and PT7.3.x.lacO promoters (for more details see Figure 5a and Table 1).
Figure 5.
Redesigned promoter sequences. (a) Promoter sequences of PT7.1.x.lacO, PT7.2.x.lacOand PT7.3.x.lacO. The mutations are highlighted in red and the lacO operator in gray. The PT7pol original promoter sequence was included for comparison purposes. (b) Promoter sequences of Ptrc.x.lacO, Ptrc.x.araO, Ptrc.x.tetO1and Ptrc.x.tetO2. The predicted -35 and -10 boxes are highlighted in purple, the lacO operator in gray, the lacOid operator in green, the araO operators in yellow, the tetO operator in light blue and the modified tetO operator in dark blue. The Ptrc1O original sequence was included for comparison purposes.
Table 1.
List of parts used in this study
| Designation | Plasmid | Description | References/sources |
|---|---|---|---|
| PrnpB | pSB1A2 | Promote—reference for promoter strength | (8) |
| PT7.1.x.lacO | pJ201 | Promoter variant 1 based on PT7 with a single mutation and containing a lacO operator | This study |
| PT7.2.x.lacO | pJ201 | Promoter variant 2 based on PT7 with a single mutation and containing a lacO operator | This study |
| PT7.3.x.lacO | pJ201 | Promoter variant 3 based on PT7 with a single mutation and containing a lacO operator | This study |
| PpsbA2* | pJ201 | Promoter based on Synechocystis native PpsbA2 promoter | (13) |
| PtacI | pJ201 | Hybrid promoter containing a lacO operator | (32) |
| Ptrc.x.lacO | pJ201 | Synthetic promoter containing two lacO operators | This study |
| Ptrc.x.araO | pJ201 | Synthetic promoter containing an araO operator | This study |
| Ptrc.x.tetO1 | pJ201 | Synthetic promoter containing a tetO operator | This study |
| Ptrc.x.tetO2 | pJ201 | Synthetic promoter containing a modified tetO operator | This study |
| ggpS | pSB1A2 | Synechocystis’ native ggpS gene | This study |
| PλcI | pSB1A2 | BioBrick promoter | Part: BBa_R0051 |
| PluxR | pSB1A2 | BioBrick promoter | Part: BBa_R0062 |
| PT7pol | pSB1AK8 | BioBrick promoter | Part: BBa_I712074 |
| ParaC | pSB1A2 | BioBrick promoter | Part: BBa_R0080 |
| PBADwt | pSB1A3 | BioBrick promoter | Part: BBa_I13453 |
| GFP generator | pSB1A2 | GFP transcriptional unit (RBS, CDS and double terminator) | Part: BBa_E0240 |
| RBS | pSB1A2 | Ribosome binding site | Part: BBa_B0030 |
| T7pol | pSB1C3 | RNA polymerase from phage T7 | Part: BBa_K145001 |
| cI | pSB1A2 | Regulator | Part: BBa_C0051 |
| lacI | pSB1A2 | Regulator | Part: BBa_C0012 |
| tetR | pSB1C3 | Regulator | Part: BBa_C0040 |
| araC | pSB1C3 | Regulator | Part: BBa_C0080 |
| luxR | pSB1A2 | Regulator | Part: BBa_C0062 |
The other novel synthetic promoters were based on the well-characterized hybrid promoter Ptrc (33) that was redesigned replacing the existing lacO sequence by other operators and/or including additional operators such as, the lacOid, tetO or araI1 half-site (for more details see Figure 5b and Table 1). All the synthetic promoters are flanked by the prefix and suffix sequences of the BioBrick RFC[10] standard and were synthesized and cloned into pJ201 vector (DNA 2.0, Inc.).
2.4 DNA devices assembly
All DNA constructs were generated following the standard assembly protocol, and the BioBrick DNA parts used were obtained from the Registry of Standard Biological Parts (Table 1). Briefly, to test promoter constitutive expression, the GFP generator (part BBa_E0240 that comprises the RBS BBa_B0032, the coding sequence for GFP BBa_E0040, and the double terminator BBa_B0015) was excised from the delivery plasmid with XbaI and PstI and cloned downstream each promoter (plasmids digested with SpeI and PstI). To test the promoters’ regulated expression, the lacI, tetR, cI, luxR or araC regulators’ ORFs were cloned under regulation of the constitutive promoter PrnpB and the RBS BBa_B0030. The ORFs were excised with XbaI and PstI and cloned in the plasmid containing the promoter, previously digested with SpeI and PstI. Subsequently, the devices containing the promoter and GFP were excised with EcoRI and SpeI and cloned upstream of the respective PrnpB:: regulator assembly (plasmid digested with EcoRI and XbaI). For the T7 polymerase (T7pol), a similar assembly was performed.
For generation of the GG device used in the proof of concept, the ggpS ORF was amplified by PCR using Synechocystis genomic DNA as template with primers flanked by the BioBrick prefix or suffix (see Supplementary Table S1). Amplification was performed using Phusion high-fidelity DNA polymerase (Thermo Scientific), according to the manufacturer’s instructions. The PCR product was purified using the NZYGelpure kit (NZYTech), digested with XbaI and PstI and cloned downstream of Ptrc.x.lacOand BBa_B0030 RBS (plasmid digested with SpeI and PstI).
All the generated constructions were transferred to pSEVA251 or pSEVA351 shuttle vectors (obtained from the ‘Standard European Vector Architecture’ repository) (34) and the construct harboring the RNA polymerase from phage T7 (PrnpB::T7pol) was cloned into the pSN15K vector (13). The correct assembly of the generated constructs was confirmed by PCR, restriction analysis and sequencing (STAB VIDA).
2.5 Generation of the Synechocystis ggpS deletion mutant
The construction of integrative plasmids for the deletion of the ggpS gene was performed as described previously (35). Briefly, the plasmids were based on pGEM-T® Easy (Promega) and contain the Synechocystis chromosomal regions flanking the ggpS gene. The 5′- and 3′-flanking regions were amplified from the cyanobacterium’s genome using Pfu DNA polymerase and the primer pairs 5-O/5-I and 3-O/3-I (Supplementary Table S1), respectively. Subsequently, the purified PCR fragments were fused by ‘overlap PCR’ using primers 5-O/3-O and 80 ng of each amplicon (Supplementary Table S1). The resulting product was purified and cloned into the vector pGEM-T® Easy, according to the manufacturer’s instructions, originating the pGDggpS plasmid. A selection cassette, containing the nptII gene (conferring resistance to neomycin and kanamycin), obtained from the plasmid pK18mobsacB (36), was cloned in the AgeI restriction site of pGDggpS using the T4 DNA ligase to generate the plasmid pGDggpS.K (Table 2). All the constructs were confirmed by DNA sequencing (STAB VIDA).
Table 2.
List of plasmids used to transform Synechocystis
| Designation | Plasmid | Description | References/sources |
|---|---|---|---|
| pGDggpS.K | pGEM-T® Easy | pGEM-T easy vector containing the kanamycin resistance cassette flanked by the two regions for double homologous recombination on the ggpS locus | This study |
| pSEVA251 | pSEVA251 | Replicative plasmid/shuttle vector, KmR, ori RSF1010 | (34) |
| pSEVA351 | pSEVA351 | Replicative plasmid/shuttle vector, CmR, ori RSF1010 | (34) |
| pSEVA451 | pSEVA451 | Replicative plasmid/shuttle vector, SpR/SmR, ori RSF1010 | (34) |
| PrnpB::gfp | pSEVA251/pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the native constitutive promoter PrnpB | (8) |
| PλcI::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PλcIpromoter (Part: BBa_ R0051) | This study |
| PλcI::gfp-PrnpB::cI | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PλcIpromoter (Part: BBa_ R0051), upstream of the CI regulator coding region (Part: BBa_ C0051) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| ParaC::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the ParaCpromoter (Part: BBa_ R0080) | This study |
| ParaC::gfp PrnpB::araC | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the ParaC promoter (Part: BBa_R0080), upstream of the AraC regulator coding sequence (Part: BBa_C0080) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PluxR::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PluxRpromoter (Part: BBa_ R0062) | This study |
| PluxR::gfp-PrnpB::luxR | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PluxRpromoter (Part: BBa_R0062), upstream of the LuxR regulator coding sequence (Part: BBa_C0062) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PBADwt::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PBADwtpromoter (Part: BBa_ I13453) | This study |
| PBADwt::gfp PrnpB::araC | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PBADwtpromoter (Part: BBa_I13453) upstream of the AraC regulator coding sequence (Part: BBa_C0080) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PT7pol::gfp-PrnpB::T7pol | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the PT7pol promoter (Part: BBa_I712074), upstream of the T7 polymerase coding sequence (Part: BBa_K145001) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PrnpB::T7pol | pSN15K | T7 polymerase under the control of the native constitutive promoter PrnpBand the BBa_B0030 RBS | This study |
| PT7.1.x.lacO::gfp | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.1.x.lacO variant | This study |
| PT7.2.x.lacO::gfp | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.2.x.lacO variant | This study |
| PT7.3.x.lacO::gfp | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.3.x.lacO variant | This study |
| PT7.1.x.lacO::gfp-PrnpB::lacI | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.1.x.lacO variant upstream of the LacI regulator coding region (Part: BBa_C0012) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PT7.2.x.lacO::gfp-PrnpB::lacI | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.2.x.lacO variant upstream of the LacI regulator coding region (Part: BBa_C0012) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PT7.3.x.lacO:: gfp-PrnpB:: lacI | pSEVA351 | GFP generator (Part: BBa_E0240) under the control of the PT7.3.x.lacO variant upstream of the LacI regulator coding region (Part: BBa_C0012) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PtacI::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the hybrid promoter PtacI | (32) |
| Ptrc.x.tetO1::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.tetO1 | This study |
| Ptrc.x.tetO1::gfp-PrnpB::tetR | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.tetO1, upstream of the TetR regulator coding region (Part: BBa_C0040) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| Ptrc.x.araO::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.araO | This study |
| Ptrc.x.araO::gfp-PrnpB::araC | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.araO, upstream of the AraC regulator coding region (Part: BBa_C0080) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| Ptrc.x.tetO2::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.tetO2 | This study |
| Ptrc.x.tetO2::gfp-PrnpB::tetR | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.tetO2, upstream of the TetR regulator coding region (Part: BBa_C0040) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| Ptrc.x.lacO::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.lacO | This study |
| Ptrc.x.lacO::gfp-PrnpB::lacI | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the synthetic promoter Ptrc.x.lacO, upstream of the LacI regulator coding region(Part: BBa_C0012) under the control of the native PrnpB and the BBa_B0030 RBS | This study |
| PpsbA2*::gfp | pSEVA251 | GFP generator (Part: BBa_E0240) under the control of the modified psbA2 promoter (PpsbA2*) | (13) |
| Ptrc.x.lacO::ggpS | pSEVA351 | Native ggpS coding region under the control of the synthetic promoter Ptrc.x.lacO and the BBa_B0030 RBS | This study |
For the generation of the ΔggpS mutant, Synechocystis was transformed via natural transformation based on the procedure described previously (37). Briefly, Synechocystis was grown in standard conditions to an OD730 ≈ 0.5. Cells were harvested by centrifugation (10 min at 3850 g) and resuspended in BG11 to a final OD730 ≈ 2.5. The pGDggpS.K plasmid was incubated for 5 h with 500 μl of those cells (final plasmid DNA concentration of 20 μg/ml), in light and at 25°C. Cells were then spread onto Immobilon™-NC membranes (0.45 μm pore size, 82 mm, Millipore) resting on solid BG11 plates, grown in standard growth conditions and transferred to selective plates after 24 h (Km, 10 μg/ml). Transformants were observed after 2 weeks. For complete segregation, Km-resistant colonies were grown at increasing Km concentrations (25 and 50 μg/ml), then transferred to liquid medium and the antibiotic concentration was increased up to 500 μg/ml. The full segregation of the mutant was confirmed by Southern blot with a probe covering the 5′ flanking region of the ggpS gene labeled with digoxigenin using the DIG DNA labeling kit (Roche Molecular Biochemicals) and following the manufacturer’s instructions. The Southern blot was carried out using the Synechocystis strains gDNA (4 μg) that was digested with AvaII Fast-Digest® for 45 min at 37°C, followed by an agarose gel electrophoresis. The remaining protocol was performed according to the DIG High Prime DNA Detection Starter kit (Roche Molecular Biochemicals) instructions. The final results were observed with a ChemiDocTM XRS+ Imager (Bio-Rad).
2.6 Synechocystis transformation and mutants/transformants confirmation
The three replicative plasmids pSEVA251 (KmR), pSEVA351 (CmR) and pSEVA451 (Sp/SmR), as well as the derived plasmids (listed in Table 2) were introduced into Synechocystis by electroporation, adapting the protocol previously described (38). Briefly, Synechocystis cultures were grown at 25°C under a continuous light regimen until an OD730 ≈ 0.5 was reached. Cells were collected by centrifugation at 4190 g for 10 min and washed three times with 10 ml of 1 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic (HEPES) acid buffer, pH 7.5. The cells were then suspended in 1 ml of HEPES buffer and 60 μL aliquots were mixed with 1 μg of plasmid DNA (eluted in water) and electroporated using a Gene Pulser™ (Bio-Rad). The capacitor was set to 25 μF and the resistor to 400 Ω, for a time constant of 9 ms with an electric field of 12 kV/cm. Immediately after the electric pulse, cells were transferred to 400 μL of fresh BG11 medium and spread onto Immobilon-NC membranes (0.45 μm pore size, 82 mm, Merck Millipore) resting on solid BG11 plates incubated at 25°C, under a 16 h light/8 h dark regimen for 24 h. Then, the membranes were transferred to solid BG11 plates supplemented with the appropriate antibiotic and incubated in the same conditions described above. Colonies were observed after 1–2 weeks.
The integrative plasmid pSN15K (KmR) and the pSEVA replicative plasmids were introduced into Synechocystis by natural transformation, as described previously (37). Additionally, the replicative plasmid pSEVA451 was introduced into Synechocystis by conjugation, as described previously (39).
The presence of replicative plasmids or mutant segregation was confirmed by PCR using specific primers (Supplementary Table S1). For this purpose, Synechocystis DNA was extracted using 2 ml of culture that were centrifuged at 14 100 g for 3 min and washed with 1 ml of distilled water. Then, cell pellet was resuspended in 200 μL of distilled water and, 1 μL of RNase solution (20 mg/ml, Sigma) and 0.1 g of 0.2 mm-diameter glass beads (acid washed, Sigma) were added. Cells were disrupted by two cycles of vortexing for 1 min with incubation on ice for 1 min in between. Subsequently, the mixture was centrifuged at 14 100 g for 2 min, and the DNA containing supernatant was transferred to a new tube. The PCR reaction mixtures (20 µl) were setup using GoTaq® G2 Flexi DNA Polymerase (Promega), according to manufacturer’s instructions, and 2 µl of DNA containing supernatant.
2.7 Growth experiments
Synechocystis cells (wild-type and mutants harboring pSEVA251, pSEVA351 and pSEVA451) were inoculated into 25 ml of BG11 medium in 50 ml Erlenmeyer flasks to an initial OD730 ≈ 0.5, and grown in batch cultures at 30°C, 100 rpm, under a 12 h light (20 μE/m2/s)/12 h dark regimen or continuous light (non-enriched air). Cell growth was monitored by measuring the OD730 for a 16 days period. All the growth experiments were performed in the absence of selective pressure and included three biological replicates with technical duplicates.
For the halotolerance growth experiments, pre-cultures of Synechocystis wild-type (WT), ΔggpS mutant and the complemented ΔggpS mutant (ΔggpS_GG device) were inoculated in BG11 medium (supplemented with 10 μg/ml Cm and/or 25 μg/ml Km, when appropriate), and grown in an orbital shaker (150 rpm), at 30°C under a 12 h light (25 μE/m2/s)/12 h dark regimen (non-enriched air). The cultures were grown until an OD730 ≈ 2 was reached and, subsequently diluted in fresh BG11 medium without antibiotic to a final OD730 ≈ 0.5. Fifty milliliter of the dilution were transferred to 100 ml Erlenmeyer flasks (previously sterilized) containing NaCl, providing the cultures with the following final NaCl concentrations: 0, 3, 5 and 7% (w/v). These cultures were maintained in the same conditions as the pre-cultures and growth was monitored as described above. Each experiment was performed in triplicate.
2.8 Flow cytometry analysis
Cells of Synechocystis wild-type and mutants harboring pSEVA251, pSEVA251 PrnpB::gfp and pSEVA251 Ptrc.x.lacO::gfp were cultivated in liquid BG11 medium without or with kanamycin (Km, 25 µg/ml) under a 12 h light (20 μE/m2/s)/12 h dark regimen as described above (Section 2.7). At Days 0, 4, 9 and 16 of cultivation, cells were diluted to OD730 ≈ 0.1 and 50 000 events per sample were acquired after cell debris exclusion [forward scatter (FSC) versus side scatter (SSC) plot] using a FACSCaliburTM flow cytometer (BD Biosciences). FSC, SSC and GFP fluorescence signal (acquired in the FL1 channel) were plotted in a logarithmic scale, and data were analyzed with Cellquest software. Average GFP fluorescence signal and cell population percentages from three biological replicates with technical duplicates were calculated using the FlowJo® software vX10.0.7. Fluorescence signal of wild-type cells and cells harboring pSEVA251 were used as negative controls to establish the GFP+ gates that allow discriminating the cell population that emits GFP fluorescence (GFP+ cells).
2.9 GFP fluorescence analysis
For the evaluation of constitutive or regulated expression, cultures of Synechocystis carrying the promoter or promoter + regulator devices were grown to a final OD730 ≈ 1 in 25 ml flasks, under continuous light at 30°C. The cultures were diluted to a final OD730 ≈ 0.5 and 200 μL aliquots were distributed in Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (Thermo Fisher Scientific). For regulated expression experiments, the inducers were used in the following final concentrations: 1, 2 or 20 mM IPTG; 2 or 5 mM lactose; 1 nM, 1 µM or 1 mM AHL; 0.2% or 1% (wt/vol) L-arabinose and 1, 2 or 10 μg/ml aTc. For the latter, a control was performed growing cells in 1% (vol/vol) ethanol since the aTc stock was prepared in 50% (vol/vol) ethanol. The 96-wells plates were incubated at 30°C under continuous light, with the exception of experiments using aTc that were maintained in the dark and cells were grown in mixotrophic conditions, adding glucose to a final concentration of 5 mM and providing a daily light pulse during the measurement procedure (10–15 min). Each experiment was repeated three times, including three technical replicates and the measurements were carried out in duplicate 0, 24, 48 and 72 h after plate setup. GFP fluorescence and OD790 were detected using the Synergy 2 Multi-Mode Microplate Reader and the Gen5™ software (BioTek Instruments, Inc.). For fluorescence detection, an excitation filter of 485/20 nm and an emission filter of 528/20 nm were used (sensibility set for 110). For data analysis, the background fluorescence and absorbance of the BG11 medium was subtracted from the values obtained for the samples and, the fluorescence values were normalized by optical density. Then, the normalized fluorescence obtained for the cells harboring the empty plasmids were subtracted from the normalized fluorescence of all samples.
2.10 Confocal microscopy
Synechocystis cells harboring the pSEVA251 or the promoter::GFP generator devices were inoculated in 20 ml of BG11 medium to a final OD730 ≈ 0.5 and grown under continuous light at 30°C until an OD730 ≈ 1.0 was reached. Five microliter of each culture were added to 20 μL of 1% (wt/vol) low-melting point agarose beds (dissolved in BG11 medium) and covered with a coverslip. The cultures were observed using a Leica TCS SP5 confocal microscope (Leica Microsystems). Initially, the microscopy settings were adjusted in order to minimize the autofluorescence of Synechocystis wild-type harboring the pSEVA251 and were the same throughout the experiment: GFP emission (collected between 500 and 540 nm) was observed when cells were exposed to a laser beam at 488 nm, and cyanobacterial autofluorescence was collected between 640 and 680 nm after excitation at 633 nm. Images were analyzed with the Leica Application Suite X software (Leica Microsystems).
2.11 SDS-PAGE and western blot
To confirm the presence of the LacI in Synechocystis LacI-expressing strains, cell-free extracts were obtained by sonication as described by Pinto et al. (13) from Synechocystis wild-type and mutants harboring pSEVA251, pSEVA251 Ptrc.x.lacO::gfp-PrnpB::lacI, pSEVA351, pSEVA351 PT7.1.x.lacO::gfp-PrnpB::lacI, PT7.2.x.lacO::gfp-PrnpB::lacI and PT7.3.x.lacO::gfp-PrnpB::lacI with the T7 RNA polymerase under the control of PrnpB and RBS BBa_B0030 integrated into the chromosomal neutral site N15. Samples were denaturated in reducing SDS sample buffer for 6 min at 95°C and later separated by electrophoresis on 12% (wt/vol) SDS-polyacrylamide gels. The proteins were visualized with colloidal Coomassie Brilliant Blue (Sigma) or blotted onto nitrocellulose membrane pore size 0.45 µm (AmershamTM ProtanTM, GE Healthcare). Western blots were performed using the rabbit anti-LacI polyclonal antibody (Abnova) and the HRP-conjugated goat anti-rabbit IgG antibody (Sigma). The blot was developed using ClarityTM Western ECL Substrate (Bio-Rad) on a ChemiDocTM XRS+ Imager (Bio-Rad). In this experiment, three independent biological replicates were performed.
2.12 Transcription analysis by RT-qPCR
Pre-cultures of Synechocystis wild-type and the mutant harboring the pSEVA351 Ptrc.x.lacO::ggpS (GG device) were grown at 30°C, with rotary shaking (150 rpm), under a 12 h light/12 h dark regimen, until an OD730 ≈ 2 was reached. Subsequently, cultures were diluted in 50 ml of fresh BG11 medium supplemented or not with 3% (wt/vol) NaCl to a final OD730 ≈ 0.5. One-hundred milliliter of culture were collected 3 days post-inoculation (OD730 ≈ 1), centrifuging cells for 10 min at 3850 g, pellets were treated with RNAprotect Bacteria Reagent (Qiagen) according to instructions and stored at −80°C. Total RNA was extracted using the TRIzol® Reagent (Ambion) in combination with the PurelinkTM RNA Mini Kit (Ambion). Briefly, the cells were disrupted in TRIzol containing 0.2 g of 0.2 mm-diameter glass beads (acid washed, Sigma) using FastPrep®-24 (MP Biomedicals), the following extraction steps included the On-column PureLink® DNase treatment. All steps were performed according to the manufacturer’s instructions except DNase treatment, in which a 90 min incubation period at 25°C was used. RNA was quantified on a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies, Inc.) and the quality and integrity was checked using the Experion™ RNA StdSens Analysis Kit (Bio-Rad). The absence of genomic DNA contamination was checked by PCR, in reaction mixtures containing: 0.5 U of GoTaq® G2 Flexi DNA Polymerase (Promega), 1x Green GoTaq Flexi buffer, 200 µM of each deoxyribonucleotide triphosphate (dNTP), 1.5 mM MgCl2, 0.25 µM of each rnpB primer (Supplementary Table S1) and 1 µl total RNA. The PCR profile was: 2 min at 95°C followed by 25 cycles of 30 s at 95°C, 30 s at 56°C and 30 s at 72°C and a final extension at 72°C for 7 min. The PCR reactions were run on agarose gel electrophoresis.
For cDNA synthesis, 1 µg of total RNA was transcribed with the iScriptTM Reverse Transcription Supermix for RT-qPCR (Bio-Rad) in a final volume of 20 µl, following the manufacturer’s instructions. A control PCR was performed using 1 µl of cDNA as template and the same reaction conditions and PCR program described above. Five-fold standard dilutions of the cDNAs were made (1/5, 1/25, 1/125 and 1/625) and stored at −20°C.
RT-qPCRs were performed as described in Pinto et al. (35) using 2 µl of template cDNA (dilution 1/25) and the primers pairs are listed in Supplementary Table S1. The genes 16S, petB and rnpB were validated as reference genes for data normalization using the geNorm application included in the qbase+ software v3.1 (Biogazelle). All RT-qPCR parameters in these experiments were in agreement with the MIQE (minimum information for publication of quantitative real-time PCR experiments) guidelines. RT-qPCR data from three biological replicates and three technical replicates were analyzed using the IQ5™ Optical System Software v2.1 (Bio-Rad).
2.13 Compatible solutes detection
Pre-cultures of Synechocystis wild-type, ΔggpS mutant and the complemented ΔggpS mutant harboring the pSEVA351 Ptrc.x.lacO::ggpS (GG device), were grown as described for the RT-qPCR experiments. Five hundred milliliter of culture were cultivated in presence or absence of 3% (wt/vol) NaCl (distributed in ten 50 ml cultures). Three days after inoculation (OD730 ≈ 1) cells were harvested as described in Santos et al. (40) with modifications. Cell cultures were centrifuged at 4470 g for 10 min at room temperature and the supernatant discarded. Cells were washed adding 100 ml of cold deionized water or 3% (wt/vol) NaCl solution (identical to the growth medium). Centrifugation was repeated and the cell pellet was suspended in 20 ml of the respective solution. Two 1 ml aliquots were removed for protein quantification, centrifuged and stored at −20°C until further use. The remaining cell suspension was centrifuged at 4°C and the cell pellet was stored at −20°C. Ethanol-chloroform extraction of solutes was performed according to Santos et al. (40) with adaptations. Briefly, cell pellets were suspended in 50 ml of 80% (vol/vol) ethanol and the cell suspension was transferred to a 250 ml round flask containing a magnetic stir bar. The flask was then connected to a coil condenser (circulating cold water), and the flask was heated in a boiling water bath with stirring for 10 min. The suspension was transferred to a 50 ml tube and centrifuged at 8000 g for 10 min at RT. The supernatant was kept in a clean 250 ml round flask and the pellet suspended again in 50 ml of 80% (vol/vol) ethanol for a new extraction; this step was repeated three times. The remainder protocol was performed as described. Detection, identification and quantification of compatible solutes were carried out by Proton NMR (ITQB magnetic resonance center, CERMAX).
2.14 Statistical analysis
The statistical analysis was performed by means of a one-way ANOVA, using GraphPad Prism v6.01 (GraphPad software Inc.).
3. Results and discussion
For the efficient use of the unicellular cyanobacterium Synechocystis sp. PCC 6803 as a photoautotrophic chassis, a well-characterized portfolio of tools/biological parts is mandatory. Therefore, three SEVA replicative plasmids and an array of heterologous and redesigned promoters were tested in this organism.
3.1 SEVA plasmids
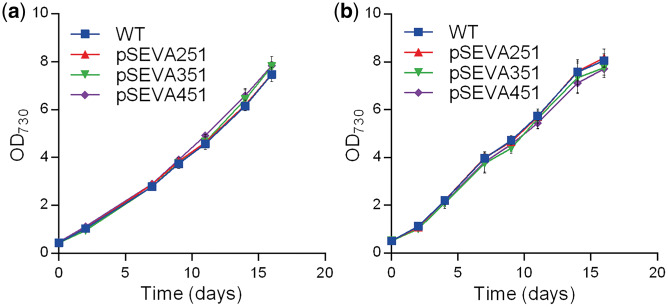
The SEVA repository (http://seva.cnb.csic.es/) comprises plasmids that are formed by three variable modules—cargo, replication origin and antibiotic marker—separated by three permanent regions, the T0 and T1 terminators and the oriT conjugation origin (34). In this study, the vectors that include the default multiple cloning site, the RSF1010 broad-host-range replicon, and the antibiotic markers conferring resistance to kanamycin (pSEVA251), chloramphenicol (pSEVA351) or spectinomycin/streptomycin (pSEVA451), were tested. The plasmids were successfully transformed into Synechocystis by three different methods: natural transformation, electroporation and conjugation (data not shown). Transformation was confirmed by the acquisition of resistance to the respective antibiotic, by PCR, and by further sequencing of the respective fragment. The presence of the plasmid did not lead to an evident phenotype or hinder Synechocystis mutants’ growth compared to the wild-type (Figure 1). The replication of these vectors in the cyanobacterium was expected since RSF1010 is the origin of replication present in most of the self-replicating plasmids currently used to transform Synechocystis [e.g. pAWG1.1 (41) or pPMQAK1 (8)]. In comparison with these vectors that are over 8 kb (8), the pSEVAs are relatively small (pSEVA251: 5275 bp, pSEVA351: 5120 bp and pSEVA451: 5334 bp), making handling and transference easier. In addition, the possibility to use electroporation reduces the amount of DNA and time required to transform Synechocystis cells and the transformant colonies are obtained faster (about 1 week compared to at least 2 weeks for natural transformation and 4 weeks for conjugation).
Figure 1.
Growth curves of Synechocystis wild-type (WT) and mutants harboring the replicative plasmids pSEVA251, pSEVA351 and pSEVA451. Cultures were grown at 30°C with rotary shaking (100 rpm) under (a) 12 h light (20 µE/m2/s)/12 h dark or (b) continuous light (20 µE/m2/s). Growth was monitored by measuring optical density at 730 nm (OD730). Error bars correspond to standard deviations from three biological replicates with technical duplicates.
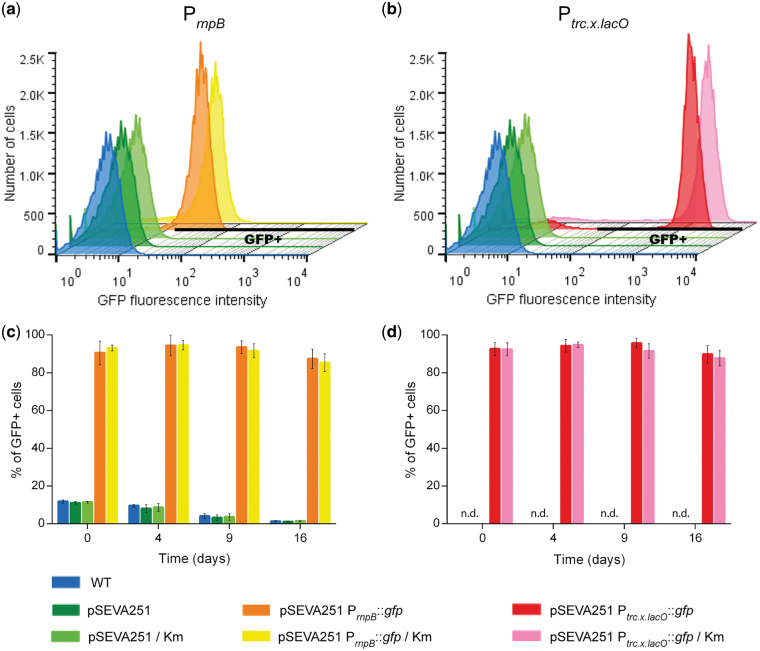
To evaluate the capacity of the Synechocystis mutants to retain the replicative plasmid pSEVA251 during cultivation in medium without the selective pressure, a flow cytometry analysis was performed. Cells transformed with the empty vector pSEVA251, or with the same vector containing the GFP generator BBa_E0240 under the control of either PrnpB (8) (the cyanobacterial reference promoter) or Ptrc.x.lacO (one of the redesigned promoters characterized in this study, see Section 3.3) (for details see Figure 3a), were grown in BG11 medium with or without Km. The mean GFP fluorescence intensity in cells of Synechocystis harboring the Ptrc.x.lacO was 34-fold higher compared to the cells harboring the PrnpB (Figure 2a and b). The results also showed that the percentage of cells emitting fluorescence in the GFP detecting channel determined by the GFP+ gates (hereafter named GFP+ cells) was similar for the two promoters tested (∼92%, Figure 2c and d). In addition, the percentage of GFP+ cells cultivated in medium with selective pressure was not significantly different from that of cells grown in medium without antibiotic (Figure 2c and d). These results were further confirmed by observing the cells with a confocal microscope. Overall, it is possible to determine that at least 90% of the cells grown for 16 days without selective pressure were able to retain the replicative plasmid, and that this flow cytometry methodology can accurately be used to follow the fraction of the population that keeps carrying a replicative plasmid for a given period of time.
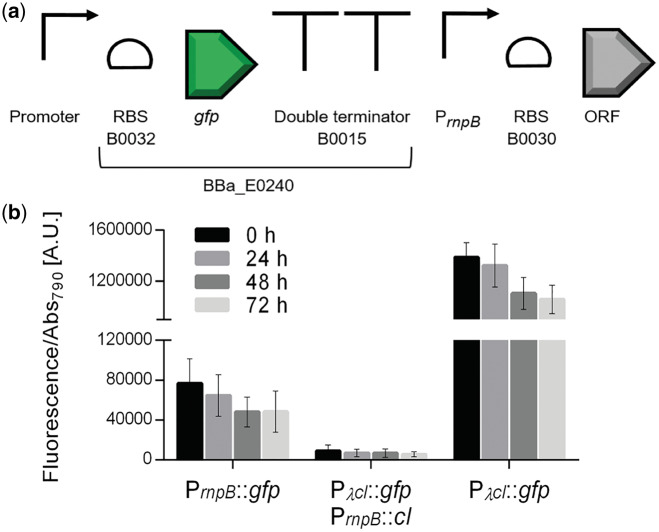
Figure 3.
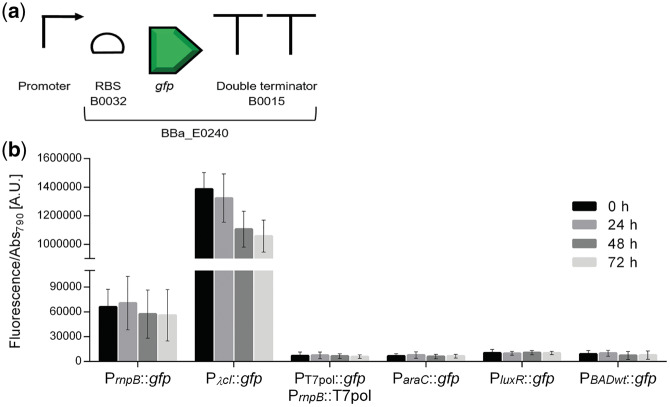
Characterization of heterologous promoters in Synechocystis. (a) Schematic representation of the promoters and the GFP generator (BBa_E0240) assembly. (b) Normalized GFP fluorescence of Synechocystis cultures harboring the PλcI, PT7pol, ParaC, PluxR and PBADwt promoters. Measurements were performed up to three days (0, 24, 48 and 72 h) and the fluorescence was normalized to Abs790. Fluorescence of the control strain harboring the pSEVA251 was subtracted from each sample. Error bars correspond to standard deviations from three biological replicates with technical triplicates (measured in duplicate). PrnpB::gfp was included for comparison purposes.
Figure 2.
Flow cytometry analysis of GFP fluorescence intensity in cells of Synechocystis wild-type (WT) and Synechocystis mutants harboring the empty plasmid pSEVA251 or the GFP generator BBa_E0240 under the control of two different promoters PrnpB (reference promoter) and Ptrc.x.lacO (later characterized in this study)—pSEVA251 PrnpB::gfp (a and c) or pSEVA251 Ptrc.x.lacO::gfp (b and d). The cells were grown in BG11 without or with kanamycin (Km). Histograms of GFP fluorescence intensities acquired in FL1 channel show the establishment of the GFP+ gates (black solid line) in Synechocystis mutants harboring the promoter PrnpB (a) or Ptrc.x.lacO (b) to evaluate the percentage of GFP+ cells (c and d). Histograms are representative of fluorescence intensity of cells analyzed after 16 days of cultivation; the error bars correspond to standard deviations from three biological replicates with technical duplicates. n.d. indicates non-detectable percentage of GFP+ cells for the WT and the strains harboring empty plasmid (d).
3.2 Heterologous promoters
A set of heterologous promoters well described and routinely used in E. coli either from bacterial origin (PluxR, ParaC and PBADwt) or bacteriophage-derived (PλcI and PT7pol) were characterized in Synechocystis. For this purpose, the promoters were assembled with the GFP generator BBa_E0240 (Figure 3a), subsequently each module was transferred to pSEVA251 and transformed into Synechocystis by electroporation. For promoter characterization, GFP expression was determined by measuring fluorescence of the cultures and normalizing it to cell density. The promoter of the RNase P gene (PrnpB) was used as reference (8).
The heterologous promoters PT7pol, ParaC, PluxR and PBADwt exhibited lower activity compared to the reference promoter PrnpB (Figure 3b), supporting earlier observations that promoters with strong activity in E. coli may perform differently in cyanobacteria (8). Beyond the genomic context, the transcriptional machinery of Synechocystis has several differences compared to E. coli, namely the RNA polymerase subunits (42). In contrast, 20-fold higher fluorescence levels were detected when PλcI was used. This result can be explained by the high similarity of the PλcI-10 and -35 consensus boxes with the cyanobacterial type I promoters (42). Nevertheless, Huang et al. (8) reported no activity for this promoter (referred to as PR) in Synechocystis, using two different fluorescence detection systems. The only difference between our constructs and those of Huang et al. (8) is the RBS (here BBa_B0032 was used, while Huang et al. (8) used BBa_B0034). However, this should not have any influence on the output since it has been reported that these RBS have similar translation efficiencies in Synechocystis (43).
To evaluate whether the ParaC, PBADwt, PluxR and PλcI promoters could be regulated in Synechocystis, the genes encoding the respective regulatory proteins (araC—BBa_C0080; luxR—BBa_C0062 or cI—BBa_C0051) were cloned under the control of PrnpB with a strong RBS (BBa_B0030) and placed downstream of the respective promoter::gfp assembly (Figure 4a). The AraC regulated promoters, ParaC and PBADwt were not repressed in presence of the protein, probably due to the absence of the araO2 operator region that has been shown to be essential for promoter repression in E. coli (44, 45). For the PBADwt, the presence of the AraC regulatory protein led to a 6-fold increase in GFP fluorescence independently of the presence of L-arabinose (Supplementary Figure S1). In the native system, the PBADwt is induced by the binding of an AraC dimer to the araI1 and araI2 operators in an L-arabinose dependent manner (46). However, our results indicate that in Synechocystis AraC binds to the araI1 and araI2 operators in the absence of L-arabinose, as previously suggested for E. coli (47) and that can be explained by the high affinity of AraC to these operators. The PluxR promoter from Vibrio fischeri promotes weak constitutive expression of downstream genes and can be induced by the binding of LuxR complexed with signaling molecules from the acyl-homoserine lactone class (AHLs) (48). In Synechocystis, an 8-fold increase in fluorescence was observed in presence of LuxR compared to constitutive levels (Supplementary Figure S2), and the induction was shown to be independent of AHLs. Similarly, in E. coli, it was shown that backwards transcription from the PluxR can occur by binding of the LuxR to the promoter in the absence of AHLs (49). The regulated expression of the heterologous PλcI promoter was also tested in Synechocystis. The presence of the CI protein in the system resulted in 99% repression compared to constitutive levels (Figure 4b), displaying a similar behavior to the previously reported (50). A tight and stable repression was registered during the 3 days of the experiment demonstrating that PλcI, together with the regulating protein CI can be used to efficiently control transcription. Therefore, regulation of CI expression levels would allow the implementation of a controlled circuit using the PλcI promoter.
Figure 4.
Characterization of the PλcI regulated expression in Synechocystis. (a) Schematic representation of the promoter GFP generator (BBa_E0240) assembly followed by PrnpB::ORF encoding the regulatory protein. (b) Normalized GFP fluorescence of Synechocystis cultures harboring the promoter PλcI in presence (PλcI::gfp-PrnpB::cI) or in absence (PλcI::gfp) of the CI repressor. Measurements were performed up to three days (0, 24, 48 and 72 h) and the fluorescence was normalized to Abs790. The fluorescence of the control strain harboring the pSEVA251 was subtracted from each sample. Error bars correspond to standard deviations from three biological replicates with technical triplicates (measured in duplicate). PrnpB::gfp was included for comparison purposes.
3.3 Redesigned promoters
In addition to the heterologous promoters, the use of orthogonal regulatory parts based on the T7 RNA polymerase promoter was also explored. For this purpose, three existing variants of the PT7pol, differing only in one nucleotide (meant to obtain promoters with different strengths) (30, 31), and based on Novagen® pET expression vectors, were redesigned to include an operator region for the LacI regulator originating the PT7.1.x.lacO, PT7.2.x.lacO and PT7.3.x.lacO promoters (Figure 5a, for details see Table 1).
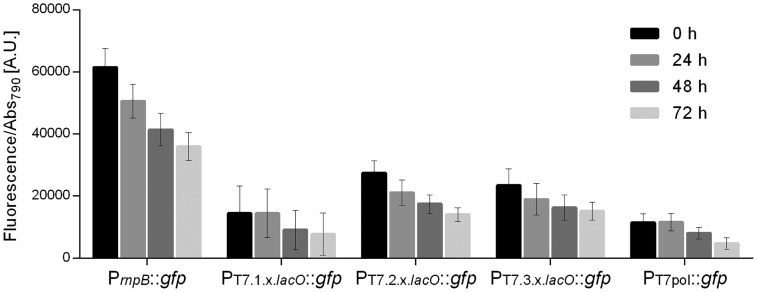
These redesigned T7 promoters were assembled with the GFP generator BBa_E0240 (depicted in Figure 3a, see also Section 2.4), cloned in pSEVA351 and transformed into a Synechocystis mutant with the T7 RNA polymerase under the control of the PrnpBand RBS BBa_B0030 integrated into the chromosomal neutral site N15 (13).
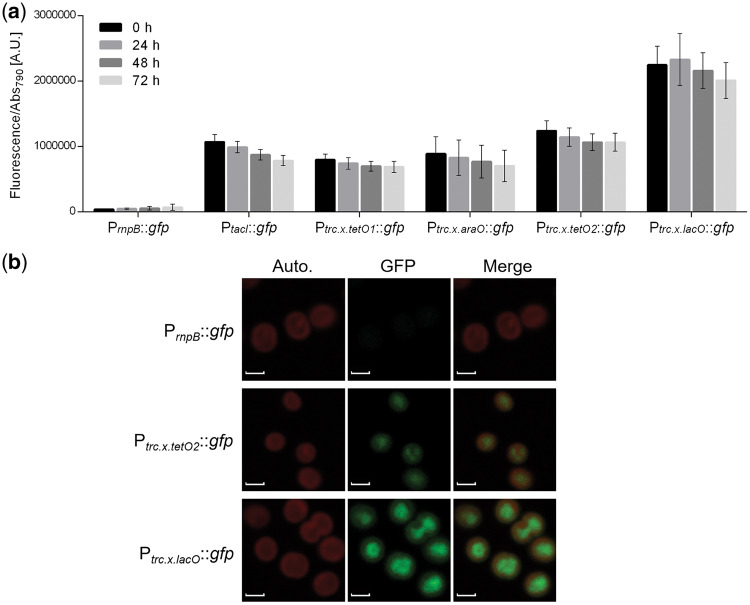
All the redesigned PT7pol variants showed lower GFP expression levels compared to the reference promoter PrnpB; while the comparison of the expression levels of the variants with the PT7pol revealed significant differences for PT7.2.x.lacO and PT7.3.x.lacO, P-value <0.0001 (Figure 6). Furthermore, our results showed that the GFP expression levels driven by the PT7pol were similar when the T7 polymerase was integrated into the genome (Figure 6, PT7pol::gfp) or in a self-replicating plasmid (see Figure 3, PT7pol::gfp-PrnpB::T7pol). The regulation of these promoters by the LacI protein was also tested. The Lac repressor from E. coli is one of the most well-characterized transcription factors and is orthogonal to cyanobacteria (51), hence a low risk of cross-talking was expected. However, the results revealed that the redesigned PT7pol variants were not repressed in the presence of the co-expressed LacI (data not shown). The presence of LacI, in the cells harboring the three variants was confirmed by western blot (Supplementary Figure S3), implying that the lack of repression was not due to the lack of the regulatory protein. One possible explanation is that LacI is not binding to the operator region. The lack of repression was also observed in PtacI (data not shown), a hybrid promoter that contains the -35 sequence from Ptrp (tryptophan operon promoter from E. coli) and the PlacUV5 sequence that includes the -10 box and the lacO operator (32). Interestingly, Huang et al. (8) reported LacI-mediated repression of the Ptrc1O promoter, which differs from the PtacI only in the spacer size between the -35 and -10 boxes, with 17 bp instead of 16 bp. This change in the core promoter also influenced the constitutive expression levels: PtacI is 17-fold stronger than PrnpB, while Ptrc1O is 83-fold stronger (8). From the promoters characterized in Synechocystis and referred herein, the Ptrc1O is the strongest and was, therefore, redesigned including the operator sequences for the regulatory proteins LacI, AraC and TetR (promoter sequences shown in Figure 5b). The constitutive GFP expression driven by the redesigned promoters Ptrc.x.tetO1, Ptrc.x.araO, Ptrc.x.tetO2 and Ptrc.x.lacO was evaluated being respectively 13-, 14-, 21- and 41-fold stronger than the one registered for PrnpB (Figure 7a). However, the strength of the promoters was lower than expected since the core promoter from the Ptrc1O was maintained in these elements. The fluorescence of Synechocystis cells harboring the constructs with the promoters PrnpB, Ptrc.x.tetO2and Ptrc.x.lacO was also analyzed by confocal microscopy, showing that the fluorescence signal was evenly distributed within the cells (Figure 7b). Moreover, differences in GFP expression driven by the three promoters could be readily observed (Figure 7b) and are consistent with the differences in promoter strength previously determined (Figure 7a).
Figure 6.
Characterization of the PT7pol and redesigned PT7pol promoter variants in Synechocystis constitutively expressing the T7 polymerase. Normalized GFP fluorescence of Synechocystis cultures harboring the PT7pol, PT7.1.x.lacO, PT7.2.x.lacO and PT7.3.x.lacO. Measurements were performed up to three days (0, 24, 48 and 72 h) and the fluorescence was normalized to Abs790. The fluorescence of the control strain harboring the pSEVA251 was subtracted from each sample. Error bars correspond to standard deviations from three biological replicates with technical triplicates (measured in duplicate). PrnpB::gfp was included for comparison purposes.
Figure 7.
Characterization of the hybrid PtacI and the redesigned trc promoters in Synechocystis. (a) Normalized GFP fluorescence of Synechocystis cultures harboring the PtacI, Ptrc.x.tetO1, Ptrc.x.araO, Ptrc.x.tetO2 and Ptrc.x.lacO. Measurements were performed up to three days (0, 24, 48 and 72 h) and the fluorescence was normalized to Abs790. The fluorescence of the control strain harboring the pSEVA251 was subtracted from each sample. Error bars correspond to standard deviations from three biological replicates with technical triplicates (measured in duplicate). PrnpB::gfp was included for comparison purposes. (b) Confocal micrographs of Synechocystis cells harboring the PrnpB, Ptrc.x.tetO2or Ptrc.x.lacO promoters assembled with the GFP generator. Autofluorescence is depicted in the left column, GFP signal in the middle column and the merged signals in the right column. Scale bar, 2 µm.
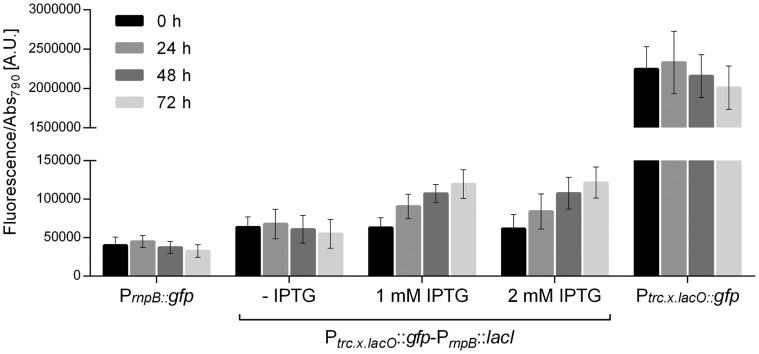
The regulated expression of the redesigned promoters based on Ptrc1O was also evaluated, cloning the genes encoding the regulatory proteins LacI, AraC or TetR downstream of the respective promoter::gfp assembly, in a similar approach to that described for the heterologous promoters (Figure 4a). The promoter Ptrc.x.lacO was designed to include a second lac operator—the lacOid that has a modified sequence known to increase its affinity to the LacI in E. coli—16 bp upstream of the -35 box (52). In Synechocystis, characterization of the Ptrc.x.lacO promoter in the presence of LacI, revealed a 97% reduction in fluorescence compared to constitutive levels (Figure 8). This is in agreement with previous work in E. coli demonstrating that the presence of two spatially separated LacI-binding operators in phase can confer a tight repression of Plac through the formation of a DNA loop (53). In Synechocystis, similar repression levels were obtained for a Plac based promoter with two lacOid operators separated by 50 bp (51), the same distance between the operators of the Ptrc.x.lacO. In E. coli, the LacI mediated repression can be reverted in presence of isopropyl β-D-1-thiogalactopyranoside (IPTG) or lactose. Therefore, the derepression of the Ptrc.x.lacO by IPTG was tested in Synechocystis. Three days (72 h) after the addition of 1 or 2 mM of IPTG, the expression levels increased 2.2-fold compared to repression conditions (-IPTG) (Figure 8). However, increasing the IPTG concentration up to 20 mM or using lactose did not increase the efficiency of derepression, which could be due to: high amounts of LacI, tight repression by DNA looping, high affinity of the lacOid operator and/or lack of an import system for IPTG or lactose. This limited derepression was also previously reported by Huang et al. (8) Recently, the permeability of Synechocystis to small organic nutrients was shown to be >20-fold lower than that of E. coli (54), which could account for the poor promoter derepression response to IPTG. Nevertheless, the introduction of the E. coli’s lactose permease (lacY) in Synechocystis did not result in increased levels of derepression (20).
Figure 8.
Characterization of the regulated expression of the Ptrc.x.lacO in Synechocystis. Normalized GFP fluorescence was determined for cell cultures harboring the redesigned trc promoter and expressing the LacI repressor (Ptrc.x.lacO::gfp-PrnpB::lacI) without (-) or with addition (1 or 2 mM) of IPTG. Measurements were performed up to three days (0, 24, 48 and 72 h) and the fluorescence was normalized to Abs790. The fluorescence of the control strain harboring the pSEVA251 was subtracted from each sample. Error bars correspond to standard deviations from three biological replicates with technical triplicates (measured in duplicate). Data from PrnpB::gfp and Ptrc.x.lacO::gfp were included for comparison purposes.
Ptrc.x.araO was designed to be efficiently repressed by the AraC. For this purpose, two araI1 half-sites from the E. coli’s PBAD promoter (55) were included. However, only a 15% reduction in fluorescence was observed in the presence of AraC compared to the constitutive levels (data not shown), which may be due to the inefficient formation of the DNA loop using two araI1 operator regions. This is unexpected, since the 111 bp distance between the centers of the araI1 operators should be sufficient to enable the formation of the DNA loop through the binding of the AraC homodimer, and also, the operators should be in phase since 11.1 bp is the periodicity of PBAD repression (45).
Finally, the Ptrc promoter was redesigned replacing the original operator sequence by the tetO operator that was included 8 bp downstream of the -10 box – Ptrc.x.tetO1. In Synechocystis, evaluation of the expression regulated by TetR revealed a significant repression with 89% reduction in fluorescence compared to constitutive levels (Supplementary Figure S4). The derepression of this promoter using anhydrotetracycline hydrochloride (aTc) was tested, and a 2.3-fold increase in GFP expression levels was registered 72 h after the addition of 1 µg/ml aTc, compared to repression conditions (-aTc) (Supplementary Figure S4). Increasing the aTc concentration up to 10 µg/ml did not result in higher derepression levels (data not shown), which cannot be related to the light-sensitivity of aTc (10) since these experiments were conducted in the dark. However, the use of aTc was previously demonstrated to derepress TetR regulated promoters but this occurred in the presence of two operator regions (56) instead of the single operator used in this study. The presence of two operators seems to be essential to regulate the system. A second version of a TetR regulated Ptrc promoter was designed harboring a modified tetO operator that differs from the original sequence in four nucleotides (57). When tested in Synechocystis, the Ptrc.x.tetO2 was not repressed in the presence of TetR (data not shown), which is in agreement with the results obtained in E. coli (57).
Another promoter tested in this study was PpsbA2*, a synthetic regulatory element based on one of the strongest Synechocystis native promoters (PpsbA2) from which the elements for light dependent regulation were removed (13). The functionality of the PpsbA2* was previously demonstrated in Synechocystis (13), and its characterization here revealed that it is 6-fold stronger than the reference promoter PrnpB (Supplementary Figure S5).
3.4 The expanded toolbox at work: restoring the production of a compatible solute
From the set of novel and validated molecular tools described above, the synthetic constitutive promoter Ptrc.x.lacO was used in the assembly of a synthetic device meant to restore the production the compatible solute GG in a Synechocystis deletion mutant (ΔggpS). Compatible solutes are low molecular weight organic molecules that can be accumulated intracellularly without interfering with essential processes or metabolism and allow the organism to cope with environmental stresses such as temperature, salt or drought (58). Synechocystis is a moderately halotolerant cyanobacterium that is known to produce sucrose and GG as major compatible solutes (58). GG is synthesized via the glucosylglycerol-phosphate synthase (GgpS) and glucosylglycerol-phosphate phosphatase (GgpP) pathway (58). The synthetic GG producing device, including the ggpS ORF preceded by Ptrc.x.lacO and the strong RBS BBa_B0030 (Supplementary Figure S6a), was cloned in pSEVA351 and transformed into Synechocystis ΔggpS via electroporation.
The growth of Synechocystis wild-type (WT), the ΔggpS mutant, and the complemented ΔggpS mutant (ΔggpS_GG device) in BG11 medium containing 0, 3 or 5% (wt/vol) NaCl was monitored (Supplementary Figure S6b). The results suggest that, in absence of salt, the presence of the GG device imposes a metabolic burden due to increased ggpS gene transcription (Supplementary Figure S7). However, in the presence of 3% (wt/vol) NaCl no significant differences between the three strains were observed, while with 5% (wt/vol) NaCl the growth of the ΔggpS mutant complemented with the GG device is similar to the wild-type, pointing out to the functionality of our device/tools.
In addition, the transcript levels of ggpS and ggpP were analyzed by RT-qPCR (Supplementary Figure S7). As shown in Supplementary Figure S7, cultivation of Synechocystis wild-type in medium supplemented with NaCl leads to an increase in relative transcript levels of both ggpS and ggpP, in agreement with previous studies (59, 60). In the ΔggpS mutant, the transcription profile of ggpP was similar to the one exhibited by the wild-type. When the ΔggpS mutant is complemented by the introduction of the GG device, a 45-fold increase in transcription could be observed for ggpS in 5% (wt/vol) NaCl compared to the wild-type, clearly demonstrating the functionality of the synthetic GG device at the transcriptional level.
Moreover, the presence of salt-stress related compatible solutes was evaluated using H-NMR (Supplementary Figure S8). In the wild-type, GG is detected only in presence of salt while glutamate is present independently of the stress. As expected, GG was not detected in the ΔggpS mutant, but since sucrose and glutamate could be detected under 3% (wt/vol) NaCl, and the ΔggpS mutant was able to grow similarly to the wild-type, we postulate that sucrose and glutamate can compensate for the absence of GG. The complementation of the ΔggpS mutant with the GG device restored the production of this compatible solute in presence of NaCl (Supplementary Figure S8, lower right panel). This salt-dependent synthesis of GG is in agreement with previous works, showing that the GgpS enzyme is inhibited by electrostatic interaction with nucleic acids and that its activation occurs only in the presence of salt (60, 61). Thus, the synthetic GG device assembled using the promoters developed herein was shown to be fully functional.
4. Conclusions
In this study, we expanded the synthetic biology toolbox for the photoautotrophic cyanobacterial chassis Synechocystis sp. PCC 6803: three self-replicating plasmids available at the SEVA repository were validated and 13 heterologous and redesigned promoters based on PT7pol and Ptrc were characterized, exhibiting a wide range of activities varying from ∼0.13- to ∼41-fold compared to PrnpB. From this set of promoters, three could be efficiently repressed (PλcI, Ptrc.x.lacO and Ptrc.x.tetO1). Derepression of the LacI- and TetR-regulated redesigned promoters using IPTG or aTc was also shown, although a modest effect (about ∼2-fold increase) was observed. This study constitutes a solid basis for the development and implementation of more efficient and tightly regulated elements required by applications using cyanobacteria as cell factories. Moreover, the tools developed and presented here allowed the assembly of a synthetic module with ggpS gene and its introduction into the chassis—a Synechocystis ggpS deficient mutant, restoring the production of the compatible solute GG.
Supplementary Material
Acknowledgements
The authors wish to thank the Parasite Disease group (i3S/IBMC) for the use of Synergy 2 Multi-Mode Microplate Reader. We are also grateful to Prof. Helena Santos and Sara Rebelo (ITQB, Universidade Nova de Lisboa) for the collaboration and technical assistance regarding compatible solutes extraction and quantification, and Prof. Peter Lindblad (Uppsala University) for providing the initial aliquot of the anti-LacI polyclonal antibody.
Funding
European Commission through the 7th Framework Programme FP7-ENERGY-2012-1-2STAGE-308518 project CyanoFactory [308518] and 6th Framework Programme EU FP6-NEST-2005-Path-SYN project BioModularH2 [043340]; National Portuguese Funds through FCT—Fundação para a Ciência e a Tecnologia scholarships SFRH/BD/117508/2016 (E.A.F.), SFRH/BD/36378/2007 (F.P.), SFRH/BPD/64095/2009 (C.C.P.) and SFRH/BPD/74894/2010 (P.O.) and by the FCT Investigator Programme (IF/00256/2015) (P.O.); and by project NORTE-01-0145-FEDER-000012—Structured Programme on Bioengineering Therapies for Infectious Diseases and Tissue Regeneration, supported by Norte Portugal Regional Operational Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF).
Conflict of interest statement. None declared.
References
- 1. Knoll A.H. (2008) Cyanobacteria and earth history In: Flores FG, Herrero A (eds). The Cyanobacteria: Molecular Biology, Genomics, and Evolution. Caister Academic Press, Norfolk, UK, pp. 1–19. [Google Scholar]
- 2. Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hirosawa M., Sugiura M., Sasamoto S.. et al. (1996) Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC 6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res., 3, 109–136. [DOI] [PubMed] [Google Scholar]
- 3. Montagud A., Zelezniak A., Navarro E., de Córdoba P.F., Urchueguía J.F., Patil K.R. (2011) Flux coupling and transcriptional regulation within the metabolic network of the photosynthetic bacterium Synechocystis sp. PCC 6803. Biotechnol. J., 6, 330–342. [DOI] [PubMed] [Google Scholar]
- 4. Fu P. (2009) Genome-scale modeling of Synechocystis sp. PCC 6803 and prediction of pathway insertion. J. Chem. Technol. Biotechnol., 84, 473–483. [Google Scholar]
- 5. Branco dos Santos F., Du W., Hellingwerf K.J. (2014) Synechocystis: not just a plug-bug for CO2, but a green E. coli. Front. Bioeng. Biotechnol., 2, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Tsai C.-S., Kwak S., Turner T.L., Jin Y.-S. (2015) Yeast synthetic biology toolbox and applications for biofuel production. FEMS Yeast Res., 15, 1–15. [DOI] [PubMed] [Google Scholar]
- 7. Xu P., Vansiri A., Bhan N., Koffas M.A.G. (2012) ePathBrick: a synthetic biology platform for engineering metabolic pathways in E. coli. ACS Synth. Biol., 1, 256–266. [DOI] [PubMed] [Google Scholar]
- 8. Huang H.-H., Camsund D., Lindblad P., Heidorn T. (2010) Design and characterization of molecular tools for a Synthetic Biology approach towards developing cyanobacterial biotechnology. Nucleic Acids Res., 38, 2577–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cardinale S., Arkin A.P. (2012) Contextualizing context for synthetic biology—identifying causes of failure of synthetic biological systems. Biotechnol. J., 7, 856–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huang H.-H., Lindblad P. (2013) Wide-dynamic-range promoters engineered for cyanobacteria. J. Biol. Eng., 7, 10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Englund E., Liang F., Lindberg P. (2016) Evaluation of promoters and ribosome binding sites for biotechnological applications in the unicellular cyanobacterium Synechocystis sp. PCC 6803. Sci. Rep., 6, 36640.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Immethun C.M., DeLorenzo D.M., Focht C.M., Gupta D., Johnson C.B., Moon T.S. (2017) Physical, chemical, and metabolic state sensors expand the synthetic biology toolbox for Synechocystis sp. PCC 6803. Biotechnol. Bioeng., 114, 1561–1569. [DOI] [PubMed] [Google Scholar]
- 13. Pinto F., Pacheco C.C., Oliveira P., Montagud A., Landels A., Couto N., Wright P.C., Urchueguía J.F., Tamagnini P. (2015) Improving a Synechocystis-based photoautotrophic chassis through systematic genome mapping and validation of neutral sites. DNA Res., 22, 425–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Liu D., Pakrasi H.B. (2018) Exploring native genetic elements as plug-in tools for synthetic biology in the cyanobacterium Synechocystis sp. PCC 6803. Microb. Cell Fact. 17, 48.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lindberg P., Park S., Melis A. (2010) Engineering a platform for photosynthetic isoprene production in cyanobacteria, using Synechocystis as the model organism. Metab. Eng., 12, 70–79. [DOI] [PubMed] [Google Scholar]
- 16. Zhou J., Zhang H., Meng H., Zhu Y., Bao G., Zhang Y., Li Y., Ma Y. (2015) Discovery of a super-strong promoter enables efficient production of heterologous proteins in cyanobacteria. Sci. Rep., 4, 4500.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Peca L., Kós P., Vass I. (2007) Characterization of the activity of heavy metal-responsive promoters in the cyanobacterium Synechocystis PCC 6803. Acta Biol. Hung., 58, 11–22. [DOI] [PubMed] [Google Scholar]
- 18. Muramatsu M., Hihara Y. (2007) Coordinated high-light response of genes encoding subunits of photosystem I is achieved by AT-rich upstream sequences in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol., 189, 2750–2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Abe K., Miyake K., Nakamura M., Kojima K., Ferri S., Ikebukuro K., Sode K. (2014) Engineering of a green-light inducible gene expression system in Synechocystis sp. PCC 6803. Microb. Biotechnol., 7, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Albers S.C., Gallegos V.A., Peebles C.A.M. (2015) Engineering of genetic control tools in Synechocystis sp. PCC 6803 using rational design techniques. J. Biotechnol., 216, 36–46. [DOI] [PubMed] [Google Scholar]
- 21. Taton A., Unglaub F., Wright N.E., Zeng W.Y., Paz-Yepes J., Brahamsha B., Palenik B., Peterson T.C., Haerizadeh F., Golden S.S.. et al. (2014) Broad-host-range vector system for synthetic biology and biotechnology in cyanobacteria. Nucleic Acids Res., 42, e136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ng A.H., Berla B.M., Pakrasi H.B. (2015) Fine-tuning of photoautotrophic protein production by combining promoters and neutral sites in the cyanobacterium Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol., 81, 6857–6863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ducat D.C., Way J.C., Silver P.A. (2011) Engineering cyanobacteria to generate high-value products. Trends Biotechnol., 29, 95–103. [DOI] [PubMed] [Google Scholar]
- 24. Lindblad P., Lindberg P., Oliveira P., Stensjö K., Heidorn T. (2012) Design, engineering, and construction of photosynthetic microbial cell factories for renewable solar fuel production. Ambio, 41, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang B., Wang J., Zhang W., Meldrum D.R. (2012) Application of synthetic biology in cyanobacteria and algae. Front. Microbiol., 3, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kanesaki Y., Shiwa Y., Tajima N., Suzuki M., Watanabe S., Sato N., Ikeuchi M., Yoshikawa H. (2012) Identification of substrain-specific mutations by massively parallel whole-genome resequencing of Synechocystis sp. PCC 6803. DNA Res., 19, 67–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Trautmann D., Voss B., Wilde A., Al-Babili S., Hess W.R. (2012) Microevolution in cyanobacteria: re-sequencing a motile substrain of Synechocystis sp. PCC 6803. DNA Res., 19, 435–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Stanier R.Y., Kunisawa R., Mandel M., Cohen-Bazire G. (1971) Purification and properties of unicellular blue-green algae (order Chroococcales). Bacteriol. Rev., 35, 171–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sambrook J., Russel D. (2001) Molecular Cloning: A Laboratory Manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York, USA. [Google Scholar]
- 30. Imburgio D., Rong M., Ma K., McAllister W.T. (2000) Studies of promoter recognition and start site selection by T7 RNA polymerase using a comprehensive collection of promoter variants. Biochemistry, 39, 10419–10430. [DOI] [PubMed] [Google Scholar]
- 31. Diaz G.A., Raskin C.A., McAllister W.T. (1993) Hierarchy of base-preference in the binding domain of the bacteriophage T7 promoter. J. Mol. Biol., 229, 805–811. [DOI] [PubMed] [Google Scholar]
- 32. de Boer H.A., Comstock L.J., Vasser M. (1983) The tac promoter: a functional hybrid derived from the trp and lac promoters. Proc. Natl. Acad. Sci. U. S. A., 80, 21–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Brosius J., Erfle M., Storella J. (1985) Spacing of the -10 and -35 regions in the tac promoter. Effect on its in vivo activity. J. Biol. Chem., 260, 3539–3541. [PubMed] [Google Scholar]
- 34. Silva-Rocha R., Martínez-García E., Calles B., Chavarría M., Arce-Rodríguez A., de las Heras A., Páez-Espino A.D., Durante-Rodríguez G., Kim J., Nikel P.I.. et al. (2013) The Standard European Vector Architecture (SEVA): a coherent platform for the analysis and deployment of complex prokaryotic phenotypes. Nucleic Acids Res., 41, D666–D675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pinto F., Pacheco C.C., Ferreira D., Moradas-Ferreira P., Tamagnini P. (2012) Selection of suitable reference genes for RT-qPCR analyses in cyanobacteria. PLoS One, 7, e34983.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schäfer A., Tauch A., Jäger W., Kalinowski J., Thierbach G., Pühler A. (1994) Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene, 145, 69–73. [DOI] [PubMed] [Google Scholar]
- 37. Williams J.G. (1988) Construction of specific mutations in photosystem II photosynthetic reaction center by genetic engineering methods in Synechocystis 6803. Methods Enzymol., 167, 766–778. [Google Scholar]
- 38. Ludwig A., Heimbucher T., Gregor W., Czerny T., Schmetterer G. (2008) Transformation and gene replacement in the facultatively chemoheterotrophic, unicellular cyanobacterium Synechocystis sp. PCC 6714 by electroporation. Appl. Microbiol. Biotechnol., 78, 729–735. [DOI] [PubMed] [Google Scholar]
- 39. Elhai J., Wolk C.P. (1988) Conjugal transfer of DNA to cyanobacteria. Methods Enzymol., 167, 747–754. [DOI] [PubMed] [Google Scholar]
- 40. Santos H., Lamosa P., Borges N. (2006) Characterization and quantification of compatible solutes in (hyper)thermophilic microorganisms In: Rainey FA and Oren A (eds.) Methods in Microbiology. Academic Press, Amsterdam, Netherlands, pp. 173–199. [Google Scholar]
- 41. Dyczmons N.G. (2006) Expression and regulation of membrane proteins: special focus on cytochrome bd-oxidase from Synechocystis sp. PCC 6803, Ph. D. Thesis. Ruhr-Universität Bochum, Bochum, Germany. [Google Scholar]
- 42. Camsund D., Lindblad P. (2014) Engineered transcriptional systems for cyanobacterial biotechnology. Front. Bioeng. Biotechnol., 2, 40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Heidorn T., Camsund D., Huang H.-H., Lindberg P., Oliveira P., Stensjö K., Lindblad P. (2011) Synthetic biology in cyanobacteria: engineering and analyzing novel functions. Methods Enzymol., 497, 539–579. [DOI] [PubMed] [Google Scholar]
- 44. Hamilton E.P., Lee N. (1988) Three binding sites for AraC protein are required for autoregulation of araC in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A., 85, 1749–1753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lee D.H., Schleif R.F. (1989) In vivo DNA loops in araCBAD: size limits and helical repeat. Proc. Natl. Acad. Sci. U. S. A., 86, 476–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Lee N., Francklyn C., Hamilton E.P. (1987) Arabinose-induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. U. S. A., 84, 8814–8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schleif R. (2010) AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev., 34, 779–796. [DOI] [PubMed] [Google Scholar]
- 48. Nasser W., Reverchon S. (2007) New insights into the regulatory mechanisms of the LuxR family of quorum sensing regulators. Anal. Bioanal. Chem., 387, 381–390. [DOI] [PubMed] [Google Scholar]
- 49. Pearson B., Lau K.H., DeLoache W., Penumetcha P., Rinker V.G., Allen A., Cool R., Feeney E., Igo J., Eckdahl T.T.. et al. (2011) Bacterial hash function using DNA-based XOR logic reveals unexpected behavior of the luxR promoter. Interdiscip. Bio Central, 3, 1. [Google Scholar]
- 50. Lewis D., Le P., Zurla C., Finzi L., Adhya S. (2011) Multilevel autoregulation of λ repressor protein CI by DNA looping in vitro. Proc. Natl. Acad. Sci. U. S. A., 108, 14807–14812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Camsund D., Heidorn T., Lindblad P. (2014) Design and analysis of LacI-repressed promoters and DNA-looping in a cyanobacterium. J. Biol. Eng., 8, 4.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Müller J., Oehler S., Müller-Hill B. (1996) Repression of lac promoter as a function of distance, phase and quality of an auxiliary lac operator. J. Mol. Biol., 257, 21–29. [DOI] [PubMed] [Google Scholar]
- 53. Schleif R. (1992) DNA looping. Annu. Rev. Biochem., 61, 199–223. [DOI] [PubMed] [Google Scholar]
- 54. Kowata H., Tochigi S., Takahashi H., Kojima S. (2017) Outer membrane permeability of cyanobacterium Synechocystis sp. strain PCC 6803: studies of passive diffusion of small organic nutrients reveal the absence of classical porins and intrinsically low permeability. J. Bacteriol., 199, e00371–e00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Seabold R.R., Schleif R.F. (1998) Apo-AraC actively seeks to loop. J. Mol. Biol., 278, 529–538. [DOI] [PubMed] [Google Scholar]
- 56. Yao L., Cengic I., Anfelt J., Hudson E.P. (2016) Multiple gene repression in cyanobacteria using CRISPRi. ACS Synth. Biol., 5, 207–212. [DOI] [PubMed] [Google Scholar]
- 57. Krueger M., Scholz O., Wisshak S., Hillen W. (2007) Engineered Tet repressors with recognition specificity for the tetO-4C5G operator variant. Gene, 404, 93–100. [DOI] [PubMed] [Google Scholar]
- 58. Hagemann M. (2013) Genomics of salt acclimation: synthesis of compatible solutes among cyanobacteria In: Franck Chauvat CC-C. (ed.). Advances in Botanical Research. Academic Press, Amsterdam, Netherlands, pp. 27–55. [Google Scholar]
- 59. Hagemann M., Richter S., Mikkat S. (1997) The ggtA gene encodes a subunit of the transport system for the osmoprotective compound glucosylglycerol in Synechocystis sp. strain PCC 6803. J. Bacteriol., 179, 714–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Marin K., Huckauf J., Fulda S., Hagemann M. (2002) Salt-dependent expression of glucosylglycerol-phosphate synthase, involved in osmolyte synthesis in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol., 184, 2870–2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Novak J.F., Stirnberg M., Roenneke B., Marin K. (2011) A novel mechanism of osmosensing, a salt-dependent protein-nucleic acid interaction in the cyanobacterium Synechocystis species PCC 6803. J. Biol. Chem., 286, 3235–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.