Abstract
Background
Ticks are hematophagous arthropods responsible for maintenance and transmission of several pathogens of veterinary and medical importance. Current knowledge on species diversity and pathogens transmitted by ticks infesting camels in Nigeria is limited. Therefore, the aim of this study was to unravel the status of ticks and tick-borne pathogens of camels in Nigeria.
Methods
Blood samples (n = 176) and adult ticks (n = 593) were collected from one-humped camels (Camelus dromedarius) of both sexes in three locations (Kano, Jigawa and Sokoto states) in north-western Nigeria and screened for the presence of Rickettsia spp., Babesia spp., Anaplasma marginale, Anaplasma spp. and Coxiella-like organisms using molecular techniques. All ticks were identified to species level using a combination of morphological and molecular methods.
Results
Ticks comprised the three genera Hyalomma, Amblyomma and Rhipicephalus. Hyalomma dromedarii was the most frequently detected tick species (n = 465; 78.4%) while Amblyomma variegatum (n = 1; 0.2%) and Rhipicephalus evertsi evertsi (n = 1; 0.2%) were less frequent. Other tick species included H. truncatum (n = 87; 14.7%), H. rufipes (n = 19; 3.2%), H. impeltatum (n = 18; 3.0%) and H. impressum (n = 2; 0.3%). The minimum infection rates of tick-borne pathogens in 231 tick pools included Rickettsia aeschlimannii (n = 51; 8.6%); Babesia species, (n = 4; 0.7%) comprising of B. occultans (n = 2), B. caballi (n = 1) and Babesia sp. (n = 1); Coxiella burnetii (n = 17; 2.9%); and endosymbionts in ticks (n = 62; 10.5%). We detected DNA of “Candidatus Anaplasma camelli” in 40.3% of the blood samples of camels. Other tick-borne pathogens including Anaplasma marginale were not detected. Analysis of risk factors associated with both tick infestation and infection with Anaplasma spp. in the blood indicated that age and body condition scores of the camels were significant (P < 0.05) risk factors while gender was not.
Conclusions
This study reports low to moderate prevalence rates of selected tick-borne pathogens associated with camels and their ticks in north-western Nigeria. The presence of zoonotic R. aeschlimannii emphasizes the need for a concerted tick control programme in Nigeria.
Keywords: Ticks, Tick-borne pathogens, Piroplasms, “Candidatus Anaplasma camelli”, Camels, Nigeria
Background
Ticks are responsible for substantial economic losses to farmers in livestock-keeping tropical regions of the world. Tick infestations cause wounds and inflammations due to tick bites, blood loss and potential diseases through transmission of pathogens [1]. The tick fauna infesting livestock in Africa is diverse with species belonging to the genera Hyalomma, Rhipicephalus and Amblyomma, having the highest impact on the productivity and health of these animals [2]. Tick-borne pathogens (TBPs) include viruses, bacteria, protozoans, and helminths afflicting humans’ and animals’ health worldwide [1]. Complex and dynamic interactions occur inside ticks with multiple microbes ranging from pathogens to endosymbionts [3]. The former is responsible for diseases, while the latter play a crucial role in maintaining fitness to the vector.
Tick-borne rickettsioses are caused by intracellular bacteria of the genus Rickettsia. Clinical manifestations include high fever, rash, myalgia, headache and lymphadenitis [4]. Rickettsia africae and R. aeschlimannii belong to the zoonotic spotted fever group (SFG) rickettsiae and have been reported from feeding hard ticks collected from livestock in Nigeria [5, 6].
Members of the genera Anaplasma and Ehrlichia (family Anaplasmataceae) can infect both animals and humans [7]. Limited studies have been conducted regarding the infection of camels with Anaplasmataceae. For example, Anaplasma marginale has been detected in camels using serological tests [8, 9]. However, other studies found no evidence of DNA of this bacterium [10, 11]. On the other hand, DNA of a novel species of Anaplasma, “Candidatus Anaplasma camelii” has been confirmed by sequencing in the blood of camels in various countries [12–14]. Infected animals may present clinical signs like anorexia, respiratory distress, edema of the sternum and xiphoid or even sudden death [13].
Coxiella burnetii, the causative agent of Q fever, is a zoonotic pathogen of vertebrates which is distributed worldwide [3]. Clinical manifestations are self-limiting febrile conditions in the majority of the cases and reproductive disorder in some animals [15]. Interestingly, strains of Coxiella burnetii have their origin from the diverse group of Coxiella-like endosymbionts, which are descendants of a Coxiella-like progenitor hosted by ticks [3].
Apicomplexan protozoans of the genus Babesia are transmitted by hard ticks [16]. Dromedaries are no exception to infection with Babesia although very few published reports exist so far [17, 18]. The pathogenicity differs according to the Babesia species. Babesia caballi causes severe clinical disease in equines characterized by fever, anemia, hemoglobinuria, and edema in some cases [19], while B. occultans is of lower pathogenicity in animals as previously reported in cattle with no visible clinical signs [20]. In camels, reported clinical signs of babesiosis includes anemia, fever, icterus, hemoglobinuria, and gastro-intestinal stasis [21].
Current estimates in Nigeria on the one-humped camel (Camelus dromedaries) population are at about 283,395 heads [22]. Pastoralists primarily keep these animals for transportation and as source of meat. The carcass yield from camels are higher under cheap management system. Recent estimates show that the consumption of camel meat in Nigeria has increased substantially due to its nutritional value and for health reasons [23]. Camel meat has relatively less fat compared to cattle and sheep and is acclaimed to cure diseases like hypertension, hyperacidity, and cardiovascular disease [24]. On the other hand, researchers in Nigeria consider the dromedary camel as a ‘foreign animal’ and this has led to research apathy on this animal species in recent past [25]. As desertification continues to encroach into sub-Saharan Africa, renewed interest is also gradually building up in northern Nigeria, as the camel is resilient to the arid land conditions and seems certainly the best option to mitigate the effects of environmental conditions on livestock production among the pastoralist in northern Nigeria [26]. In Nigeria, dromedary camels are raised in semi-arid conditions grazing on poor pastures for most of the year where they are exposed to a wide variety of vectors including ticks. This shows the need to ascertain this source of potential disease. In order to be better prepared to raise this animal species successfully without the debilitating effects of ticks and tick-borne disease on their health and productivity, this study was carried out to assess (i) the species diversity of ticks on camels, (ii) the occurrence of selected tick-borne pathogens in ticks collected and blood taken from camels and (iii) the risk factors associated with infection of camels with tick-borne pathogens in Nigeria.
Methods
Study area
The North-West region is a semi-arid zone and the largest region in Nigeria with a combined human population of 35,786,944 [27]. This region has a savannah type of vegetation favorable to camel husbandry because they are easily predisposed to foot rot associated with wetland and this hence the concentration of camels at this region [28]. The temperature ranges from 18 °C to 45 °C with a mean temperature of 27 °C. There is a single rainy season from May to October with mean annual rainfall of 508–1016 mm. Three states (Sokoto, Jigawa and Kano) were selected for sampling (Fig. 1).
Fig. 1.
Map of Nigeria with insert of the northwestern region showing the study areas where samples were collected (Maps were created using ArcGIS version 10.6 by ESRI, Redlands, CA, USA)
Study design and sampling locations
A cross sectional study was carried out from September to November 2017. Additional samples were collected in November 2018. Non-probability sampling, combining both convenient and snowball sampling techniques were employed. Blood and tick samples were collected from several sampling points across the three study areas comprising of abattoirs, livestock markets and herders/pastoralists. All samples from Kano (n = 92) were collected from the Kano metropolitan abattoir (12.0123540N, 8.520795E) located in the city of Kano. For Sokoto state, all samples (n = 55) were collected from herders/pastoralists at several locations within the state. The geographical coordinates for the state of Sokoto are 12.1358N, 4.8654E. Finally, the livestock market located in Maigatari local government of Jigawa state (12.8125483N, 9.444303E) as well as adjoining local villages within this area were used for sampling in Jigawa state (n = 29). Sampled animals from all study areas were raised under the traditional nomadic (extensive) management system typical of camel husbandry in Africa with little access to veterinary care. Information such as age (< 5 years/> 5 years), sex (male/female) and presence/absence of ticks were collected for each animal to assess possible risk factors associated with tick-borne pathogen infection. Body condition score were classified into any of the three classes (poor, moderate and good) based on the fat storage at the back and flank region using visual inspection. All samples were collected from animals that were apparently healthy without any clinical signs of infection after seeking the owner’s consent and approval.
Blood and tick sample collection
About 5 ml of whole blood were collected from the jugular vein and in some cases from the lateral abdominal vein of clinically healthy animals and in the case of slaughtered animals, from severed jugular blood vessels. All collected blood samples were transferred into labelled EDTA coated tubes and transported to the laboratory on ice packs within 4 h. In the laboratory, 125 µl of blood was dispensed on the marked spot within the Classic FTA card (Whatman ® GE Healthcare, Buckinghamshire, UK). All cards were labeled, air dried and stored at room temperature for further analysis. The skin of the camels covering known predilection sites for ticks including the perineum region, abdomen and thigh, ear, neck, and dewlap were carefully examined for the presence of ticks. Ticks were collected using tweezers into labelled tubes plugged with cotton. Ticks from each animal were kept in separate tubes. The labelled tubes contained information on the identity of the animal including their location.
Morphological identification of ticks
All tick samples collected from infested animals were identified to species level based on standard keys using a stereomicroscope (Olympus®, Tokyo, Japan) separately by two of the co-authors [29]. Specimens were separated based on species, life stage and sex. All tick specimens were preserved in 70% ethanol and kept at 4 °C after identification.
Washing and homogenization of ticks
Individual ticks were washed twice with double distilled water after the removal of ethanol in individual Eppendorf tubes as described by Silaghi et al. [30]. A 5 mm sterile stainless-steel bead and 100 µl of sterile PBS were added to each tube. Ticks were homogenized using Tissue Lyser II (Qiagen, Hilden, Germany) for 60 s twice with 30 s break in between at an oscillation frequency of 30 Hz. After centrifugation at 2500× rpm for 3 min, the supernatant was removed.
Pooling of supernatant and extraction of genomic DNA from tick homogenates and FTA cards
Prior to extraction, the supernatants from ticks of the same species and the same animal were pooled with a maximum of 5 ticks per pool. A maximum of 80 µl of the homogenate (supernatant) was used for DNA extraction (individual tick contributing a maximum of 16 µl). For partially-fed ticks, supernatants were either pooled or used individually (engorged ticks were used individually). Extraction of DNA from FTA cards (blood) was performed from approximately a 6 mm punch from the dried blood spot on the card. The spot was carefully excised into a sterile 2 ml labelled Eppendorf tube containing one 4 mm sterile stainless-steel bead. The samples were then lysed using Tissue Lyser II (Qiagen) for 60 s twice. Isolation of genomic DNA was carried out with QIAamp DNA Mini Kit (Qiagen) according to manufacturer’s instruction. Genomic DNA was stored at − 20 °C until use.
Tick species identification using PCR
For the molecular identification of tick species, three different genes (12S rRNA, 16S rRNA and cox1) were targeted using primer pairs shown in Table 1. Genetic identification of ticks was carried out using DNAs extracted from a single tick representative of each species. The reaction was performed in total volume of 25 μl using the GoTaq® G2 Flexi DNA Polymerase Kit (Promega, Madison, WI, USA). The PCR mix consisted of 5 μl GoTaq® 5× Flexi Buffer (green), 3 μl 25 mM MgCl2 solution, 0.5 μl 10 mM dNTPs, 1 μl of each primer (both forward and reverse) (10 µM), 0.1 μl of GoTaq® DNA Polymerase (5 u/μl), 9.4 µl nuclease-free water (NFW) and 5 µl template DNA. A thermal cycler C1000 (Bio-Rad, Munich, Germany) was used for amplification and the cycling conditions are provided in Table 1.
Table 1.
Primer sets used for DNA amplification and sequencing of ticks and tick-borne pathogens in ticks and camels from north-western Nigeria
| Target | Method | Gene target | Primer sequence (5′-3′) | Product size (bp) | Positive control (DNA) from ticks | Reference |
|---|---|---|---|---|---|---|
| Tick identification | PCR | 12S rRNA | T1B: AAACTAGGATTAGATACCCT | 360 | Hyalomma dromedarii | [35] |
| T2A: AATGAGAGCGACGGGCGATGT | ||||||
| Tick identification | PCR | 16S rRNA | 16S + 1: CTGCTCAATGATTTTTTAAATTGCTGTGG | 456 | H. dromedarii | [36] |
| 16S − 1: CCGGTCTGAACTCAGATCAAGTA | ||||||
| Tick identification | PCR | cox1 | Cox1F: GGAACAATATATTTAATTTTTGG | 360 | H. dromedarii | [37] |
| Cox1R: ATCTATCCCTACTGTAAATATATG | ||||||
| Rickettsia spp. | PCR | gltA | Rsfg877: GGGGGCCTGCTCACGGCGG | 381 | Rickettsia helvetica | [38] |
| Rfsg1258: ATTGCAAAAAGTACAGTGAACA | ||||||
| Rickettsia spp. | PCR | ompA | Rr190.70p: ATGGCGAATATTTCTCCAAAA | 631 | R. helvetica | [39] |
| Rr190.701n: GTTCCGTTAATGGCAGCATCT | ||||||
| Rickettsia spp. | PCR | ompB | 120–2788: AAACAATAATCAAGGTACTGT | 765 | R. helvetica | [40] |
| 120–3599: TACTTCCGGTTACAGCAAAGT | ||||||
| Babesia/Theileria | PCR | 18S rRNA | BJ1: GTCTTGTAATTGGAATGATGG | 411–452 | Babesia spp. | [41] |
| BN2: TAGTTTATGGTTAGGACTACG | ||||||
| BabsppF1: GTTTCTGMCCCATCAGCTTGAC | 422–440 | [42] | ||||
| BabsppR: CAAGACAAAAGTCTGCTTGAAAC | ||||||
| Anaplasma marginale | qPCR | msp1ß | AM-forward: TTGGCAAGGCAGCAGCTT | 95 | Anaplasma marginale | [31] |
| AM-reverse: TTCCGCGAGCATGTGCAT | ||||||
| AM-probe: FAM TCGGTCTAACATCTCCAGGCTTTCAT BHQ1 | ||||||
| Anaplasma/Ehrlichia | PCR | 16S rRNA | EHR16SD: GGTACCYACAGAAGAAGTCC | 345 | A. marginale | [43] |
| EHR16SR: TAGCACTCATCGTTTACAGC | ||||||
| Anaplasma spp. | Semi-nested PCR | 16S rRNA | fD1: AGAGTTTGATCCTGGCTCAG | 760 | A. marginale | [44] |
| EHR16SR: TAGCACTCATCGTTTACAGC | ||||||
| fD1: AGAGTTTGATCCTGGCTCAG | 426 | [45] | ||||
| GA1UR: GAGTTTGCCGGGACTTCTTCT | ||||||
| Coxiella-like organisms | Semi-nested PCR | 16S rDNA | Cox16SF1: CGTAGGAATCTACCTTRTAGWGG | 1321–1416 | Coxiella burnetii | [46] |
| Cox16SR2: GCCTACCCGCTTCTGGTACAATT | ||||||
| Cox16SF2: TGAGAACTAGCTGTTGGRRAGT | 624–625 | |||||
| Cox16SR2: GCCTACCCGCTTCTGGTACAATT |
Molecular detection of pathogens using PCR
For pathogen detection, PCRs were used for amplification of DNA of Rickettsia spp., Anaplasma/Ehrlichia spp., A. marginale, C. burnetii and Babesia/Theileria spp. from tick DNA, while A. marginale, “Ca. A. camelii” and Babesia/Theileria spp. were screened from blood DNA. All reactions were performed in total volume of 25 μl using the GoTaq® G2 Flexi DNA Polymerase Kit (Promega) The PCR mix contained 5 μl GoTaq® 5× Flexi Buffer (green), 3 μl 25 mM MgCl2 solution, 0.5 μl 10 mM dNTPs, 400 nM of each primer (forward and reverse), 0.1 μl of GoTaq® DNA Polymerase (5 u/µl), 9.4 µl NFW and 5 µl of template DNA. Every reaction set had a positive and negative control (molecular grade water). Table 1 summarizes the PCR cycling conditions.
Gel electrophoresis and sequencing
Agarose gel electrophoresis at a concentration of 1.5% was used for the separation of PCR products with 2 μl GelRed™ (1×; equivalent to 1 µl/10 ml) (Biotium Fremont, CA, USA). Bands were visualized using a ChemiDoc™ MP imaging system (Bio-Rad). Amplicons were purified with NucleoSEQ® columns (Macherey-Nagel, Düren, Germany), according to the manufacturer’s instructions, and then sequenced in one direction using an ABI PRISM® 3130 sequencer (Applied Biosystem, California, USA) at the Institute of Diagnostic Virology, Friedrich Loeffler Institute, Germany. The nucleotide sequences were viewed and edited using Geneious 9.1 software (Biomatters, Auckland, New Zealand) and analyzed against sequences deposited in GenBank using BLASTn (National Centre for Biotechnology Information; www.blast.ncbi.nlm.nih.gov/Blast) for high similarity sequences.
Real-time PCR for the amplification of A. marginale
The msp1ß gene of A. marginale was targeted in DNA samples from ticks and camel blood using species-specific primers and probe (Table 1) as previously described [31, 32]. The PCR was carried out using a CFX-96 Real-Time System (Bio-Rad) with the cycling conditions described in Table 1. PCR amplification was carried out using iTaq™ Universal Probes Supermix (Bio-Rad) in a total volume of 25 µl comprising of 200 nM of forward and reverse primers, 100 nM of probe (Table 1), 12.5 µl (2×) iTaq™ Supermix, 0.9 µl RNase free water and 10 µl template DNA. Each reaction run included a positive and negative control.
Statistical analysis
For pooled tick samples, the prevalence was estimated using the minimum infection rate (MIR). MIR assumes that only one tick is infected in a positive pool [33]. Results are also presented with a 95% confidence interval (CI: lower and upper) for the infection rates and MIR for the detected pathogens. MIR was expressed in simple percentages (only one tick was considered as positive, in a pool of adult ticks). The calculation was carried out thus MIR = (P/N) × 100%, where P is the number of positive pools, N is the total number of ticks tested. Chi-square test was used to test for statistical significance between the various risk factors. The odds ratio was used to test the association/likelihood of the presence of ticks and infection with Anaplasma spp. The level of significance was set as P < 0.05. Statistical significance was carried out using GraphPad Prism version 5.0 (GraphPad Software, La Jolla California USA; www.graphpad.com).
Phylogenetic analysis
The nucleotide sequences were viewed and edited using Geneious 11.1.5 and analyzed against references in GenBank using BLASTn (www.blast.ncbi.nlm.nih.gov/Blast) for high similarity sequences to confirm identity for the ticks as well as for pathogens. Sequences were added to alignment explorer in MEGA 7 and aligned with ClustalW [33]. Reference sequences were also added to the aligned datasets. Model test was run in MEGA 7 prior to the tree construction in order to select the suitable model. The phylogenetic tree was constructed using the Maximum Likelihood method based on the Kimura 2-parameter model [34] with 1000 replicates. Median joining network was constructed using PopART (http://popart.otago.ac.na) to examine the haplotype distribution and relationships.
Results
Morphological and molecular identification of tick species
Of the 176 camels examined, 92 (52.3%) were infested with ticks from a total collection of 593. All ticks collected were identified as adult with no immature stages comprising of 440 (74.2%) males and 153 (25.8%) females. The largest number of ticks was collected from Kano (396; 66.8%) followed by Jigawa (145; 24.5%) and Sokoto (52; 8.8%) state (Table 2).
Table 2.
Demography of adult tick species infesting 176 camels in north-western Nigeria
| Parameter/Tick species | Total number (%) | Study locations | |||||
|---|---|---|---|---|---|---|---|
| Kano (n = 396) | Jigawa (n = 145) | Sokoto (n = 52) | |||||
| Male | Female | Male | Female | Male | Female | ||
| Hyalomma dromedarii | 465 (78.41) | 254 | 75 | 87 | 27 | 12 | 10 |
| H. truncatum | 87 (14.67) | 22 | 18 | 15 | 6 | 22 | 4 |
| H. rufipes | 19 (3.20) | 3 | 5 | 7 | 1 | 3 | 0 |
| H. impeltatum | 18 (3.03) | 12 | 4 | 1 | 0 | 1 | 0 |
| H. impressum | 2 (0.34) | 0 | 2 | 0 | 0 | 0 | 0 |
| Amblyomma variegatum | 1 (0.17) | 1 | 0 | 0 | 0 | 0 | 0 |
| Rhipicephalus evertsi evertsi | 1 (0.17) | 0 | 0 | 0 | 1 | 0 | 0 |
| Total | 593 (100.0) | 292 (49.2) | 104 (17.5) | 110 (18.5) | 35 (5.9) | 38 (6.4) | 14 (2.4) |
Altogether, 7 species of ticks were identified: Hyalomma dromedarii (n = 465; 78.4%), H. truncatum (n = 87; 14.7%), H. rufipes (n = 19; 3.2%), H. impressum (n = 2; 0.3%), H. impeltatum (n = 18; 3.0%), Amblyomma variegatum (n = 1; 0.2%) and Rhipicephalus evertsi evertsi (n = 1; 0.2%) (Table 2). Three tick species (H. dromedarii, H. truncatum and H. rufipes) were found in all three locations while H. impeltatum was found only in Kano and Sokoto. Amblyomma variegatum was collected in Kano state and R. e. evertsi in Jigawa state only and lastly, H. impressum in Kano state (Table 2). To confirm the identity of these species of ticks, molecular identification was carried out. A BLASTn query of the obtained sequences for the 16S rRNA gene revealed a high identity match ranging from 98.9% to 100% for all tick species except for H. rufipes. Due to ambiguity of the 16S rRNA gene for H. rufipes, the 12S rRNA gene was amplified and a BLASTn query of the obtained sequences still could not clear this ambiguity. Lastly, we amplified the cox1 gene followed by sequencing. The sequence analysis gave 99.8% homology with H. rufipes (GenBank: KX000641.1). The newly generated sequences were deposited in the GenBank database under the accession numbers MN394427-MN39444 (16S rRNA gene), MN394457-MN394461 (12S rRNA gene) and MN601291-MN601294 (cox1).
Risk factors associated with tick infestations of camels
Tick infestation rates were slightly higher in male camels across the three different locations with 65.6% (40 /61) in Kano, 63.2% (12/19) in Jigawa and 27.8% (10/36) in Sokoto compared with female camels with 64.5% (20/31), 60.0% (6/10) and 21.1% (4/19), respectively. No significant difference was observed between sexes across the study locations (P > 0.05) (Table 3). The infestation rate was significantly higher in camels > 5 years-old compared with those < 5 years-old and differed significantly (P < 0.05) (Table 3). The odds of infestation with ticks were higher in camels > 5 years-old from Kano (OR: 1.44, 95% CI: 0.49–4.18) compared with Jigawa (OR: 0.76, 95% CI: 0.15–4.30) and Sokoto states (OR: 0.49, 95% CI: 0.14–1.73) (Table 3). Finally, camels with a good body condition score were significantly less infested with ticks across the three study locations compared with those with either poor or moderate body condition score (P < 0.05) (Table 3).
Table 3.
Risk factors associated with infestation of 176 camels with ticks in north-western Nigeria
| Risk factor | n | Total no. infested with ticks (%) | Study location | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Kano | Jigawa | Sokoto | |||||||||
| No. examined | No. infested (%) | Odds ratio (95% CI) | No. examined | No. infested (%) | Odds ratio (95% CI) | No. examined | No. infested (%) | Odds ratio (95% CI) | |||
| Sex | |||||||||||
| Male | 116 | 62 (53.5) | 61 | 40 (65.6) | Ref | 19 | 12 (63.2) | Ref | 36 | 10 (27.8) | Ref |
| Female | 60 | 30 (50.0) | 31 | 20 (64.5) | 1.05 (0.42–2.59) | 10 | 6 (60.0) | 1.14 (0.24–5.50) | 19 | 4 (21.1) | 1.44 (0.38–5.41) |
| Total | 176 | 92 (52.3) | 92 | 60 (65.2) | 29 | 18 (62.2) | 55 | 14 (25.5) | |||
| Age | |||||||||||
| < 5 years | 66 | 27 (40.9) | 21 | 15 (71.4) | Ref | 7 | 4 (57.1) | Ref | 38 | 8 (21.1) | Ref |
| > 5 years | 110 | 65 (64.5) | 71 | 45 (47.9) | 1.44 (0.49–4.18) | 22 | 14 (63.6) | 0.76 (0.15–4.30) | 17 | 6 (42.9) | 0.49 (0.14–1.73) |
| Total | 176 | 92 (52.3) | 92 | 60 (65.2) | 29 | 18 (62.1) | 55 | 14 (25.5) | |||
| Body condition | |||||||||||
| Poor | 10 | 7 (70.0) | 7 | 4 (57.1) | Ref | 3 | 2 (66.7) | Ref | 3 | 1 (33.3) | |
| Moderate | 158 | 81 (51.3) | 81 | 55 (67.9) | 0.63 (0.13–3.03) | 25 | 16 (64.0) | 0.63 (0.05–8.44) | 47 | 10 (21.3) | 1.85 (0.15–22.55) |
| Good | 8 | 4 (50.0) | 4 | 1 (25.0) | 4.00 (0.27–60.37) | 1 | 0 (0) | 1.22 (0.0915.12) | 5 | 3 (60.0) | 0.33 (0.02–6.66) |
| Total | 176 | 92 (52.3) | 92 | 60 | 29 | 18 (62.1) | 55 | 14 (25.5) | |||
Abbreviation: n, total number; Ref, reference value
Molecular detection of tick-borne pathogens in ticks
Rickettsia spp.
Altogether, 67 out of 231 tick pools (comprising of 593 ticks) produced bands of the correct length in the gltA PCR and all were sequenced. Out of those, 51 resulted in good quality sequences and could be evaluated as Rickettsia spp. Therefore, the minimum infection rate (MIR) for tick pools for Rickettsia spp. was 8.6%. Across the study locations, 31 pools were positive (MIR 7.8%) in Kano state, 14 pools (MIR 9.7%) in Jigawa state and 6 pools (MIR 11.5%) in Sokoto state (Table 4). No significant difference (P > 0.05) was observed between the different study locations.
Table 4.
Minimum infection rates of tick-borne pathogens detected in tick pools from different study locations in north-western Nigeria
| Study location | Total no. of ticks tested (n) | Minimum infection rate % (number of positives) [95% CI] | ||
|---|---|---|---|---|
| Rickettsia spp.(n) | Babesia spp. (n) | Coxiella burnetii (n) | ||
| Kano | 396 (150) | 7.8 (31) [5.4–10.9] | 0.8 (3) [0.2–2.2] | 3.0 (12) [1.6–5.2] |
| Jigawa | 145 (57) | 9.7 (14) [5.4–15.7] | 0.7 (1) [0.0–3.8] | 2.1 (3) [0.4–5.9] |
| Sokoto | 52 (24) | 11.5 (6) [4.4–23.4] | 0 (0) | 3.8 (2) [0.5–13.2] |
| Total | 593 (231) | 8.6 (51) [6.5–11.2] | 0.7 (4) [0.2–1.7] | 2.9 (17) [1.7–4.6] |
Abbreviation: n, number of pools
Rickettsia spp. was detected in four different tick species with H. rufipes having the highest MIR (36.8%) and H. impeltatum with the lowest MIR (5.6%). Others include H. truncatum (16.1%) and H. dromedarii (6.2%) (Table 5).
Table 5.
Minimum infection rate of tick-borne pathogens in tick pools in relation to tick species collected from camels in north-western Nigeria
| Study location | Total no. of ticks tested (n) | Minimum infection rate (no. of positives) [95% CI] | ||
|---|---|---|---|---|
| Rickettsia spp. (n) | Babesia spp. (n) | Coxiella burnetii (n) | ||
| Hyalomma dromedarii | 465 (154) | 6.2 (29) [4.2–8.8] | 0.6 (3) [0.1–1.9] | 3.4 (16) [2.0–5.5] |
| H. truncatum | 87 (46) | 16.1 (14) [9.1–25.5] | 0 (0) | 1.1 (1) [0.0–6.2] |
| H. rufipes | 19 (16) | 36.8 (7) [16.3–61.6] | 0 (0) | 0 (0) |
| H. impeltatum | 18 (11) | 5.6 (1) [0.1–27.3] | 5.6 (1) [0.1–27.3] | 0 (0) |
| H. impressum | 2 (2) | 0 (0) | 0 (0) | 0 (0) |
| Amblyomma variegatum | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Rhipicephalus evertsi evertsi | 1 (1) | 0 (0) | 0 (0) | 0 (0) |
| Total | 593(231) | 8.6 (51) [6.5–11.2] | 0.7 (4) [0.2–1.7] | 2.9 (17) [1.7–4.6] |
Abbreviation: n, number of pools
Following a BLASTn query on the NCBI database, 45 of these sequences showed 100% identity with R. aeschlimannii (GenBank: MH267736.1) and 6 showed high similarity scores ranging between 98.7–99.7% (GenBank: MH267736.1). BLASTn analysis of one of the sequences obtained from the gltA gene of Rickettsia spp., gave 100% homology with C. burnetii (GenBank: CP035112.1).
To further confirm the genotypes of Rickettsia spp., ompA and ompB genes were partially amplified. We tested all gltA-positive samples with good quality sequences (n = 51) for both ompA and ompB. For ompA and ompB, 39 and 43 tick pools, respectively were positive, from which 13 and 16 amplicons were selected for sequencing and gave good quality sequences. BLASTn analysis of these sequences obtained for both genes showed 99.9–100% similarity with R. aeschlimannii on GenBank. All newly generated sequences were deposited in the GenBank database under the accession numbers MN601304-MN601344 (gltA), MT126809-MT126818 (ompA) and MN601295-MN601303 (ompB).
Anaplasma/Ehrlichia spp.
The PCR targeting the 16S rRNA gene of Anaplasma/Ehrlichia spp. was positive in 62 out of 231 tick pools with a MIR of 10.5%. Based on location, the MIR in Sokoto state was 15.4% followed by Kano state with 10.1% while Jigawa state had the lowest MIR of 9.7%. No significant difference (P > 0.05) was observed across the different sampling locations (Table 4).
Five different tick species were positive: H. dromedarii (MIR, 8.0%), H. truncatum (MIR, 16.1%), H. rufipes (MIR, 36.8%), H. impeltatum (MIR, 11.1%) and H. impressum (MIR, 100.0%) (Table 5).
BLASTn analysis of sequences obtained from positive samples of the Anaplasma/Ehrlichia PCR had sizes of 240 bp with 98–100% similarity to Peptoniphilus spp. (GenBank: LC145547.1), “Candidatus Midichloria mitochondrii” (GenBank: KU559921.11) and a Rickettsiales bacterium (GenBank: DQ379964.1).
Anaplasma marginale
Anaplasma marginale DNA was not detected in DNA from ticks.
Coxiella burnetii
The DNA of C. burnetii was detected in 17 out of 231 tick pools with a MIR of 2.9%. The MIR for the states of Sokoto, Kano and Jigawa was 3.8%, 3.0% and 2.1%, respectively (Table 4). No significant difference (P > 0.05) was observed across the different study locations. Most C. burnetii positive tick pools were H. dromedarii (MIR 3.4%) and H. truncatum (MIR 1.1%) (Table 5). Only 1 out of 87 (1.1%) H. truncatum ticks in the pools were positive to C. burnetti (Table 5). Sequences had similarity scores ranging between 99.2–100% to C. burnetii (GenBank: CP035112.1). The newly generated sequences were deposited in the GenBank database under the accession numbers MN396571-MN396578.
Babesia spp.
The MIR for Babesia spp. was 0.7% (4/593) across the study locations with 3 positives in Kano state (MIR 0.8%) and one in Jigawa state (MIR 0.7%) (Table 4). No significant difference (P > 0.05) was observed across the different sampling locations. Three out of the four positive pools were detected in H. dromedarii (MIR 0.6%) and one was detected in H. impeltatum (MIR 5.6%) (Table 5).
BLASTn analysis showed that 2 sequences showed 100% identity to B. occultans (GenBank: MG920540.1), 1 with 100% identity to B. caballi (GenBank: MG052892.1) and 1 showed 98.5% similarity to Babesia spp. (GenBank: KC249945.1). A further attempt to characterize the undifferentiated species of Babesia using a different primer pair [41] showed 100% homology with Babesia spp. (GenBank: KC249946.1). The newly generated sequences were deposited in the GenBank database under the accession numbers MN394378-MN394381.
Co-detection of tick-borne pathogens in ticks
A low co-detection rate was observed in the study with all co-detections occurring in Kano state only. Co-detection was observed for Rickettsia spp. + Babesia spp. in one tick pool as well as for Rickettsia spp. + C. burnetii in another tick pool.
Molecular detection of tick-borne pathogens in the blood of camels
“Candidatus Anaplasma camelii”
The overall prevalence of “Ca. A. camelii” from the three study locations was 40.3% (71/176). Kano state had the highest prevalence of 59.8% (55/92), followed by Jigawa with 37.9% (11/29) and Sokoto state with 9.1% (5/55) (Table 6).
Table 6.
Risk factors associated with infection with “Candidatus Anaplasma camelli” in blood collected from camels in north-western Nigeria
| Parameter | n | Total no. infected (%) | Study location | |||||
|---|---|---|---|---|---|---|---|---|
| Kano | Jigawa | Sokoto | ||||||
| No. examined | No. infected (%) | No. examined | No. infected (%) | No. examined | No. infected (%) | |||
| Sex | ||||||||
| Male | 116 | 41 (35.34) | 61 | 31 (50.82) | 19 | 7 (36.84) | 36 | 3 (8.33) |
| Female | 60 | 30 (50.00) | 31 | 24 (77.41) | 10 | 4 (40.0) | 19 | 2 (10.53) |
| Total | 176 | 71 (40.34) | 92 | 55 (59.78) | 29 | 11 (37.93) | 55 | 5 (9.09) |
| Age | ||||||||
| < 5 years | 66 | 20 (30.30) | 21 | 15 (71.43) | 7 | 2 (28.57) | 38 | 3 (7.89) |
| > 5 years | 110 | 51 (46.36) | 71 | 40 (56.34) | 22 | 9 (40.91) | 17 | 2 (11.76) |
| Total | 176 | 71 (40.34) | 92 | 55 (59.78) | 29 | 11 (37.93) | 55 | 5 (9.09) |
| Body condition | ||||||||
| Poor | 10 | 4 (40.00) | 7 | 3 (42.86) | 3 | 1 (33.33) | 3 | 0 (0) |
| Moderate | 158 | 66 (41.77) | 81 | 52 (64.19) | 25 | 10 (40.0) | 47 | 4 (8.51) |
| Good | 8 | 1 (12.50) | 4 | 0 (0) | 1 | 0 (0) | 5 | 1 (20.0) |
| Total | 176 | 71 (40.34) | 92 | 55 (59.78) | 29 | 11 (37.93) | 55 | 5 (9.09) |
| Presence of ticks | ||||||||
| Yes | 92 | 42 (45.65) | 60 | 35 (58.33) | 18 | 6 (33.33) | 14 | 1 (7.14) |
| No | 84 | 29 (34.52) | 32 | 20 (62.5) | 11 | 5 (45.45) | 41 | 4 (9.76) |
| Total | 176 | 71 (40.34) | 92 | 55 (59.78) | 29 | 11 (37.93) | 55 | 5 (9.09) |
Abbreviation: n, total number
GenBank analysis of representative sequences (n = 15) selected from all the study locations with a product size of 345 bp showed 99.6–100% similarity to 16S rDNA of Anaplasma platys (GenBank: MH762081.1) and “Ca. A. camelii” (GenBank: KF843827.1). In the attempt to differentiate these two species, semi-nested PCR targeting the 16S rRNA gene of Anaplasma spp. was used, generating a PCR product of 426 bp. BLASTn analysis of the sequences yielded “Ca. A. camelii” (GenBank: KF843825.1) with the highest identity score of 100% (GenBank: KF843825.1). The newly generated sequences were deposited in the GenBank database under the accession numbers MN396629-MN396638.
Babesia spp. and Anaplasma marginale
DNA of neither pathogen was amplified in the blood of camels.
Risk factors associated with “Candidatus A. camelii” infection in blood of camels
A higher number of female camels were infected as compared to males, although no significant difference (P > 0.05) was observed (Table 6). Furthermore, the prevalence was higher in camels > 5 years-old across the three study areas compared with those < 5 years-old old. A significant difference was observed between age groups (P < 0.05) (Table 6).
Camels with poor or moderate body condition had higher infection rates with “Ca. A. camelii” compared to those with a good body condition with a significant difference (P < 0.05). Only one camel (20.0%, 1/5) with a good body condition score was infected in Sokoto state (Table 6). Finally, camels infested with ticks were two times more likely to be infected with “Ca. A. camelii” compared with those without ticks (OR: 1.59, 95% CI: 0.9–2.9).
Phylogenetic and haplotype analysis of “Ca. A. camelii”
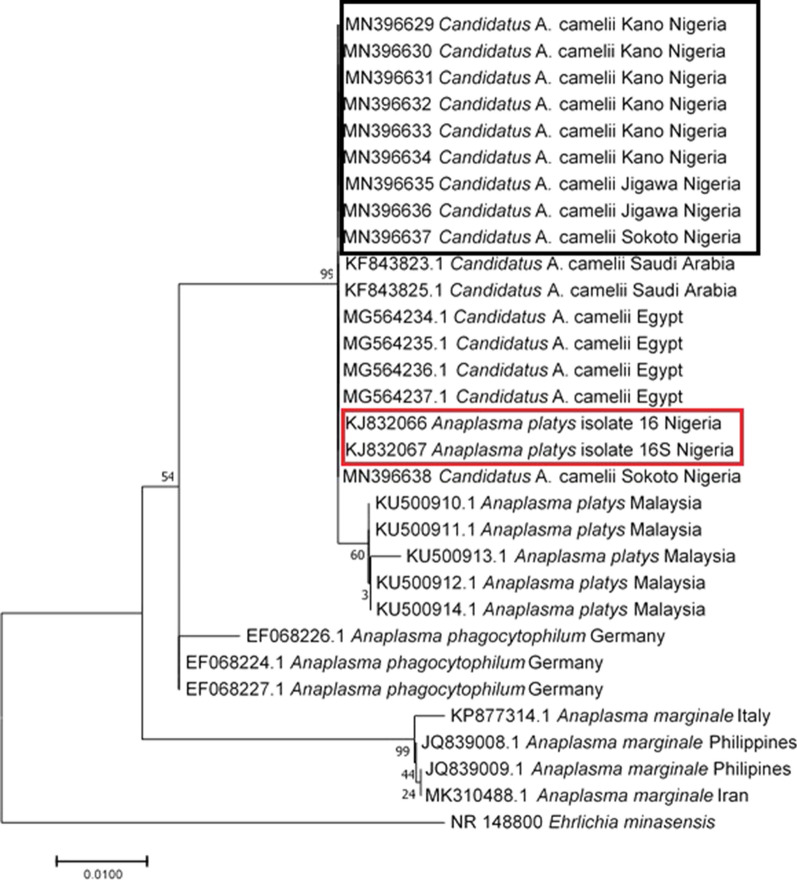
“Candidatus A. camelii” nucleotide sequences from this study clustered together with all other “Ca. A. camelii” sequences from Saudi Arabia (GenBank: KF843823-KF843825) and Egypt (GenBank: MG564235-MG564237) (Fig. 2). In addition, A. platys sequences from a previous study in Nigeria clustered with the sequences from this study.
Fig. 2.
Phylogenetic tree based on 16S rDNA sequences of “Candidatus A. camelii” isolates identified in this study (indicated in the black box) and Anaplasma platys sequences from a previous study from Sokoto, Nigeria, retrieved from GenBank (indicated in the red box). The Maximum Likelihood method based on the Kimura 2-parameter model was used to construct the tree at 1000 replicates using MEGA 7. Ehrlichia minasensis was used as the outgroup
Only one haplotype was found in this study (Fig. 3), which is similar to the haplotype detected from other “Ca. A. camelii” in Egypt and Saudi Arabia based on the sequences retrieved from the NCBI database. This haplotype differs slightly by a single mutation from A. platys of dogs in Malaysia (GenBank: KU500910). Furthermore, it also differs by 3 mutations from A. phagocytophilium and by 8 mutations from A. marginale (Fig. 3).
Fig. 3.
Median joining network of “Candidatus Anaplasma camelii” haplotypes based on 16S rDNA sequences and its relatedness with other species of Anaplasma
Discussion
This study confirmed the occurrence of several tick-borne pathogens and the species diversity of ticks infesting camels in Nigeria. The overall rate of tick infestation in this study was 52.3%, lower than 80.0% reported by Abdullahi et al. [47] in Kebbi state, Nigeria. Differences between the two studies could be basically attributed to the smaller sample size in the latter study. Other factors possibly attributing to differences in tick infestations include geographical distribution, climatic factors, the management system as well as the frequency of acaricides application. We observed high numbers of male ticks compared with female ticks which is unsurprising considering the fact that the latter are known to detach from their host few days after feeding to oviposit, while the males stay longer for weeks before detaching [48].
In this study, ticks from Nigeria were morphologically identified and confirmed using molecular markers targeting several genes. So far, studies on tick identification in camels from Nigeria were based on morphology [6, 47, 49, 50]. Combining several mitochondrial markers (12S, 16S and cox1), we were able to identify several species of Hyalomma ticks. The use of these markers has been increasingly useful for tick identification in several studies [51–54].
Hyalomma dromedarii was the most frequent tick species encountered in this study. Similar observations have also been reported by other researchers on ticks of camels in Nigeria [47, 49, 50], within other African countries [55, 56] and other parts of the world [57, 58]. The dromedary camel was found to be the preferred host for this tick species, but it also infests sheep, goats, cattle and horses [29]. In another study conducted at a single site in Nigeria, Kamani et al. [6], reported H. impeltatum as the most prevalent tick species of camel. We attributed some reasons for these differences. First, we sampled three study locations in north-western Nigeria. Secondly, we sampled towards the end of rainy season while Kamani et al. [6] sampled throughout the dry season. Nonetheless, previous studies on tick abundance of camels and the influence of season in both Kano and Sokoto states reported H. dromedarii as the most prevalent tick species of dromedary which was not influenced by season as this tick species showed preponderance on camels during both dry and wet seasons [50, 59]. Most likely, abiotic factors such as temperature and humidity may play a role in this observation, but this remains highly speculative.
Hyalomma impressum was the least prevalent tick species with only two specimens collected. This corroborates the observation of previous studies on ticks of camels in Nigeria [6] and Algeria [60] where two and three specimens respectively were collected. It is likely that environment as well as sampling time could also impact the prevalence of H. impressum. All other species of the genus Hyalomma such as H. rufipes, H. truncatum and H. impeltatum as well as A. variegatum and R. evertsi evertsi have been reported infesting camels in Nigeria [6, 47, 49].
Rickettsia aeschlimannii belongs to the spotted fever group of Rickettsia and is maintained and/or transmitted primarily by ticks. The MIR of R. aeschlimannii in ticks collected from camels was 7.8–11.5% across the three study locations (Kano, Jigawa and Sokoto states) and 5.6–36.8% across tick species. This confirms the presence of this species of Rickettsia in ticks infesting camels in Nigeria. A previous study in Kano, Nigeria reported an infection rate of 23.8% in ticks from camels [6]. The prevalence of pathogens detected in ticks is shown here as the minimum infection rate, assuming that only one sample is positive in each positive pool. Of course, this approach is only approximate and prevalence rates for the identified pathogens might be higher than reported in the present study.
All isolates in this study were identified as R. aeschlimannii, suggesting that the organism is endemic and widespread among Hyalomma tick species infesting camels in Nigeria. Furthermore, this bacterium has been detected in several countries within Africa in Hyalomma ticks [60–62]. The highest prevalence of R. aeschlimannii DNA was detected in H. rufipes, which agrees with the report of previous studies in Nigeria [6], Egypt [62] and Senegal [61]. Furthermore, all species of Hyalomma ticks were positive for R. aeschlimannii DNA except H. impressum. Similar findings were registered in previous studies in Algeria and Nigeria [4, 6, 60].
Piroplasms of the genera Babesia and Theileria are tick-borne pathogens of livestock including camels. The overall MIR of piroplasms (Babesia spp.) in ticks from this study was low in addition to the non-detection of these protozoan parasites in the blood. This corroborates with previous reports on piroplasms (both Babesia spp. and Theileria spp.) of camels [63, 64]. Previously, a low prevalence of Theileria ovis was reported in blood of camels from Sokoto, Nigeria using reverse line blot (RLB) [63] and in H. dromedarii ticks in Saudi Arabia [64]. Nevertheless, a high prevalence of 74.5% has been registered in the blood of camels in Sudan [65].
In the present study, B. occultans DNA was amplified and confirmed by sequencing in Hyalomma ticks (H. impeltatum and H. dromedarii) for the first time after its first morphological description in the haemolymph of Hyalomma ticks over three decades ago in Nigeria [66]. The DNA of B. occultans has been detected in other species of ticks such as H. asiaticum in China [67], H. marginatum in Tunisia [68] and Rhipicephalus turanicus and H. marginatum rufipes in Turkey [69]. In addition, DNA of this pathogen has also been detected in the blood of cows in Italy displaying fever, anemia, and hematological alterations [20]. Furthermore, DNA of B. caballi was amplified in a H. dromedarii tick. The detection of B. caballi in our study may not be surprising considering the fact that both camels and horses are infested with similar tick species [70]. Previous studies on camel piroplasms have detected B. caballi in the blood of camels in Sudan [71], Jordan [70] and Iraq [72].
The low infection rate of C. burnetii in Hyalomma tick species reported in the present study is comparable with that reported elsewhere for Hyalomma ticks [53, 73]. Most of the positives were detected in H. dromedarii and only one in a H. truncatum tick (1.1%). In a similar study in Egypt, C. burnetii was detected in H. dromedarii exclusively [53], while in China, most of the infection was in H. asiaticum asiaticum [73]. Furthermore, while Coxiella-like bacteria have been found in ticks as endosymbionts and play a role in tick fitness, C. burnetii is responsible for Q fever in vertebrates including humans [3]. Since ticks serve as a carrier of C. burnetii in livestock, the close association between man and livestock could probably lead to human infections [74]. An epidemiological survey among veterinarians and other high-risk individuals with regular contact with animals showed a high antibody titre to C. burnetii, suggesting possible transmission [75, 76].
Anaplasmosis in camels due to Anaplasma marginale causes subclinical disease as registered in other studies [77, 78]. In our study, camels from the three study areas tested positives to a novel species of Anaplasma named “Ca. A. camelii” by Bastos et al. [79]. This species is genetically related to A. platys [10, 12, 63, 79]. An earlier study in one of the study areas (Sokoto) reported a high prevalence for A. platys in camels [63]. The overall prevalence of “Ca. A. camelii” in camels in our study was 40.3%, which is comparable to results reported by Lbacha et al. [13] in Morocco, but higher compared with data from China (7.20%) [10] and Tunisia (17.70%) [12]. The variations in prevalence rates may result from differences in husbandry practices, tick control programmes and reservoir hosts [12].
Phylogenetic analyses in our study based on DNA sequencing clusters the A. platys reported earlier in one of the study areas (Sokoto state) (GenBank: KJ832066-KJ832067) with 99.5% identity to that obtained in our study (GenBank: MN396629-MN396638). It is therefore possible that the A. platys as earlier reported by Lorusso et al. [63], could be “Ca. A. camelii”. The 16S sequences generated in the present study were identical to other “Ca. A. camelii” isolated in Saudi Arabia [79]. The haplotype analysis in our study shows that only one nucleotide differentiates A. platys with “Ca. A. camelii”, an observation similar to that observed by Sazmand et al. [14] in Iran.
Risk factors associated with “Ca. A. camelii” infection indicate that the female camels were more infected compared with the males corroborating previous studies [12, 63]. Immunosuppression associated with pregnancy and lactation has been attributed to be responsible for this observation [80]. The opposite was the case with respect to tick infestation, as more male camels were infested compared with females. It could also be that the male camels despite being more infested with ticks due to their natural behavior for space triggered by androgenic hormones, had better immunity against tick-borne infection than the females. A poor body condition score was a risk factor to both tick infestation and infection with “Ca. A. camelii” infection.
Older animals were more often infested with ticks as well as positive for “Ca. A. camelii” DNA than younger camels. According to Azmat et al. [81], the infection rate of camels with anaplasmosis increases with age. The occurrence of “Ca. A. camelii” infection in our study was positively associated with the presence of ticks. This finding confirms previous reports on anaplasmosis of camels [81, 82]. Also, A. marginale is responsible for bovine intra-erythrocytic anaplasmosis in bovines, but we did not find A. marginale in the investigated camels and ticks, an observation that has also been reported by other researchers [10, 11]. Furthermore, it has been postulated that dromedaries are not relevant reservoirs for already named Anaplasma species which include A. marginale, A. centrale, A. phagocytophilum and A. bovis [12].
Conclusions
This study revealed the occurrence of different tick species and different tick-borne pathogens in ticks infesting camels as well as in their blood in Nigeria. We identified several subspecies of Hyalomma ticks and their associated tick-borne pathogens. Pathogen DNA detected in ticks using PCR and sequencing includes R. aeschlimannii, B. caballi, Babesia spp. and C. burnetii. Furthermore, we amplified B. occultans DNA in Hyalomma ticks infesting camels in Nigeria. “Candidatus A. camelii”, a novel species variant of Anaplasma, was the only pathogen amplified in the blood of the investigated camels. The detection of the two zoonotic pathogens, R. aeschlimannii and C. burnetii, may necessitate further investigation on the role of camels in their maintenance and reservoir status.
Acknowledgements
We thank all animal owners for their co-operation during the study. The council of the University of Maiduguri, Nigeria is hereby acknowledged for granting TEO study leave to undertake this study. We thank the assistance of Adamu Aliyu, Gerald Chibuzo Agu and Andries Phukuntsi for their help during the sample collection and analysis. Special thanks to Marlene Hausner and Amira Al-Hosary for their useful suggestions during the laboratory work.
Abbreviations
- MIR
minimum infection rates
- TBP
tick-borne pathogens
- BLAST
Basic Local Alignment Search Tool
- PCR
polymerase chain reaction
- CI
confdence interval
- HM
Hyalomma marginatum
- HR
Hyalomma rufipes
- cox1
cytochrome c oxidase subunit 1
- glt
citrate synthase
- omp
outer membrane protein
Authorsʼ contributions
TEO, OT and CS conceived and organized the study. CS supported the acquisition of fellowship for OET. OET carried out field work. OET, CR, OT, NIO, SF, AV and MS performed laboratory work. OET, CR, SF, AAB, OT and CS analyzed the data. CR and CS critically reviewed the manuscript drafts. All authors read and approved the final manuscript.
Funding
The financial support of the Deutscher Akademischer Austauschdienst (DAAD) through a short-term research grant (Grant No: 91709125) made available to TEO is hereby acknowledged.
Availability of data and materials
All data generated or analyzed during this study are included in this published article. All newly generated sequences were submitted to the GenBank database.
Ethics approval and consent to participate
Permission to carry out the study was approved by the animal research ethics committee of the North-West University South Africa with ethics number NWU-01242-19-A9 in line with the guidelines of the committee.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
ThankGod E. Onyiche, Email: et.onyiche@unimaid.edu.ng
Cristian Răileanu, Email: cristian.raileanu@fli.de.
Oliver Tauchmann, Email: oliver.tauchmann@fli.de.
Susanne Fischer, Email: susanne.fischer@fli.de.
Ana Vasić, Email: ana.vasic@fli.de.
Mandy Schäfer, Email: mandy.schaefer@fli.de.
Abdullahi A. Biu, Email: biuvet@yahoo.com
Ndudim I. Ogo, Email: ogoendy@yahoo.com
Oriel Thekisoe, Email: oriel.thekisoe@nwu.ac.za.
Cornelia Silaghi, Email: cornelia.silaghi@fli.de.
References
- 1.de la Fuente J, Estrada-Pena A, Venzal JM, Kocan KM, Sonenshine DE. Overview: ticks as vectors of pathogens that cause disease in humans and animals. Front Biosci. 2008;13:6938–6946. doi: 10.2741/3200. [DOI] [PubMed] [Google Scholar]
- 2.Rajput ZI, Hu SH, Chen WJ, Arijo AG, Xiao CW. Importance of ticks and their chemical and immunological control in livestock. J Zhejiang Univ Sci. 2006;7:912–921. doi: 10.1631/jzus.2006.B0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duron O, Noël V, Mccoy KD, Bonazzi M, Sidi-Boumedine K, Morel O, et al. The recent evolution of a maternally-inherited endosymbiont of ticks led to the emergence of the Q fever pathogen, Coxiella burnetii. PLoS Pathog. 2015;11:e1004892. doi: 10.1371/journal.ppat.1004892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parola P, Paddock CD, Socolovschi C, Labruna MB, Mediannikov O, Kernif T, et al. Update on tick-borne rickettsioses around the world: a geographic approach. Clin Microbiol Rev. 2014;27:166. doi: 10.1128/CMR.00032-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reye AL, Arinola OG, Hübschen JM, Muller CP. Pathogen prevalence in ticks collected from the vegetation and livestock in Nigeria. Appl Environ Microbiol. 2012;78:2562–2568. doi: 10.1128/AEM.06686-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamani J, Baneth G, Apanaskevich DA, Mumcuoglu KY, Harrus S. Molecular detection of Rickettsia aeschlimannii in Hyalomma spp. ticks from camels (Camelus dromedarius) in Nigeria, West Africa. Med Vet Entomol. 2015;29:205–209. doi: 10.1111/mve.12094. [DOI] [PubMed] [Google Scholar]
- 7.Sharifiyazdi H, Jafari S, Ghane M, Nazifi S, Sanati A. Molecular investigation of Anaplasma and Ehrlichia natural infections in the dromedary camel (Camelus dromedarius) in Iran. Comp Clinical Pathol. 2017;26:99–103. [Google Scholar]
- 8.Wernery U, Thomas R, Syriac G, Raghavan R, Kletzka S. Seroepidemiological studies for the detection of antibodies against nine infectious diseases in dairy dromedaries (Part-I) J Camel Pract Res. 2007;14:85–90. [Google Scholar]
- 9.Al-Gharban HA, AL-Taee HS. Seroclinical diagnosis of Anaplasma marginale bacteria in carrier arabian one-humped camels. Basrah J Vet Res. 2006;15:346–359. [Google Scholar]
- 10.Li Y, Yang J, Chen Z, Qin G, Li Y, Li Q, et al. Anaplasma infection of Bactrian camels (Camelus bactrianus) and ticks in Xinjiang, China. Parasites Vectors. 2015;8:313. doi: 10.1186/s13071-015-0931-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wernery U, Pfister K, Marina R, Hakimudin F, Silaghi C. No evidence of Mycoplasma haemolamae and Anaplasma marginale in anaemic dromedaries in the United Arab Emirates. J Camel Pract Res. 2014;21:5–8. [Google Scholar]
- 12.Belkahia H, Said MB, Sayahi L, Alberti A, Messadi L. Detection of novel strains genetically related to Anaplasma platys in Tunisian one-humped camels (Camelus dromedarius) J Infect Dev Ctries. 2015;9:1117–1125. doi: 10.3855/jidc.6950. [DOI] [PubMed] [Google Scholar]
- 13.Lbacha HA, Zouagui Z, Alali S, Rhalem A, Petit E, Ducrotoy MJ, et al. “Candidatus anaplasma camelii” in one-humped camels (Camelus dromedarius) in Morocco: a novel and emerging Anaplasma species? Infect Dis Poverty. 2017;6:1. doi: 10.1186/s40249-016-0216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sazmand A, Harl J, Eigner B, Hodžić A, Beck R, Hekmatimoghaddam S, et al. Vector-borne bacteria in blood of camels in Iran: new data and literature review. Comp Immunol Microbiol Infect Dis. 2019;65:48–53. doi: 10.1016/j.cimid.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klemmer J, Njeru J, Emam A, El-Sayed A, Moawad AA, Henning K, et al. Q fever in Egypt: epidemiological survey of Coxiella burnetii specific antibodies in cattle, buffaloes, sheep, goats and camels. PLoS ONE. 2018;13:e0192188. doi: 10.1371/journal.pone.0192188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oechslin CP, Heutschi D, Lenz N, Tischhauser W, Péter O, Rais O, et al. Prevalence of tick-borne pathogens in questing Ixodes ricinus ticks in urban and suburban areas of Switzerland. Parasites Vectors. 2017;10:558. doi: 10.1186/s13071-017-2500-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khamesipour F, Doosti A, Koohi A, Chehelgerdi M, Mokhtari-Farsani A, Chengula AA. Determination of the presence of Babesia DNA in blood samples of cattle, camel and sheep in Iran by PCR. Arch Biol Sci Belgrade. 2015;63:83–90. [Google Scholar]
- 18.Faraj AA, Al-Amery AMA. Comparison of the efficacy of different methods in the different methods in the diagnosis of babesiosis in Camels (Camelus dromedarius) in Alnajaf Al-ashraf Province-Iraq of Najaf—Iraq. Basrah J Vet Res. 2018;17:593–602. [Google Scholar]
- 19.Radostits OM, Gay CC, Blood DC, Hinchliff KW. Veterinary medicine: a text book of the diseases of cattle, sheep, goats and horses. 10. London: WB Saunders; 2007. pp. 1483–1498. [Google Scholar]
- 20.Decaro N, Larocca V, Parisi A, Losurdo M, Lia RP, Greco MF, et al. Clinical bovine piroplasmosis caused by Babesia occultans in Italy. J Clin Microbiol. 2013;51:2432–2434. doi: 10.1128/JCM.00713-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Swelum AA, Ismael AB, Khalaf AF, Abouheif MA. Clinical and laboratory findings associated with naturally occurring babesiosis in dromedary camels. Bull Vet Inst Pulawy. 2014;58:229–233. [Google Scholar]
- 22.FAO. FOOSTAT. Rome: Food and Agriculture Organization of the United Nations; 2018. http://www.fao.org/faostat/en/#data/QA. Accessed 3 Mar 2020.
- 23.Akpa GN, Abbaya HY, Saley ME. Comparative evaluation of the influence of species, age and sex on carcass characteristics of camels, cattle, sheep and goats in Sahel environment. Anim Res Int. 2017;14:2588. [Google Scholar]
- 24.Kadim IT, Mahgoub O, Purchas RW. A review of the growth, and of the carcass and meat quality characteristics of the one-humped camel (Camelus dromedaries) Meat Sci. 2018;80:555–569. doi: 10.1016/j.meatsci.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 25.Mohammed I. Study of the integration of the dromedary in smallholder crop-livestock production systems in northwestern Nigeria. Göttingen: Cuvillier Verlag; 2000. [Google Scholar]
- 26.Abdussamad AM, Charruau P, Kalla DJU, Burger PA. Validating local knowledge on camels: colour phenotypes and genetic variation of dromedaries in the Nigeria-Niger corridor. Livest Sci. 2015;181:131–136. [Google Scholar]
- 27.National Population Commission. Population and housing census of the Federal Republic of Nigeria. Priority Tables, 1; 2006. http://www.nigerianstat.gov.ng. Accessed 26 June 2019.
- 28.Bourn D, Wint W, Blench R, Woolley E. Nigerian livestock resources survey. World Anim Rev. 1994;78:49–58. [Google Scholar]
- 29.Walker AR, Bouattour A, Camicas JL. Ticks of domestic animals in Africa: a guide to identification of species. Edinburgh: Bioscience Reports; 2003.
- 30.Silaghi C, Hamel D, Thiel C, Pfister K, Pfeffer M. Spotted fever group rickettsiae in ticks, Germany. Emerg Infect Dis. 2011;17:890. doi: 10.3201/eid1705.101445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carelli G, Decaro N, Lorusso A, Elia G, Lorusso E, Mari V, et al. Detection and quantification of Anaplasma marginale DNA in blood samples of cattle by real-time PCR. Vet Microbiol. 2007;124:107–114. doi: 10.1016/j.vetmic.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 32.Pothmann D, Poppert S, Rakotozandrindrainy R, Hogan B, Mastropaolo M, Thiel C, et al. Prevalence and genetic characterization of Anaplasma marginale in zebu cattle (Bos indicus) and their ticks (Amblyomma variegatum, Rhipicephalus microplus) from Madagascar. Ticks Tick Borne Dis. 2016;7:1116–1123. doi: 10.1016/j.ttbdis.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Cowling DW, Gardner IA, Johnson WO. Comparison of methods for estimation of individual-level prevalence based on pooled samples. Prev Vet Med. 1999;39:211–225. doi: 10.1016/s0167-5877(98)00131-7. [DOI] [PubMed] [Google Scholar]
- 34.Kimura M. A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 35.Beati L, Keirans JE. Analysis of the systematic relationships among ticks of the genera Rhipicephalus and Boophilus (Acari: Ixodidae) based on mitochondrial 12S ribosomal DNA gene sequences and morphological characters. J Parasitol. 2001;87:32–48. doi: 10.1645/0022-3395(2001)087[0032:AOTSRA]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 36.Black WC, Piesman J. Phylogeny of hard-and soft-tick taxa (Acari: Ixodida) based on mitochondrial 16S rDNA sequences. Proc Natl Acad Sci USA. 1994;91:10034–10038. doi: 10.1073/pnas.91.21.10034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chitimia L, Lin RQ, Cosoroaba I, Braila P, Song HQ, Zhu XQ. Molecular characterization of hard and soft ticks from Romania by sequences of the internal transcribed spacers of ribosomal DNA. Parasitol Res. 2009;105:907–911. doi: 10.1007/s00436-009-1474-1. [DOI] [PubMed] [Google Scholar]
- 38.Regnery RL, Spruill CL, Plikaytis BD. Genotypic identification of rickettsiae and estimation of intraspecies sequence divergence for portions of two rickettsial genes. J Bacteriol. 1991;173:1576–1589. doi: 10.1128/jb.173.5.1576-1589.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roux V, Fournier PE, Raoult D. Differentiation of spotted fever group rickettsiae by sequencing and analysis of restriction fragment length polymorphism of PCR-amplified DNA of the gene encoding the protein OmpA. J Clin Microbiol. 1996;34:2058–2065. doi: 10.1128/jcm.34.9.2058-2065.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roux V, Raoult D. Phylogenetic analysis of members of the genus Rickettsia using the gene encoding the outer-membrane protein rOmpB (ompB) Int J Syst Evol Microbiol. 2000;50:1449–1455. doi: 10.1099/00207713-50-4-1449. [DOI] [PubMed] [Google Scholar]
- 41.Casati S, Sager H, Gern L, Piffaretti J. Presence of potentially pathogenic Babesia sp. for human in Ixodes ricinus in Switzerland. Ann Agric Environ Med. 2006;13:65. [PubMed] [Google Scholar]
- 42.Hilpertshauser H, Deplazes P, Schnyder M, Gern L, Mathis A. Babesia spp identified by PCR in ticks collected from domestic and wild ruminants in southern Switzerland. Appl Environ Microbiol. 2006;72:6503–6507. doi: 10.1128/AEM.00823-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Parola P, Roux V, Camicas JL, Baradji I, Brouqui P, Raoult D. Detection of ehrlichiae in African ticks by polymerase chain reaction. Trans R Soc Trop Med Hyg. 2000;94:707–708. doi: 10.1016/s0035-9203(00)90243-8. [DOI] [PubMed] [Google Scholar]
- 44.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Warner CK, Dawson JF. Genus- and species-level identification of Ehrlichia species by PCR and sequencing. In: Persing DH, editor. PCR protocols for emerging infectious diseases: a supplement to diagnostic molecular microbiology: principles and applications. Washington: American Society for Microbiology; 1996. [Google Scholar]
- 46.Duron O, Jourdain E, McCoy KD. Diversity and global distribution of the Coxiella intracellular bacterium in seabird ticks. Ticks Tick Borne Dis. 2014;5:557–563. doi: 10.1016/j.ttbdis.2014.04.003. [DOI] [PubMed] [Google Scholar]
- 47.Abdullahi YM, Magami IM, Audu A, Mainasara MM. Prevalence of ticks on camels and cattle brought to Dodoru market Kebbi state, Nigeria. Traektoriâ Nauki. 2008;4:3001–3004. [Google Scholar]
- 48.Hoogstraal H. African Ixodoidea: 1. Ticks of the Sudan (with special reference to equatorial province and with preliminary reviews of the genera Boophilus, Margaropus, and Hyalomma) Washington DC: US Navy; 2008. [Google Scholar]
- 49.Biu AA, Konto M. Survey of tick species infesting the one humped camel (Camelus dromedarius) in Borno state, Nigeria. J Agric Vet Sci. 2008;4:1–4. [Google Scholar]
- 50.Alayande MO, Mayaki AM, Lawal MD, Bandi NI, Ibrahim DD, Talabi AO. Pattern of tick infestation on one humped camels (Camelus dromedarius) in Sokoto, Nigeria. Bull Anim Health Prod Afr. 2015;63:349–354. [Google Scholar]
- 51.Zhang RL, Zhang B. Prospects of using DNA barcoding for species identification and evaluation of the accuracy of sequence databases for ticks (Acari: Ixodida) Ticks Tick Borne Dis. 2014;5:352–358. doi: 10.1016/j.ttbdis.2014.01.001. [DOI] [PubMed] [Google Scholar]
- 52.Lv J, Wu S, Zhang Y, Chen Y, Feng C, Yuan X, et al. Assessment of four DNA fragments (COI, 16S rDNA, ITS2, 12S rDNA) for species identification of the Ixodida (Acari: Ixodida) Parasites Vectors. 2014;7:93. doi: 10.1186/1756-3305-7-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abdullah HH, El-Shanawany EE, Abdel-Shafy S, Abou-Zeina HA, Abdel-Rahman EH. Molecular and immunological characterization of Hyalomma dromedarii and Hyalomma excavatum (Acari: Ixodidae) vectors of Q fever in camels. Vet World. 2018;11:1109. doi: 10.14202/vetworld.2018.1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roth A, Akad F, Zonstein I, King R, Orshan L, Erster O. Molecular characterization of six Hyalomma species using mitochondrial markers. Ticks Tick-Borne Dis. 2019;10:911–917. doi: 10.1016/j.ttbdis.2019.04.015. [DOI] [PubMed] [Google Scholar]
- 55.Barghash SM, Hafez AA, Darwish AM, El-Naga TRA. Molecular detection of pathogens in ticks infesting camels in Matrouh Governorate, Egypt. J Bacteriol Parasitol. 2016;7:2. [Google Scholar]
- 56.Djerbouh A, Lafri I, Kechemir-Issad N. Bitam I. Endo-and ectoparasites (Ixodidae) of camels (Camelus dromedarius) from Southern Algeria. Livest Res Rural Dev. 2018. http://www.lrrd.org/lrrd30/8/djerb30141.html. Accessed 29 Sept 2019.
- 57.Azagi T, Klement E, Perlman G, Lustig Y, Mumcuoglu KY, Apanaskevich DA, et al. Francisella-like endosymbionts and Rickettsia species in local and imported Hyalomma ticks. Appl Environ Microbiol. 2017;83:e01302–e01317. doi: 10.1128/AEM.01302-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fard SRN, Fathi S, Asl EN, Nazhad HA, Kazeroni SS. Hard ticks on one-humped camel (Camelus dromedarius) and their seasonal population dynamics in southeast, Iran. Trop Anim Health Prod. 2012;44:197–200. doi: 10.1007/s11250-011-9909-y. [DOI] [PubMed] [Google Scholar]
- 59.Umar YA, George BDJ, Ajanusi OJ. Survey of hard ticks (Ixodidae) infesting one-humped camels (Camelus dromedaries) in Kano state—Nigeria. Nigeria J Parasitol. 2011;32:61–66. [Google Scholar]
- 60.Djerbouh A, Kernif T, Beneldjouzi A, Socolovschi C, Kechemir N, Parola P, et al. The first molecular detection of Rickettsia aeschlimannii in the ticks of camels from southern Algeria. Ticks Tick Borne Dis. 2012;3:374–376. doi: 10.1016/j.ttbdis.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 61.Mediannikov O, Diatta G, Fenollar F, Sokhna C, Trape JF, Raoult D. Tick-borne rickettsioses, neglected emerging diseases in rural Senegal. PLoS Negl Trop Dis. 2010;4:e821. doi: 10.1371/journal.pntd.0000821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Abdel-Shafy S, Allam NA, Mediannikov O, Parola P, Raoult D. Molecular detection of spotted fever group rickettsiae associated with ixodid ticks in Egypt. Vector Borne Zoonotic Dis. 2012;12:346–359. doi: 10.1089/vbz.2010.0241. [DOI] [PubMed] [Google Scholar]
- 63.Lorusso V, Wijnveld M, Latrofa MS, Fajinmi A, Majekodunmi AO, Dogo AG, et al. Canine and ovine tick-borne pathogens in camels, Nigeria. Vet Parasitol. 2016;228:90–92. doi: 10.1016/j.vetpar.2016.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Alanazi AD, Abdullah S, Helps C, Wall R, Puschendorf R, ALHarbi SA, et al. Tick-borne pathogens in ticks and blood samples collected from camels in Riyadh Province, Saudi Arabia. Int J Zool Res. 2018;14:30–36. [Google Scholar]
- 65.Ibrahim AM, Kadle AA, Nyingilili HS. Microscopic and molecular detection of camel piroplasmosis in Gadarif State, Sudan. Vet Med Int. 2017;2017:9345231. doi: 10.1155/2017/9345231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dipeolu OO, Amoo A. The presence of kinetes of a Babesia species in the haemolymph smears of engorged Hyalomma ticks in Nigeria. Vet Parasitol. 1984;17:41–46. doi: 10.1016/0304-4017(84)90063-3. [DOI] [PubMed] [Google Scholar]
- 67.Sun M, Wang J, Liu Z, Guan G, Li Y, Liu J, et al. First molecular evidence of Babesia occultans and Theileria separata infection in ticks and sheep in China. Exp Appl Acarol. 2019;78:223–229. doi: 10.1007/s10493-019-00369-1. [DOI] [PubMed] [Google Scholar]
- 68.Ros-García A, Mghirbi Y, Bouattour A, Hurtado A. First detection of Babesia occultans in Hyalomma ticks from Tunisia. Parasitology. 2011;138:578–582. doi: 10.1017/S0031182011000060. [DOI] [PubMed] [Google Scholar]
- 69.Aktas M, Vatansever Z, Ozubek S. Molecular evidence for trans-stadial and transovarial transmission of Babesia occultans in Hyalomma marginatum and Rhipicephalus turanicus in Turkey. Vet Parasitol. 2014;204:369–371. doi: 10.1016/j.vetpar.2014.05.037. [DOI] [PubMed] [Google Scholar]
- 70.Qablan MA, Sloboda M, Jirků M, Oborník M, Dwairi S, Amr ZS, et al. Quest for the piroplasms in camels: identification of Theileria equi and Babesia caballi in Jordanian dromedaries by PCR. Vet Parasitol. 2012;186:456–460. doi: 10.1016/j.vetpar.2011.11.070. [DOI] [PubMed] [Google Scholar]
- 71.Abdelrahim IA, Ismail AA, Majid AM, Mohammad AS, Ibrahim A, Allsop M, et al. Detection of Babesia caballi in the one-humped Camel (Camelius dromedarius) using the Reverse Line Block (RLB) in Sudan. Sudan J Vet Res. 2019;24:69–72. [Google Scholar]
- 72.Jasim HJ, Azzal GY, Othman RM. Conventional and molecular detection of Babesia caballi and Theileria equi parasites in infected camels in south of Iraq. Basrah J Vet Res. 2015;14:110–121. [Google Scholar]
- 73.Luo D, Yin XP, Wang AD, Tian YH. The first detection of C. burnetii in Hyaloma asiaticum asiaticum in Dzungarian gate: the boundary of China and Kazakhstan. Dis Surveill. 2016;31:814–816. [Google Scholar]
- 74.De Bruin A, Van der Plaats RQJ, De Heer L, Paauwe R, Schimmer B, Vellema P, et al. Detection of Coxiella burnetii DNA on small-ruminant farms during a Q fever outbreak in the Netherlands. Appl Environ Microbiol. 2012;78:1652–1657. doi: 10.1128/AEM.07323-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bernard H, Brockmann SO, Kleinkauf N, Klinc C, Wagner-Wiening C, Stark K, et al. High seroprevalence of Coxiella burnetii antibodies in veterinarians associated with cattle obstetrics, Bavaria, 2009. Vector Borne Zoonotic Dis. 2012;12:552–557. doi: 10.1089/vbz.2011.0879. [DOI] [PubMed] [Google Scholar]
- 76.Schimmer B, Schotten N, Van Engelen E, Hautvast JLA, Schneeberger PM, Van Duijnhoven YTHP. Coxiella burnetii seroprevalence and risk for humans on dairy cattle farms, the Netherlands, 2010–2011. Emerg Infect Dis. 2014;20:417. doi: 10.3201/eid2003.131111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alsaad KM. Clinical, hematological and biochemical studies of anaplasmosis in arabian one humped camels (Camelus dromedarius) J Anim Vet Adv. 2019;8:2106–2109. [Google Scholar]
- 78.Sudan V, Sharma RL, Borah MK. Subclinical anaplasmosis in camel (Camelus dromedarius) and its successful therapeutic management. J Parasit Dis. 2014;38:163–165. doi: 10.1007/s12639-012-0206-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bastos AD, Mohammed OB, Bennett NC, Petevinos C, Alagaili AN. Molecular detection of novel Anaplasmataceae closely related to Anaplasma platys and Ehrlichia canis in the dromedary camel (Camelus dromedarius) Vet Microbiol. 2015;179:310–314. doi: 10.1016/j.vetmic.2015.06.001. [DOI] [PubMed] [Google Scholar]
- 80.Wernery U, Kaaden OR. Infectious diseases of camelids. Berlin: Blackwell Science; 2012. p. 404. [Google Scholar]
- 81.Azmat M, Ijaz M, Farooqi SH, Ghaffar A, Ali A, Masud A, et al. Molecular epidemiology, associated risk factors, and phylogenetic analysis of anaplasmosis in camel. Microb Pathog. 2018;123:377–384. doi: 10.1016/j.micpath.2018.07.034. [DOI] [PubMed] [Google Scholar]
- 82.Ashraf QU, Khan AU, Khattak RM, Ali M, Shaikh RS, Iqbal F. A report on the high prevalence of Anaplasma sp. in buffaloes from two provinces in Pakistan. Ticks Tick Borne Dis. 2013;4:395–398. doi: 10.1016/j.ttbdis.2013.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article. All newly generated sequences were submitted to the GenBank database.