Abstract
Background
Urolithiasis is a worldwide urological problem with significant contribution of genetic factors. Pakistan, which resides within the Afro-Asian stone belt, has a high reported prevalence (12%) of urolithiasis. Osteopontin (SPP1) is a urinary macromolecule with a suggested critical role in modulating renal stone formation, genetic polymorphisms of which may determine individual risk of developing urolithiasis. However, results of previous studies regarding SPP1 polymorphisms and susceptibility to urolithiasis have apparent inconsistencies with no data available for local population.
Methods
A total of 235 urolithiasis patients and 243 healthy controls, all of Pakistani ancestry, underwent genotyping for six SPP1 genetic polymorphisms in an effort to investigate potential association with urolithiasis using indigenous candidate gene association study design. Further, a comprehensive meta-analysis following a systematic literature search was also done to ascertain an evidence based account of any existent association regarding SPP1 promoter polymorphisms and risk of developing urolithiasis.
Results
Three SPP1 promoter polymorphisms, rs2853744:G > T, rs11730582:T > C and rs11439060:delG>G, were found to be significantly associated with risk of urolithiasis in indigenous genetic association study (OR = 3.14; p = 0.006, OR = 1.78; p = 0.006 and OR = 1.60; p = 0.012, respectively). We also observed a 1.68-fold positive association of a tri-allelic haplotype of these SPP1 promoter polymorphisms (G-C-dG) with risk of urolithiasis (OR = 1.68; p = 0.0079). However, no association was evident when data were stratified according to gender, age at first presentation, stone recurrence, stone multiplicity, parental consanguinity and family history of urolithiasis. The overall results from meta-analysis, which included 4 studies, suggested a significant association of SPP1 rs2853744:G > T polymorphism with susceptibility of urolithiasis (OR = 1.37; p = 0.004), but not for other SPP1 polymorphic variants analyzed.
Conclusions
In conclusion, we report significant association of 3 SPP1 polymorphisms with urolithiasis for the first time from South Asia, however, this association persisted only for SPP1 rs2853744:G > T polymorphism after meta-analysis of pooled studies. Further studies with a larger sample size will be required to validate this association and assess any potential usefulness in diagnosis and prognosis of renal stone disease.
Keywords: Candidate Gene Association study, Meta-analysis, Pakistan, Renal calculi, SPP1, SNP
Background
Urolithiasis is a common urological problem (worldwide prevalence of 4–20%) [1] causing high patient morbidity and associated healthcare burden involving recurrence, frequent hospitalization and sometimes progression to renal failure resulting from chronic kidney disease (CKD) [2, 3]. Pakistan resides in the middle of Afro-Asian renal stone belt, characterized by relatively higher prevalence (12–15%) of urolithiasis, complicated by environmental determinants of urolithiasis risk such as chronic dehydration and nutrition [4].
The reported etiology of urolithiasis is multifactorial involving environmental and genetic risk factors with heritability of 50% [5]. Only a few Genome Wide Association Studies (GWAS) are available regarding urolithiasis (predominantly from European and Japanese cohorts) that identified common genetic variants in various genetic loci regulating calcium and phosphate metabolism, urinary transporters and macromolecules among others, as urolithiasis associated risk factors [6, 7]. The “common disease-common variant” paradigm stresses the small but significant risk contributed by common genetic variations in the development of complex genetic disorders like urolithiasis [8]. Although genome wide and global association analyses options are readily available and trending, still, candidate gene association studies, if executed appropriately, provide a practical and a cost effective way to evaluate genetic determinants in complex diseases, especially in research settings where resource may be limited [9].
Evidence that macromolecular proteins, especially osteopontin, may play an important role in the modulation and development of urolithiasis, is growing [10]. Osteopontin, also called as secreted phosphoprotein 1, is a macromolecular glycoprotein with pleotropic expression and function [11, 12]. In kidneys, renal epithelial cells produce osteopontin with subsequent secretion into the urine as a normal macromolecular constituent of it [13]. The hypothesis that osteopontin may play a critical role in modulating renal stone formation is supported by many observations such as; (1) SPP1 as organic component in the matrix of renal stones [14]; (2) SPP1 as important regulator of renal calcification [15]; (3) Changes in SPP1 expression and urinary SPP1 levels in hyperoxaluric rats and human subjects with urolithiasis, respectively [16]; (4) In vitro cell culture based studies and in vivo SPP1 knockout animal models suggesting an important role of osteopontin in various phases of renal stone formation, including crystal nucleation, aggregation, retention, adhesion to renal epithelial cells and stone formation [17, 18]; and (5) Candidate gene association studies demonstrating association of SPP1 polymorphisms and urolithiasis in different ethnic groups [19, 20].
Osteopontin gene on chromosome 4q21–25 exhibits many functional polymorphisms in the promoter/coding regions that may influence osteopontin expression/activity [21] and have been analyzed for potential association with urolithiasis in different ethnic groups [19, 20, 22, 23], but with varied results. Therefore, in this context, the current study aimed to determine any potential association between common genetic variants in osteopontin gene and the susceptibility of urolithiasis in the indigenous sample set. Further, we also applied a systematic approach by collecting and analyzing the previously available data on the osteopontin polymorphisms in association with urolithiasis susceptibility, as determined by candidate gene association studies using urolithiasis patients and healthy controls, in the form of a meta-analysis that evaluated the varying results of previous studies and provided a more comprehensive and accurate estimate of any existent association expressed as OR (95% CI) and associated p-value.
Material and methods
Case control study cohort
Study participants
We recruited 235 urolithiasis patients, based on ultrasound finding of at least one renal stone [supplemented by other renal stone diagnostic procedures including X-ray imaging or non-contrast-enhanced computed tomography (NCCT) and urine analysis in most cases], presenting at five different tertiary care hospitals of Punjab, during a period of 29 months. All urolithiasis patients provided clinical and pedigree details, with confirmation provided by their attending clinician (a urologist) in addition to relevant medical records, and whole blood samples (EDTA anticoagulated) for genetic study. In addition, 243 healthy subjects that were age and gender matched with same ethnic origin having no personal or familial antecedents of urolithiasis were also recruited as control group. Specific details pertaining to enrollment of study subjects and their baseline characteristics have been described earlier [24, 25].
Genotyping of SPP1 polymorphisms
Subjects were genotyped for six polymorphisms of SPP1 gene including five promoter polymorphisms (rs28357094:T > G, rs11439060:delG>G, rs11730582:T > C, rs2853744:G > T and T-593A) by Sanger sequencing and one coding polymorphism (rs1126616:C > T) by PCR-RFLP based approach (assay details provided in Additional file 1). For Sanger sequencing based genotyping of SPP1 promoter polymorphisms, two DNA fragments of 369 bp and 289 bp, covering SPP1 promoter region, were amplified in a standard PCR reaction, purified using ethanol precipitation method and subjected to DNA sequencing. Resultant DNA sequences were aligned and analyzed using human SPP1 reference sequence gene (NCBI accession number NG_030362.1). Two researchers independently determined the genotypes of all SPP1 polymorphisms, based on DNA sequencing or PCR-RFLP, to minimize chances of genotyping bias.
Statistical analysis
The protocol followed for statistical analysis of datasets in the present study has been described elsewhere in detail [25]. Briefly, analysis of coded study data, including allelic and genotypic frequencies expressed as counts (percentages), was accomplished using the statistical package for social sciences (SPSS) version 20 for windows and online web tool SNPstats [26]. Using a Chi-square test, Hardy-Weinberg equilibrium (HWE) was performed which served as a statistical control for systematic genotyping error and population stratification where SPP1 polymorphisms that violated HWE, as indicated by p < 0.05 for in the control group, were not processed for further data analyses. Odds ratios (ORs) with associated 95% confidence intervals (CI) were determined to assess strength of statistical association, if any, considering allelic, genotypic, recessive, dominant and log-additive models by the same SNPstats program. The pairwise linkage disequilibrium and haplotype analysis for SPP1 polymorphisms was conducted using the Haploview program [27]. Bonferroni correction for multiple testing was performed in calculating the ORs and associated p-values for genotype and haplotype associations between SPP1 polymorphisms and urolithiasis. Also, the post-hoc power of the study estimates for SPP1 polymorphisms were performed using the Power and Sample Size Program (PS) version 3.0 available at http://biostat.mc.vanderbilt.edu/PowerSampleSize [28]. A p < 0.05 in two-sided analysis was considered significant unless specified differently.
Meta-analysis of SPP1 polymorphisms and risk of urolithiasis
The current meta-analysis of selected SPP1 genetic variants and urolithiasis susceptibility was performed using a modified protocol described in previous studies [25, 29], following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines [30] for performing and reporting of meta-analysis studies including considerations for literature search, eligibility, screening and selection of studies, data extraction, characteristics of included studies, reporting of effect sizes, assessment of risk of bias, summary of evidence along with any limitations, conclusions and disclosure of funding sources. A summary description of current meta-analysis protocol employed is as follows.
Strategy for systematic literature search
The review protocol of the present study was not pre-registered. The meta-analysis investigation included published studies using a case control study format for exploring genetic association of SPP1 polymorphisms and urolithiasis. Initial systematic literature search identified such published studies available before September, 2018 from the online databases of the PubMed, Google Scholar, ScienceDirect, Embase and Cochrane library. The literature search used relevant keywords related to the urolithiasis (renal stones, urolithiasis, nephrolithiasis and kidney stones) and osteopontin gene polymorphisms (osteopontin, SPP1, OPN). The search was restricted to studies with human subjects only and limited to publications in English language. Additional relevant articles were included by screening the bibliography provided in the articles retrieved as a result of initial literature search. Three researchers, working independently, completed the literature search step and discrepancies were resolved through discussion.
Selection of studies using a defined eligibility criteria and data extraction
Selection of relevant studies for inclusion in the meta-analysis was based on the following pre-defined eligibility criteria; 1) original studies following a case-control study for determining genetic association of SPP1 polymorphisms and urolithiasis; 2) SPP1 polymorphic sites investigated in the study should include at least one of the mentioned sites (rs2853744, rs11730582 and rs11439060); 3) Data presentation is appropriate enabling the calculations of Odds Ratios, confidence intervals and p-values. In accordance with this eligibility criteria, full texts of the selected articles were used to determine the relevancy and sufficiency of the included data. To ensure the robustness of analyses performed, three researchers independently performed the screening process resolving any conflicting issues through discussion. Information about reference, publication year, region, ethnicity, total number of study subjects including number of cases and controls, source of control samples, genotyping method, Hardy-Weinberg equilibrium status and genotype frequencies of the three SPP1 polymorphisms in cases and controls was extracted from each selected study. Collection of additional information by approaching the corresponding author of any of the included studies was not required and the data in its entirety that was included in the current meta-analysis was only extracted from published articles. The studies excluded from meta-analysis were; (1) studies with insufficient data presentation; (2) studies not pertaining to urolithiasis patients or SPP1 polymorphisms; (3) studies not following a case-control study design; (4) review articles; (5) meta-analysis studies; (6) meeting abstracts with insufficient data; and (7) unpublished reports. The Newcastle-Ottawa scale (NOS) was the reference used to evaluate the quality of eligible studies where quality score of ≥6 was considered as threshold for inclusion of studies in the meta-analysis.
Statistical analysis for meta-analysis part of study
Calculations of effect sizes and, other relevant meta-analysis measures and production of visual plots (including forest and Begg’s funnel plots) was achieved using Review Manager (RevMan) version 5 [31]. Use of Mantel–Haenszel statistics enabled analysis of dichotomous /categorical data. Odds ratio (OR) with associated confidence interval (CI) represented the principal measure that reflected strength of association between SPP1 genetic variants and urolithiasis susceptibility. The calculations of between-study heterogeneity in the current meta-analysis were based on the index of heterogeneity (I2) and chi-square (χ2) tests. A two tailed approach with a statistical significance threshold of 0.05 was used for all the statistical tests employed, except for the heterogeneity test which required the use of p < 0.10 as a threshold to reflect statistical significance because of the observation that the traditional Chi-square test has limited statistical power for studies involving a relatively small sample sizes. Fixed-effect model was used by default for determination of effect size, however, for studies displaying significant heterogeneity (as suggested by I2 values of > 50%), effect size was determined using a random effect model. The estimation of between-study variance was done using tau-squared (τ2) test. To assess the effect of a single study on cumulative results, each study was removed sequentially in a sensitivity analysis. Use of R version 3.5.2 [32] enabled assessment of any potential publication bias based on the Begg’s rank correlation test [33] and Egger’s linear regression test [34] . Stratified data analysis could not be performed due to limited number of studies available for meta-analysis part of the study.
Results
Case control study cohort
The salient demographic and clinical features of the present sample set and primary information of SPP1 polymorphisms analyzed in this study are presented in Additional files 2 and 3, respectively. For urolithiasis patients included in this study, the median age was 34 years with a gender distribution of 1.6:1 (Males: Females ratio), both of which were comparable with that of control group (p = 0.77 and 0.69 for age and gender distribution, respectively). 23% of patients presented at a younger age (< 18 years). Almost half of the patients had multiple renal stones (41%), recurrent disease (49%), positive family history of urolithiasis (48%) and parental consanguinity (53%). SPP1–593 T/A polymorphism was found to be monomorphic in this study. The allelic and genotypic distribution for SPP1 rs28357094:T > G and rs1126616:C > T SNPs deviated from the HWE in control group, and as a result, were not included in the subsequent data analyses. Sanger sequencing electropherograms for representative genotypes of each SPP1 polymorphism included in final analysis are presented in Additional file 4.
Data analysis for allelic and genotypic distribution suggested no significant association between the risk of urolithiasis and any of the SPP1 polymorphisms analyzed except for rs11439060:delG>G (OR = 0.40; p = 0.002 for G/dG genotype in co-dominant model) (Table 1). Additionally, SPP1 rs2853744:G > T polymorphism showed significant associated with increased risk of urolithiasis in a dominant model (OR = 3.14; p = 0.006). While, SPP1 rs11730582:T > C and rs11439060:delG>G polymorphisms were significantly associated with the risk of urolithiasis (OR = 1.78; p = 0.006 and OR = 1.60; p = 0.012, respectively) considering a recessive genetic model (Table 2).
Table 1.
Association analysis of urolithiasis risk and SPP1 genetic variants considering their allelic and genotypic frequencies
| SPP1 genetic variants | Genotype/Allele | Urolithiasis patients n = 235, n (%) |
Healthy controls n = 243, n (%) |
OR (95% CI) | p-value (corrected)† |
|---|---|---|---|---|---|
| rs2853744:G > T | T/T | 07 (3.1%) | 21 (9.1%) | Referent | 0.024 |
| G/T | 61 (27.2%) | 62 (26.8%) | 2.95 (1.17–7.45) | ||
| G/G | 156 (69.6%) | 148 (64.1%) | 3.16 (1.31–7.66) | ||
| T | 75 (17%) | 104 (23%) | Referent | 0.035 | |
| G | 373 (83%) | 358 (77%) | 1.44 (1.03–2.01) | ||
| rs11730582:T > C | T/T | 63 (28%) | 69 (29.6%) | Referent | 0.017 |
| T/C | 87 (38.7%) | 113 (48.5%) | 0.84 (0.54–1.31) | ||
| C/C | 75 (33.3%) | 51 (21.9%) | 1.61 (0.98–2.64) | ||
| T | 213 (47%) | 251 (54%) | Referent | 0.056 | |
| C | 237 (53%) | 215 (46%) | 1.29 (1.00–1.68) | ||
| rs11439060:delG > G | G/G | 19 (8.3%) | 12 (5%) | Referent | 0.002 |
| G/dG | 65 (28.5%) | 103 (43.3%) | 0.40 (0.18–0.88) | ||
| dG/dG | 144 (63.2%) | 123 (51.7%) | 0.74 (0.35–1.58) | ||
| G | 103 (23%) | 127 (27%) | Referent | 0.170 | |
| dG | 353 (77%) | 349 (73%) | 1.24 (0.92–1.68) |
†p-values are corrected for age and gender. p-value adjustment for multiple testing using Bonferroni method was also made (p-value threshold 0.016). Statistical significance is highlighted in bold
OR Odds ratio; n (%), frequency and CI Confidence interval
Table 2.
Association analysis of urolithiasis risk and SPP1 genetic variants considering dominant, recessive and log-additive genetic models
| SPP1 polymorphisms | Model | Genotypes | Patients n = 235, n (%) | Controls value 243, n (%) | OR (95% CI) | p-value (corrected)† |
|---|---|---|---|---|---|---|
| rs2853744:G > T | Dominant | T/T | 07 (3.1%) | 21 (9.1%) | 1.00 | 0.006 |
| G/T-G/G | 217 (96.9%) | 210 (90.9%) | 3.14 (1.29–7.45) | |||
| Recessive | T/T-G/T | 68 (30.4%) | 83 (35.1%) | 1.00 | 0.210 | |
| G/G | 156 (69.4%) | 148 (64.9%) | 1.29 (0.87–1.90) | |||
| Log-additive | – | – | – | 1.38 (1.01–1.89)S | 0.040 | |
| rs11730582:T > C | Dominant | T/T | 63 (28%) | 69 (29.6%) | 1.00 | 0.700 |
| T/C-C/C | 162 (72%) | 164 (70.4%) | 1.08 (0.72–1.62) | |||
| Recessive | T/T-T/C | 150 (66.7%) | 182 (78.1%) | 1.00 | 0.006 | |
| C/C | 75 (33.3%) | 51 (21.9%) | 1.78 (1.18–2.71) | |||
| Log-additive | – | – | – | 1.26 (0.99–1.61) | 0.062 | |
| rs11439060:delG > G | Dominant | G/G | 19 (8.3%) | 12 (5%) | 1.00 | 0.150 |
| dG/G-dG/dG | 209 (91.7%) | 226 (95%) | 0.58 (0.28–1.23) | |||
| Recessive | G/G-dG/G | 84 (36.8%) | 115 (48.3%) | 1.00 | 0.012 | |
| dG/dG | 144 (63.2%) | 123 (51.7%) | 1.60 (1.11–2.60) | |||
| Log-additive | – | – | – | 1.24 (0.92–1.67) | 0.15 |
†p-values are corrected for age and gender. p-value adjustment for multiple testing using Bonferroni method was also made (p-value threshold 0.016). Statistical significance is highlighted in bold
OR Odds ratio; n (%), frequency and CI Confidence interval
Frequency of G-C-dG haplotype (SPP1 rs2853744-rs11730582-rs11439060 polymorphisms, respectively) was significantly higher in urolithiasis patients as compared to controls (OR = 1.68; p = 0.0079), suggesting an association with increased risk of urolithiasis in haplotype analysis (Table 3). However, pair wise linkage disequilibrium (LD) and haplotype plot structure analysis demonstrated lack of any substantial LD measures (based on D’ values) for each pair of SPP1 loci analyzed, suggesting that LD in this region is low (Additional file 5).
Table 3.
Association of urolithiasis risk with of SPP1 genetic variants considering haplotype analysis
| SPP1 haplotypes (rs2853744:G > T- rs11730582:T > C- rs11439060:delG > G) | Haplotype frequency† | Case, control ratios | OR (95% CI) | p-value (corrected)‡ |
|---|---|---|---|---|
| G-T-dG | 0.323 | 0.313, 0.332 | Referent | – |
| G-C-dG | 0.312 | 0.374, 0.248 | 1.68 (1.15–2.46) | 0.0079 |
| G-C-G | 0.090 | 0.086, 0.107 | 0.95 (0.57–1.58) | 0.840 |
| G-T-G | 0.078 | 0.079, 0.068 | 1.25 (0.61–2.56) | 0.550 |
| T-T-dG | 0.060 | 0.083, 0.053 | 1.43 (0.72–2.87) | 0.310 |
| T-C-dG | 0.059 | 0.025, 0.081 | 0.53 (0.24–1.17) | 0.120 |
OR Odds ratio; 95% CI 95% confidence interval
†Haplotypes with a frequency > 5% were analyzed
‡p-values are corrected for age and gender. p-value adjustment for multiple testing using Bonferroni method was also made (p-value threshold 0.0083). Statistical significance is highlighted in bold
The SPP1 polymorphisms data was also analyzed considering sub-groups of Pakistani urolithiasis patients based on demographics (gender, age at first presentation), clinical features (stone multiplicity and stone recurrence) and histories (parental consanguinity and family history of urolithiasis). But, none of these comparisons yielded any significant associations (Additional file 6).
Meta-analysis
Qualitative synthesis for association of urolithiasis and SPP1 genetic variants
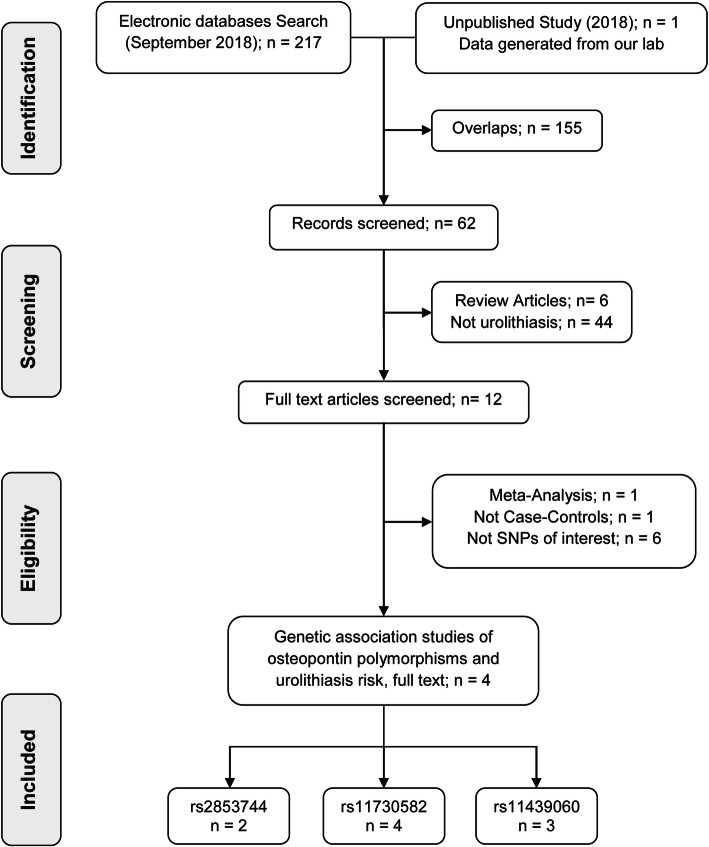
A flow diagram, reflecting the sequence of study selection for genetic association of 3 SPP1 genetic variants and susceptibility of urolithiasis, is described in Fig. 1. An initial online search of literature databases by means of defined MeSH terms concerning urolithiasis and osteopontin genetic variants, resulted in retrieval of a total of 217 articles. However, in the end, a total of 4 studies were included in the present meta-analysis, comprising of 3 previously published reports obtained after rigorous screening according to the eligibility criteria, combined with the indigenous genetic epidemiology study. There were 2 studies exploring the association of SPP1 polymorphism rs2853744, 4 for rs11730582 and 3 for rs11439060. All included studies had NOS score of 6 or better.
Fig. 1.
Flow diagram reflecting selection process of eligible studies included in meta-analysis. Initial literature search identified a total of 217 records from different online databases including Google Scholar, PubMed, ScienceDirect, Cochrane library and Embase. Of these, 155 overlapping studies (same studies that were indexed in two or more of these databases and resulted in redundant entries) were excluded, retaining 62 unique studies for further screening. Subsequent screening according to defined eligibility criteria yielded a total of 04 final studies including in this meta-analysis
The salient characteristics of the studies comprising the present meta-analysis are described in Table 4. The publication period for the selected studies ranged from 2010 to 2018. All four studies included in the meta-analysis were case-control studies, based on Asian population, and most (3/4) studies used control groups collected from general population. Also, all studies conformed to HWE with respect to their control group. Among these, three studies also analyzed the association of other polymorphisms in SPP1 gene with urolithiasis. However, data pertaining to additional polymorphisms were not included in the present meta-analysis. All studies showed least one of the analyzed SPP1 genetic variants to be positively associated with the susceptibility of urolithiasis.
Table 4.
Main characteristics and findings of the eligible studies included in this meta-analysis
| Reference (first author, year) | Region | Ethnic group | Controls source | Samples (N) | Cases | Controls | Polymorphic sites | HWE status† | Genotyping method | Findings |
|---|---|---|---|---|---|---|---|---|---|---|
| Liu, 2010 [35] | Taiwan | Asian | Hospital based | 496 | 249 | 247 | rs11730582, rs11439060 | Yes | TaqMan genotyping assay | rs11439060 of SPP1 promoter associated with risk of UL in allelic and genotypic models |
| Safarinejad, 2013 [19] | Iran | Asian | Population based | 1026 | 342 | 684 | rs2853744, rs11730582 | Yes | PCR-FRET | SPP1 SNP rs2853744 showed significant association with UL |
| Xiao, 2016 [20] | China | Asian | Population based | 480 | 230 | 250 | rs11730582, rs11439060 | Yes | TaqMan genotyping assay | rs11439060 in SPP1 promoter significantly associated with risk of UL as well as clinical characteristics in UL |
| Present study, 2018 | Pakistan | Asian | Population based | 478 | 235 | 243 | rs2853744, rs11730582 and rs11439060 | Yes | Sanger sequencing | All 3 SPP1 promoter SNPs associated with UL under different genetic models |
†Yes indicates consistence with HWE
FRET Fluorescence resonance energy transfer; HWE Hardy-Weinberg equilibrium; N Total number of samples; SPP1 Osteopontin; SNP Single-nucleotide polymorphism; UL Urolithiasis
Quantitative synthesis for association of urolithiasis and SPP1 genetic variants
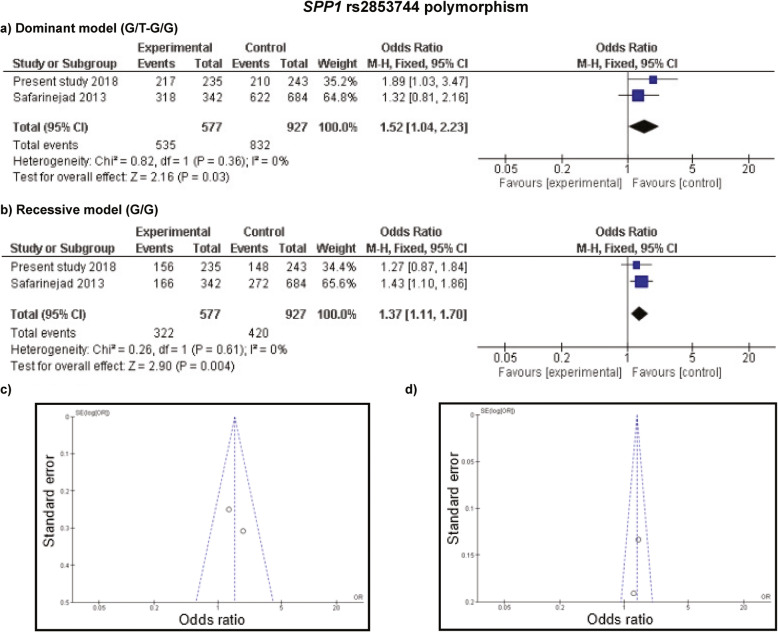
For the association of SPP1 rs2853744 polymorphism, only 2 studies were available including 577 cases and 927 controls where overall results from recessive model reflected a statistically noteworthy association with the susceptibility of urolithiasis (OR = 1.37; p = 0.004, Fig. 2b). However, no association was detected under dominant model after considering correction for multiple testing (Fig. 2a). Measures of heterogeneity in this set of studies were not significant (I2 = 0%, p = 0.61 for recessive model) therefore, fixed effect model was employed to determine the cumulative results (Fig. 2c and d).
Fig. 2.
Meta-analysis of SPP1 rs2853744:G > T polymorphism with risk of urolithiasis. a and b Forest plots of urolithiasis association with rs2853744 polymorphism assuming dominant and recessive model, respectively. c and d Funnel plots of rs2853744 polymorphism in dominant and recessive inheritance, respectively, using fixed effect model
Meta-analysis of SPP1 rs11730582 and rs11439060 polymorphisms included 4 (1056 cases and 1424 controls) and 3 (714 cases and 740 controls) studies, respectively. The summarization of all studies indicated no significant associations between rs11730582, and rs11439060 polymorphisms and urolithiasis using either a dominant or recessive model (Additional files 7 and 8, parts a and b) after correction for multiple testing. Heterogeneity analysis for SPP1 rs11730582 polymorphism was insignificant, but not for SPP1 rs11439060 polymorphism, therefor fixed and random effect models, respectively, were applied in calculation of pooled results (Additional files 7 and 8, parts c and d).
Shape of funnel plots and results of Egger’s test (as depicted in Fig. 2, and Additional files 7 and 8) rendered calculation of the publication bias where absence of any significant publication bias was evident except for the analysis of SPP1 rs11439060 polymorphism and urolithiasis (Additional file 8, parts c and d); however, the Egger’s test was not significant (p = 0.19 for dominant model and p = 0.15 for recessive model) in that case too. The outcome in sensitivity analyses, performed by removing each study at a time, indicated the reliability of the current meta-analysis results as no significant influence of an individual study was evident on the cumulative OR.
The raw dataset containing the individual phenotypic and genotypic data for each of the SPP1 polymorphisms analyzed has been provided as additional file 9 in the supplementary data.
Discussion
The role of genetic variations with low penetrance has earned special concern in urolithiasis research, which, in association with other risk factors, may define the critical threshold necessary for development of clinically significant renal stones. A number of genetic markers in different urolithiasis genes including SPP1, VDR, CaSR, urokinase, prothrombin, interleukins and others have been investigated in this regard [6]. Osteopontin has earned a particular prominence among these genetic risk factors as an importance determinant and regulator of renal calcification and stone formation [11, 13, 36]. However, the results of genetic association studies in urolithiasis are still to achieve diagnostic and translational significance. The gap in existing knowledge and inconsistent results for potential genetic associations in urolithiasis indicate a need of genetic epidemiology studies performed in diverse populations.
For the first time, we present a genetic association study investigating the role of common genetic variants in SPP1 gene using a sample set of Pakistani patients manifesting clinically significant urolithiasis where we demonstrate significant association of three SPP1 promoter polymorphisms (rs2853744:G > T, rs11730582:T > C and rs11439060:delG>G) with urolithiasis. We also demonstrated that subjects simultaneously harboring G-C-dG alleles of SPP1 rs2853744-rs11730582-rs11439060 polymorphisms, respectively, have 1.68 times increased risk of urolithiasis that is statistically significant as determined by haplotype association analysis.
Moreover, estimates of post-hoc study power showed that the levels of power associated with SPP1 rs2853744:G > T, rs11730582:T > C and rs11439060:delG>G polymorphisms were 78.4, 79.8 and 99.7%. These results reflect that the sample size of 235 provided fairly adequate power (almost 80%) in determining genetic associations between these polymorphic variants of SPP1 gene and urolithiasis in the indigenous population.
We also analyzed whether additional risk factors, including gender, early age at presentation, severe disease (multiple renal stones, recurrences), presence of familial history of urolithiasis and parental consanguinity modulated the SPP1 polymorphisms based potential genetic risk of urolithiasis. However, sub-group analyses considering these additional risk factors reflected no moderation of genetic risk for SPP1 polymorphisms analyzed by any of the additional risk factors, at least in the context of present sample set.
Considerable heterogeneity in correlation of these SPP1 polymorphisms and risk of urolithiasis have been reported by studies conducted in different ethnic groups. An Iranian study [19] reported a positive association of G allele/GG genotype of rs2853744:G > T with risk of developing urolithiasis. In contrast to our study results, no significant association of rs11730582:T > C with urolithiasis risk was reported in 3 independent studies from Taiwan, Iran and China [19, 20, 35]. For rs11439060:delG>G, significant association with urolithiasis phenotype was described in two studies, however, in consistence with our study, first study of Taiwanese origin reported dG/dG genotype as the risk genotype associated with increased susceptibilty of urolithiasis [35], while the second study found insertion allele or genotype (G allele or G/G genotype) to be more prevalent in Chinese urolithiasis patients as compared to controls [20].
The observed overall heterogeneity in the results of different studies regarding association of SPP1 polymorphisms with urolithiasis may be attributable to many factors of genetic and non-genetic (including demographic, environmental and analytical variants) in origin. With respect to genetic modulators, variations in allele/genotype distributions and LD pattern of SPP1 polymorphisms in different populations, reflecting a population specific genetic architecture, may determine substantial heterogeneity observed in genetic risk of urolithiasis. Similarly, non-genetic confounding factors may also explain a part of heterogeneity observed in the results of different genetic studies of SPP1 polymorphisms and urolithiasis risk, including; a) differential prevalence of urolithiasis in different regions home to different ethnicities [e.g. high (12–15%) vs moderate (9.6%) vs low (5.7%) prevalence of urolithiasis in Pakistan, Taiwan and Iran, respectively] [37–39]; b) environmental risk factors (most importantly a lithogenic diet and lifestyle, and chronic dehydration) owing to differences in their context and relative contribution [4]; and c) lack of standardization and consistency in study methodologies (reflecting selection bias, control source, genotyping method used, statistical analysis approach especially when it comes to HWE conformance and applying correction for multiple testing).
Meta-analysis is a powerful tool that provides evidence based comprehensive and reliable results compared to a single study when investigating association of potential risk factors and disease phenotype. Therefore, in addition to presenting results of indigenous study, we also conducted a meta-analysis to clarify the possible association between SPP1 polymorphisms and risk of urolithiasis. To the best of our knowledge, no meta-analysis has been carried out previously regarding association of SPP1 promoter polymorphisms with urolithiasis risk. The results of present meta-analysis reveal that GG genotype of SPP1 rs2853744:G > T significantly increased the risk of urolithiasis by 1.37 fold in a recessive model. However, no significant association between other SPP1 polymorphisms analyzed (rs11730582:T > C and rs11439060:delG>G) was observed after correction for multiple testing. Several indicators of the robustness of analyses done and results generated in the current meta-analysis can be identified including; a) all the studies included in the meta-analysis were in HWE, b) no publication bias or heterogeneity was observed except for rs11439060:delG>G, c) a single study did not influenced the cumulative results as suggested by sensitivity analysis, and d) correction for multiple testing was applied. Inclusion of Pakistani dataset in the overall meta-analysis also reinforced the results obtained in this study. However, the results should still be interpreted with caution because of the limited number of primary studies available for present meta-analysis.
Currently, there is only one meta-analysis available on the subject which revealed positive association of SPP1 coding region genetic variant (rs1126616:C > T) and lower serum and urine osteopontin levels with increased risk of developing urolithiasis [40], however, they did not include any other SPP1 polymorphism (including SPP1 promoter polymorphisms investigated in this study) in the analysis, which limits the usefulness and broader applicability of that study.
Despite the efforts made to generate evidence based and robust statistical results through current case control and meta-analysis study, a number of limitations should be acknowledged. First, a comprehensive investigation and correlation of biochemical parameters of renal stone disease (including stone analysis and serum/urine osteopontin levels) could not be done due to limited resources available. Second, all the eligible studies, including our own data, could not address all the known risk factors involved. Keeping in view the multifactorial nature of the urolithiasis phenotype, a more comprehensive and precise analysis should be based on adjusted estimates considering covariates such as gender, age, lifestyle, dietary habits including fluid intake, and other genetic factors, thus also investigating gene-gene and gene-environment interactions. Third, sub-group analysis based on ethnicity, source of control samples and other factors, could not be performed due to limited number of published studies available for current meta-analysis. Further, we did not include other SPP1 polymorphisms because we could find only a couple of studies with limited sample size.
Conclusions
In conclusion, the current study provides first account of a modest but statistically significant association of three SPP1 promoter polymorphisms and their tri-allelic haplotype with increased risk of urolithiasis from South-Asian region under the indicated genetic model. In addition, evidence from meta-analysis part of the study supports the positive association of rs2853744:G > T SPP1 SNP and susceptibility of urolithiasis. Further studies using larger sample sizes and analyses of gene-gene and gene-environment factors in diverse populations are suggested to validate and determine usefulness and broader relevance of SPP1 and other genetic polymorphisms in accessing risk and prognosis of urolithiasis.
Supplementary information
Additional file 1. Oligonucleotide sequences, PCR conditions and restriction enzyme used for the genotyping of SPP1 gene polymorphisms.
Additional file 2. Basic characteristics of the study participants in the case-control part of the study.
Additional file 3. Basic information and HWE analysis for SPP1 gene polymorphisms analyzed in this study.
Additional file 4. Representative electropherograms for each genotype of three SPP1 polymorphisms. (A) rs11730582:T > C, (B) rs2853744:G > T and (C) rs11439060:delG>G.
Additional file 5. Pairwise linkage disequilibrium (LD) map, based on D´ values, of SPP1 polymorphic markers analyzed in the present study. No significant LD was apparent in any of the SPP1 polymorphic pairs analyzed.
Additional file 6. A Association of SPP1 rs2853744:G > T with different clinical characteristics of urolithiasis. B Association of SPP1 rs11730582:T > C with different clinical characteristics of urolithiasis. C Association of SPP1 rs11439060:delG>G with different clinical characteristics of urolithiasis.
Additional file 7. Meta-analysis of SPP1 rs11730582:T > C polymorphism with risk of urolithiasis. a) and b) Forest plots of urolithiasis association with rs11730582 polymorphism using dominant and recessive model, respectively. c) and d) Funnel plots of rs11730582 polymorphism assuming dominant and recessive inheritance, respectively, using fixed effect model.
Additional file 8. Meta-analysis of SPP1 rs11439060:delG>G polymorphism with susceptibility of urolithiasis. a) and b) Forest plots of urolithiasis association with rs11439060 polymorphism following dominant and recessive model, respectively. c) and d) Funnel plots of rs11439060 polymorphism using dominant and recessive inheritance, respectively, by random effect model.
Additional file 9. Raw dataset with details of genotypes and phenotypes of every study subject analyzed for SPP1 polymorphisms in this study.
Acknowledgements
We thank patients who participated in the study and hospital staff who facilitated the research team in data and sample collection. We also acknowledge the facilities and support provided by university staff.
Abbreviations
- CaSR
Calcium sensing receptor
- CI
Confidence interval
- CKD
Chronic kidney disease
- EDTA
Ethylenediaminetetraacetic acid
- GWAS
Genome wide association studies
- HWE
Hardy-weinberg equilibrium
- I2
Index of heterogeneity test
- LD
Linkage disequilibrium
- MeSH
Medical subject headings
- NCCT
Non-contrast-enhanced computed tomography
- NOS
Newcastle-ottawa scale
- OPN
Osteopontin
- OR
Odds ratio
- PCR-RFLP
Polymerase chain reaction-restriction fragment length polymorphism
- PRISMA
Preferred reporting items for systematic reviews and meta-analyses
- PS
Power and sample size program
- SPP1
Secreted phosphoprotein 1
- VDR
Vitamin D receptor
- τ2
Tau-squared test
- χ2
Chi-square test
Authors’ contributions
AA1 performed data curation, genetic and statistical analysis, wrote the original manuscript and contributed in review/editing of final manuscript. AA2 contributed towards data curation, genetic analysis and manuscript review/editing. AH and MA contributed in patient recruitment, clinical data acquisition and analysis and manuscript review/editing. HN contributed towards genetic analysis. ARK and AA3 performed the meta-analysis and were significant contributors towards manuscript review/editing. SK designed and supervised the study, acquired funding and contributed significantly towards manuscript review/editing. All authors read and approved the final manuscript.
Funding
This work was supported by Higher Education Commission of Pakistan (grant number NRPU-1987 to Shagufta Khaliq). The funding body had no role in the design of the study and collection, analysis, and interpretation of data. The meta-analysis part of the study did not receive any specific funding.
Availability of data and materials
The raw sequence data used and/ analyzed in this study has been deposited in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) and is publically available under the accession numbers MT450478 to MT450678. Sequencing data of all the cases and control samples was aligned and analyzed on the platform of SPP1 reference sequence gene using NCBI GenBank accession number NG_030362.1 available at https://www.ncbi.nlm.nih.gov/nuccore/353249890. The datasets supporting the findings reported in this article are included in this published article and its supplementary information files. Details of individual phenotype and genotypes for each of the SPP1 polymorphisms analyzed have been provided as additional file 9 in the supplementary data.
Ethics approval and consent to participate
The Ethical Review Committee for Medical and Biomedical Research, University of Health Sciences Lahore, which adheres to the latest guidelines provided in Declaration of Helsinki, approved the study protocol. All the study participants (urolithiasis patients and healthy controls) provided written informed consent before enrollment in the study. In case of minor subjects (< 16 years old), written informed consent was provided by their parents or legal guardians.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12881-020-01101-2.
References
- 1.Sorokin I, Mamoulakis C, Miyazawa K, Rodgers A, Talati J, Lotan Y. Epidemiology of stone disease across the world. World J Urol. 2017;35(9):1301–1320. doi: 10.1007/s00345-017-2008-6. [DOI] [PubMed] [Google Scholar]
- 2.Rule AD, Bergstralh EJ, Melton LJ, Li X, Weaver AL, Lieske JC. Kidney stones and the risk for chronic kidney disease. Clin J Am Soc Nephrol. 2009;4(4):804–811. doi: 10.2215/CJN.05811108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miller DC, Saigal CS, Litwin MS. The demographic burden of urologic diseases in America. Urol Clin N Am. 2009;36(1):11–27. doi: 10.1016/j.ucl.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rizvi SH, Naqvi SA, Hussain Z, Hashmi A, Hussain M, Zafar MN, et al. The management of stone disease. BJU Int. 2002;89(Suppl 1):62–68. doi: 10.1046/j.1465-5101.2001.134.x. [DOI] [PubMed] [Google Scholar]
- 5.Goldfarb DS, Avery AR, Beara-Lasic L, Duncan GE, Goldberg J. A twin study of genetic influences on nephrolithiasis in women and men. Kidney Int Rep. 2019;4(4):535–540. doi: 10.1016/j.ekir.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayer JA. Progress in understanding the genetics of calcium-containing nephrolithiasis. J Am Soc Nephrol. 2017;28(3):748–759. doi: 10.1681/ASN.2016050576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanikawa C, Kamatani Y, Terao C, Usami M, Takahashi A, Momozawa Y, et al. Novel risk loci identified in a genome-wide association study of Urolithiasis in a Japanese population. J Am Soc Nephrol. 2019;30(5):855–864. doi: 10.1681/ASN.2018090942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritchard JK, Cox NJ. The allelic architecture of human disease genes: common disease-common variant…or not? Hum Mol Genet. 2002;11(20):2417–2423. doi: 10.1093/hmg/11.20.2417. [DOI] [PubMed] [Google Scholar]
- 9.Foulkes AS. Applied statistical genetics with R: for population-based association studies. New York: Springer-Verlag; 2009. p. 2009. [Google Scholar]
- 10.Ertan P, Tekin G, Oger N, Alkan S, Horasan GD. Metabolic and demographic characteristics of children with urolithiasis in Western Turkey. Urol Res. 2011;39(2):105–110. doi: 10.1007/s00240-010-0306-1. [DOI] [PubMed] [Google Scholar]
- 11.Mazzali M, Kipari T, Ophascharoensuk V, Wesson JA, Johnson R, Hughes J. Osteopontin--a molecule for all seasons. QJM. 2002;95(1):3–13. doi: 10.1093/qjmed/95.1.3. [DOI] [PubMed] [Google Scholar]
- 12.Kazanecki CC, Uzwiak DJ, Denhardt DT. Control of osteopontin signaling and function by post-translational phosphorylation and protein folding. J Cell Biochem. 2007;102(4):912–924. doi: 10.1002/jcb.21558. [DOI] [PubMed] [Google Scholar]
- 13.Watts RWE. Idiopathic urinary stone disease: possible polygenic aetiological factors. QJM. 2005;98(4):241–246. doi: 10.1093/qjmed/hci041. [DOI] [PubMed] [Google Scholar]
- 14.Kohri K, Nomura S, Kitamura Y, Nagata T, Yoshioka K, Iguchi M, et al. Structure and expression of the mRNA encoding urinary stone protein (osteopontin) J Biol Chem. 1993;268(20):15180–15184. [PubMed] [Google Scholar]
- 15.Konya E, Umekawa T, Iguchi M, Kurita T. The role of osteopontin on calcium oxalate crystal formation. Eur Urol. 2003;43(5):564–571. doi: 10.1016/S0302-2838(03)00088-5. [DOI] [PubMed] [Google Scholar]
- 16.Khan SR, Johnson JM, Peck AB, Cornelius JG, Glenton PA. Expression of osteopontin in rat kidneys: induction during ethylene glycol induced calcium oxalate nephrolithiasis. J Urol. 2002;168(3):1173–1181. doi: 10.1016/S0022-5347(05)64621-6. [DOI] [PubMed] [Google Scholar]
- 17.Kleinman JG, Wesson JA, Hughes J. Osteopontin and calcium stone formation. Nephron Physiol. 2004;98(2):p43–p47. doi: 10.1159/000080263. [DOI] [PubMed] [Google Scholar]
- 18.Tsuji H, Shimizu N, Nozawa M, Umekawa T, Yoshimura K, De Velasco MA, et al. Osteopontin knockdown in the kidneys of hyperoxaluric rats leads to reduction in renal calcium oxalate crystal deposition. Urolithiasis. 2014;42(3):195–202. doi: 10.1007/s00240-014-0649-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Safarinejad MR, Shafiei N, Safarinejad S. Association between polymorphisms in osteopontin gene (SPP1) and first episode calcium oxalate. Urolithiasis. 2013;41(4):303–313. doi: 10.1007/s00240-013-0582-7. [DOI] [PubMed] [Google Scholar]
- 20.Xiao X, Dong Z, Ye X, Yan Y, Chen X, Pan Q, et al. Association between OPN genetic variations and nephrolithiasis risk. Biomed Rep. 2016;5(3):321–326. doi: 10.3892/br.2016.724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giacopelli F, Marciano R, Pistorio A, Catarsi P, Canini S, Karsenty G, et al. Polymorphisms in the osteopontin promoter affect its transcriptional activity. Physiol Genomics. 2004;20(1):87–96. doi: 10.1152/physiolgenomics.00138.2004. [DOI] [PubMed] [Google Scholar]
- 22.Gao B, Yasui T, Itoh Y, Li Z, Okada A, Tozawa K, et al. Association of osteopontin gene haplotypes with nephrolithiasis. Kidney Int. 2007;72(5):592–598. doi: 10.1038/sj.ki.5002345. [DOI] [PubMed] [Google Scholar]
- 23.Gögebakan B, Igci YZ, Arslan A, Igci M, Erturhan S, Oztuzcu S, et al. Association between the T-593A and C6982T polymorphisms of the osteopontin gene and risk of developing nephrolithiasis. Arch Med Res. 2010;41(6):442–448. doi: 10.1016/j.arcmed.2010.08.014. [DOI] [PubMed] [Google Scholar]
- 24.Amar A, Majmundar AJ, Ullah I, Afzal A, Braun DA, Shril S, et al. Gene panel sequencing identifies a likely monogenic cause in 7% of 235 Pakistani families with nephrolithiasis. Hum Genet. 2019. [DOI] [PMC free article] [PubMed]
- 25.Amar A, Afzal A, Hussain SA, Hameed A, Khan AR, Shakoor M, et al. Association of vitamin D receptor gene polymorphisms and risk of urolithiasis: results of a genetic epidemiology study and comprehensive meta-analysis. Urolithiasis. 2019.. [DOI] [PubMed]
- 26.Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. doi: 10.1093/bioinformatics/btl268. [DOI] [PubMed] [Google Scholar]
- 27.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 28.Dupont WD, Plummer WD. Power and sample size calculations. A review and computer program. Control Clin Trials. 1990;11(2):116–128. doi: 10.1016/0197-2456(90)90005-M. [DOI] [PubMed] [Google Scholar]
- 29.Abid A, Ajaz S, Khan AR, Zehra F, Hasan AS, Sultan G, et al. Analysis of the glutathione S-transferase genes polymorphisms in the risk and prognosis of renal cell carcinomas. Case-control and meta-analysis. Urol Oncol. 2016;34(9):419.e1–419e12. doi: 10.1016/j.urolonc.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 30.Moher D, Liberati A, Tetzlaff J, Altman DG, Group P Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6(7):e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deeks JJ, Higgins JPT. Statistical algorithms in review manager 5. Stat Methods Group Cochrane Collaboration. 2010:1–11..
- 32.Team RC. R: a language and environment for statistical computing. 2013.
- 33.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50(4):1088–1101. doi: 10.2307/2533446. [DOI] [PubMed] [Google Scholar]
- 34.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315(7109):629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu C-C, Huang S-P, Tsai L-Y, Wu W-J, Juo S-HH, Chou Y-H, et al. The impact of osteopontin promoter polymorphisms on the risk of calcium urolithiasis. Clin Chim Acta. 2010;411(9–10):739–743. doi: 10.1016/j.cca.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 36.Mo L, Liaw L, Evan AP, Sommer AJ, Lieske JC, Wu X-R. Renal calcinosis and stone formation in mice lacking osteopontin, Tamm-Horsfall protein, or both. Am J Physiol Renal Physiol. 2007;293(6):F1935–F1943. doi: 10.1152/ajprenal.00383.2007. [DOI] [PubMed] [Google Scholar]
- 37.Talati J, Khan F, Drago H, Lall E, Khan NZ, Talati A, et al. Epidemiology of urolithiasis in Pakistan. In: Talati J, RAL S, Moazam F, Ahmed M, et al., editors. The Management of Lithiasis. Developments in Nephrology. Netherlands: Springer; 1997. pp. 21–33. [Google Scholar]
- 38.Safarinejad MR. Adult urolithiasis in a population-based study in Iran: prevalence, incidence, and associated risk factors. Urol Res. 2007;35(2):73–82. doi: 10.1007/s00240-007-0084-6. [DOI] [PubMed] [Google Scholar]
- 39.Lee Y-H, Huang W-C, Tsai J-Y, Lu C-M, Chen W-C, Lee M-H, et al. Epidemiological studies on the prevalence of upper urinary calculi in Taiwan. Urol Int. 2002;68(3):172–177. doi: 10.1159/000048445. [DOI] [PubMed] [Google Scholar]
- 40.Li X, Liu K, Pan Y, Zhang J, Lv Q, Hua L, et al. Roles of Osteopontin gene polymorphism (rs1126616), Osteopontin levels in urine and serum, and the risk of Urolithiasis: a meta-analysis. Biomed Res Int. 2015;2015:e315043. doi: 10.1155/2015/315043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1. Oligonucleotide sequences, PCR conditions and restriction enzyme used for the genotyping of SPP1 gene polymorphisms.
Additional file 2. Basic characteristics of the study participants in the case-control part of the study.
Additional file 3. Basic information and HWE analysis for SPP1 gene polymorphisms analyzed in this study.
Additional file 4. Representative electropherograms for each genotype of three SPP1 polymorphisms. (A) rs11730582:T > C, (B) rs2853744:G > T and (C) rs11439060:delG>G.
Additional file 5. Pairwise linkage disequilibrium (LD) map, based on D´ values, of SPP1 polymorphic markers analyzed in the present study. No significant LD was apparent in any of the SPP1 polymorphic pairs analyzed.
Additional file 6. A Association of SPP1 rs2853744:G > T with different clinical characteristics of urolithiasis. B Association of SPP1 rs11730582:T > C with different clinical characteristics of urolithiasis. C Association of SPP1 rs11439060:delG>G with different clinical characteristics of urolithiasis.
Additional file 7. Meta-analysis of SPP1 rs11730582:T > C polymorphism with risk of urolithiasis. a) and b) Forest plots of urolithiasis association with rs11730582 polymorphism using dominant and recessive model, respectively. c) and d) Funnel plots of rs11730582 polymorphism assuming dominant and recessive inheritance, respectively, using fixed effect model.
Additional file 8. Meta-analysis of SPP1 rs11439060:delG>G polymorphism with susceptibility of urolithiasis. a) and b) Forest plots of urolithiasis association with rs11439060 polymorphism following dominant and recessive model, respectively. c) and d) Funnel plots of rs11439060 polymorphism using dominant and recessive inheritance, respectively, by random effect model.
Additional file 9. Raw dataset with details of genotypes and phenotypes of every study subject analyzed for SPP1 polymorphisms in this study.
Data Availability Statement
The raw sequence data used and/ analyzed in this study has been deposited in the NCBI GenBank database (https://www.ncbi.nlm.nih.gov/genbank/) and is publically available under the accession numbers MT450478 to MT450678. Sequencing data of all the cases and control samples was aligned and analyzed on the platform of SPP1 reference sequence gene using NCBI GenBank accession number NG_030362.1 available at https://www.ncbi.nlm.nih.gov/nuccore/353249890. The datasets supporting the findings reported in this article are included in this published article and its supplementary information files. Details of individual phenotype and genotypes for each of the SPP1 polymorphisms analyzed have been provided as additional file 9 in the supplementary data.