Abstract
PURPOSE
The evolution of precision oncology increasingly requires oncologists to incorporate genomic testing into practice. Yet, providers’ confidence with genomic testing is poorly documented. This article describes medical oncologists’ confidence with genomic testing and the association between genomic confidence and test use.
METHODS
We used data from the 2017 National Survey of Precision Medicine in Cancer Treatment to characterize oncologists’ confidence with genomic testing. Genomic confidence was examined separately by type of test user: next-generation sequencing (NGS) only, gene expression (GE) only, both NGS and GE, or nonuser. Predictors of genomic confidence were examined with multinomial logistic regression. The association between genomic confidence and test use was examined with multivariable linear regression.
RESULTS
More than 75% of genomic test users were either moderately or very confident about using results from multimarker tumor panel tests to guide patient care. Confidence with using multimarker tumor panel tests was highest among both NGS and GE test users, with 60.1% very confident in using test results, and lowest among NGS-only test users, with 38.2% very confident in using test results. Oncologists were most confident in using single-gene tests and least confident in using whole-genome or -exome sequencing to guide patient care. Genomic confidence was positively associated with self-reported test use. In adjusted models, training in genomics, larger patient volume, and treating patients with solid tumors predicted higher genomic confidence. Onsite pathology services and receipt of electronic medical record alerts for genomic testing predicted lower genomic confidence.
CONCLUSION
Oncologists’ confidence varies by testing platform, patient volume, genomic training, and practice infrastructure. Research is needed to identify modifiable factors that can be targeted to enhance provider confidence with genomic testing.
INTRODUCTION
The landscape of cancer care has changed dramatically over the past decade as therapies are increasingly personalized to the molecular characteristics of a patient’s tumor.1 Whereas genomic testing in oncology started with testing for single mutations in specific genes, advances in molecular profiling has spurred the development and now widespread use of multimarker tumor panels.2,3 These panels assess different types of genomic alterations commonly detected through next-generation sequencing (NGS) and gene expression (GE) profiles. Such testing can identify therapeutic targets that can be treated with particular anticancer agents and determine when individuals are at higher risk for recurrence and may need therapy.3,4 Although not standard practice as of yet, whole-genome or whole-exome sequencing is a newer technology that shows promise in informing patient care.4
CONTEXT
Key Objective
This article provides unique data about oncologists in the United States and their confidence in using results from commercially available multimarker tumor panel tests to guide decisions about patient treatment and management.
Knowledge Generated
Oncologists who used both next-generation sequencing (NGS) and gene expression tests reported the highest confidence in using multimarker tumor panel results to guide patient care; oncologists who only used NGS tests had the lowest confidence. Confidence was higher among oncologists who received training in genomics and varied by provider specialty, patient volume, and practice infrastructure. Oncologists with higher confidence were more likely to report using results from multimarker tumor panels to inform patient care.
Relevance
These findings underscore the importance of genomic training and suggest other provider- and practice-level factors that can be leveraged to build oncologists’ confidence in using genomic testing.
The rapid proliferation of genomic testing has outpaced the development of practice guidelines on their appropriate use.5,6 Multimarker tumor panel tests generate vast data about the biology of a patient’s tumor. In cases where the tumor can be successfully sequenced, an actionable target is identified for 39%-90% of patients, a wide range that varies by patient population and the size of the panel test.7-10 Although multimarker panel testing reports typically highlight potentially beneficial therapies and relevant clinical trials, individual providers must navigate the process of communicating results to their patients and selecting an appropriate targeted therapy. Furthermore, multimarker panel testing frequently identifies variants of unknown significance or germline variants, which can further complicate interpretation and care management.11-13
Very few studies have examined providers’ attitudes about genomic testing. There is some evidence that providers lack confidence in using new genomic tests, which could limit the extent to which new tests are appropriately applied in practice.5,6,14,15 For example, providers report less confidence in using multimarker tumor panels compared with single-gene tests, which are well established for certain patient populations.6 In addition, providers vary in their understanding of how to interpret and act upon results from whole-genome sequencing, which is not available outside of clinical trials.16 Given the potential of precision oncology, there is a need to better understand oncologists’ attitudes and experiences with genomic testing and to identify factors that foster genomic testing confidence.
This article builds upon previous studies by describing oncologists’ confidence with genomic testing and the association between confidence and genomic test use over the past 12 months. The analysis primarily focuses on oncologists’ confidence in using commercially available multimarker tumor panel tests. Oncologists’ confidence in using multimarker tumor panels was first examined in relation to their confidence in using single-gene somatic tests and whole-genome or -exome sequencing. Oncologists’ confidence was then further characterized by exploring the individual- and practice-level characteristics associated with confidence and assessing whether their confidence was associated with their use of multimarker tumor panels in practice.
METHODS
Data and Study Population
This study uses data from the National Survey of Precision Medicine in Cancer Treatment, a nationally representative study of how oncologists use genomic testing in practice. Eligible providers were identified from the American Medical Association Physician Masterfile and selected using multistage probability sampling. Eligible medical oncologists, hematologists, and hematologists/oncologists completed the 20-minute survey by mail. Each participant was sent up to three mailed surveys and could have received up to two e-mail reminders and a follow-up phone call. The survey was administered from February to May 2017, with completed surveys returned by 1,281 eligible individuals (cooperation rate, 38%). Additional information about the study design and methods have been published elsewhere.17 The survey is available upon request.
Measures
The National Survey of Precision Medicine in Cancer Treatment collected detailed data about oncologists’ confidence in using genomic testing; their genomic test use; and demographics, specialty, and practice characteristics. Genomic confidence was measured with a series of questions that captured oncologists’ level of confidence in using the results from genomic tests to guide treatment decisions. Confidence was assessed for genomic tests for individual genes or chromosomal alterations (referred to as single-gene tests), commercially available multimarker tumor panel tests, and whole-genome or -exome sequencing. Response options were very confident, moderately confident, a little confident, or not at all confident.
Type of genomic test user was determined on the basis of oncologists’ reported use of 18 specific multimarker tumor panels in the past 12 months. The multimarker tumor panel tests included commercially available GE tests (eg, Oncotype DX Breast; Genomic Health, Redwood City, CA), commercially available NGS tests (eg, FoundationOne; Foundation Medicine, Cambridge, MA), and noncommercial panels performed at an academic medical center. Oncologists who used either commercially available NGS panels or noncommercial panels performed at an academic medical center were considered NGS users. For this analysis, oncologists were categorized into 1 of 4 groups: those who only used NGS tests were defined as NGS-only users, those who only used GE tests were defined as GE-only users, those who used both NGS and GE tests were defined as NGS and GE users, and those who used neither NGS nor GE tests in the past 12 months were defined as genomic test nonusers.
Use of genomic test results was defined as the percentage of patients receiving multimarker panel tests for whom test results guided patient care decisions in the past 12 months. Oncologists were instructed to exclude Oncotype DX when answering this question because it is the most commonly used GE test for which there is clear guidance for interpreting and acting upon test results.18 Frequency of genomic test use was categorized into quartiles corresponding to < 5%, 5%-10%, 11%-49% and ≥ 50% of patients.
Provider characteristics were assessed with questions about provider specialty (treated solid tumors, hematologic malignancies, or both), years since medical school graduation (≤ 10, 11-20, 21-30, > 30), census region (Midwest, northeast, south, west), sex, and race (White, Asian, other). Training in genomics was also captured and defined as any formal training (eg, instruction during residency/fellowship, professional lectures or seminars, symposiums, conferences, continuing medical education) in the use of genomic testing.
Practice setting information was assessed with questions about the number of unique patients with cancer treated per month. Responses were categorized as < 49, 50-100, 100-199, ≥ 200. Respondents were also asked about the different settings where they saw patients for treatment and evaluation. Responses were categorized as academic medical center or medical school and nonacademic setting.
Infrastructure to support precision medicine was assessed with a series of questions about whether the respondent’s primary practice had the following genomic testing services: onsite pathology, internal policies or protocols for use of genomic and biomarker testing, an electronic medical record (EMR) that alerts when a genomic test is recommended, or a genomic/molecular tumor board. Response options were categorized as yes and no or do not know.
Analysis
Weighted percentages were calculated to describe the sample and provider confidence in using single-gene tests, multimarker tumor panel tests, and whole-genome or -exome sequencing. Confidence was analyzed separately by type of genomic test user and compared using χ2 tests of independence. Multinomial logistic regression was used to identify factors associated with confidence in using multimarker tumor panel test results to guide treatment decisions. Two sets of models were examined: one that adjusted for type of genomic test user and a second that simultaneously adjusted for type of genomic test user and the other physician and practice characteristics in the model. Results are presented as odds ratios and adjusted percentages, also referred to as predicted margins.19 Multivariable linear regression was then used to assess whether confidence was associated with the percentage of patients for whom multimarker panel test results guided patient care in the past 12 months. Analyses were limited to NGS users. Multivariable models adjusted for years since graduation, sex, race, census region, specialty, training in genomics, practice setting, patient volume, and infrastructure for precision medicine. Results are presented as adjusted percentages.19 All analyses were conducted using SUDAAN release 11.0 statistical software (RTI International, Research Triangle Park, NC) and weighted to account for the complex sampling approach and survey nonresponse.
RESULTS
Sample Characteristics
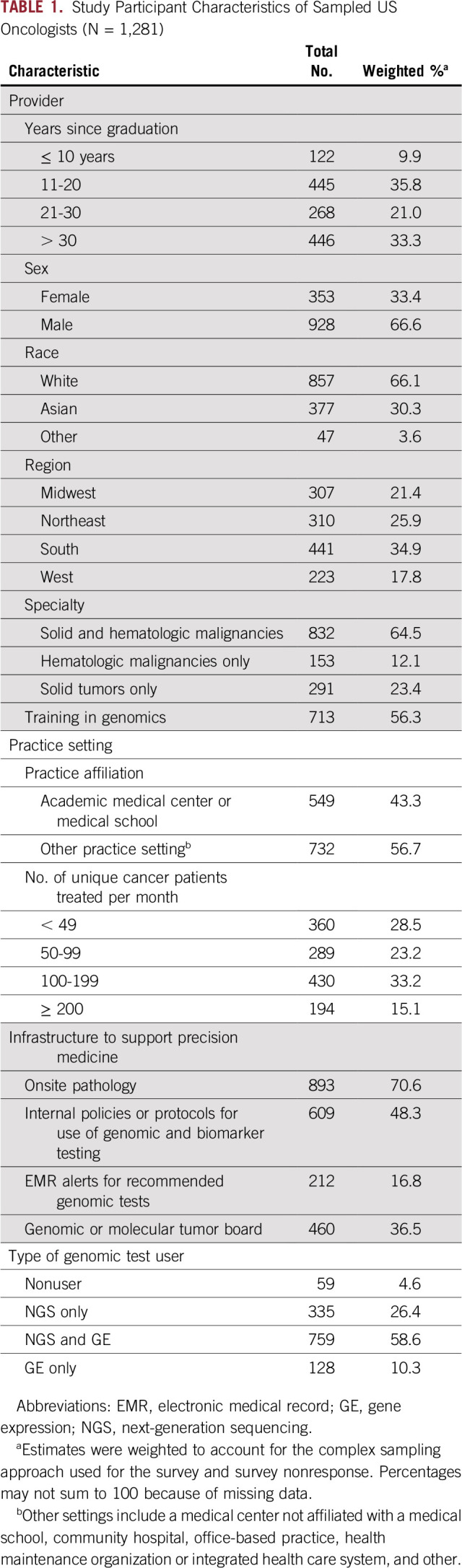
The sample comprised 1,281 oncologists (Table 1). Participants were predominately White (66.1%) and male (66.6%), and 45.7% graduated from medical school within the past 20 years. Most participants (56.7%) practiced outside an academic medical center or medical school, and approximately half (48.9%) treated ≥ 100 unique patients per month. Most treated a mix of solid tumors and hematologic malignancies (64.5%) and reported training in genomics (56.3%). Participants reported a range of institutional resources to support precision medicine, with onsite pathology and internal policies or protocols for use of genomic and biomarker testing reported most frequently. Genomic test use was reported by 95.4% of the sample, with 58.6% using both NGS and GE tests, 26.4% using only NGS tests, and 10.3% using only GE tests.
TABLE 1.
Study Participant Characteristics of Sampled US Oncologists (N = 1,281)
Confidence Using Genomic Tests in Practice
Confidence using multimarker tumor panel tests to guide decisions about patient treatment and management was highest among NGS and GE users, with 60.1% very confident and 35.6% moderately confident in using test results. Confidence was lowest among NGS-only users, with 38.2% very confident and 44.5% moderately confident in using test results. Even among oncologists who did not use multimarker tumor panel tests in the past 12 months, 27.1% were very confident and 37.8% were moderately confident in using test results (Fig 1B).
FIG 1.
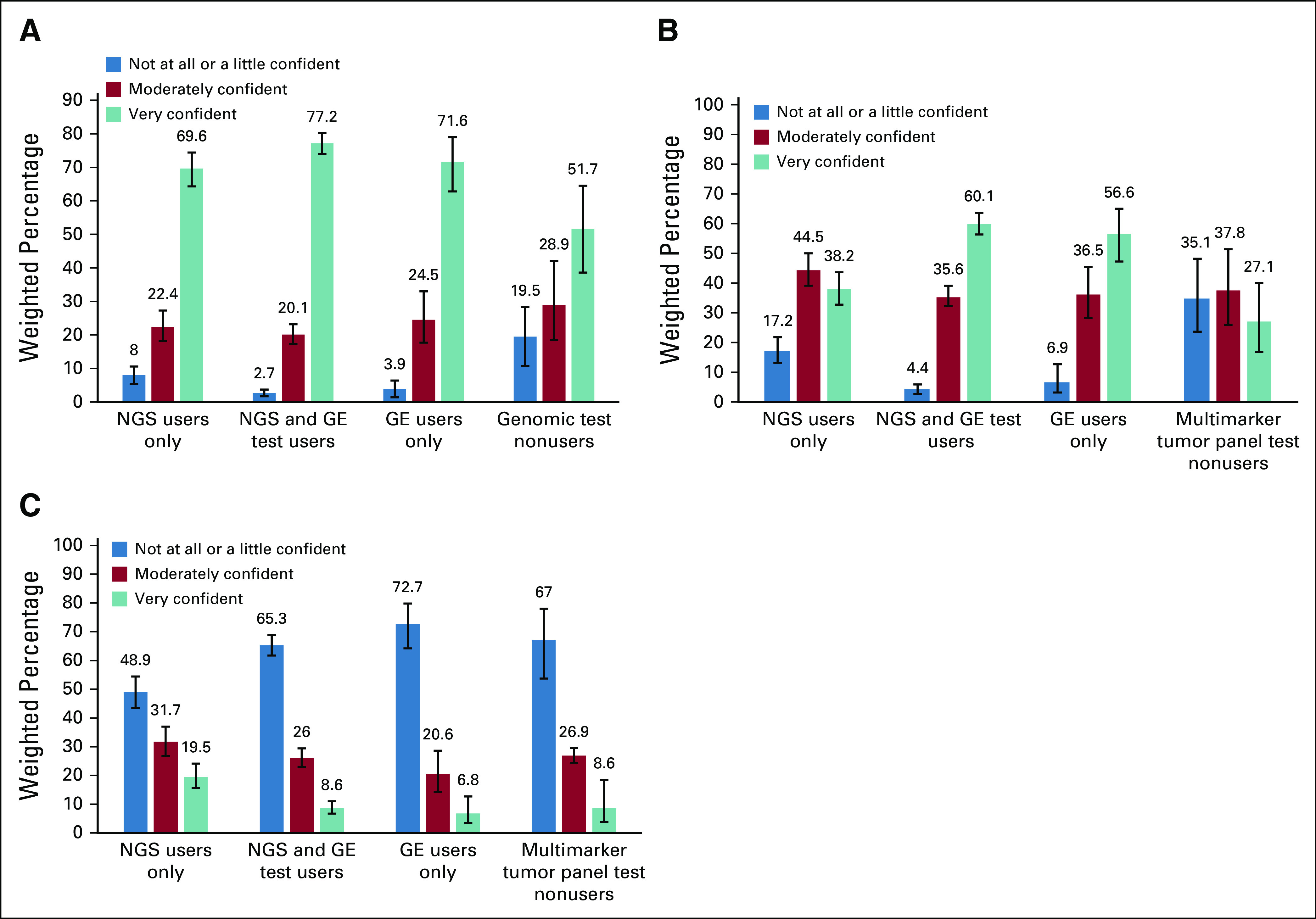
Oncologists’ confidence in using results from (A) single-gene tests, (B) commercially available multimarker somatic panels, and (C) whole-genome or -exome sequencing to guide decisions about patient treatment and management (N = 1,281). On the basis of χ2 tests for independence, overall differences in confidence among 4 types of genomic test users were statistically significant at P < .001 for single-gene tests, multimarker tumor panels, and whole-genome or -exome sequencing.
In contrast to multimarker tumor panel tests, oncologists were more confident in using somatic single-gene tests and less confident in using whole-genome or -exome sequencing (Figs 1A and 1C). For example, 77.2% of NGS and GE users, 71.6% of GE-only users, and 69.6% of NGS-only users were very confident in using results from single-gene somatic tests (Fig 1A). For whole-genome or -exome sequencing, only 19.5% of NGS users, 8.6% of NGS and GE users, and 6.9% of GE users were very confident using test results to guide patient care.
Physician- and System-Level Characteristics and Confidence Using Multimarker Panel Tests
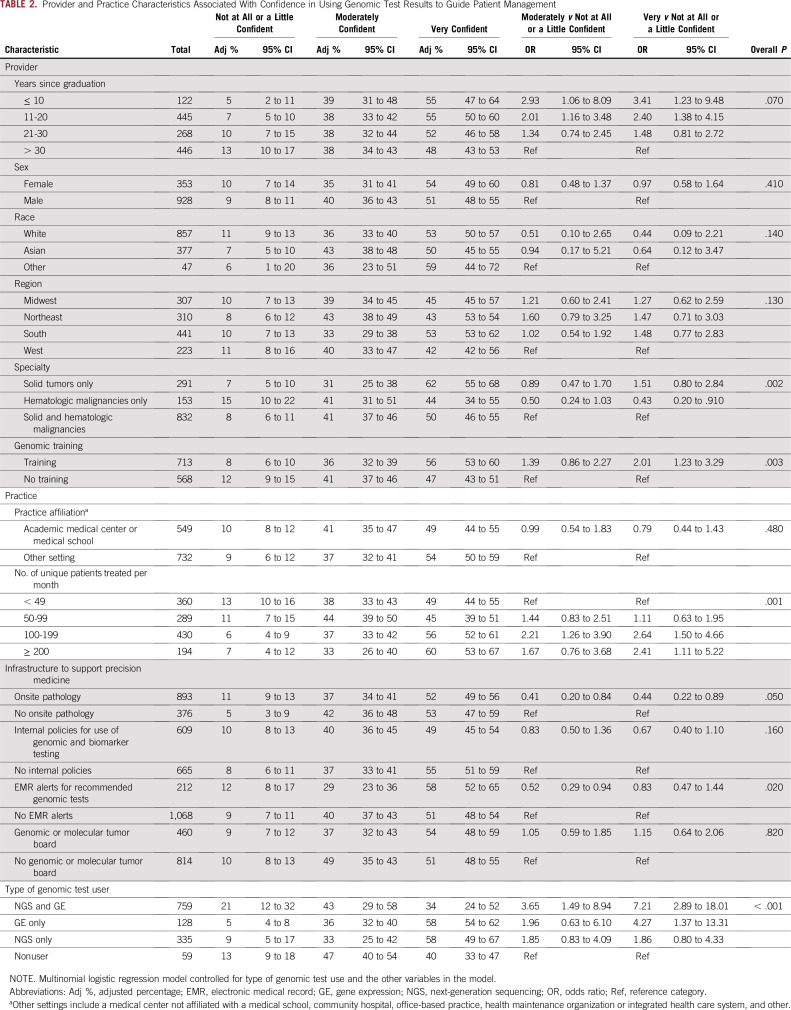
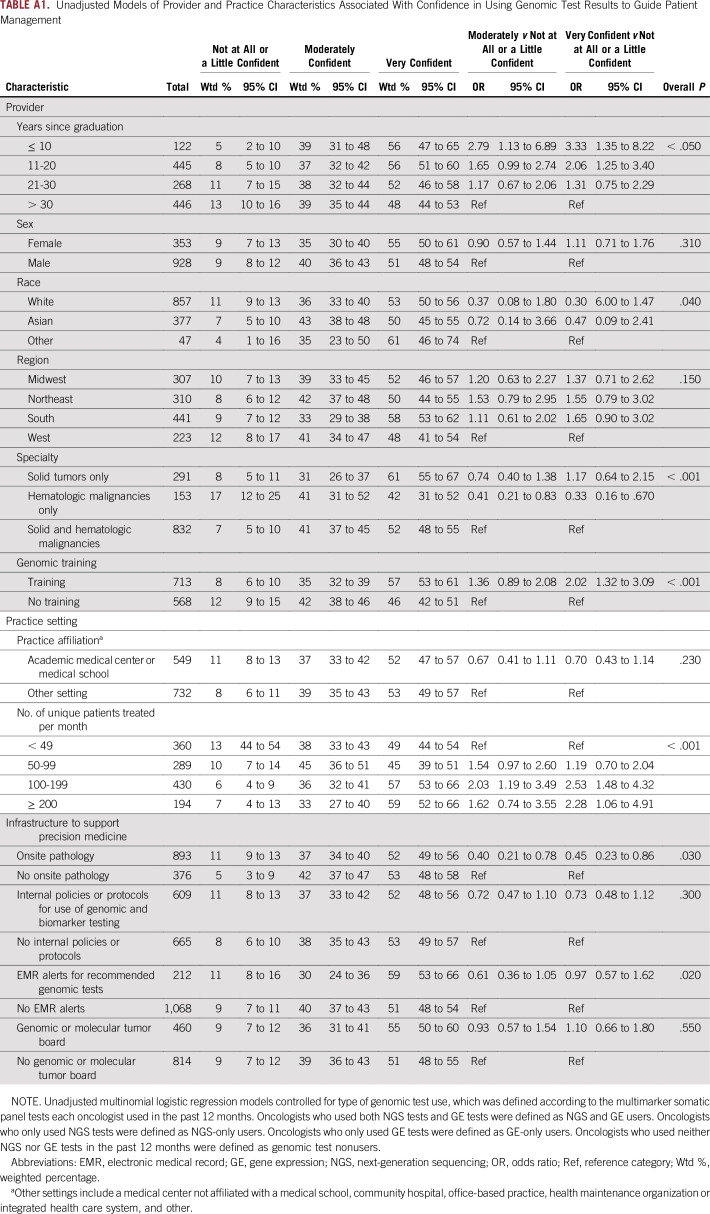
Physician- and system-level factors were associated with confidence using multimarker tumor panel test results to guide decisions about patient treatment and management. In the fully adjusted models (Table 2) and in models that only adjusted for the type of genomic test user (Appendix Table A1), oncologists with training in genomics had higher confidence than those who did not. Likewise, oncologists who treated ≥ 100 patients per month had higher confidence than those who treated < 49 patients per month. Compared with oncologists who treated both solid and hematologic malignancies, those who only treated solid tumors were more confident, whereas those who treated only hematologic malignancies were less confident in using results of multimarker panel tests to guide patient care. Oncologists who practice in institutions with onsite pathology or EMR alerts for recommended genomic tests had lower confidence than those who practice in institutions without onsite pathology or EMR alerts. Years since graduation were marginally associated with confidence such that more-recent graduates had higher confidence than oncologists who graduated > 20 years ago.
TABLE 2.
Provider and Practice Characteristics Associated With Confidence in Using Genomic Test Results to Guide Patient Management
Level of Genomic Confidence and Use of Multimarker Panel Test Results to Inform Patient Care Decisions
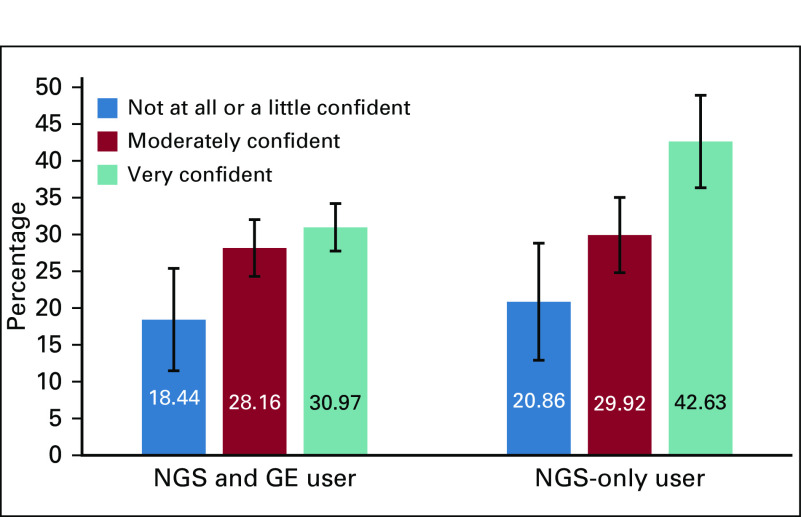
Compared with NGS and GE users who were not at all or a little confident, NGS and GE users who were moderately or very confident were more likely to use the results from multimarker panel tests to inform patient care (P < .01; Fig 2). For example, NGS and GE users who were very confident used multimarker panel test results to inform care for 30.97% of their patients (who had received testing) compared with 18.44% among NGS and GE users who were not at all or a little confident. Findings were similar for NGS-only users. Those who were very confident used multimarker panel test results to inform care for 42.36% of their patients (who had received testing) compared with 20.86% among NGS-only users who were not at all or a little confident (P < .01).
FIG 2.
Percentage of patients for whom multimarker tumor panel test results informed patient care decisions by level of genomic confidence. Presented are adjusted percentages from multivariable linear regression models that were adjusted for years since graduation, sex, race, census region, specialty, training in genomics, practice setting, and infrastructure for precision medicine. Pairwise comparisons with oncologists who were not at all or a little confident were statistically significant at P < .01.
DISCUSSION
The current study adds to the literature on provider attitudes about genomic testing. We found that oncologists’ confidence with genomic testing varied greatly by the types of tests they use in their practice. When asked about multimarker tumor panel tests, GE-only users and users of both NGS and GE tests reported the highest levels of confidence. Most GE panel tests are used to inform the treatment of patients with breast cancer, and there are clear guidelines for testing and the interpretation of test results.20,21 For users of other multimarker tumor panel tests, the application of test results to patient management is guideline endorsed for a limited set of clinical indications.
Oncologists’ confidence also varied by test type, which is consistent with previous research.6 Single-gene tests have been used for several decades as companion diagnostics to targeted therapies; thus, clinical appropriateness and utility are well prescribed.22 Even among genomic test nonusers, more than half reported being very confident in using single-gene tests to guide patient care. Using multimarker tumor panel tests to inform care is more complicated than using single-gene tests. At the time the survey was conducted, there were only consensus recommendations for multimarker tumor panel testing in treating non–small-cell lung cancer.23 Despite this, providers’ confidence using multimarker tumor panel tests was relatively high, with > 75% of genomic test users reporting that they were either moderately or very confident in using results to guide patient care. Providers who reported higher levels of confidence also tended to use test results more frequently to inform patient care compared with providers who reported lower levels of confidence. These findings are consistent with other studies of provider confidence.15,24-26
Multimarker tumor panel tests are increasingly being used in practice,17 yet they are still at an early phase of implementation. Thus, we conducted additional analyses to understand provider and practice characteristics associated with confidence using multimarker panel tests that could inform provider-level interventions. We found that confidence was relatively low among providers who only treated hematologic malignancies. Historically, providers who treat patients with a hematologic malignancy have relied on different technologies (eg, flow cytometry for immunophenotyping) or single-gene somatic tests to identify relevant targeted therapies.27 In contrast, use of multimarker tumor panel tests has been more common in the treatment of solid tumors.23
The importance of experience was also highlighted in our findings that providers who treat > 100 patients per month had higher confidence than providers who treat < 50. Providers in high-volume clinics have more opportunity to encounter patients for whom multimarker tumor panel tests are warranted. Indeed, patient volume has been associated with better treatment outcomes in other settings.28 Providers in lower-volume clinics may benefit from additional resources, including clinical decision support, to strengthen evidence-based use of genomic testing in treatment planning.29
Our findings underscore the importance of genomic training to increase provider confidence in using multimarker panel tests to guide patient care. Training was defined broadly and included any instruction during residency or fellowship, professional lectures, conferences, or continuing medical education in the use of genomic testing. Additional research should identify the specific competencies needed by different types of oncologists and be tailored to support providers who demonstrate relatively low confidence in using genomic testing in practice. Several categories of training could be pursued, including revision of the curricula of medical and nursing schools to provide training in genomics; advanced training of physicians and other practitioners in genomics; and continuing medical education to help practicing clinicians to stay up to date on advances in the field.30,31 Strategies such as case-based learning, clinical decision-support tools, administrative support for prior authorization, and clinical trial matching may be helpful in strengthening provider confidence around genomic testing at the point of care and supporting successful implementation of precision medicine programs.29
Genomic testing services available in an oncologist’s primary practice were either not associated with provider confidence or, paradoxically, associated with lower confidence. Internal policies for use of genomic and biomarker testing and availability of a genomic or molecular tumor board were not associated with provider confidence. Resources such as onsite pathology and EMR alerts for recommended genomic or biomarker tests were associated with lower confidence. However, the pattern of confidence between oncologists in settings with and without EMR alerts was less consistent than between oncologists with and without onsite pathology. Oncologists who reported that their practice had onsite pathology spent a lower percentage of time in delivering patient care (data not shown), which could have influenced their confidence in using genomic test results to guide treatment decisions. Oncologists in settings with onsite pathology could also have been more likely to have a research focus outside of genomics, although that is not something we can explore with our data.
In addition, it is possible that other features of the practice setting or availability of appropriate training were more important drivers of provider confidence. More research is needed to identify practice- and system-level factors that can be applied to build provider confidence in using genomic testing. This work should address both the knowledge and the skills to use genomic testing appropriately and to act on test results as well as system-level resources to support precision oncology.
Our study results should be considered in light of a few limitations. First, providers were asked generally about their confidence in using genomic test results to guide patient care; we do not know how confidence with a particular testing approach may vary on the basis of specific patient subgroups or clinical scenarios. To address this limitation, we stratified our sample by the type of test user to better understand the variability in provider confidence. Second, the survey cooperation rate was low. However, participants are representative of practicing oncologists in the United States in terms of age, sex, and geographic location, and all analyses were weighted to adjust estimates for survey nonresponse. That said, 43% of our sample had an academic affiliation, which may be higher than the general population of practicing oncologists. Third, our data on test use were self-reported and limited to the estimated percentage of patients for whom testing informed care. We do not know exactly how test results were used and whether care was guideline concordant and clinically appropriate. Finally, it is possible that genomic confidence differs between oncologists who responded to the survey and those who did not. Thus, our results may over- or underestimate genomic confidence levels among oncologists in the United States.
In conclusion, the use of multimarker tumor panel testing has increased dramatically over the past decade.2 Since the National Survey of Precision Medicine in Cancer Treatment was conducted, the US Food and Drug Administration (FDA) approved the FoundationOne CDx test, and the Centers for Medicare & Medicaid Services issued a national coverage determination for NGS testing with the FoundationOne genomic profiling assay.32 In addition, the FDA approved Oncomine Dx Target Test (Thermo Fisher Scientific, Waltham, MA), an NGS-based companion diagnostic to identify lung cancer mutations.33 Relatedly, there has been a move to develop tissue-agnostic therapies, which will further drive the use of NGS testing.34,35 Additional evidence is needed to better understand the impact of NGS on outcomes, including disease response and cost.36 Continued research is also needed to determine how best to support oncologists and other cancer care providers, as well as patients and families, in fully harnessing these advances and addressing the challenges associated with the rapid evolution of tools for precision oncology to optimize patient care.3,37
Appendix
TABLE A1.
Unadjusted Models of Provider and Practice Characteristics Associated With Confidence in Using Genomic Test Results to Guide Patient Management
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Andrew N. Freedman
Administrative support: Andrew N. Freedman
Provision of study material or patients: Andrew N. Freedman
Collection and assembly of data: Janet S. de Moor, Carrie N. Klabunde, Andrew N. Freedman
Data analysis and interpretation: Janet S. de Moor, Stacy W. Gray, Sandra A. Mitchell, Carrie N. Klabunde, Andrew N. Freedman
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Janet S. de Moor
Employment: Biogen (I)
Travel, Accommodations, Expenses: Biogen (I)
Stacy W. Gray
Stock and Other Ownership Interests: Magenta Therapeutics (I)
Consulting or Advisory Role: Grail Industries, Magenta Therapeutics (I)
Expert Testimony: Riley and Associates
No other potential conflicts of interest were reported.
REFERENCES
- 1.Kalia M. Biomarkers for personalized oncology: Recent advances and future challenges. Metabolism. 2015;64:S16–S21. doi: 10.1016/j.metabol.2014.10.027. [DOI] [PubMed] [Google Scholar]
- 2.Morganti S, Tarantino P, Ferraro E, et al. Complexity of genome sequencing and reporting: Next generation sequencing (NGS) technologies and implementation of precision medicine in real life. Crit Rev Oncol Hematol. 2019;133:171–182. doi: 10.1016/j.critrevonc.2018.11.008. [DOI] [PubMed] [Google Scholar]
- 3.El-Deiry WS, Goldberg RM, Lenz HJ, et al. The current state of molecular testing in the treatment of patients with solid tumors, 2019. CA Cancer J Clin. 2019;69:305–343. doi: 10.3322/caac.21560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakagawa H, Wardell CP, Furuta M, et al. Cancer whole-genome sequencing: Present and future. Oncogene. 2015;34:5943–5950. doi: 10.1038/onc.2015.90. [DOI] [PubMed] [Google Scholar]
- 5.Miller FA, Krueger P, Christensen RJ, et al. Postal survey of physicians and laboratories: Practices and perceptions of molecular oncology testing. BMC Health Serv Res. 2009;9:131. doi: 10.1186/1472-6963-9-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blazer KR, Nehoray B, Solomon I, et al. Next-generation testing for cancer risk: Perceptions, experiences, and needs among early adopters in community healthcare settings. Genet Test Mol Biomarkers. 2015;19:657–665. doi: 10.1089/gtmb.2015.0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tan O, Shrestha R, Cunich M, et al. Application of next-generation sequencing to improve cancer management: A review of the clinical effectiveness and cost-effectiveness. Clin Genet. 2018;93:533–544. doi: 10.1111/cge.13199. [DOI] [PubMed] [Google Scholar]
- 8.Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol. 2015;33:2753–2762. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson DB, Dahlman KH, Knol J, et al. Enabling a genetically informed approach to cancer medicine: A retrospective evaluation of the impact of comprehensive tumor profiling using a targeted next-generation sequencing panel. Oncologist. 2014;19:616–622. doi: 10.1634/theoncologist.2014-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwaederle M, Daniels GA, Piccioni DE, et al. On the road to precision cancer medicine: Analysis of genomic biomarker actionability in 439 patients. Mol Cancer Ther. 2015;14:1488–1494. doi: 10.1158/1535-7163.MCT-14-1061. [DOI] [PubMed] [Google Scholar]
- 11.Hoffman-Andrews L. The known unknown: The challenges of genetic variants of uncertain significance in clinical practice. J Law Biosci. 2018;4:648–657. doi: 10.1093/jlb/lsx038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ngeow J, Eng C. Precision medicine in heritable cancer: When somatic tumour testing and germline mutations meet. NPJ Genom Med. 2016;1:15006. doi: 10.1038/npjgenmed.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Statz CM, Patterson SE, Mockus SM. Barriers preventing the adoption of comprehensive cancer genomic profiling in the clinic. Expert Rev Mol Diagn. 2017;17:549–555. doi: 10.1080/14737159.2017.1319280. [DOI] [PubMed] [Google Scholar]
- 15.Gray SW, Hicks-Courant K, Cronin A, et al. Physicians’ attitudes about multiplex tumor genomic testing. J Clin Oncol. 2014;32:1317–1323. doi: 10.1200/JCO.2013.52.4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weipert CM, Ryan KA, Everett JN, et al. Physician experiences and understanding of genomic sequencing in oncology. J Genet Couns. 2018;27:187–196. doi: 10.1007/s10897-017-0134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Freedman AN, Klabunde CN, Wiant K, et al: Use of next-generation sequencing tests to guide cancer treatment: Results from a nationally representative survey of oncologists in the United States. JCO Precis Oncol 10.1200/PO.18.00169. [DOI] [PMC free article] [PubMed]
- 18.McVeigh TP, Kerin MJ. Clinical use of the Oncotype DX genomic test to guide treatment decisions for patients with invasive breast cancer. Breast Cancer (Dove Med Press) 2017;9:393–400. doi: 10.2147/BCTT.S109847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graubard BI, Korn EL. Predictive margins with survey data. Biometrics. 1999;55:652–659. doi: 10.1111/j.0006-341x.1999.00652.x. [DOI] [PubMed] [Google Scholar]
- 20.Duffy MJ, Harbeck N, Nap M, et al. Clinical use of biomarkers in breast cancer: Updated guidelines from the European Group on Tumor Markers (EGTM) Eur J Cancer. 2017;75:284–298. doi: 10.1016/j.ejca.2017.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Nicolini A, Ferrari P, Duffy MJ. Prognostic and predictive biomarkers in breast cancer: Past, present and future. Semin Cancer Biol. 2018;52:56–73. doi: 10.1016/j.semcancer.2017.08.010. [DOI] [PubMed] [Google Scholar]
- 22.Agarwal A, Ressler D, Snyder G. The current and future state of companion diagnostics. Pharm Genomics Pers Med. 2015;8:99–110. doi: 10.2147/PGPM.S49493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ettinger DS, Wood DE, Akerley W, et al. NCCN guidelines insights: Non-small cell lung cancer, version 4.2016. J Natl Compr Canc Netw. 2016;14:255–264. doi: 10.6004/jnccn.2016.0031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. West HJ, Miller G: Genomic Testing and Precision Medicine in Cancer Care, 2017. https://www.medscape.com/slideshow/genomics-and-oncology-report-6008655.
- 25.Godin G, Bélanger-Gravel A, Eccles M, et al. Healthcare professionals’ intentions and behaviours: A systematic review of studies based on social cognitive theories. Implement Sci. 2008;3:36. doi: 10.1186/1748-5908-3-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson LM, Valdez JM, Quinn EA, et al. Integrating next-generation sequencing into pediatric oncology practice: An assessment of physician confidence and understanding of clinical genomics. Cancer. 2017;123:2352–2359. doi: 10.1002/cncr.30581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hochhaus A, Saussele S, Rosti G, et al. Chronic myeloid leukaemia: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2018;29:iv261. doi: 10.1093/annonc/mdy159. [DOI] [PubMed] [Google Scholar]
- 28.Amato L, Fusco D, Acampora A, et al. Volume and health outcomes: Evidence from systematic reviews and from evaluation of Italian hospital data. Epidemiol Prev. 2017;41:1–128. doi: 10.19191/EP17.5-6S2.P001.100. [DOI] [PubMed] [Google Scholar]
- 29.Levit LA, Kim ES, McAneny BL, et al. Implementing precision medicine in community-based oncology programs: Three models. J Oncol Pract. 2019;15:325–329. doi: 10.1200/JOP.18.00661. [DOI] [PubMed] [Google Scholar]
- 30.McGrath S, Ghersi D. Building towards precision medicine: Empowering medical professionals for the next revolution. BMC Med Genomics. 2016;9:23. doi: 10.1186/s12920-016-0183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ha VTD, Frizzo-Barker J, Chow-White P. Adopting clinical genomics: A systematic review of genomic literacy among physicians in cancer care. BMC Med Genomics. 2018;11:18. doi: 10.1186/s12920-018-0337-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Allegretti M, Fabi A, Buglioni S, et al. Tearing down the walls: FDA approves next generation sequencing (NGS) assays for actionable cancer genomic aberrations. J Exp Clin Cancer Res. 2018;37:47. doi: 10.1186/s13046-018-0702-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dy GK, Nesline MK, Papanicolau-Sengos A, et al. Treatment recommendations to cancer patients in the context of FDA guidance for next generation sequencing. BMC Med Inform Decis Mak. 2019;19:14. doi: 10.1186/s12911-019-0743-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garber K. In a major shift, cancer drugs go ‘tissue-agnostic’. Science. 2017;356:1111–1112. doi: 10.1126/science.356.6343.1111. [DOI] [PubMed] [Google Scholar]
- 35.Lemery S, Keegan P, Pazdur R. First FDA Approval agnostic of cancer site - When a biomarker defines the indication. N Engl J Med. 2017;377:1409–1412. doi: 10.1056/NEJMp1709968. [DOI] [PubMed] [Google Scholar]
- 36. Pennell NA, Mutebi A, Zhou Z-Y, et al: Economic impact of next-generation sequencing versus single-gene testing to detect genomic alterations in metastatic non–small-cell lung cancer using a decision analytic model. JCO Precis Oncol 10.1200/PO.18.00356. [DOI] [PubMed]
- 37.Bradbury AR. Implementation of precision cancer medicine: Progress and the path to realizing the promise of tumor sequencing. J Oncol Pract. 2019;15:297–299. doi: 10.1200/JOP.19.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]