Abstract
PURPOSE
The development and use of predictive biomarkers to guide treatment decisions are paramount not only for improving survival in patients with metastatic colorectal cancer (mCRC), but also for sparing them from unnecessary toxicity and reducing the economic burden of expensive treatments. We conducted a systematic review of published studies and evaluated the predictive biomarker landscape in the mCRC setting from a molecular and clinical viewpoint.
METHODS
Studies analyzing predictive biomarkers for approved therapies in patients with mCRC were identified systematically using electronic databases. Preclinical studies and those providing no relevant information were excluded.
RESULTS
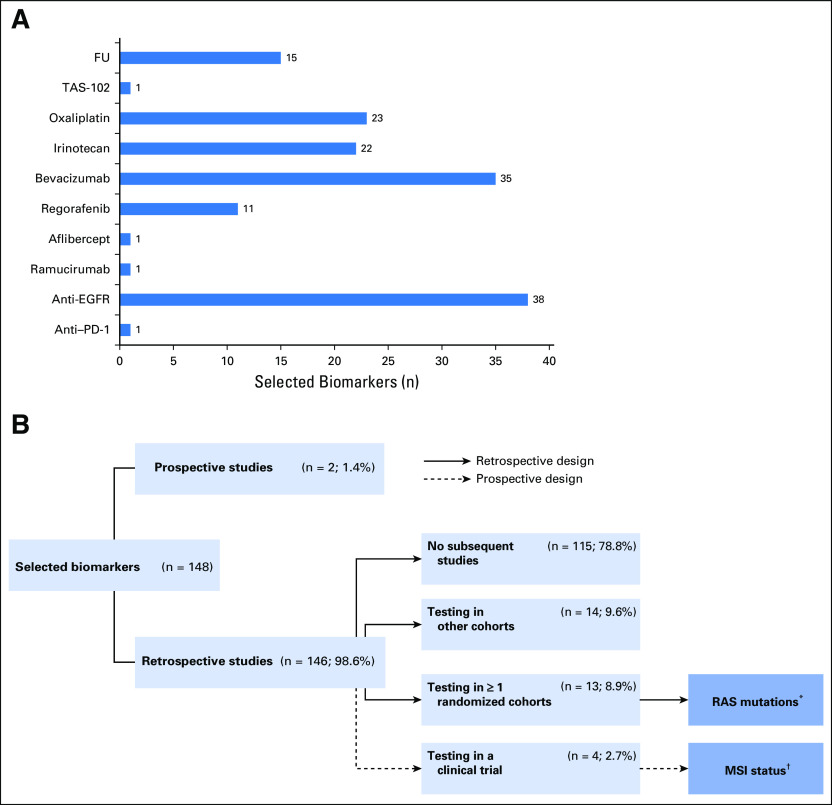
A total of 173 studies comprising 148 biomarkers were selected for final analysis. Of all the biomarkers analyzed, 1.4% (two of 148) were explored in a prospective manner, whereas 98.6% (146 of 148) were evaluated in retrospective studies. Of the latter group, 78.8% (115 of 146) were not tested in subsequent phases, 9.6% (14 of 146) were tested in other retrospective cohorts, 8.9% (13 of 146) were retrospectively tested in at least one or more randomized cohorts, and only 2.7% (four of 146) were prospectively tested in a clinical trial. Finally, only 1.4% (two of 148) were validated sufficiently and are recognized as biomarkers for guiding treatment decision making in patients with mCRC. These markers were RAS mutational status for anti-EGFR antibodies and microsatellite instability status for anti–programmed cell death-1 drugs.
CONCLUSION
Despite notable efforts to identify predictive biomarkers for various therapies used in the mCRC setting, because of a lack of data beyond retrospective studies and successful biomarker-driven approaches, only two molecular biomarkers have thus far found their translation into the clinic, highlighting the imperative need for implementing novel strategies and additional research in this clinically important field.
INTRODUCTION
Colorectal cancer (CRC) remains the third leading cause of cancer-related deaths in the western world. Despite ongoing efforts aimed at increased population screening and improved early detection strategies, approximately 20% of patients still present with metastatic disease at diagnosis, and approximately 35% of those who undergo curative surgeries for a localized disease relpase.1 During the past three decades, the median overall survival (OS) of patients with metastatic CRC (mCRC) has gradually increased because of the implementation of combined chemotherapy regimens as well as targeted molecular therapies against EGFR and angiogenic factors.2 Since the identification of RAS mutations as a negative predictive marker, anti-EGFR therapy has had the greatest impact on the management of patients with mCRC; nonetheless, the response rates of these treatments remain only approximately 40% to 60%. In addition, the recognition of immunotherapy in the treatment landscape for patients with microsatellite instability-high (MSI-H) or DNA deficient mismatch-repair (dMMR) mCRC has been encouraging for this subset of patients. In this review focused on mCRC, we systematically summarize the most relevant milestones achieved in the field of predictive biomarkers for various treatments and discuss methodologic aspects, current trends, and future directions in this exciting area.
METHODS
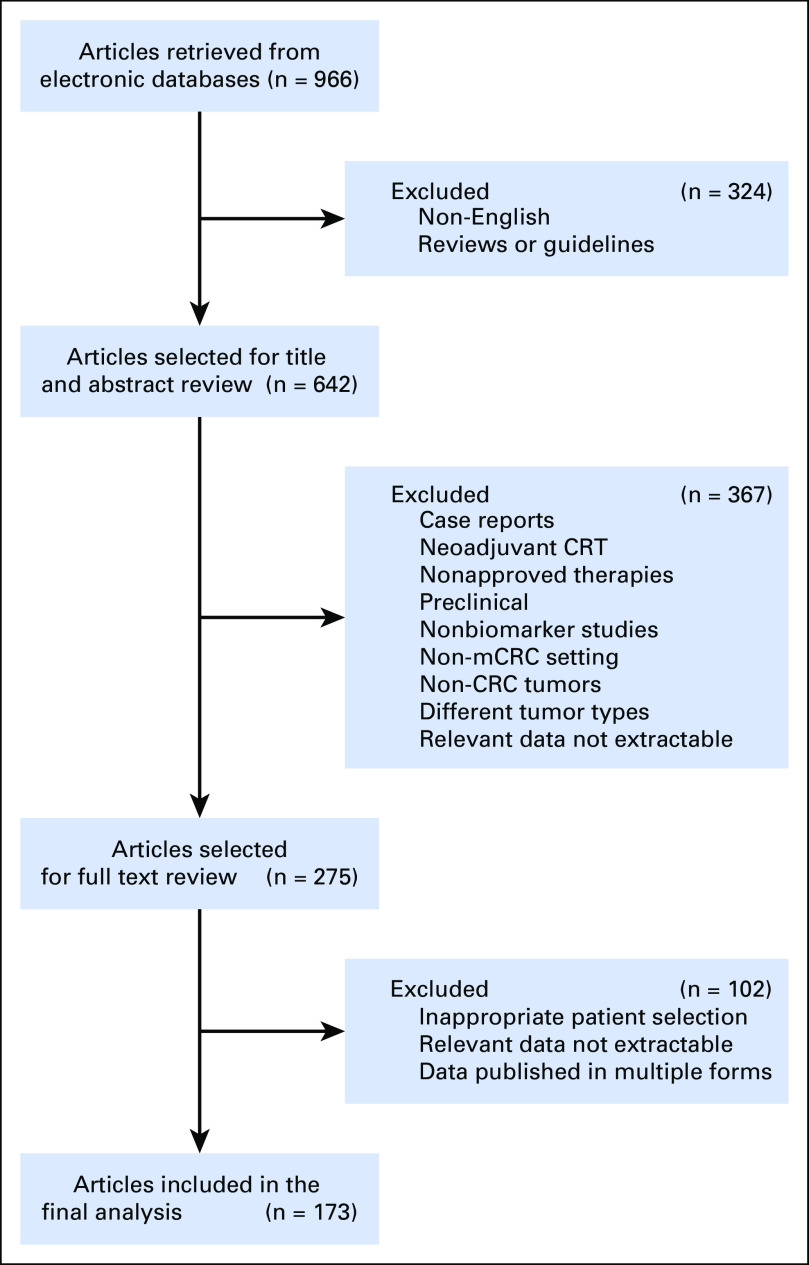
A systematic literature search was conducted using PubMed, EMBASE, and Web of Science up to January 2018. The query was developed and executed in PubMed (Appendix Fig A1) and was subsequently translated to other databases. For all selected articles, titles and abstracts were examined to exclude review articles and studies lacking evidence-based data. All remaining articles were screened carefully, and the bibliographies from these publications were also screened for other relevant studies. Duplicate studies or articles that did not meet these criteria on full review were subsequently excluded. The most pertinent articles were selected for inclusion in this review (Fig 1). The results were reviewed by a multidisciplinary team of medical oncologists and translational research scientists with longstanding expertise in this biomarker field. Critical issues regarding the biomarker study design were identified (Data Supplement), and key findings from the selected studies are summarized succinctly in this systematic review on this important clinical topic.
FIG 1.
Flow diagram of the study selection process. CRC, colorectal cancer; mCRC, metastatic colorectal cancer; Neoadjuvant-CRT, neoadjuvant chemoradiotherapy.
RESULTS
A total of 173 studies comprising 148 biomarkers, individually or as a panel, were selected for this review article. The detailed strategy for study and biomarker selection is illustrated in Fig 2.
FIG 2.
Selected biomarkers for the final analysis. (A) Distribution of biomarkers by individual treatment group. (B) Distribution of biomarkers by study design. Anti–PD-1, anti–programmed cell death-1 drugs; FU, fluoropyrimidine-based chemotherapy; Irinotecan, irinotecan-based chemotherapy; MSI, microsatellite instability; Oxaliplatin, oxaliplatin-based chemotherapy; TAS-102, trifluridine/tipiracil. (*) Predictive biomarker for anti-EGFR drugs (cetuximab/panitumumab); (†) predictive biomarker for anti–PD-1 drugs (pembrolizumab and nivolumab alone or in combination with ipilimumab).
Conventional Chemotherapy and Trifluridine/Tipiracil
The backbone of treatment in patients with mCRC has historically been chemotherapy, and several chemotherapeutic agents are now approved in this setting: fluoropyrimidines (fluorouracil [FU] and capecitabine), oxaliplatin, irinotecan, and since 2015, trifluridine/tipiracil (TAS-102).
Fluoropyrimidine-based chemotherapy and TAS-102.
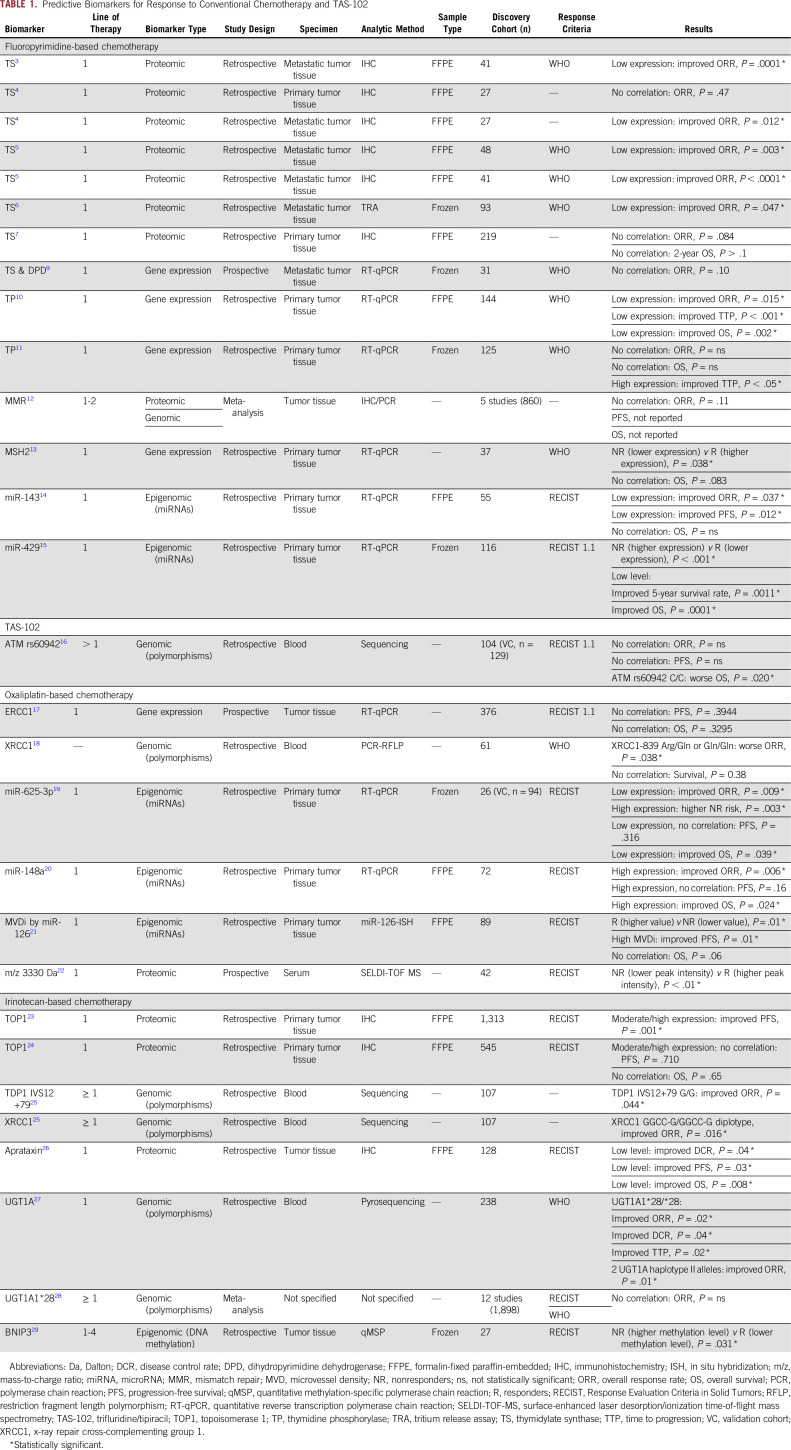
Over the years, several studies have investigated the predictive role of fluoropyrimidine metabolic pathway enzymes in response to FU-based therapies. Studies of the role of thymidylate synthase (TS) in fluoropyrimidine-based therapy (primarily FU plus leucovorin) in various retrospective and prospective studies have yielded discordant results. In this regard, multiple studies have shown that low levels of TS expression in metastatic tumor tissues generally correlate with higher overall response rate (ORR).3-6 Surprisingly, such a correlation was not evident when TS levels were measured in primary tumor tissues.4,7 Similarly, low levels of TS and dihydropyrimidine dehydrogenase in metastatic tumor tissues were associated with a favorable response to FU in patients with mCRC8; however, a subsequent report in 2006 did not validate these findings.9 Likewise, the role of thymidine phosphorylase as a predictive biomarker was also investigated, but the results remain inconclusive.10,11
In 2009, a meta-analysis of five studies examining a total of 861 patients with mCRC concluded that compared with microsatellite-stable patients, MSI-H patients did not achieve a statistically significant better response rate to FU-based chemotherapy.12 Similarly, while investigating the relationship between MSH2 gene expression and capecitabine efficacy in patients with mCRC, Jensen et al13 observed that a higher MSH2 expression was associated with a better response. In an attempt to identify noncoding RNA-based predictive biomarkers, a low expression of miR-143 was shown to be associated with improved ORR and progression-free survival (PFS) in patients treated with capecitabine.14 Likewise, a low expression of miR-429 correlated with improved 5-year disease-free survival and OS in patients with mCRC treated with FU-based chemotherapy.15 Other predictive biomarkers for fluoropyrimidine-based chemotherapy are summarized in Table 1 and in the Data Supplement.
TABLE 1.
Predictive Biomarkers for Response to Conventional Chemotherapy and TAS-102
On the basis of the results of the phase III RECOURSE (Study of TAS-102 in Patients With Metastatic Colorectal Cancer Refractory to Standard Chemotherapies) trial, the US Food and Drug Administration approved TAS-102 for patients with mCRC. In 2015, Hamauchi et al30 found that patients who develop grade 3 or 4 neutropenia during the first cycle of TAS-102 treatment had a smaller risk of disease progression. More recently, another study reported improved OS rates in patients harboring any G allele at the ATM rs609429 locus, when compared with those with a C/C variant.16
Oxaliplatin-based chemotherapy.
In examining the mechanism of action of oxaliplatin, several key genes involved in the nucleotide excision repair pathway have been explored as potential predictive biomarkers. The most notable attempts in this regard have been for the ERCC1 gene, and as reported from the MAVERICC (Marker Evaluation for Avastin Research in CRC) trial, intratumoral ERCC1 gene expression failed to predict response to oxaliplatin treatment.17 Another gene evaluated in this setting was the x-ray repair cross-complementing group 1 (XRCC1) gene, a base excision repair modulator, wherein a polymorphism in this gene (XRCC1-839 Arg/Gln or Gln/Gln) correlated with worse ORR to FU/oxaliplatin.18
Interestingly, several microRNAs (miRNAs) have been explored for their predictive response potential to FU, leucovorin, and oxaliplatin (FOLFOX)– or capecitabine plus oxaliplatin (CAPEOX)–based regimens. In this treatment setting, high miR-625-3p and low miR-148a expression were associated with poor response,19,20 whereas high miR-126 microvessel density was associated with improved PFS.21
In addition to studies of messenger RNA- and miRNA-based markers, two independent studies reported that evaluation of serum protein expression patterns is implicated in predicting response to CAPEOX and FOLFOX, respectively.22,31 Other potential predictive biomarkers for oxaliplatin-based chemotherapy are summarized in Table 1 and in the Data Supplement.
Irinotecan-based chemotherapy.
In terms of predictive biomarkers for irinotecan treatment, the most notable marker studied in this setting is topoisomerase 1 (TOP1). The first large study in which TOP1 predictive power was evaluated used samples from the FOCUS (Fluorouracil, Oxaliplatin, CPT-11: Use and Sequencing) trial and reported that a moderate or high expression of TOP1 was associated with a significant gain in survival after irinotecan-based therapy.23 Unfortunately, these findings were not confirmed subsequently by analyzing samples from 545 patients involved in the CAIRO (Capecitabine, Irinotecan, Oxaliplatin) study, despite similar treatment regimens and analytic approaches.24
While studying the role of genetic polymorphisms within the TDP1 and XRCC1 genes in response to irinotecan-based regimens, a positive correlation with improved ORR was observed in patients with the TDP1 IVS12+79G>T and XRCC1 GGCC-G/GGCC-G genotypes.25 Aprataxin, a protein member of the histidine triad superfamily, has also shown a potential ability to discriminate responders from nonresponders. In a retrospective cohort of 128 patients treated with irinotecan-based chemotherapy, a low expression of aprataxin correlated with improved disease control rate, PFS, and OS.26 Although studies of UGT1A gene polymorphisms and their predictive value for response to irinotecan treatment have yielded conflicting results,27,28 higher methylation levels of the BNIP3 gene in patients treated with irinotecan plus S1 correlated with lower response rates.29 Additional predictive biomarkers for irinotecan-based chemotherapy are summarized in Table 1 and in the Data Supplement.
Antiangiogenic Drugs
Bevacizumab.
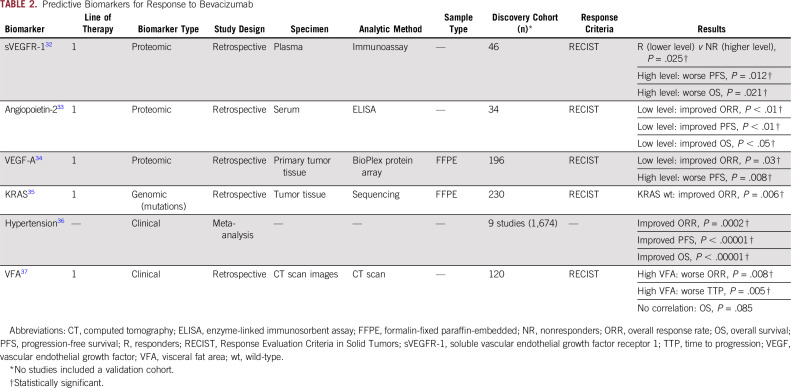
Since the introduction of bevacizumab for the management of patients with mCRC, substantial efforts have been made to discover predictive biomarkers for this antiangiogenic drug. Using a proteomic approach in 2010, Aoyagi et al32 reported that lower levels of plasma soluble vascular endothelial growth factor (VEGF) receptor 1 were associated with improved disease control in a subset of 46 patients treated with modified FU, leucovorin, and oxaliplatin (mFOLFOX6) plus bevacizumab. In the same year, another study identified a significant correlation between low angiopoetin-2 serum levels and better survival outcomes in a cohort of patients with mCRC treated with bevacizumab-based therapy.33
Conversely, the correlation between VEGF-A levels and the clinical benefit of bevacizumab-based chemotherapy is still under evaluation. In 2014, a study by Bruhn et al34 reported higher ORR and improved PFS in patients with a low expression of VEGF-A in primary tumor specimens. Perhaps in the near future, MAVERICC trial results will offer more insights into the usefulness of plasma VEGF-A levels in this setting.17 With regard to the predictive role of RAS, in a subset of 230 patients with mCRC treated in a phase III randomized clinical trial with either irinotecan, FU, and leucovorin (IFL) plus placebo, or IFL plus bevacizumab, only the wild-type KRAS subset of patients obtained a significantly higher response rate in the bevacizumab arm.35
In addition to the molecular markers listed previously, a few clinical factors have been studied in relation to bevacizumab response. Perhaps the best documented feature in this context is the association between bevacizumab-induced hypertension and a better outcome in terms of response rate, PFS, and OS.36 On the contrary, another clinical factor such as a high visceral fat area has been associated with poor response rate, time to progression, and OS.37 Other potential predictive biomarkers for bevacizumab-based therapy are summarized in Table 2 and in the Data Supplement.
TABLE 2.
Predictive Biomarkers for Response to Bevacizumab
Regorafenib.
Two retrospective studies based on the data from the CORRECT (Patients With Metastatic Colorectal Cancer Treated With Regorafenib or Placebo After Failure of Standard Therapy) and CONSIGNA (Regorafenib in Subjects With Metastatic Colorectal Cancer Who Have Progressed After Standard Therapy) trials have evaluated the usefulness of different computed tomography scan–based parameters in predicting the clinical benefit of regorafenib therapy. One of the first studies, in 2016, reported a correlation between improved PFS and lung metastases cavitation before therapy initiation and even at week 8.38 In this study, baseline lung metastases cavitation and changes in the sum of target lesion diameters were deemed to be predictors for improved OS in the multivariate analysis. A year later, the CORRECT trial also demonstrated a significant association between survival (PFS and OS) and several radiologic parameters such as response or stable disease in size and density of lung metastases.39 Likewise, highlighting the use of dynamic contrast-enhanced magnetic resonance imaging, a recent study reported that a > 70% decrease in KeF (the product of the median values of volume transfer constant and enhancing fraction) correlated positively with an improved disease control rate and longer PFS and OS.40 Conversely, tissue-based molecular markers such as the downregulation of p53 and phosphorylated-proline–rich AKT substrate have been shown to correlate with higher PFS and metabolic response, respectively.41 Regarding clinical variables and on the basis of the REBECCA (Regorafenib in Metastatic Colorectal Cancer: A Cohort Study in the Real-Life Setting) study, a prognostic score was developed that included the following several parameters independently associated with poorer OS: high Eastern Cooperative Oncology Group performance status, a shorter time from initial diagnosis of metastases, an initial regorafenib dose of < 160 mg, more than three metastatic sites, liver metastases, and KRAS mutations.42 Other potential biomarkers for regorafenib therapy are summarized in the Data Supplement.
Aflibercept and ramucirumab.
In 2015, 87 patients with mCRC enrolled as part of the phase II AFFIRM (Study of Aflibercept and Modified FOLFOX6 as First-Line Treatment in Patients With Metastatic Colorectal Cancer) trial, who were treated with aflibercept plus mFOLFOX6, were analyzed, and it was reported that high plasma levels of interleukin-8 at baseline, together with their increase, were correlated with shorter PFS.43 In 2017, Tabernero et al44 described, in a translational research study based on the RAISE (Ramucirumab Versus Placebo in Combination With Second-Line FOLFIRI in Patients With Metastatic Colorectal Carcinoma That Progressed During or After First-Line Therapy With Bevacizumab, Oxaliplatin, and a Fluoropyrimidine) trial, that high VEGF-D basal levels correlated with a better PFS and OS in patients treated in the ramucirumab arm (Data Supplement).
Anti-EGFR Drugs: Cetuximab and Panitumumab
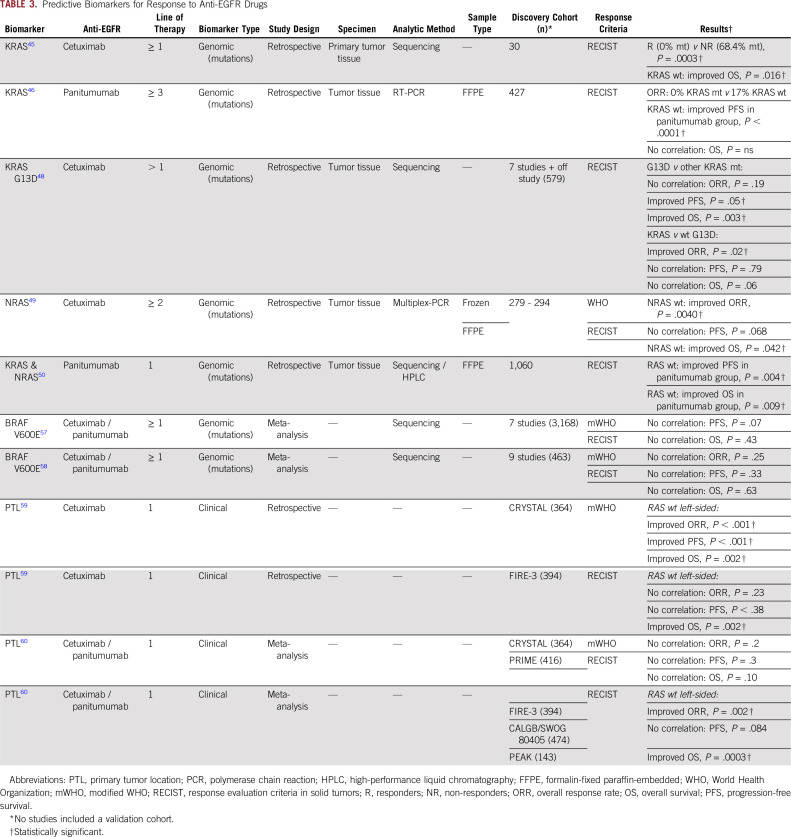
KRAS mutations in tumor tissues were the first predictive biomarker approved to guide decision making for determining eligibility for anti-EGFR therapy in patients with mCRC. One of the first studies comprising a series of 30 patients with mCRC treated with cetuximab-based regimens noted a significant correlation between the presence of KRAS mutations and the lack of response to anti-EGFR therapy,45 an observation that was validated subsequently in a cohort of 427 patients with mCRC treated with panitumumab.46 Similarly, the role of NRAS mutation status as a negative response predictor for panitumumab and cetuximab was later confirmed in various clinical trials and meta-analyses.47-51 Interestingly, in addition to the lack of efficacy in patients with RAS-mutant mCRC, retrospective data analysis from two phase III clinical trials reported a detrimental effect when panitumumab or cetuximab was given in combination with FOLFOX50,52; such a negative effect for these drugs when given in conjunction with FU, leucovorin, and irinotecan (FOLFIRI) has not been confirmed to date.53 In an attempt to translate these tissue-based predictive biomarkers into circulating tumor DNA (ctDNA)–based liquid biopsy assays, two studies in 2017 used an innovative beads, emulsions, amplification, and magnetics (BEAMing) assay and reported promising agreement rates of 89.7% and 93% for the mutational status of RAS between tissue and ctDNA.54,55
Regarding the role of the mutant BRAF gene, the presence of V600E mutation within this gene often reflects a poor prognosis in patients with CRC.56 In addition, two meta-analyses reported a lack of benefit in terms of PFS, OS, and ORR when anti-EGFR therapies were combined with standard chemotherapy in the subset of patients harboring BRAF mutations.57,58 Despite these early data suggesting that the presence of the BRAF V600E mutation may dictate a lack of response to anti-EGFR–based therapies, there is still not enough clinical evidence to consider BRAF mutational status as a predictive biomarker in patients with advanced disease.
In 2016, in a retrospective analysis of the CRYSTAL (Cetuximab Combined With Irinotecan in First-Line Therapy for Metastatic Colorectal Cancer) and FIRE-3 (FOLFIRI Plus Cetuximab Versus FOLFIRI Plus Bevacizumab as First-Line Treatment for Patients With Metastatic Colorectal Cancer) trials, Tejpar et al59 highlighted the significance of tumor location within the colorectum as a predictor of treatment response to anti-EGFR drugs. In these trials, the researchers showed substantially better ORR and a corresponding increase in PFS and OS in patients with wild-type RAS and left-sided tumors.59 However, a subsequent meta-analysis of results from the PRIME (Panitumumab Randomized Trial in Combination With Chemotherapy for Metastatic Colorectal Cancer to Determine Efficacy) and CRYSTAL trials in 2017 failed to show a significant correlation between primary tumor location and ORR, PFS, or OS.60 Nonetheless, a meta-analysis of the FIRE-3/AIO KRK0306, CALGB/SWOG 80405, and PEAK (Panitumumab Efficacy in Combination With mFOLFOX6 Against Bevacizumab Plus mFOLOFOX6 in mCRC Subjects With Wild-Type KRAS Tumors) trials still demonstrated a statistically significant benefit in terms of ORR and OS in patients with wild-type RAS and left-sided tumors.60 In addition to molecular markers, the secondary effects of anti-EGFR treatments, including skin rash, xerosis, or hypomagnesemia, have been postulated as serving as early response predictors. These and other potential predictive biomarkers for anti-EGFR therapy are summarized in Table 3 and in the Data Supplement.
TABLE 3.
Predictive Biomarkers for Response to Anti-EGFR Drugs
Anti–Programmed Cell Death-1 Drugs: Pembrolizumab and Nivolumab
In May and July of 2017, the US Food and Drug Administration approved pembrolizumab and nivolumab for the treatment of patients with MSI-H mCRC in whom the disease has progressed after treatment with fluoropyrimidine, oxaliplatin, and irinotecan therapies. Almost a year later, in July 2018, a nivolumab plus ipilimumab combined regimen was approved, which opened up three novel treatment options for patients with MSI-H or dMMR mCRC, who represent approximately 5% of all patients with mCRC.23 The belief is that, despite their worse prognosis, a large proportion of lymphocytic infiltration and the presence of mutation-associated neoantigens61 confer to patients with MSI-positive mCRC the clinical benefit they derive from anti–programmed cell death-1 (PD-1) therapy.62-65 This exciting discovery has now led to universal MSI testing for the management of patients with mCRC.
Disease Monitoring by Liquid Biopsies
Recently, liquid biopsies have emerged as powerful tools for monitoring disease evolution and therapeutic response through the analysis of cell-free DNA and RNA biomarkers in bodily fluids. One of the first studies, in 1979, which reported that a gradual decrease in carcinoembryonic antigen (CEA) levels during chemotherapy was significantly associated with better survival rates, was the basis for this concept.66 Such a correlation between CEA flare and improved PFS and OS was confirmed a few years later in a subset of 670 patients with mCRC undergoing first-line chemotherapy.67 Although CEA is not a CRC-specific biomarker, CEA monitoring in blood, alone or in addition to CA 19-9,68 is still performed commonly in routine clinical practice. In this context, the accuracy of CEA change in predicting disease progression has been demonstrated recently in a study involving 2,828 patients from seven first-line clinical trials.69 In addition to this, other analyses of circulating tumor cells70 and endothelial cells71 in mCRC have been undertaken by several groups. In 2015, Hansen et al72 reported that circulating levels of miRNA-126 in a subset of 68 patients with mCRC were predictive of tumor response to bevacizumab-based chemotherapy. Two years later, another study reported an association between increasing levels of vasoactive peptides and better treatment outcomes.73 The role of ctDNA in genotyping CRCs and tracking clonal evolution during and after treatment with anti-EGFR–based schedules was first evaluated by Siravegna et al74 in 2015. Since then, multiple studies have reported distinct genetic alterations in ctDNA from patients with primary disease or acquired resistance to anti-EGFR–based therapies in genes such as KRAS, NRAS, MET, ERBB2, FLT3, EGFR, and MAP2K1 by droplet digital polymerase chain reaction, BEAMing, and next-generation sequencing methodologies.74,75 Using a massively parallel sequencing-based assay in a prospective cohort of 53 patients with mCRC, it was shown that early changes in ctDNA during first-line standard chemotherapy can also predict subsequent radiologic response.76 Similarly, in 2017, a study demonstrated a significant correlation between the decrease in RAS mutant clones in blood after 8 weeks of therapy and improved PFS and OS in a cohort of patients treated with regorafenib.40 Intriguingly, clonal evolution is a dynamic process, yet the emergence of drug-resistant clones in circulation increases during treatment, whereas drug withdrawal results in a decrease of such clones. The understanding of this fact has paved a path for novel treatment strategies that are already under evaluation as part of the RASINTRO (RAS Mutations in ctDNA and Anti-EGFR Reintroduction in mCRC) study (ClinicalTrials.gov identifier: NCT03259009) and the CHRONOS (Rechallenge With Panitumumab Driven by RAS Dynamic of Resistance) trial (ClinicalTrials.gov identifier: NCT03227926), which are evaluating the predictive impact of ctDNA RAS mutations on the efficacy of anti-EGFR monotherapy rechallenge in patients with RAS wild-type mCRC whose disease has progressed after anti-EGFR–free chemotherapy. In addition, a five-gene methylation panel for monitoring tumor burden in liquid biopsies using a methyl-BEAMing assay was described recently77 in 182 patients with mCRC treated with chemotherapy and/or targeted therapy, in which the authors discovered a significant correlation between the dynamics of methylation markers and ORR and PFS.
DISCUSSION
Despite the tremendous body of effort devoted to the identification of predictive biomarkers for various treatments used in patients with mCRC, thus far only two of such markers have been translated into routine clinical practice. The first one, the mutations in the RAS gene, serves as a negative predictive biomarker that is present in approximately 55% of patients with mCRC78 and correlates with the lack of efficacy of anti-EGFR antibody treatments. The second marker is the tumor MSI status, which has emerged as a predictive marker for anti–PD-1 drugs. The exciting result of the association between MSI-H and response to nivolumab in the first-in-human clinical trial (ClinicalTrials.gov identifier: NCT00441337) led to two subsequent phase II clinical trials, which were instrumental in the approval of anti–PD-1 drugs (pembrolizumab or nivolumab) alone or in combination with ipilimumab (nivolumab plus ipilimumab) as a treatment option for patients with MSI-H or dMMR mCRC.63-65,79
Other well-described predictive biomarkers used in the management of several tumor types have shown promising usefulness in selecting patients with mCRC for various targeted therapy-based regimens. Results from two clinical trials in patients with BRAF V600E–positive mCRC have highlighted this mutation as a predictive biomarker for BRAF inhibitor–based regimens (Data Supplement).80,81 Regarding the role of human epidermal growth factor receptor 2 (HER2) amplification or overexpression as a predictive biomarker for anti-HER2–based therapies, the results of two phase II clinical trials evaluating the dual HER2 blockade in a biomarker-selected subset of heavily pretreated patients with mCRC, with either trastuzumab plus lapatinib (HERACLES [HER2 Amplification for Colorectal Cancer Enhanced Stratification] trial) or with pertuzumab and trastuzumab (MyPathway trial), demonstrate an impressive ORR of approximately 30% to 40% (Data Supplement).82,83
Nonetheless, the discovery and validation of novel predictive biomarkers that can assist in decision making has been a challenging endeavor, resulting in a long list of failed predictive markers. As highlighted in this article, this task seems even more daunting in terms of conventional chemotherapy and antiangiogenic drugs. In CRC, because the use of single-agent chemotherapeutic regimens has shown limited efficacy, and the majority of current treatment options include various combinations of drugs, biomarker discovery for specific drugs is more complicated, not surprisingly, because of the interactions among different cytotoxic agents.84 Similar concerns remain regarding developing predictive biomarkers for therapeutic response to bevacizumab, because (1) it is also not used as a single agent in the clinic,84 (2) its mechanisms of action are poorly understood,85 and (3) angiogenesis is an intriguingly adaptive process that involves numerous factors.86 Presumably, the inherent complexity of angiogenesis has been a substantial hurdle in the attempts to develop response-predictive biomarkers for other multitargeted antiangiogenic drugs such as aflibercept or regorafenib. Additional insights into the tumor microenvironment, including the role of tumor-associated stromal cells, could possibly shed light on this tortuous process in the future. The gap between the discovery phase and subsequent biomarker development is evident, highlighting the necessity for the implementation of robust worldwide platforms to move predictive biomarker validation algorithms forward.
Another important question worthy of discussion in any biomarker discovery effort is the origin of tumor tissue samples: primary tumor tissue or metastatic lesions? An interesting example of this important concept is TS expression as a predictive biomarker for FU-based chemotherapy, because its efficacy has been discordant depending on the tumor tissue origin.4,87 This concept is highly congruent with tumor heterogeneity, which is a possible source of discrepancy even when the molecular marker is analyzed in a different region of the same source.88 Besides their spatial heterogeneity, tumors are dynamic entities that continue to evolve over time, especially if they are under selective pressure.89 For this reason, the time from sample acquisition to biomarker analysis is of clinical relevance; however, this is an issue that is overlooked in most studies. Because only approximately 20% of patients with CRC present with metastatic disease at the time of diagnosis, it is often the practice or only option available to analyze archival tissues from the primary tumor to identify biomarkers, which is not always optimal.90
Patient selection is gaining importance, which is evidenced by the recent initiative, the US National Cancer Institute’s Exceptional Responder Program.91 Consideration of extreme phenotypes such as long-term responders and extremely early progressors for biomarker discovery can facilitate successful identification of molecular alterations that better correlate with clinical phenotypes. For instance, in the majority of studies presented in this article, there was no consideration of PFS as a selection criterion, and many studies included in the nonresponders patients with stable disease. In general, improved ORR and longer PFS are superior indicators of the true efficacy of any drug intervention, whereas inclusion of gain in OS as a selection feature may inadvertently introduce bias. In addition, new biomarker-driven study designs such as basket or umbrella trials, which assign a treatment according to tumor molecular characteristics, not only are going to improve clinical drug development, but also will facilitate improved biomarker validation. Besides the examples already described previously in the text, new drugs that target tyrosine kinase fusions in genes such as NRTK1/2/3, RET, ALK, and ROS1 are emerging with promising preliminary results in phase I and II clinical trials that include patients with CRC (Data Supplement).92-97
Although analysis of clinical specimens with robust follow-up data from retrospective series or randomized trials are of tremendous value, subsequent prospective clinical cohorts using longitudinally collected specimens are much needed to establish clinically translatable predictive biomarkers. In addition, although many surgical specimens are of suitable quality, needle biopsy–derived metastatic lesions often yield lower amounts of DNA and RNA than that required for robust sequencing experiments98,99; having access to liquid biopsy–based predictive markers would be transformative in overcoming this limitation in patients with mCRC. Furthermore, liquid biopsy biomarkers will improve patient compliance and eliminate the concerns surrounding intratumor heterogeneity associated with tumor or biopsy specimens and may also help in disease monitoring as well as in predicting secondary resistance.
The international community has to consolidate initiatives to improve biomarker development studies and, more importantly, undertake conscious efforts to validate the results gathered from retrospective studies in prospective randomized multicenter cohorts. Such efforts will guarantee improved success and will decrease the economic burden by allowing precision treatment of patients with cancer. Last, the implementation of novel high-throughput molecular analytic techniques and the integration of multiomic approaches with clinical and epidemiologic data using machine-learning algorithms will definitely hasten biomarker development in the coming years.100
Despite many attempts over the past decades, there remain only two well-established predictive biomarkers, mutations in the RAS gene and MSI status, that currently guide treatment decisions in patients with mCRC. Although past efforts in this context may not have been as rewarding, we currently are at a frontier where the future looks promising. It is just a matter of time until we have access to robust predictive biomarkers for response to cancer therapeutics, as we usher in the new era of precision oncology.
Appendix
FIG A1.
PubMed search query.
Footnotes
Supported by Grant Nos. CA181572, CA184792, CA187956, and CA214254 from the National Cancer Institute, National Institutes of Health; by Grant No. RP140784 from the Cancer Prevention Research Institute of Texas; and by grants from the Baylor Foundation and Baylor Scott & White Research Institute, Dallas, TX.
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Financial support: Ajay Goel
Collection and assembly of data: Juan Ruiz-Bañobre, Raju Kandimalla
Data analysis and interpretation: Juan Ruiz-Bañobre
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS’ DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST AND DATA AVAILABILITY STATEMENT
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Juan Ruiz-Bañobre
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Merck Sharp & Dohme, Ipsen, PharmaMar
No other potential conflicts of interest were reported.
REFERENCES
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 2.Kopetz S, Chang GJ, Overman MJ, et al. Improved survival in metastatic colorectal cancer is associated with adoption of hepatic resection and improved chemotherapy. J Clin Oncol. 2009;27:3677–3683. doi: 10.1200/JCO.2008.20.5278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cascinu S, Catalano V, Aschele C, et al. Immunohistochemical determination of p53 protein does not predict clinical response in advanced colorectal cancer with low thymidylate synthase expression receiving a bolus 5-fluorouracil-leucovorin combination. Ann Oncol. 2000;11:1053–1056. doi: 10.1023/a:1008362511552. [DOI] [PubMed] [Google Scholar]
- 4.Aschele C, Debernardis D, Tunesi G, et al. Thymidylate synthase protein expression in primary colorectal cancer compared with the corresponding distant metastases and relationship with the clinical response to 5-fluorouracil. Clin Cancer Res. 2000;6:4797–4802. [PubMed] [Google Scholar]
- 5.Aschele C, Debernardis D, Bandelloni R, et al. Thymidylate synthase protein expression in colorectal cancer metastases predicts for clinical outcome to leucovorin-modulated bolus or infusional 5-fluorouracil but not methotrexate-modulated bolus 5-fluorouracil. Ann Oncol. 2002;13:1882–1892. doi: 10.1093/annonc/mdf327. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Grimaldi M-C, Formento J-L, Francoual M, et al. K-Ras mutations and treatment outcome in colorectal cancer patients receiving exclusive fluoropyrimidine therapy. Clin Cancer Res. 2008;14:4830–4835. doi: 10.1158/1078-0432.CCR-07-4906. [DOI] [PubMed] [Google Scholar]
- 7.Johnston PG, Benson AB, III, Catalano P, et al. Thymidylate synthase protein expression in primary colorectal cancer: Lack of correlation with outcome and response to fluorouracil in metastatic disease sites. J Clin Oncol. 2003;21:815–819. doi: 10.1200/JCO.2003.07.039. [DOI] [PubMed] [Google Scholar]
- 8.Salonga D, Danenberg KD, Johnson M, et al. Colorectal tumors responding to 5-fluorouracil have low gene expression levels of dihydropyrimidine dehydrogenase, thymidylate synthase, and thymidine phosphorylase. Clin Cancer Res. 2000;6:1322–1327. [PubMed] [Google Scholar]
- 9.Smorenburg CH, Peters GJ, van Groeningen CJ, et al. Phase II study of tailored chemotherapy for advanced colorectal cancer with either 5-fluouracil and leucovorin or oxaliplatin and irinotecan based on the expression of thymidylate synthase and dihydropyrimidine dehydrogenase. Ann Oncol. 2006;17:35–42. doi: 10.1093/annonc/mdj046. [DOI] [PubMed] [Google Scholar]
- 10.Gustavsson B, Kaiser C, Carlsson G, et al. Molecular determinants of efficacy for 5-FU-based treatments in advanced colorectal cancer: mRNA expression for 18 chemotherapy-related genes. Int J Cancer. 2009;124:1220–1226. doi: 10.1002/ijc.23852. [DOI] [PubMed] [Google Scholar]
- 11.Lindskog EB, Derwinger K, Gustavsson B, et al. Thymidine phosphorylase expression is associated with time to progression in patients with metastatic colorectal cancer. BMC Clin Pathol. 2014;14:25. doi: 10.1186/1472-6890-14-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Des Guetz G, Uzzan B, Nicolas P, et al. Microsatellite instability: A predictive marker in metastatic colorectal cancer? Target Oncol. 2009;4:57–62. doi: 10.1007/s11523-008-0103-8. [DOI] [PubMed] [Google Scholar]
- 13.Jensen LH, Danenberg KD, Danenberg P V, et al. Predictive value of MSH2 gene expression in colorectal cancer treated with capecitabine. Clin Colorectal Cancer. 2007;6:433–435. doi: 10.3816/CCC.2007.n.012. [DOI] [PubMed] [Google Scholar]
- 14.Simmer F, Venderbosch S, Dijkstra JR, et al. MicroRNA-143 is a putative predictive factor for the response to fluoropyrimidine-based chemotherapy in patients with metastatic colorectal cancer. Oncotarget. 2015;6:22996–23007. doi: 10.18632/oncotarget.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dong S-J, Cai X-J, Li S-J. The clinical significance of MiR-429 as a predictive biomarker in colorectal cancer patients receiving 5-fluorouracil treatment. Med Sci Monit. 2016;22:3352–3361. doi: 10.12659/MSM.900674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suenaga M, Schirripa M, Cao S, et al. Genetic variants of DNA repair-related genes predict efficacy of TAS-102 in patients with refractory metastatic colorectal cancer. Ann Oncol. 2017;28:1015–1022. doi: 10.1093/annonc/mdx035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lenz H-J, Lee F-C, Yau L, et al. MAVERICC, a phase 2 study of mFOLFOX6-bevacizumab (BV) vs FOLFIRI-BV with biomarker stratification as first-line (1L) chemotherapy (CT) in patients (pts) with metastatic colorectal cancer (mCRC) J Clin Oncol. 2016;34:493. [Google Scholar]
- 18.Stoehlmacher J, Ghaderi V, Iobal S, et al. A polymorphism of the XRCC1 gene predicts for response to platinum based treatment in advanced colorectal cancer. Anticancer Res. 2001;21:3075–3079. [PubMed] [Google Scholar]
- 19.Rasmussen MH, Jensen NF, Tarpgaard LS, et al. High expression of microRNA-625-3p is associated with poor response to first-line oxaliplatin based treatment of metastatic colorectal cancer. Mol Oncol. 2013;7:637–646. doi: 10.1016/j.molonc.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi M, Cuatrecasas M, Balaguer F, et al. The clinical significance of MiR-148a as a predictive biomarker in patients with advanced colorectal cancer. PLoS One. 2012;7:e46684. doi: 10.1371/journal.pone.0046684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TF, Nielsen BS, Jakobsen A, et al. Visualising and quantifying angiogenesis in metastatic colorectal cancer: A comparison of methods and their predictive value for chemotherapy response. Cell Oncol (Dordr) 2013;36:341–350. doi: 10.1007/s13402-013-0139-3. [DOI] [PubMed] [Google Scholar]
- 22.Helgason HH, Engwegen JYM, Zapatka M, et al. Identification of serum proteins as prognostic and predictive markers of colorectal cancer using surface enhanced laser desorption ionization-time of flight mass spectrometry. Oncol Rep. 2010;24:57–64. doi: 10.3892/or_00000828. [DOI] [PubMed] [Google Scholar]
- 23.Braun MS, Richman SD, Quirke P, et al. Predictive biomarkers of chemotherapy efficacy in colorectal cancer: Results from the UK MRC FOCUS trial. J Clin Oncol. 2008;26:2690–2698. doi: 10.1200/JCO.2007.15.5580. [DOI] [PubMed] [Google Scholar]
- 24.Koopman M, Knijn N, Richman S, et al. The correlation between topoisomerase-I (Topo1) expression and outcome of treatment with capecitabine and irinotecan in advanced colorectal cancer (ACC) patients (pts) treated in the CAIRO study of the Dutch Colorectal Cancer Group (DCCG) EJC Suppl. 2009;7:321–322. [Google Scholar]
- 25.Hoskins JM, Marcuello E, Altes A, et al. Irinotecan pharmacogenetics: Influence of pharmacodynamic genes. Clin Cancer Res March. 2008;14:1788–1796. doi: 10.1158/1078-0432.CCR-07-1472. [DOI] [PubMed] [Google Scholar]
- 26.Dopeso H, Mateo-Lozano S, Elez E, et al. Aprataxin tumor levels predict response of colorectal cancer patients to irinotecan-based treatment. Clin Cancer Res. 2010;16:2375–2382. doi: 10.1158/1078-0432.CCR-09-3275. [DOI] [PubMed] [Google Scholar]
- 27.Cecchin E, Innocenti F, D’Andrea M, et al. Predictive role of the UGT1A1, UGT1A7, and UGT1A9 genetic variants and their haplotypes on the outcome of metastatic colorectal cancer patients treated with fluorouracil, leucovorin, and irinotecan. J Clin Oncol. 2009;27:2457–2465. doi: 10.1200/JCO.2008.19.0314. [DOI] [PubMed] [Google Scholar]
- 28.Dias MM, McKinnon RA, Sorich MJ. Impact of the UGT1A1*28 allele on response to irinotecan: A systematic review and meta-analysis. Pharmacogenomics. 2012;13:889–899. doi: 10.2217/pgs.12.68. [DOI] [PubMed] [Google Scholar]
- 29.Hiraki M, Kitajima Y, Nakafusa Y, et al. CpG island methylation of BNIP3 predicts resistance against S-1/CPT-11 combined therapy in colorectal cancer patients. Oncol Rep. 2010;23:191–197. [PubMed] [Google Scholar]
- 30.Hamauchi S, Yamazaki K, Masuishi T, et al. Neutropenia as a predictive factor in metastatic colorectal cancer treated with TAS-102. Clin Colorectal Cancer. 2017;16:51–57. doi: 10.1016/j.clcc.2016.07.005. [DOI] [PubMed] [Google Scholar]
- 31.Yuan Y, Tan C-W, Shen H, et al. Identification of the biomarkers for the prediction of efficacy in first-line chemotherapy of metastatic colorectal cancer patients using SELDI-TOF-MS and artificial neural networks. Hepatogastroenterology. 2012;59:2461–2465. doi: 10.5754/hge12127. [DOI] [PubMed] [Google Scholar]
- 32.Aoyagi Y, Iinuma H, Horiuchi A, et al. Association of plasma VEGF-A, soluble VEGFR-1 and VEGFR-2 levels and clinical response and survival in advanced colorectal cancer patients receiving bevacizumab with modified FOLFOX6. Oncol Lett. 2010;1:253–259. doi: 10.3892/ol_00000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goede V, Coutelle O, Neuneier J, et al. Identification of serum angiopoietin-2 as a biomarker for clinical outcome of colorectal cancer patients treated with bevacizumab-containing therapy. Br J Cancer. 2010;103:1407–1414. doi: 10.1038/sj.bjc.6605925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruhn MA, Townsend AR, Khoon Lee C, et al. Proangiogenic tumor proteins as potential predictive or prognostic biomarkers for bevacizumab therapy in metastatic colorectal cancer. Int J Cancer. 2014;135:731–741. doi: 10.1002/ijc.28698. [DOI] [PubMed] [Google Scholar]
- 35.Hurwitz HI, Yi J, Ince W, et al. The clinical benefit of bevacizumab in metastatic colorectal cancer is independent of K-ras mutation status: Analysis of a phase III study of bevacizumab with chemotherapy in previously untreated metastatic colorectal cancer. Oncologist. 2009;14:22–28. doi: 10.1634/theoncologist.2008-0213. [DOI] [PubMed] [Google Scholar]
- 36.Chen C, Sun P, Ye S, et al. Hypertension as a predictive biomarker for efficacy of bevacizumab treatment in metastatic colorectal cancer: A meta-analysis. J BUON. 2014;19:917–924. [PubMed] [Google Scholar]
- 37.Guiu B, Petit JM, Bonnetain F, et al. Visceral fat area is an independent predictive biomarker of outcome after first-line bevacizumab-based treatment in metastatic colorectal cancer. Gut. 2010;59:341–347. doi: 10.1136/gut.2009.188946. [DOI] [PubMed] [Google Scholar]
- 38.Ricotta R, Verrioli A, Ghezzi S, et al. Radiological imaging markers predicting clinical outcome in patients with metastatic colorectal carcinoma treated with regorafenib: Post hoc analysis of the CORRECT phase III trial (RadioCORRECT study) ESMO Open. 2017;1:e000111. doi: 10.1136/esmoopen-2016-000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vanwynsberghe H, Verbeke X, Coolen J, et al. Predictive value of early tumor shrinkage and density reduction of lung metastases in patients with metastatic colorectal cancer treated with regorafenib. Clin Colorectal Cancer. 2017;16:377–380. doi: 10.1016/j.clcc.2017.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Khan K, Rata M, Cunningham D, et al. Functional imaging and circulating biomarkers of response to regorafenib in treatment-refractory metastatic colorectal cancer patients in a prospective phase II study. Gut. 2018;67:1484–1492. doi: 10.1136/gutjnl-2017-314178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wong ALA, Lim JSJ, Sinha A, et al. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J Transl Med. 2015;13:57. doi: 10.1186/s12967-015-0405-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Adenis A, de la Fouchardiere C, Paule B, et al. Survival, safety, and prognostic factors for outcome with regorafenib in patients with metastatic colorectal cancer refractory to standard therapies: Results from a multicenter study (REBECCA) nested within a compassionate use program. BMC Cancer. 2016;16:412. doi: 10.1186/s12885-016-2440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lambrechts D, Thienpont B, Thuillier V, et al. Evaluation of efficacy and safety markers in a phase II study of metastatic colorectal cancer treated with aflibercept in the first-line setting. Br J Cancer. 2015;113:1027–1034. doi: 10.1038/bjc.2015.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tabernero J, Hozak RR, Yoshino T, et al. Analysis of angiogenesis biomarkers for ramucirumab efficacy in patients with metastatic colorectal cancer from RAISE, a global, randomized, double-blind, phase III study. Ann Oncol. 2018;29:602–609. doi: 10.1093/annonc/mdx767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lièvre A, Bachet J-B, Le Corre D, et al. KRAS mutation status is predictive of response to cetuximab therapy in colorectal cancer. Cancer Res. 2006;66:3992–3995. doi: 10.1158/0008-5472.CAN-06-0191. [DOI] [PubMed] [Google Scholar]
- 46.Amado RG, Wolf M, Peeters M, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:1626–1634. doi: 10.1200/JCO.2007.14.7116. [DOI] [PubMed] [Google Scholar]
- 47.Peeters M, Oliner KS, Parker A. 2010. Use of massively parallel, next-generation sequencing to identify gene mutations beyond ERAS that predict response to panitumumab in a randomized, phase 3, monotherapy study of metastatic colorectal cancer (mCRC) [Google Scholar]
- 48.De Roock W, Jonker DJ, Nicolantonio F Di, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304:1812–1820. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 49.De Roock W, Claes B, Bernasconi D, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- 50.Douillard J-Y, Oliner KS, Siena S, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369:1023–1034. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]
- 51.Sorich MJ, Wiese MD, Rowland A, et al. Extended RAS mutations and anti-EGFR monoclonal antibody survival benefit in metastatic colorectal cancer: A meta-analysis of randomized, controlled trials. Ann Oncol. 2015;26:13–21. doi: 10.1093/annonc/mdu378. [DOI] [PubMed] [Google Scholar]
- 52.Bokemeyer C, Köhne C-H, Ciardiello F, et al. FOLFOX4 plus cetuximab treatment and RAS mutations in colorectal cancer. Eur J Cancer. 2015;51:1243–1252. doi: 10.1016/j.ejca.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stintzing S, Miller-Phillips L, Modest DP, et al. Impact of BRAF and RAS mutations on first-line efficacy of FOLFIRI plus cetuximab versus FOLFIRI plus bevacizumab: Analysis of the FIRE-3 (AIO KRK-0306) study. Eur J Cancer. 2017;79:50–60. doi: 10.1016/j.ejca.2017.03.023. [DOI] [PubMed] [Google Scholar]
- 54.Grasselli J, Elez E, Caratù G, et al. Concordance of blood- and tumor-based detection of RAS mutations to guide anti-EGFR therapy in metastatic colorectal cancer. Ann Oncol. 2017;28:1294–1301. doi: 10.1093/annonc/mdx112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vidal J, Muinelo L, Dalmases A, et al. Plasma ctDNA RAS mutation analysis for the diagnosis and treatment monitoring of metastatic colorectal cancer patients. Ann Oncol. 2017;28:1325–1332. doi: 10.1093/annonc/mdx125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yuan Z-X, Wang X-Y, Qin Q-Y, et al. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: A meta-analysis. PLoS One. 2013;8:e65995. doi: 10.1371/journal.pone.0065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rowland A, Dias MM, Wiese MD, et al. Meta-analysis of BRAF mutation as a predictive biomarker of benefit from anti-EGFR monoclonal antibody therapy for RAS wild-type metastatic colorectal cancer. Br J Cancer. 2015;112:1888–1894. doi: 10.1038/bjc.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: A meta-analysis. Eur J Cancer. 2015;51:587–594. doi: 10.1016/j.ejca.2015.01.054. [DOI] [PubMed] [Google Scholar]
- 59.Tejpar S, Stintzing S, Ciardiello F, et al. Prognostic and predictive relevance of primary tumor location in patients with RAS wild-type metastatic colorectal cancer: Retrospective analyses of the CRYSTAL and FIRE-3 trials. JAMA Oncol. doi: 10.1001/jamaoncol.2016.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Holch JW, Ricard I, Stintzing S, et al. The relevance of primary tumour location in patients with metastatic colorectal cancer: A meta-analysis of first-line clinical trials. Eur J Cancer. 2017;70:87–98. doi: 10.1016/j.ejca.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 61.Giannakis M, Mu XJ, Shukla SA, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Reports. 2016;15:857–865. doi: 10.1016/j.celrep.2016.03.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Le DT, Uram JN, Wang H, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372:2509–2520. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Le DT, Durham JN, Smith KN, et al. Mismatch repair deficiency predicts response of solid tumors to PD-1 blockade. Science. 2017;357:409–413. doi: 10.1126/science.aan6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Overman MJ, McDermott R, Leach JL, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): An open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18:1182–1191. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Overman MJ, Lonardi S, Wong KYM, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36:773–779. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 66.Al-Sarraf M, Baker L, Talley RW, et al. The value of serial carcinoembryonic antigen (CEA) in predicting response rate and survival of patients with gastrointestinal cancer treated with chemotherapy. Cancer. 1979;44:1222–1225. doi: 10.1002/1097-0142(197910)44:4<1222::aid-cncr2820440409>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 67.Strimpakos AS, Cunningham D, Mikropoulos C, et al. The impact of carcinoembryonic antigen flare in patients with advanced colorectal cancer receiving first-line chemotherapy. Ann Oncol. 2010;21:1013–1019. doi: 10.1093/annonc/mdp449. [DOI] [PubMed] [Google Scholar]
- 68.Stiksma J, Grootendorst DC, van der Linden PWG. CA 19-9 as a marker in addition to CEA to monitor colorectal cancer. Clin Colorectal Cancer. 2014;13:239–244. doi: 10.1016/j.clcc.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 69.Gulhati P, Yin J, Pederson L, et al. Change in CEA as an early predictor of progression to first-line systemic therapy in metastatic colorectal cancer. J Clin Oncol. 2018;36 doi: 10.1093/jnci/djaa020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Cohen SJ, Punt CJA, Iannotti N, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26:3213–3221. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 71.Ronzoni M, Manzoni M, Mariucci S, et al. Circulating endothelial cells and endothelial progenitors as predictive markers of clinical response to bevacizumab-based first-line treatment in advanced colorectal cancer patients. Ann Oncol. 2010;21:2382–2389. doi: 10.1093/annonc/mdq261. [DOI] [PubMed] [Google Scholar]
- 72.Hansen TF, Carlsen AL, Heegaard NHH, et al. Changes in circulating microRNA-126 during treatment with chemotherapy and bevacizumab predicts treatment response in patients with metastatic colorectal cancer. Br J Cancer. 2015;112:624–629. doi: 10.1038/bjc.2014.652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hagman H, Bendahl P-O, Melander O, et al. Vasoactive peptides associate with treatment outcome ofbevacizumab-containing therapy in metastatic colorectal cancer. Acta Oncol. 2017;56:653–660. doi: 10.1080/0284186X.2017.1302098. [DOI] [PubMed] [Google Scholar]
- 74.Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med. 2015;21:795–801. doi: 10.1038/nm.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Van Emburgh BO, Arena S, Siravegna G, et al. Acquired RAS or EGFR mutations and duration of response to EGFR blockade in colorectal cancer. Nat Commun. 2016;7:13665. doi: 10.1038/ncomms13665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tie J, Kinde I, Wang Y, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015;26:1715–1722. doi: 10.1093/annonc/mdv177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barault L, Amatu A, Siravegna G, et al. Discovery of methylated circulating DNA biomarkers for comprehensive non-invasive monitoring of treatment response in metastatic colorectal cancer. Gut. 2018;67:1995–2005. doi: 10.1136/gutjnl-2016-313372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Peeters M, Kafatos G, Taylor A, et al. Prevalence of RAS mutations and individual variation patterns among patients with metastatic colorectal cancer: A pooled analysis of randomised controlled trials. Eur J Cancer. 2015;51:1704–1713. doi: 10.1016/j.ejca.2015.05.017. [DOI] [PubMed] [Google Scholar]
- 79.Lipson EJ, Sharfman WH, Drake CG, et al. Durable cancer regression off-treatment and effective reinduction therapy with an anti-PD-1 antibody. Clin Cancer Res. 2013;19:462–468. doi: 10.1158/1078-0432.CCR-12-2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kopetz S, McDonough SL, Lenz H-J, et al. Randomized trial of irinotecan and cetuximab with or without vemurafenib in BRAF-mutant metastatic colorectal cancer (SWOG S1406) J Clin Oncol. 2017;35:3505. doi: 10.1200/JCO.20.01994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Corcoran RB, Andre T, Atreya CE, et al. Combined BRAF, EGFR, and MEK inhibition in patients with BRAFV600E-mutant colorectal cancer. Cancer Discov. 2018;8:428–443. doi: 10.1158/2159-8290.CD-17-1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS codon 12/13 wild-type, HER2-positive metastatic colorectal cancer (HERACLES): A proof-of-concept, multicentre, open-label, phase 2 trial. Lancet Oncol. 2016;17:738–746. doi: 10.1016/S1470-2045(16)00150-9. [DOI] [PubMed] [Google Scholar]
- 83.Hainsworth JD, Meric-Bernstam F, Swanton C, et al. Targeted therapy for advanced solid tumors on the basis of molecular profiles: Results from MyPathway, an open-label, phase IIa multiple basket study. J Clin Oncol. 2018;36:536–542. doi: 10.1200/JCO.2017.75.3780. [DOI] [PubMed] [Google Scholar]
- 84.National Comprehensive Cancer Network 2017 https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- 85.Ellis LM, Hicklin DJ. VEGF-targeted therapy: Mechanisms of anti-tumour activity. Nat Rev Cancer. 2008;8:579–591. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 86.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yamada H, Ichikawa W, Uetake H, et al. Thymidylate synthase gene expression in primary colorectal cancer and metastatic sites. Clin Colorectal Cancer. 2001;1:169–173, discussion 174. doi: 10.3816/CCC.2001.n.017. [DOI] [PubMed] [Google Scholar]
- 88.Sottoriva A, Kang H, Ma Z, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47:209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Melo FDSE, Vermeulen L, Fessler E, et al. Cancer heterogeneity—a multifaceted view. EMBO Rep. 2013;14:686–695. doi: 10.1038/embor.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Khattak MA, Martin HL, Beeke C, et al. Survival differences in patients with metastatic colorectal cancer and with single site metastatic disease at initial presentation: Results from South Australian clinical registry for advanced colorectal cancer. Clin Colorectal Cancer. 2012;11:247–254. doi: 10.1016/j.clcc.2012.06.004. [DOI] [PubMed] [Google Scholar]
- 91.Chang DK, Grimmond SM, Evans TRJ, et al. Mining the genomes of exceptional responders. Nat Rev Cancer. 2014;14:291–292. doi: 10.1038/nrc3723. [DOI] [PubMed] [Google Scholar]
- 92.Drilon A, Laetsch TW, Kummar S, et al. Efficacy of larotrectinib in TRK fusion-positive cancers in adults and children. N Engl J Med. 2018;378:731–739. doi: 10.1056/NEJMoa1714448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Drilon A, Siena S, Ou S-HI, et al. Safety and antitumor activity of the multitargeted Pan-TRK, ROS1, and ALK inhibitor entrectinib: Combined results from two phase I trials (ALKA-372-001 and STARTRK-1) Cancer Discov. 2017;7:400–409. doi: 10.1158/2159-8290.CD-16-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Amatu A, Somaschini A, Cerea G, et al. Novel CAD-ALK gene rearrangement is drugable by entrectinib in colorectal cancer. Br J Cancer. 2015;113:1730–1734. doi: 10.1038/bjc.2015.401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Sartore-Bianchi A, Ardini E, Bosotti R, et al. Sensitivity to entrectinib associated with a novel LMNA-NTRK1 gene fusion in metastatic colorectal cancer. J Natl Cancer Inst. 2015;108:djv306. doi: 10.1093/jnci/djv306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Drilon A, Nagasubramanian R, Blake JF, et al. A next-generation TRK kinase inhibitor overcomes acquired resistance to prior TRK kinase inhibition in patients with TRK fusion-positive solid tumors. Cancer Discov. 2017;7:963–972. doi: 10.1158/2159-8290.CD-17-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pietrantonio F, Di Nicolantonio F, Schrock AB, et al. ALK, ROS1, and NTRK rearrangements in metastatic colorectal cancer. J Natl Cancer Inst. 2017;109(12):djx089. doi: 10.1093/jnci/djx089. [DOI] [PubMed] [Google Scholar]
- 98.Kris MG, Johnson BE, Berry LD, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. doi: 10.1001/jama.2014.3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meric-Bernstam F, Brusco L, Shaw K, et al. Feasibility of large-scale genomic testing to facilitate enrollment onto genomically matched clinical trials. J Clin Oncol. 2015;33:2753–2762. doi: 10.1200/JCO.2014.60.4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhang W, Chien J, Yong J, et al Network-based machine learning and graph theory algorithms for precision oncology. npj Precis Oncol. [DOI] [PMC free article] [PubMed]