INTRODUCTION
After a patient dies, the medical history stops unfolding and genetic evaluation typically ceases. However, newer techniques may resolve uncertain diagnoses for patients and families, even years after a cancer diagnosis and including after the death of the affected individual.
Lynch syndrome is a highly penetrant, autosomal dominant, cancer predisposition syndrome caused by germline pathogenic variants in one of the following mismatch repair (MMR) genes: MSH6, MSH2, MLH1, PMS2, or EPCAM. Individuals with Lynch syndrome have a high lifetime risk of cancer, including colorectal cancer (CRC; risk range, 10% to 80%), endometrial cancer (risk range, 16% to 60%), and others.1 Clinical suggestion for Lynch syndrome is subjectively based on personal and family history and objectively on tumor screening.2-6 Tumor screening assesses MMR deficiency by identifying DNA replication errors in microsatellite repeats (microsatellite instability-high) or by immunohistochemical (IHC) staining with decreased or absent expression in one or more MMR proteins.1,5,6
Greater than 85% of Lynch-associated tumors have a high level of microsatellite instability or abnormal findings by IHC staining.3,4 However, tumor screening is not diagnostic: 10% to 15% of sporadic CRC tumors are also MMR deficient.2,4,7,8 Screening of abnormal tumors should include assessment of sporadic causes of tumor MMR deficiency, including MLH1 hypermethylation or BRAF codon 600 mutations, and germline testing.1,5,8,9 Probands with an MMR-deficient tumor but negative somatic or germline testing present a diagnostic and clinical management challenge.4 Preventive recommendations for these individuals are not well defined and vary widely in practice.3,4
Sequencing MMR-deficient tumors in individuals with unexplained, abnormal tumor screening is a powerful tool for refining the risk of Lynch syndrome. Tumor sequencing identifies double-somatic MMR deficiency in 75% of patients and previously missed germline variants in 5% to 10% of patients.7,8,10,11 Double-somatic MMR mutations that explain tumor MMR deficiency significantly reduce clinical suggestion for Lynch syndrome, allowing cancer risk reassessment.8,10-12 For surviving relatives who may be following intensive screening guidelines based on family history, paired germline and tumor sequencing can clarify an uncertain diagnosis of Lynch syndrome and influence medical management.
CASE REPORT
All postmortem clinical requests for ColoSeq Tumor (University of Washington Genetics and Solid Tumors Laboratory, Seattle, WA) ordered between test introduction in 2014 to August 2016 were included. Tumor samples were received as formalin-fixed paraffin-embedded blocks. Germline DNA was obtained from banked samples or extracted from nonadjacent tissue. Tumor content was maximized using manual macrodissection.10 To rule out low-level mosaicism and missed germline variants, 1 μg of tumor and germline DNA was concurrently analyzed. ColoSeq Tumor is performed in the College of American Pathologists-accredited, Clinical Laboratory Improvement Amendments–certified University of Washington Genetics and Solid Tumors Laboratory. Data are processed by the University of Washington Next-Generation Sequencing Laboratory and Analytics group using a custom pipeline to detect single nucleotide variants, insertions and deletions, structural variations, microsatellite instability, and copy number alterations in 25 genes previously implicated in colorectal and other cancers, including MLH1, MSH2, MSH6, PMS2, and EPCAM (http://tests.labmed.washington.edu/COLOSEQ).7,10,12-14
Variants were categorized according to International Society for Gastrointestinal Hereditary Tumors (http://insight-group.org/variants/database/) and American College of Medical Genetics and Genomics classification criteria and expert consensus review.15,16 Tumor loss of heterozygosity was determined as previously described.10
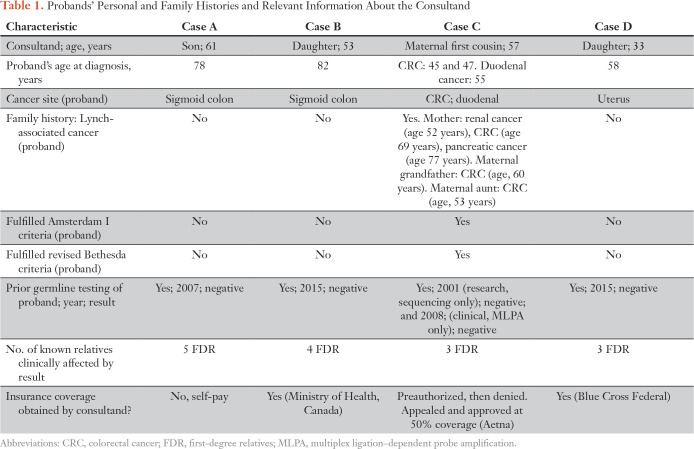
From a total of 381 clinical requests for ColoSeq Tumor, four requests were ordered on postmortem samples. In each, MMR deficiency was established by IHC staining. Germline testing was negative for all probands. ColoSeq Tumor was ordered on behalf of a surviving relative to clarify an uncertain diagnosis of Lynch syndrome and for an associated need for preventive surveillance and/or prophylactic surgery. Tumor and germline DNA was successfully extracted in all cases. Table 1 describes the personal and family history of each proband and relevant information about the consultand.
Table 1.
Probands’ Personal and Family Histories and Relevant Information About the Consultand
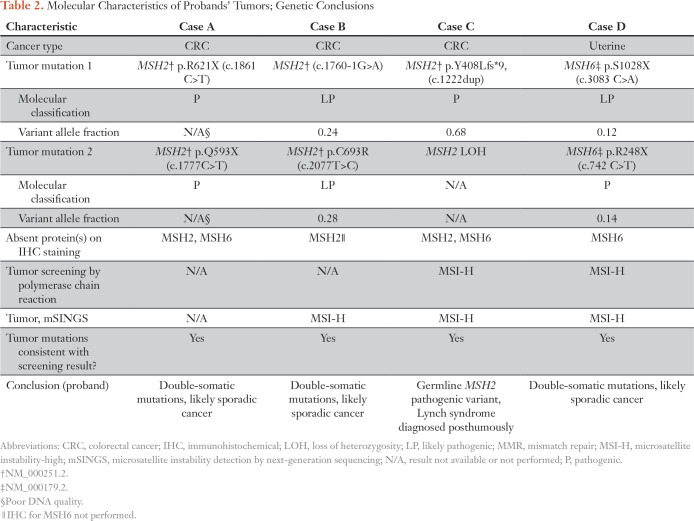
Patient A was diagnosed with CRC at age 78 years and died at age 82 years. Tumor IHC staining indicated MSH2 and MSH6 were absent. The consultand, the proband’s 61-year old son, was following high-risk surveillance recommendations based on his mother’s presumptive diagnosis of Lynch syndrome. At time of sequencing, he had already undergone 12 to 15 colonoscopies. ColoSeq Tumor revealed two somatic pathogenic mutations, MSH2 p.R621X and MSH2 p.Q593X, consistent with findings on IHC staining and a likely explanation for tumor MMR deficiency (Table 2). Postmortem sequencing results reduced the consultand’s cancer surveillance from annually to every 5 years. The consultand’s sister was considering prophylactic total hysterectomy and bilateral salpingectomy-oophorectomy (BSO). Sequencing signaled she was not at increased risk for Lynch-associated cancer and prophylactic surgery was not indicated; her surveillance schedule was revised to every 10 years. The recommendation for early, high-risk surveillance for the consultand’s children was rescinded.
Table 2.
Molecular Characteristics of Probands’ Tumors; Genetic Conclusions
Patient B was diagnosed with colorectal cancer at age 82 years and died at age 87 years. Frequent colonoscopy was recommended for her four children based on the proband’s MMR-deficient tumor. Formal genetic counseling was received at the proband’s death and the family desired additional risk assessment. ColoSeq Tumor testing revealed two likely pathogenic somatic mutations in the tumor, MSH2 c.1760-1G>A and MSH2 p.C693R.16 The cancer surveillance plan was reassessed for the consultand and her siblings; recommendation for colonoscopy was reduced to every 5 years.
Patient C was diagnosed with metachronous CRC at ages 45 and 47 years and duodenal cancer at age 55 years. High-risk surveillance was recommended for all surviving relatives given the proband’s MMR-deficient tumor and meeting Amsterdam I criteria. The proband’s 59-year-old, maternal first cousin requested ColoSeq Tumor to clarify possible Lynch syndrome. Tumor sequencing identified a pathogenic variant MSH2 c.1222dup (p.Y408Lfs*9) with associated loss of heterozygosity. This variant was present in the germline sample (banked DNA extracted from whole blood). Thus, Lynch syndrome was posthumously diagnosed in the proband. Cascade testing was initiated for surviving family members. For years, the consultand underwent colonoscopy every 1 to 2 years for high-risk surveillance. His screening schedule was revised after he was molecularly confirmed negative for the familial mutation. The proband’s daughters were considering prophylactic total hysterectomy with BSO and currently await cascade testing results to guide this decision.
Patient D was diagnosed with endometrial cancer at age 58 years and died within a year. ColoSeq Tumor was ordered on behalf of the proband’s 33-year-old daughter, who was considering initiation of high-risk cancer surveillance and prophylactic hysterectomy. ColoSeq Tumor revealed two somatic pathogenic variants, MSH6 p.S1028X and MSH6 p.R248X. Results were consistent with a likely sporadic cause of tumor MMR deficiency. The consultand and her two brothers were advised to initiate colonoscopy screening at age 50 years. Prophylactic hysterectomy and BSO were no longer recommended for proband’s daughter.
DISCUSSION
Clinical judgment often supports high-risk surveillance on the basis of an MMR-deficient tumor and/or personal and family history, even in the absence of a pathogenic germline variant. Indeed, individuals with an MMR-deficient tumor and negative germline testing have a higher recurrence rate of CRC compared with individuals with known sporadic cancer, leading to concern for a missed germline variant.4,7,8 Despite universal tumor screening recommendations, compliance is inconsistent and may be preferentially performed with high clinical suspicion of Lynch syndrome.17,18 Preventive cancer screening carries cost and risk; clarification of an uncertain diagnosis should be sought when clinically feasible.2
Genetic testing is unique; a result potentially provides information about a proband and relatives. When a consultand requests a risk assessment and a germline sample from the proband is unavailable, it is reasonable to offer genetic testing to the unaffected consultand. However, surrogate testing is not equivalent to testing the affected person and carries its own risks.19 Whenever possible, it is preferred to test the affected individual, even postmortem.
Genetic testing of deceased patients warrants special consideration and regulation.20 To our knowledge, this is the first report of clinical, postmortem, somatic sequencing used to actively change surviving relative management. As such, literature exploring this issue is limited. Information considered ethical to disclose during a patient’s lifetime, such as imminent harm to identifiable individuals and/or potential benefit for at-risk individuals or public health, is considered ethical to disclose postmortem with the family’s consent.20 Resolving an uncertain but clinically suspected diagnosis of Lynch syndrome falls within this paradigm.
Despite the clinical utility of resolving an uncertain diagnosis, several factors may hinder clinical uptake. First, misinformation abounds.21,22 Many individuals with MMR-deficient tumors and negative germline testing are confused by the diagnostic nuances. Patients may misunderstand the uncertainty of this result and erroneously believe they have Lynch syndrome or not understand the results.20 ColoSeq Tumor will likely only be offered if diagnosis is recognized as uncertain.5,21,22 Next, test implementation requires significant genetic literacy, clinical judgment, and expertise that is not universally available.17 Clinical pathology laboratories are required by the College of American Pathologists to retain patient-tissue blocks for 10 years. Thus, obtaining samples for postmortem testing may not be feasible if requested > 10 years after a patient’s tissue procurement. Last, insurance coverage for postmortem testing is challenging and, if denied, out-of-pocket cost is a deterrent.5 In this report, insurance coverage was obtained for three of four patients. These numbers are small and may reflect ascertainment bias of those who expected coverage or could self-pay.
This case series has several important limitations, including comprising few cases and that patients receiving tumor screening may be a selected group, which could skew the high diagnostic rate and significant downstream benefits for surviving family members.18 Nevertheless, this case series sufficiently demonstrates feasibility and clinical utility of resolving an uncertain Lynch syndrome diagnosis in a deceased proband.
In conclusion, postmortem paired tumor and germline sequencing is feasible, reliable, and offers a high diagnostic yield. It is a powerful tool to resolve uncertain diagnoses, inform risk assessment, and prevent unnecessary cancer surveillance. Families following intensive cancer surveillance based on unexplained tumor MMR deficiency should be offered tumor sequencing for diagnostic resolution.
Footnotes
Supported by Grant NIH5T32GM007454 to H.M.B. from the National Institutes of Health.
AUTHOR CONTRIBUTIONS
Conception and design: Heather M. Byers, Angela Jacobson, Cigdem H. Ussakli, Stephanie More, Jonathan F. Tait, Colin C. Pritchard, Eric Q. Konnick, Christina M. Lockwood
Financial support: Heather M. Byers, Colin C. Pritchard, Christina M. Lockwood
Administrative support: Angela Jacobson
Provision of study material or patients: Angela Jacobson, Anna Newlin, Amanda Hamblett, Brian Shirts, Stephanie More, Eric Q. Konnick
Collection and assembly of data: Heather M. Byers, Angela Jacobson, Cigdem H. Ussakli, Anna Newlin, Stephanie More, Amanda Hamblett, Jonathan F. Tait, Brian Shirts, Colin C. Pritchard, Eric Q. Konnick, Christina M. Lockwood
Data analysis and interpretation: Heather M. Byers, Angela Jacobson, Andrew S. McFadden, Cigdem H. Ussakli, Peter Stanich, Stephanie More, Colin C. Pritchard, Eric Q. Konnick, Christina M. Lockwood
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Heather M. Byers
Stock and Other Ownership Interests: HCA Healthcare, Spark Therapeutics, Illumina
Angela Jacobson
No relationship to disclose
Andrew S. McFaddin
No relationship to disclose
Cigdem H. Ussakli
No relationship to disclose
Anna Newlin
Employment: OPKO Health/GeneDx
Travel, Accommodations, Expenses: OPKO Health/GeneDx
Peter P. Stanich
Patents, Royalties, Other Intellectual Property: I am an author for UpToDate and receive royalties.
Stephanie More
No relationship to disclose
Amanda Hamblett
Stock and Other Ownership Interests: Edge Therapeutics, Achillion Pharmaceuticals, Cerecor, Delcath Systems, OPKO Health, Sonoma Pharmaceuticals, Sierra Oncology, Synthetic Biologics, Trevena, Oncogenix, Corbus Pharmaceuticals, Waveguide, Osiris Therapeutics, Bristol-Myers Squibb, Gilead Sciences, Cellectar Biosciences, Chimerix, Contravir, Stellar Biotechnologies, Innovate Biopharmaceuticals, Verastem
Jonathan F. Tait
Stock and Other Ownership Interests: Merck (I), Amgen (I), GlaxoSmithKline (I)
Research Funding: EviCore (Inst)
Patents, Royalties, Other Intellectual Property: EviCore copyrights (Inst)
Brian Shirts
No relationship to disclose
Colin C. Pritchard
No relationship to disclose
Eric Q. Konnick
Honoraria: Ventana Medical Systems
Travel, Accommodations, Expenses: Ventana Medical Systems
Christina M. Lockwood
Travel, Accommodations, Expenses: Cambridge Health Institute
REFERENCES
- 1.Bonadona V, Bonaïti B, Olschwang S, et al. Cancer risks associated with germline mutations in MLH1, MSH2, and MSH6 genes in Lynch syndrome. JAMA. 2011;305:2304–2310. doi: 10.1001/jama.2011.743. [DOI] [PubMed] [Google Scholar]
- 2.Palomaki GE, McClain MR, Melillo S, et al. EGAPP supplementary evidence review: DNA testing strategies aimed at reducing morbidity and mortality from Lynch syndrome. Genet Med. 2009;11:42–65. doi: 10.1097/GIM.0b013e31818fa2db. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodríguez-Soler M, Pérez-Carbonell L, Guarinos C, et al. Risk of cancer in cases of suspected Lynch syndrome without germline mutation. Gastroenterology. 2013;144:926–932.e1. doi: 10.1053/j.gastro.2013.01.044. [DOI] [PubMed] [Google Scholar]
- 4.Win AK, Buchanan DD, Rosty C, et al. Role of tumour molecular and pathology features to estimate colorectal cancer risk for first-degree relatives. Gut. 2015;64:101–110. doi: 10.1136/gutjnl-2013-306567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weissman SM, Burt R, Church J, et al. Identification of individuals at risk for Lynch syndrome using targeted evaluations and genetic testing: National Society of Genetic Counselors and the Collaborative Group of the Americas on Inherited Colorectal Cancer joint practice guideline. J Genet Couns. 2012;21:484–493. doi: 10.1007/s10897-011-9465-7. [DOI] [PubMed] [Google Scholar]
- 6.Moreira L, Balaguer F, Lindor N, et al. Identification of Lynch syndrome among patients with colorectal cancer. JAMA. 2012;308:1555–1565. doi: 10.1001/jama.2012.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sourrouille I, Coulet F, Lefevre JH, et al. Somatic mosaicism and double somatic hits can lead to MSI colorectal tumors. Fam Cancer. 2013;12:27–33. doi: 10.1007/s10689-012-9568-9. [DOI] [PubMed] [Google Scholar]
- 8.Geurts-Giele WR, Leenen CH, Dubbink HJ, et al. Somatic aberrations of mismatch repair genes as a cause of microsatellite-unstable cancers. J Pathol. 2014;234:548–559. doi: 10.1002/path.4419. [DOI] [PubMed] [Google Scholar]
- 9.Evaluation of Genomic Applications in Practice and Prevention (EGAPP) Working Group Recommendations from the EGAPP Working Group: Genetic testing strategies in newly diagnosed individuals with colorectal cancer aimed at reducing morbidity and mortality from Lynch syndrome in relatives. Genet Med. 2009;11:35–41. doi: 10.1097/GIM.0b013e31818fa2ff. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haraldsdottir S, Hampel H, Tomsic J, et al. Colon and endometrial cancers with mismatch repair deficiency can arise from somatic, rather than germline, mutations. Gastroenterology. 2014;147:1308–1316.e1. doi: 10.1053/j.gastro.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mensenkamp AR, Vogelaar IP, van Zelst-Stams WA, et al. Somatic mutations in MLH1 and MSH2 are a frequent cause of mismatch-repair deficiency in Lynch syndrome-like tumors. Gastroenterology. 2014;146:643–646.e8. doi: 10.1053/j.gastro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 12.Konnick EQ, Pritchard CC. Germline, hematopoietic, mosaic, and somatic variation: Interplay between inherited and acquired genetic alterations in disease assessment. Genome Med. 2016;8:100. doi: 10.1186/s13073-016-0350-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pritchard CC, Smith C, Salipante SJ, et al. ColoSeq provides comprehensive Lynch and polyposis syndrome mutational analysis using massively parallel sequencing. J Mol Diagn. 2012;14:357–366. doi: 10.1016/j.jmoldx.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salipante SJ, Scroggins SM, Hampel HL, et al. Microsatellite instability detection by next generation sequencing. Clin Chem. 2014;60:1192–1199. doi: 10.1373/clinchem.2014.223677. [DOI] [PubMed] [Google Scholar]
- 15.Thompson BA, Spurdle AB, Plazzer JP, et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat Genet. 2014;46:107–115. doi: 10.1038/ng.2854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shirts BH, Casadei S, Jacobson AL, et al. Improving performance of multigene panels for genomic analysis of cancer predisposition. Genet Med. 2016;18:974–981. doi: 10.1038/gim.2015.212. [DOI] [PubMed] [Google Scholar]
- 17.Beamer LC, Grant ML, Espenschied CR, et al. Reflex immunohistochemistry and microsatellite instability testing of colorectal tumors for Lynch syndrome among US cancer programs and follow-up of abnormal results. J Clin Oncol. 2012;30:1058–1063. doi: 10.1200/JCO.2011.38.4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallego CJ, Perez ML, Burt A, et al. Next generation sequencing in the clinic: A patterns of care study in a retrospective cohort of subjects referred to a genetic medicine clinic for suspected Lynch syndrome. J Genet Couns. 2016;25:515–519. doi: 10.1007/s10897-015-9902-0. [DOI] [PubMed] [Google Scholar]
- 19. doi: 10.1016/j.mayocp.2016.08.008. Ackerman JP, Bartos DC, Kapplinger JD, et al: The promise and peril of precision medicine: Phenotyping still matters most. Mayo Clin Proc. [epub ahead of print on October 8, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tassé AM. The return of results of deceased research participants. J Law Med Ethics. 2011;39:621–630. doi: 10.1111/j.1748-720X.2011.00629.x. [DOI] [PubMed] [Google Scholar]
- 21.Menko FH, Aalfs CM, Henneman L, et al. Informing family members of individuals with Lynch syndrome: A guideline for clinical geneticists. Fam Cancer. 2013;12:319–324. doi: 10.1007/s10689-013-9636-9. [DOI] [PubMed] [Google Scholar]
- 22.Katz LH, Burton-Chase AM, Advani S, et al. Screening adherence and cancer risk perceptions in colorectal cancer survivors with Lynch-like syndrome. Clin Genet. 2016;89:392–398. doi: 10.1111/cge.12653. [DOI] [PMC free article] [PubMed] [Google Scholar]