Abstract
PURPOSE
To evaluate the impact of targeted DNA sequencing on selection of cancer therapy for patients with metastatic breast cancer (MBC).
PATIENTS AND METHODS
In this prospective, single-center, single-arm trial, patients with MBC were enrolled within 10 weeks of starting a new therapy. At enrollment, tumor samples underwent next-generation sequencing for any of 315 cancer-related genes to high depth (> 500×) using FoundationOne CDx. Sequencing results were released to providers at the time of disease progression, and physician treatment recommendations were assessed via questionnaire. We evaluated three prespecified questions to assess patients’ perceptions of genomic testing.
RESULTS
In all, 100 patients underwent genomic testing, with a median of five mutations (range, 0 to 13 mutations) detected per patient. Genomic testing revealed one or more potential therapies in 98% of patients (98 of 100), and 60% of patients (60 of 100) had one or more recommended treatments with level I/II evidence for actionability. Among the 94 genomic text reports that were released, there was physician questionnaire data for 87 patients (response rate, 92.6%) and 31.0% of patients (27 of 87) had treatment change recommended by their physician. Of these, 37.0% (10 of 27) received the treatment supported by genomic testing. We did not detect a statistically significant difference in time-to-treatment failure (log-rank P = .87) or overall survival (P = .71) among patients who had treatment change supported by genomic testing versus those who had no treatment change. For patients who completed surveys before and after genomic testing, there was a significant decrease in confidence of treatment success, specifically among patients who did not have treatment change supported by genomic testing (McNemar’s test of agreement P = .001).
CONCLUSION
In this prospective study, genomic profiling of tumors in patients with MBC frequently identified potential treatments and resulted in treatment change in a minority of patients. Patients whose therapy was not changed on the basis of genomic testing seemed to have a decrease in confidence of treatment success.
INTRODUCTION
Among patients diagnosed with breast cancer, approximately 30% eventually develop metastatic breast cancer (MBC),1 and the estimated 150,000 patients living in the United States with stage IV breast cancer are relatively understudied.2-7 Evaluating multiple somatic mutations in MBC may identify mechanisms of resistance, prognostic genomic biomarkers, or molecular targets to exploit.8-10 Personalized cancer medicine—tailoring medical decisions by integrating clinical features and demographic factors with genetic information—is promising but its value remains unclear for malignancies such as breast cancer, which harbor few truly predictive genomic alterations.8,10,11
Most of our understanding of somatic genomic alterations in breast cancer comes from studies of primary breast cancer,12-14 but our understanding of genomic data on MBC are not nearly as well developed.15 In the largest clinico-genomic analysis of MBC to date, 1,918 tumors prospectively underwent targeted DNA panel sequencing.15 Mutations in frequently altered genes were similar to those in primary cancers, such as TP53 in triple-negative breast cancer (TNBC) and PIK3CA and CDH1 in hormone receptor (HR)–positive, human epidermal growth factor receptor 2 (HER2)–negative patients.15 In HR-positive/HER2-negative patients, the total number of alterations was only moderately higher in MBC tumors compared with primary tumors.15 Recent evaluation of genomic targets in breast cancer suggests there are nine alterations with level I/II evidence for actionability16: ERBB2 amplifications (level IA), germline pathogenic BRCA1 or BRCA2 mutations (IA), PIK3CA hotspot-activating missense mutations (IA), microsatellite instability (IC), NTRK fusions (IC), ESR1 hotspot-activating missense mutations (IIA), PTEN loss (IIA), AKT1 mutations (IIB), and ERBB2 hotspot-activating missense mutations (IIB).16
CONTEXT
Key Objective
DNA sequencing is increasingly incorporated into clinical care for metastatic breast cancer (MBC). How targeted panel DNA sequencing impacts cancer therapy selection and patient perception of care remains unclear. This study provides a prospective analysis of physician decision making and outcomes of patients receiving tumor sequencing as part of MBC clinical care.
Knowledge Generated
Genomic testing revealed that most patients (60%) had at least one recommended treatment with level I/II evidence for actionability; however, only a subset of patients were given a recommendation for a change in treatment, and even fewer actually changed therapy on the basis of genomic testing results. Patients whose therapy did not change on the basis of genomic testing had a decrease in confidence of treatment success.
Relevance
Even though standard of care is improving, relatively few patients with MBC changed therapy on the basis of genomic testing in this prospective study. This suggests a need to better understand barriers to implementation of mutation-directed therapy as well as a need for improved understanding of patient perception of somatic genomic testing.
Although genomic testing is promising for improving treatment efficacy, it is unclear how frequently it changes choice of treatment.17-19 Complex interactions between genomic testing and patient perception of care make it imperative to understand the impact of genetic sequencing in clinical oncology.17,20,21 To date, we have little understanding of how patients perceive the value or accuracy of genomic testing, and whether changes in treatment recommendation based on genomic testing affect patients’ perceptions of care. There are discrepancies between cancer patient and physician expectations of therapy efficacy; patients demonstrate greater optimism about therapy efficacy.22-25 Expectations of patients with MBC and their motivations for undergoing somatic genetic testing have not yet been explored.
Clinical tumor genomic analyses typically rely on targeted next-generation sequencing (NGS) of a panel of specific, typically actionable cancer-related genes to analyze somatic gene alterations from biopsy specimens. Most commercial targeted panel sequencing approaches, including FoundationOne CDx (Foundation Medicine, Cambridge, MA) in this study, provide treatment suggestions that are based on genomic alterations and available clinical literature.26 We hypothesized that prospective implementation of FoundationOne CDx testing into clinical care for patients with MBC will identify patient-specific approaches that offer improved patient outcomes and perceptions of care. In this study, patients with MBC received FoundationOne CDx testing when a new treatment was initiated and genomic testing results released to the provider at the next progression event. The primary objectives of this study were to assess the proportion of patients whose subsequent cancer-related therapy was based on the FoundationOne CDx test results (FoundationOne CDx-supported treatment change), to evaluate time-to-treatment failure (TTF) and overall survival (OS) in patients whose therapy was genomically directed versus not, and to assess patient perceptions of genomic testing.
PATIENTS AND METHODS
Study Population
This was a prospective, single-site, single-arm trial at a National Cancer Institute–designated cancer center. Patients with MBC who were within 10 weeks of starting their current line of therapy and who had an estimated survival of 3 or more months were included in this study. Included patients needed to have a tumor sample (primary or metastatic) available for genomic testing. The study was approved by The Ohio State University Institutional Review Board and informed consent was obtained from all patients.
NGS
Testing was performed by using FoundationOne CDx, which used NGS on patients’ formalin-fixed paraffin-embedded tumor tissue. The test provides information on alterations in 315 cancer-related genes and 28 selected rearrangements (Data Supplement). The selected genes sequenced by FoundationOne CDx have implications in cancer biology, prognosis, and availability of therapy that targets gene products. Genes with more than one alteration were counted once. The FoundationOne CDx report, which is based on detected genomic alterations, includes potential treatment options, such as therapies approved to treat specific tumor types, therapies approved in another tumor type, or therapies evaluated in a clinical trial that target a specific alteration. Patients were observed until disease progression or intolerance of therapy. The FoundationOne CDx results were then released to the provider.
Survey Measures
Physicians received a five-item questionnaire after the FoundationOne CDx report was released to assess whether their treatment recommendation should be changed on the basis of those results. The questionnaire asked how the physician used the FoundationOne CDx test (multiple choice) and what else they would like to see in the report (multiple choice plus an open-ended “other” option). Patients completed a pretest (before genomic testing) questionnaire at study entry and a post-test questionnaire at the end of the study. Attitudes toward FoundationOne CDx testing were assessed with three items: (1) “By having the test, I will feel more confident of my treatment’s success”, (2) “I will trust the test results”, and (3) “I think the test results will be accurate”. Patient motivation and expectations for study participation were assessed with 15 questions using a five-point Likert scale that ranged from strongly disagree to strongly agree (Data Supplement). Because of small numbers in each group, these questions were assessed as a three-level outcome (agree v neutral v disagree) and as a binary outcome (agree v neutral and disagree).
Statistical and Survival Analyses
McNemar’s test of agreement (2 level or 3 level) was used to compare pre- and post-test survey responses and pre- versus post-test survey agreement by FoundationOne CDx–supported treatment change results from the physician questionnaire. We used McNemar’s nonparametric test because of the limited sample size and non-normal data. TTF was defined as the time from the release of the FoundationOne CDx report to the off-study date. The off-study date was defined as the date of the patient’s second progression (first while on study), second treatment change (first while on study), death, or loss to follow-up. Patients for whom the off-study date was missing were excluded from the TTF analysis. OS was defined as time from the date of study enrollment to death or loss to follow-up. Patients still alive on December 18, 2018, were censored as of this date. All statistical analyses were performed using SAS version 9.4 (SAS Institute, Cary, NC) and R version 3.4.1, and survival curves were created using the packHV package.27
RESULTS
Study Population
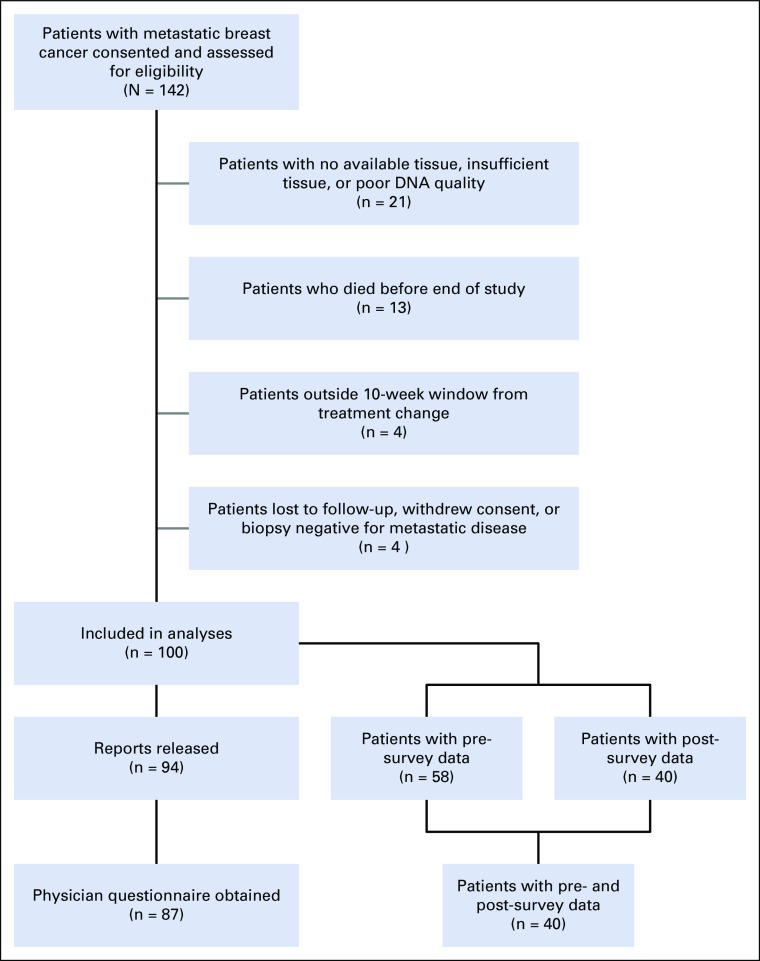
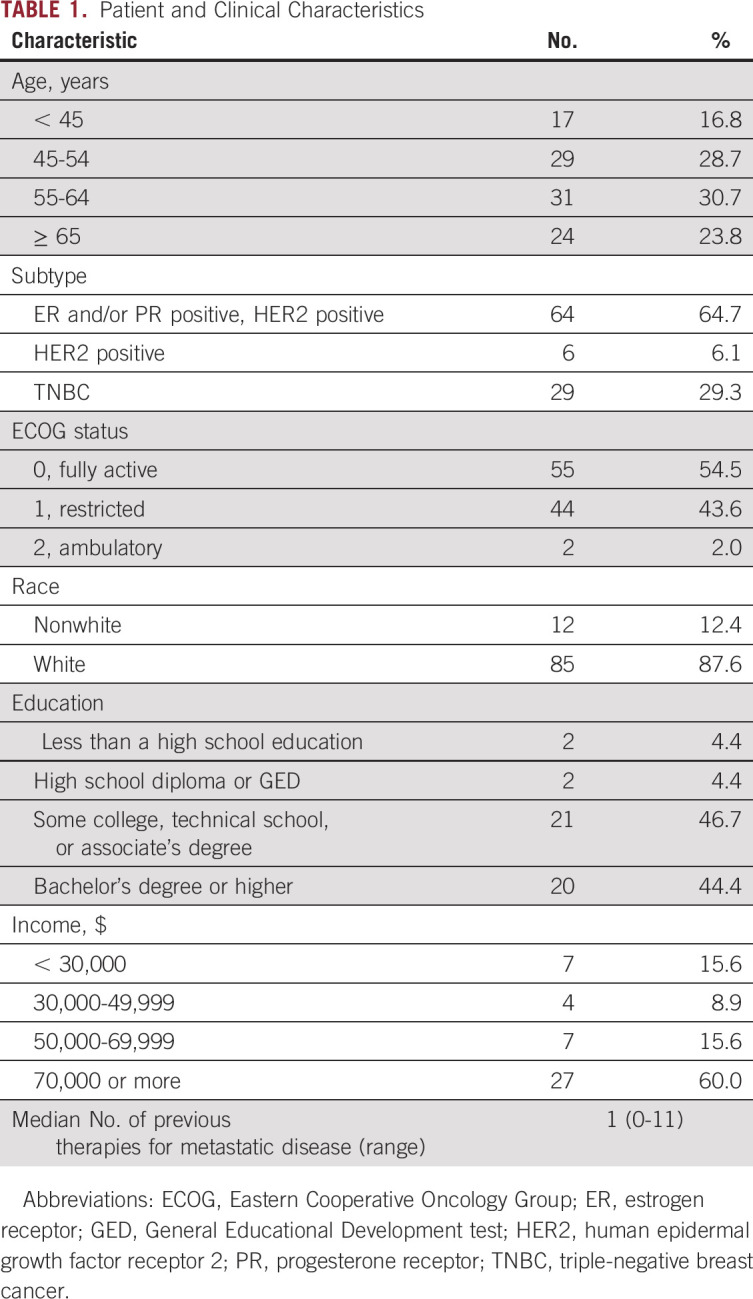
A total of 142 patients with MBC provided consent and were assessed for eligibility (Fig 1). Patients were excluded from analysis if there was no available tissue, insufficient tissue, or poor DNA quality (n = 21), died before the FoundationOne CDx report was released (n = 13), did not receive FoundationOne CDx testing within 10 weeks of starting their current line of therapy (n = 4), were lost to follow-up (n = 2), withdrew consent (n = 1), or had a biopsy that revealed no metastatic disease (n = 1). A total of 100 patients had successful FoundationOne CDx NGS testing and were evaluable for analysis. Of these samples, 71% (71 of 100) were metastatic biopsies and 29% (29 of 100) were primary tumors (breast or axillary lymph node). The most common sites of metastatic biopsy were liver (n = 24), bone (n = 12), and skin/chest wall (n = 10). Most patients’ tumors (63%) were positive for estrogen receptor (ER) and/or progesterone receptor (PR) and negative for HER2. Genomic testing was performed from September 2013 through October 2015, and FoundationOne CDx reports were released to treating physicians starting in December 2013. The study population was predominantly age 55 years or older, white, and college-educated with a high income (Table 1).
FIG 1.
CONSORT diagram.
TABLE 1.
Patient and Clinical Characteristics
FoundationOne CDx Genomic Testing Results
Among the 100 patients with successful FoundationOne CDx NGS testing, the number of mutations identified per patient ranged from 0 to 13 with a median of five mutations (Data Supplement). As anticipated, the most common mutations were missense or frameshift mutations in TP53 (n = 49) and PIK3CA (n = 40) or amplification of MYC (n = 20; Fig 2A). Mutations in TP53 were most common among patients with TNBC. Among ER-positive/HER2-negative patients, 10 (16.7%) of 63 harbored an ESR1 mutation.
FIG 2.
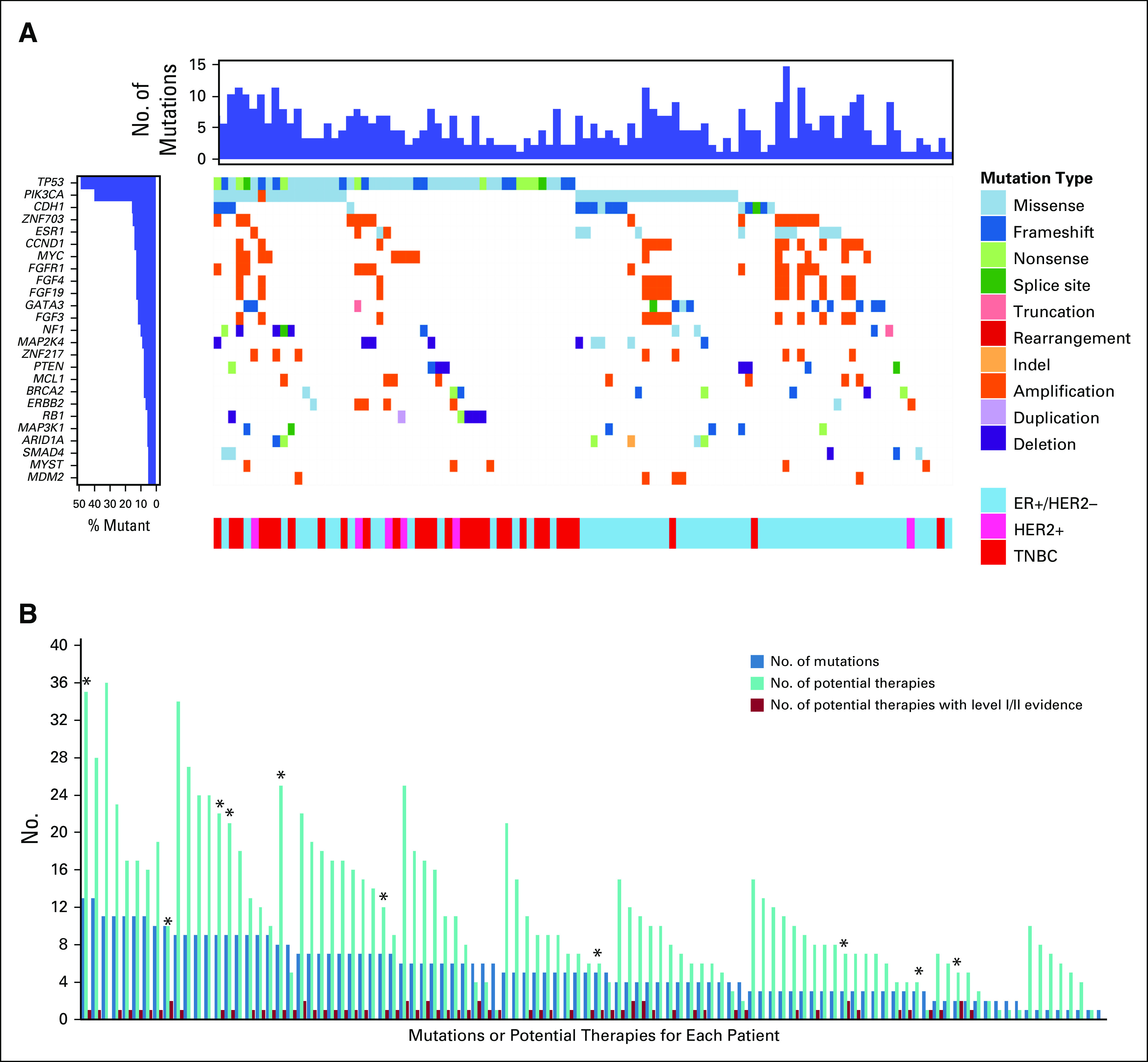
Genomic alterations and therapeutic implications. (A) Patient-level (n = 100) mutation plot of the 25 most frequently altered genes in the study population. Left: frequency of the alteration in each gene; top: number of detected mutations per patient; bottom: receptor subtype. CoMut plot created by using the GenVisR package.47 (B) For each patient (n = 100), the graph shows number of detectable mutations, number of potential therapies identified, and number of potential therapies with level I/II evidence for actionability in breast cancer based on Condorelli, et al.16 (*) Indicates patients whose treatment was changed based on genomic testing results.
In terms of therapy recommendations, 98 (98.0%) of 100 patients had at least one potential therapy identified on FoundationOne CDx testing, with a median of nine potential treatments (range, 0 to 35 potential treatment; Fig 2B). The most commonly recommended therapy based on a genomic alteration was an mTOR inhibitor (everolimus or temsirolimus) for PIK3CA mutations (Data Supplement). Among all patients, 60 (60%) of 100 had at least one recommended treatment with level I/II evidence (not including ERBB2 amplifications),16 with a median of one potential treatment with level I/II evidence (range, 0 to 2 potential treatments; Fig 2B).
Potential Germline Alterations in Somatic Tumor Testing
A known challenge in somatic tumor sequencing is potential identification of germline alterations.28-32 Among the 100 total patients, 14 had alterations identified in BRCA1 (n = 4), BRCA2 (n = 8), or PALB2 (n = 2). Of these, eight (57.1%) of 14 had previous knowledge of germline alterations but six (42.8%) of 14 did not. Of patients with no prior knowledge of germline alterations, five of six had no first-degree relatives with breast or ovarian cancer (Data Supplement). Of these six patients with no prior knowledge of germline mutations, two had germline alterations confirmed (one BRCA1 and one BRCA2) and one underwent germline testing in which no alterations were identified (presumed somatic BRCA2 mutation). Three patients did not undergo germline genetic testing: two patients were notified of FoundationOne CDx results and received a referral for genetic testing; one patient died before release of the FoundationOne CDx results. After consultation with the medical ethics committee, that patient’s family was notified of the results.
Clinical Importance of NGS Results
Of the 100 evaluable patients, 94 had FoundationOne CDx reports released, and 87 had corresponding physician questionnaire data (physician response rate, 92.6%). The six patients whose FoundationOne CDx reports were not released remained on their initial therapy without progression as of September 1, 2018; five of these patients had ER-positive/HER2-negative and one had ER-positive/HER2-positive breast cancer. Of the 87 patients with physician data, 27 (31.0%) were given a recommendation to change treatment as a result of the FoundationOne CDx results. Of these, 10 (37.0%) of 27 underwent this recommended treatment change. Across the study population, 10 (10.0%) of 100 total patients or 10 (11.5%) of 87 patients with physician questionnaire data experienced a treatment change supported by FoundationOne CDx data. Physicians described not changing treatment because the patient could not be enrolled on a clinical trial (ie, the recommended trial closed, the patient did not want to enroll, or the patient was ineligible; n = 10), patient hospitalization (n = 3), insurance denied treatment charges (n = 2), patient transferred care to other provider (n = 1), or patient pursued a nonrecommended trial (n = 1; Data Supplement).
Genomic Alterations and Survival
Among 76 patients evaluable for TTF analysis, there was no significant difference in TTF (log-rank P = .87) by FoundationOne CDx-supported treatment change status (Fig 3A). Among the 87 patients evaluable for OS analysis, the median follow-up time was 14.3 months; there was no significant difference in OS (log-rank P = .71) by FoundationOne CDx-supported treatment change status (Fig 3B). By subtype, there were significant differences in OS (log-rank P = .01); patients who had TNBC had worse OS than ER-positive/HER2-negative patients, as anticipated (Data Supplement).
FIG 3.
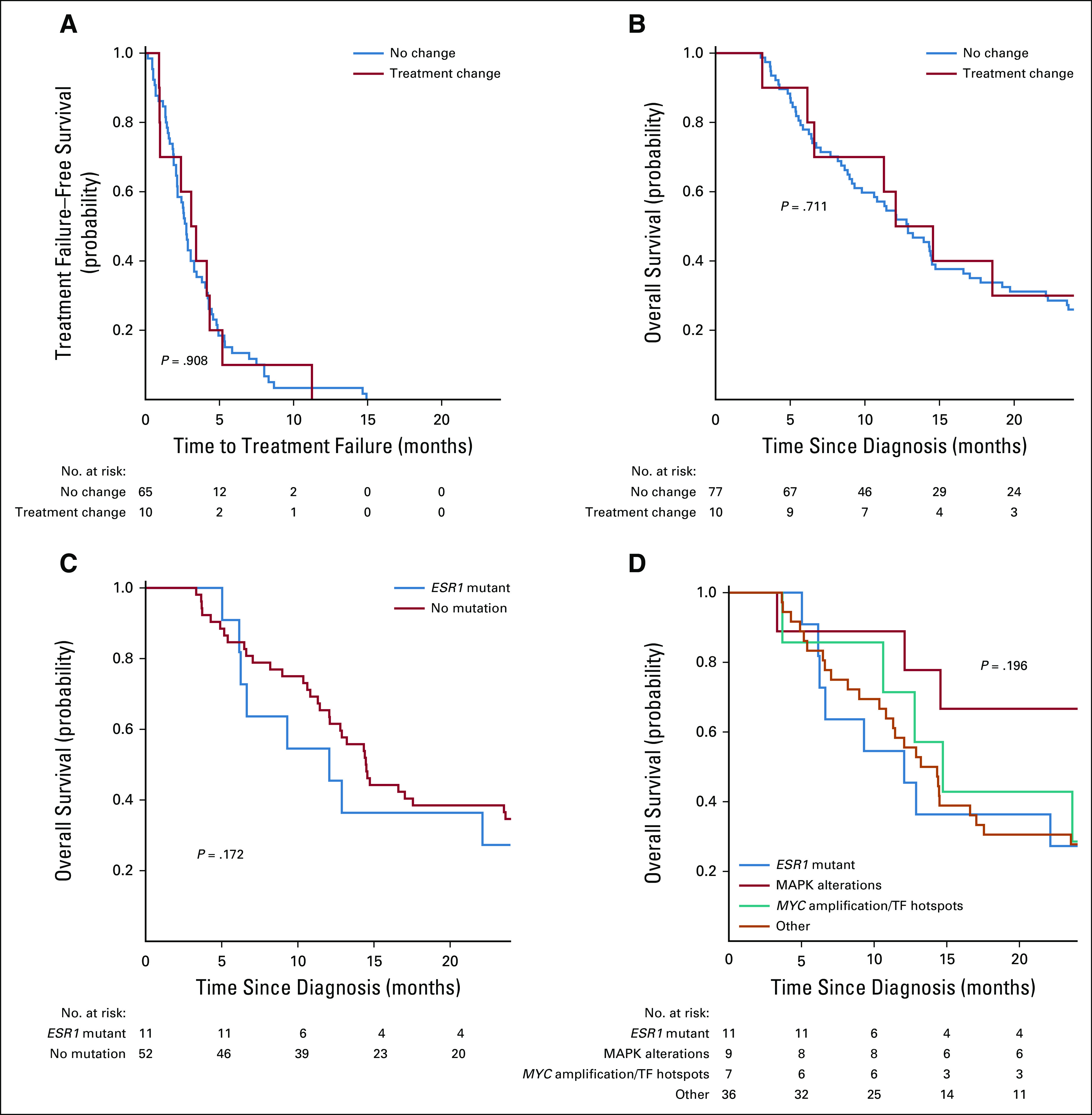
Cohort survival analyses by receptor and genomic features. Kaplan-Meier plots with log-rank test P value. (A) Kaplan-Meier plot of time to treatment failure stratified by patients with treatment change based on genomic testing (red line) v no treatment change. Time to treatment failure defined as time from release of FoundationOne CDx results to the off-study date, defined as date of second progression or treatment change (first during the study period), death, or loss to follow-up. (B) Kaplan-Meier plot of overall survival by receptor status. Overall survival is defined as the period of time from the date of study enrollment to death or loss to follow-up. Patients who were still alive on December 18, 2018, were censored on this date. (C) Kaplan-Meier plot of overall survival for patients with estrogen receptor (ER)–positive/human epidermal growth factor receptor 2 (HER2)–negative metastatic breast cancer, stratified by presence or absence of mutation in ESR1. (D) Kaplan-Meier plot of overall survival for patients with ER-positive/HER2-negative metastatic breast cancer stratified by presence or absence of ESR1 mutation, MAPK pathway alteration, MYC amplification, transcription factor (TF) alteration, or other, as defined by Razavi, et al.15
Among genomic alterations, we evaluated several mutations known to be prognostic (ESR1 mutations) or potentially predictive of outcomes (PIK3CA). In MSK-IMPACT (a targeted tumor-sequencing assay developed at Memorial Sloan Kettering Cancer Center [MSK] termed “Integrated Mutation Profiling of Actionable Cancer Targets” [IMPACT]), there was evidence that patient response to aromatase inhibitor therapy was significantly worse for patients whose tumors harbored one of several alterations.15 Among ER-positive/HER2-negative patients, we noted that the presence of an ESR1 mutation was not significantly associated with OS (Fig 3C). Despite a high frequency of mutation, there was no significant difference in OS by PIK3CA mutation status among ER-positive/HER2-negative patients (P = .18) or TNBC patients (P = .66), or when evaluating alterations that could activate the PI3K pathway (PIK3CA mutation, PTEN loss, or AKT1 mutation) among ER-positive/HER2-negative patients (P = .60) or TNBC (P = .81) patients (Data Supplement). In the MSK-IMPACT study (ClinicalTrials.gov identifier: NCT01775072), HR-positive/HER2-negative patients with detected tumor mutations in ESR1, MAP kinase pathway, or MYC or other transcription factors genes had a worse response to aromatase inhibitor therapy than patients without these alterations.15 We evaluated the association of these alterations with OS and found no significant difference among the four groups (Fig 3D).
Patients’ Perceptions of Genetic Testing
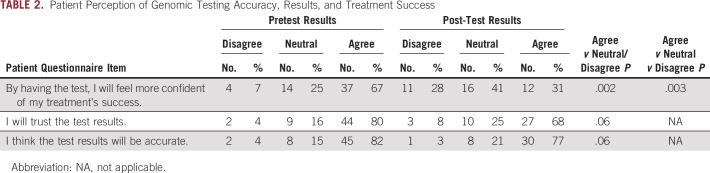
A total of 58 (58.0%) of the 100 evaluable patients completed at least a portion of the survey at enrollment (pretest), and 40 (40.0%) completed at least a portion of the survey at study conclusion (post-test). We evaluated three prespecified questions to assess patients’ perceptions of treatment success, trust of genomic testing results, and accuracy of the results (Table 2). In the pretest survey, most patients strongly agreed or somewhat agreed that by having the genetic testing they would feel more confident in their treatment’s success (36 [65.5%] of 55), whereas at study completion, a minority of patients (12 [30.8%] of 39) felt more confident in their treatment’s success by having the genetic testing. We used a paired test among patients who completed both the pretest and post-test and found a statistically significant decrease in confidence for both 3-level (agree v neutral v disagree; McNemar’s test of agreement P = .003) and 2-level (agree v neutral/disagree; McNemar’s test of agreement P = .002) analyses. There was no significant difference in patients’ trust of test results or thinking the results were accurate (McNemar’s P = .06 for both questions).
TABLE 2.
Patient Perception of Genomic Testing Accuracy, Results, and Treatment Success
We then assessed whether pretest and post-test survey results were associated with treatment change on the basis of FoundationOne CDx results in patients who completed both the pretest and post-test survey questions of interest. For treatment confidence, we found that patients who did not have a treatment change supported by FoundationOne CDx data were significantly more likely to change their response from agree to neutral or disagree (McNemar’s test of agreement P = .001; Fig 4). There was no significant change in perceptions of the group who had treatment change supported by FoundationOne CDx data (McNemar’s test of agreement P = .32).
FIG 4.
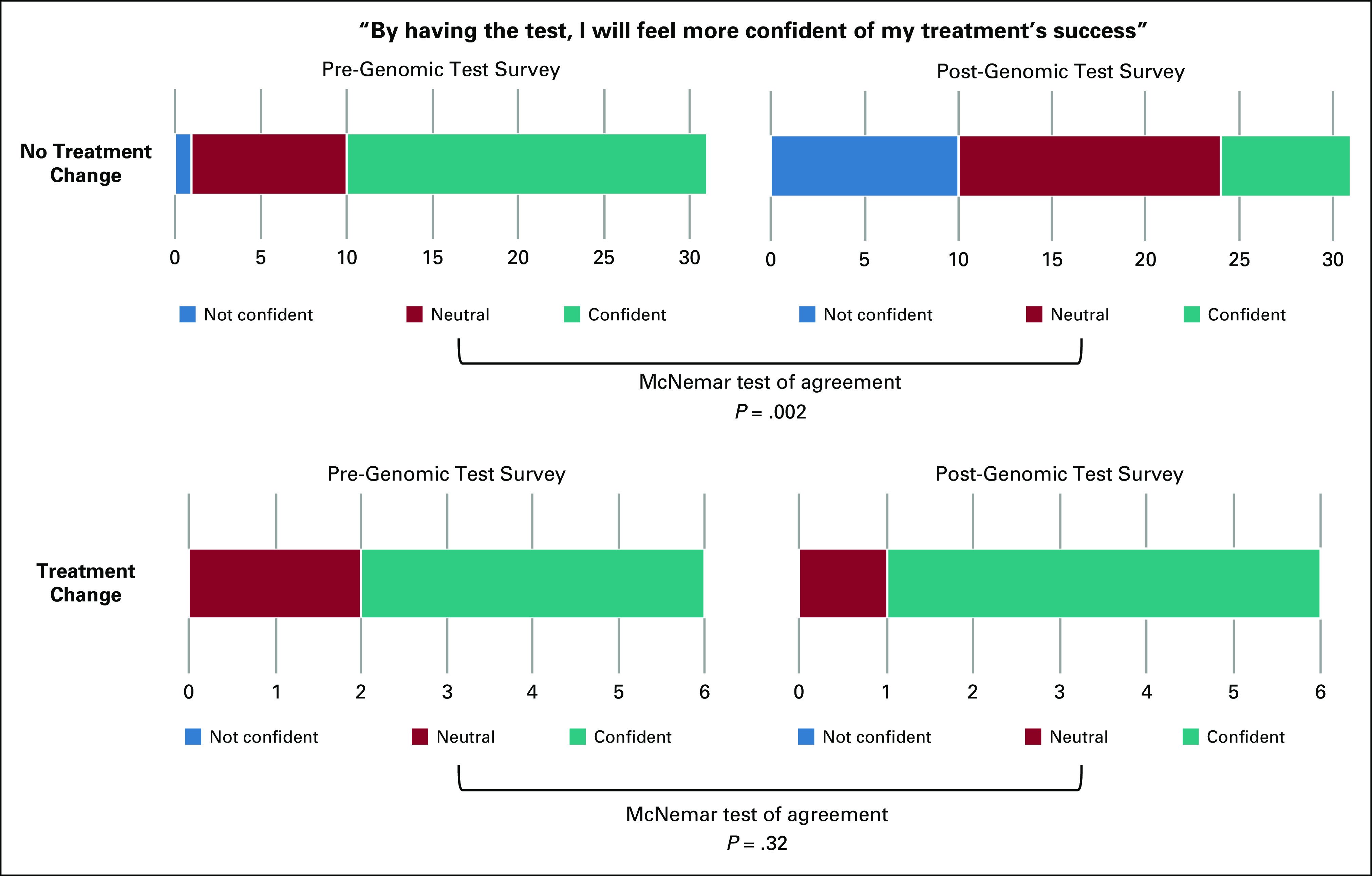
Patient confidence in treatment success before and after genomic testing. Enrolled patients were surveyed regarding their perception of genomic testing: “By having the test, I will feel more confident of my treatment’s success.” A total of 37 patients answered the question both before (pre-genomic test survey) and after (post-genomic test survey) genomic test results were shared with physicians. Number of patients who were not confident, neutral, or confident are indicated.
To understand patients’ motivations and expectations, we assessed 15 questions from the survey administered before treatment (Data Supplement). A total of 58 patients completed most or all of these questions. Most patients (61.8%) did not believe the FoundationOne CDx test would have a negative impact on their family. Most patients (80.0%) believed the FoundationOne CDx test would give them a better understanding of the chance of success for their treatment option, and 59.3% believed the test would tell them about their genetic future. About half the patients (49.1%) believed the test would tell them what medications to take. Most patients (67.9%) believed the test results would tell them their children’s risk of disease. Most patients (91.1%) believed that their participation would help researchers. To evaluate potential nonresponse bias, given the response rates, we compared patients who completed the pretest survey with those who did not. We found no significant differences between these two groups in age, cancer subtype, Eastern Cooperative Oncology Group status, race, and number of genetic mutations. Similarly, we did not find significant differences in these demographics between patients who completed both surveys and those who completed only the presurvey, suggesting that nonresponse bias is minimal.
DISCUSSION
NGS is increasingly common in MBC care and has significant potential value for physicians and patients when making cancer treatment decisions.33-35 Despite this evidence, the role of NGS for patients with MBC remains fluid, and patient perception, motivation, and expectations are poorly understood. We prospectively evaluated the impact of NGS on MBC treatment decisions and potential associations with patient outcomes and their perceptions of genomic testing.
In this study, more than 60% of patients had a genomic alteration associated with level I or II evidence for actionability (not including HER2 amplification) based on a recent consensus panel16—an impressive number that emphasizes the potential for widespread use of NGS. The 31% of evaluable patients in this prospective study for whom a change in treatment was recommended on the basis of somatic NGS findings is similar to the frequency seen in large retrospective cohorts.15 Of the patients for whom a different treatment was recommended by their provider on the basis of the FoundationOne CDx results, only 37% (11.5% of total) proceeded with the recommendation to change treatment, which implies that there are significant barriers to pursuing NGS-supported treatments. This is consistent with a previous study that identified barriers such as trial ineligibility, distance to trial, and physical/emotional exhaustion.18 Treatment change was not significantly associated with TTF or OS; however, the small number of patients experiencing treatment change limits the conclusions that can be drawn from these analyses.
This is the first study to our knowledge to prospectively evaluate the influence of somatic NGS on patient perception of care and it provides important initial data, but the lower patient survey response rate limits definitive conclusions. When comparing study surveys at admission to surveys at conclusion, patients seemed less confident in their treatment, particularly those whose treatment was not changed by FoundationOne CDx results. This is consistent with other studies that suggest genetic testing may increase negative emotions in patients with metastatic cancer,18 although most studies evaluated effects from germline testing.36-40 Our data preliminarily suggests that there may be ramifications of somatic genomic testing for patients despite its known utility in selecting therapy for MBC. Because negative emotions are associated with decreased quality of life and potentially with survival,41-44 the possible negative implications of broadly integrating NGS into clinical practice should be considered.
Furthermore, pretest survey data suggest that patients have misconceptions about what somatic mutation testing can inform. We found that patients had overly optimistic expectations for somatic tumor NGS: almost half believed the test would tell them which medication to take whereas only 11.5% of the total patients switched therapy on the basis of FoundationOne CDx reports. This is similar to previous studies that demonstrate that patients have greater optimism regarding treatment efficacy than physicians do,24 possibly because physicians provide patients with more optimistic information than their true assessment.45
In addition, most patients incorrectly believed the test would predict future genetic mutations, assess children’s disease risk, or estimate treatment success, despite the informed consent stating that participants would rarely benefit personally or therapeutically from this study. Therefore, this supports further exploration of the readability of consent forms, an area currently under investigation.46 We hypothesize that patients’ limited knowledge of genetics may explain their decreased confidence after genomic testing,17-19 and we also hypothesize that improved decision support for genomic testing for patients will improve confidence in their treatment, particularly in the context of increases in available targeted treatments for MBC.
There are known limitations to our study that prevent us from making definitive conclusions. Among survey data, we had a high physician survey response rate (> 90%) but a substantially lower response rate among evaluable participants: the response rate was 58% (58 of 100) for the pretest and 40% (40 of 100) for both pretest and post-test. Despite this, our findings provide preliminary insight into critical aspects of MBC care that warrant evaluation in a larger study. Furthermore, unlike the methodologies in the existing literature, we used a unique prospective and longitudinal methodology to evaluate patient perceptions. Future studies may benefit by including patient interviews to assess the experience of somatic NGS. The use of nonvalidated survey items and single site design in our study also support the need for further research. In addition, among all consented patients, 14.8% had no available tissue, inadequate tissue, or inadequate DNA quality. This is a realistic barrier to NGS that is infrequently captured in retrospective analyses. Furthermore, the potential treatments identified by FoundationOne CDx test results reflect drugs approved for any indication, not necessarily for MBC, and therapies available at the time of the test. For example, both everolimus and temsirolimus were reported as potential therapies for patients with alterations in NF1, PTEN, AK1, PIK3R1, RPTOR, AKT3, FBXW7, KIT, PDGFRA, STK11, and VHL; however, only everolimus is approved by the US Food and Drug Administration for MBC. In addition, since this study was completed, multiple new therapies approved by the US Food and Drug Administration have not been considered, suggesting that clinical implications of NGS should be assessed regularly.
In conclusion, our findings suggest that NGS by tools such as FoundationOne CDx may provide valuable insight for physicians and patients when determining the best treatment option for patients with MBC upon disease progression. There are clear clinical challenges involved with integrating NGS into the framework of MBC care, including barriers to preferred treatment options and complex interactions between genomic testing and patient perceptions, which warrant further study.
Footnotes
Presented in part at the Annual San Antonio Breast Cancer Symposium, San Antonio, TX, December 9-13, 2014, and the Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 3-7, 2016.
Supported by grants from Pelotonia (D.G.S, M.B.L.), the Stephanie Spielman Fund (D.G.S.), and Foundation Medicine.
ClinicalTrials.gov identifier: NCT01987726
AUTHOR CONTRIBUTIONS
Conception and design: Daniel G. Stover, Katlyn Tolliver, Cynthia D. Timmers, Sagar Sardesai, Erin R. Macrae, Bhuvaneswari Ramaswamy, Maryam B. Lustberg
Administrative support: Susan Gillespie, Maryam B. Lustberg
Provision of study materials or patients: Mathew A. Cherian, Anne M. Noonan, Sagar Sardesai, Robert Wesolowski, Charles L. Shapiro, Bhuvaneswari Ramaswamy, Maryam B. Lustberg
Collection and assembly of data: Daniel G. Stover, Raquel E. Reinbolt, Katlyn Tolliver, Mahmoud Abdel-Rasoul, Cynthia D. Timmers, Susan Gillespie, Mathew A. Cherian, Anne M. Noonan, Jeffrey VanDeusen, Robert Wesolowski, Nicole Williams, Charles L. Shapiro, Erin R. Macrae, Maryam B. Lustberg
Data analysis and interpretation: Daniel G. Stover, Raquel E. Reinbolt, Elizabeth J. Adams, Sarah Asad, Katlyn Tolliver, Mahmoud Abdel-Rasoul, Cynthia D. Timmers, James L. Chen, Siraj Mahamed Ali, Katharine A. Collier, Robert Wesolowski, Clara N. Lee, Erin R. Macrae, Bhuvaneswari Ramaswamy, Maryam B. Lustberg
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Cynthia D. Timmers
Stock and Other Ownership Interests: Array BioPharma, Seattle Genetics
James L. Chen
Consulting or Advisory Role: Novartis, Immune Design, Syapse
Speakers' Bureau: Novartis, Foundation Medicine
Research Funding: Eisai
Patents, Royalties, Other Intellectual Property: MatchTX, a genomics software package that helps researchers, oncologists, and clinical trial managers identify the full set of biomarkers that collectively predict the outcome of patients with cancer to treatment
Siraj Mahamed Ali
Employment: Foundation Medicine
Leadership: Incysus
Stock and Other Ownership Interests: Exelixis, Blueprint Medicines, Agios, Genocea Biosciences
Consulting or Advisory Role: Revolution Medicines, Azitra (I), Princeps Tx (I)
Patents, Royalties, Other Intellectual Property: Patents via Foundation Medicine and via Seres Health on microbiome in non-neoplastic disease (I)
Anne M. Noonan
Consulting or Advisory Role: Helsinn Healthcare, QED Therapeutics
Sagar Sardesai
Consulting or Advisory Role: Novartis
Speakers' Bureau: Immunomedics
Travel, Accommodations, Expenses: Novartis
Jeffrey VanDeusen
Stock and Other Ownership Interests: Immunomedics
Robert Wesolowski
Consulting or Advisory Role: Pfizer (Inst), Puma Biotechnology
Research Funding: Acerta Pharma, AstraZeneca
Travel, Accommodations, Expenses: Pfizer, Puma BiotechnologyBhuvaneswari Ramaswamy
Consulting or Advisory Role: Pfizer
Maryam B. Lustberg
Consulting or Advisory Role: Tempus, PledPharma
Other Relationship: Hologic/Cynosure
No other potential conflicts of interest were reported.
REFERENCES
- 1.O’Shaughnessy J. Extending survival with chemotherapy in metastatic breast cancer. Oncologist. 2005;10:20–29. doi: 10.1634/theoncologist.10-90003-20. [DOI] [PubMed] [Google Scholar]
- 2.Mayer M, Grober SE. Silent voices: Women with advanced (metastatic) breast cancer share their needs and preferences for information, support and practice resources. Living Beyond Breast Cancer; 2006. https://www.lbbc.org/sites/default/files/LBBCsilentvoices.pdf [Google Scholar]
- 3.Centers for Disease Control and Prevention https://www.cdc.gov/cancer/uscs/dataviz/download_data.htm United States Cancer Statistics. Based on November 2017 submission data (1999-2015).
- 4.Chu KC, Tarone RE, Kessler LG, et al. Recent trends in U.S. breast cancer incidence, survival, and mortality rates. J Natl Cancer Inst. 1996;88:1571–1579. doi: 10.1093/jnci/88.21.1571. [DOI] [PubMed] [Google Scholar]
- 5.Parker SL, Tong T, Bolden S, et al. Cancer statistics, 1997. CA Cancer J Clin. 1997;47:5–27. doi: 10.3322/canjclin.47.1.5. [DOI] [PubMed] [Google Scholar]
- 6.MBC Alliance Changing the Landscape for People Living With Metastatic Breast Cancer. 2014 https://www.mbcalliance.org/research/landscape-analysis Metastatic Breast Cancer Landscape Analysis: Research Report 2.
- 7.Mariotto AB, Etzioni R, Hurlbert M, et al. Estimation of the number of women living with metastatic breast cancer in the United States. Cancer Epidemiol Biomarkers Prev. 2017;26:809–815. doi: 10.1158/1055-9965.EPI-16-0889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin L, Andersen JN, Futreal PA. Cancer genomics: From discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- 9.Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonzalez-Garay ML. The road from next-generation sequencing to personalized medicine. Per Med. 2014;11:523–544. doi: 10.2217/pme.14.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vogenberg FR, Isaacson Barash C, Pursel M. Personalized medicine: Part 1. Evolution and development into theranostics. P T. 2010;35:560–576. [PMC free article] [PubMed] [Google Scholar]
- 12.Pereira B, Chin SF, Rueda OM, et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat Commun. 2016;7:11479. doi: 10.1038/ncomms11479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nik-Zainal S, Davies H, Staaf J, et al. Landscape of somatic mutations in 560 breast cancer whole-genome sequences. Nature. 2016;534:47–54. doi: 10.1038/nature17676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banerji S, Cibulskis K, Rangel-Escareno C, et al. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature. 2012;486:405–409. doi: 10.1038/nature11154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Razavi P, Chang MT, Xu G, et al. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34:427–438.e6. doi: 10.1016/j.ccell.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condorelli R, Mosele F, Verret B, et al. Genomic alterations in breast cancer: Level of evidence for actionability according to ESMO Scale for Clinical Actionability of Molecular Targets (ESCAT) Ann Oncol. 2019;30:365–373. doi: 10.1093/annonc/mdz036. [DOI] [PubMed] [Google Scholar]
- 17.Gray SW, Park ER, Najita J, et al. Oncologists’ and cancer patients’ views on whole-exome sequencing and incidental findings: Results from the CanSeq study. Genet Med. 2016;18:1011–1019. doi: 10.1038/gim.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miller FA, Hayeems RZ, Bytautas JP, et al. Testing personalized medicine: Patient and physician expectations of next-generation genomic sequencing in late-stage cancer care. Eur J Hum Genet. 2014;22:391–395. doi: 10.1038/ejhg.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heshka JT, Palleschi C, Howley H, et al. A systematic review of perceived risks, psychological and behavioral impacts of genetic testing. Genet Med. 2008;10:19–32. doi: 10.1097/GIM.0b013e31815f524f. [DOI] [PubMed] [Google Scholar]
- 20.Gollust SE, Gray SW, Carere DA, et al. Consumer perspectives on access to direct-to-consumer genetic testing: Role of demographic factors and the testing experience. Milbank Q. 2017;95:291–318. doi: 10.1111/1468-0009.12262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gray SW, Gagan J, Cerami E, et al. Interactive or static reports to guide clinical interpretation of cancer genomics. J Am Med Inform Assoc. 2018;25:458–464. doi: 10.1093/jamia/ocx150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee SJ, Fairclough D, Antin JH, et al. Discrepancies between patient and physician estimates for the success of stem cell transplantation. JAMA. 2001;285:1034–1038. doi: 10.1001/jama.285.8.1034. [DOI] [PubMed] [Google Scholar]
- 23.Weeks JC, Catalano PJ, Cronin A, et al. Patients’ expectations about effects of chemotherapy for advanced cancer. N Engl J Med. 2012;367:1616–1625. doi: 10.1056/NEJMoa1204410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meropol NJ, Weinfurt KP, Burnett CB, et al. Perceptions of patients and physicians regarding phase I cancer clinical trials: Implications for physician-patient communication. J Clin Oncol. 2003;21:2589–2596. doi: 10.1200/JCO.2003.10.072. [DOI] [PubMed] [Google Scholar]
- 25.Lux MP, Bayer CM, Loehberg CR, et al. Shared decision-making in metastatic breast cancer: Discrepancy between the expected prolongation of life and treatment efficacy between patients and physicians, and influencing factors. Breast Cancer Res Treat. 2013;139:429–440. doi: 10.1007/s10549-013-2557-3. [DOI] [PubMed] [Google Scholar]
- 26.Foundation Medicine FoundationOne CDX. 2018 https://www.foundationmedicine.com/genomic-testing/foundation-one-cdx?gclid=Cj0KCQjw9fntBRCGARIsAGjFq5G3gjYuLVvGLfNSasiVl5PmSS3_cl366NsQVNT2nlkKRuLADSIUoCQaAsJLEALw_wcB
- 27.Booth FV, Hassett JM. Staff communications and credentialing in a multisite institution. Ann N Y Acad Sci. 1992;670:211–214. doi: 10.1111/j.1749-6632.1992.tb26092.x. [DOI] [PubMed] [Google Scholar]
- 28.Meric-Bernstam F, Brusco L, Daniels M, et al. Incidental germline variants in 1000 advanced cancers on a prospective somatic genomic profiling protocol. Ann Oncol. 2016;27:795–800. doi: 10.1093/annonc/mdw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngeow J, Eng C. Precision medicine in heritable cancer: When somatic tumour testing and germline mutations meet. NPJ Genom Med. 2016;1:15006. doi: 10.1038/npjgenmed.2015.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsons DW, Roy A, Plon SE, et al. Clinical tumor sequencing: An incidental casualty of the American College of Medical Genetics and Genomics recommendations for reporting of incidental findings. J Clin Oncol. 2014;32:2203–2205. doi: 10.1200/JCO.2013.54.8917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Funchain P, Sohal D, Khorana AA, et al. Hereditary implications of somatic tumor testing. J Clin Oncol. 2015;33 doi: 10.1200/jco.2015.33.15_suppl.1523. (suppl; asbstr 1523) [DOI] [Google Scholar]
- 32.Barton LV, Walko CM, Pal T, et al. Potential germline relevance of tumor testing and the need for genetic referral. J Clin Oncol. 2015;33 doi: 10.1200/jco.2015.33.15_suppl.e12552. (suppl; abstr e12552) [DOI] [Google Scholar]
- 33.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A hybridization capture-based next-generation sequencing clinical assay for solid tumor molecular oncology. J Mol Diagn. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shibuya Y, Tokunaga H, Saito S, et al. Identification of somatic genetic alterations in ovarian clear cell carcinoma with next generation sequencing. Genes Chromosomes Cancer. 2018;57:51–60. doi: 10.1002/gcc.22507. [DOI] [PubMed] [Google Scholar]
- 35.Tanioka M, Fan C, Parker JS, et al. Integrated analysis of RNA and DNA from the phase III trial CALGB 40601 identifies predictors of response to trastuzumab-based neoadjuvant chemotherapy in HER2-positive breast cancer. Clin Cancer Res. 2018;24:5292–5304. doi: 10.1158/1078-0432.CCR-17-3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bakos AD, Hutson SP, Loud JT, et al. BRCA mutation-negative women from hereditary breast and ovarian cancer families: A qualitative study of the BRCA-negative experience. Health Expect. 2008;11:220–231. doi: 10.1111/j.1369-7625.2008.00494.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay JL, Meischke HW, Bowen DJ, et al. Anticipating dissemination of cancer genomics in public health: A theoretical approach to psychosocial and behavioral challenges. Ann Behav Med. 2007;34:275–286. doi: 10.1007/BF02874552. [DOI] [PubMed] [Google Scholar]
- 38.Scheuner MT, Sieverding P, Shekelle PG. Delivery of genomic medicine for common chronic adult diseases: A systematic review. JAMA. 2008;299:1320–1334. doi: 10.1001/jama.299.11.1320. [DOI] [PubMed] [Google Scholar]
- 39.Collins VR, Meiser B, Ukoumunne OC, et al. The impact of predictive genetic testing for hereditary nonpolyposis colorectal cancer: Three years after testing. Genet Med. 2007;9:290–297. doi: 10.1097/gim.0b013e31804b45db. [DOI] [PubMed] [Google Scholar]
- 40.Hendriks KS, Hendriks MM, Birnie E, et al. Familial disease with a risk of sudden death: A longitudinal study of the psychological consequences of predictive testing for long QT syndrome. Heart Rhythm. 2008;5:719–724. doi: 10.1016/j.hrthm.2008.01.032. [DOI] [PubMed] [Google Scholar]
- 41.Chida Y, Hamer M, Wardle J, et al. Do stress-related psychosocial factors contribute to cancer incidence and survival? Nat Clin Pract Oncol. 2008;5:466–475. doi: 10.1038/ncponc1134. [DOI] [PubMed] [Google Scholar]
- 42.Giese-Davis J, Collie K, Rancourt KM, et al. Decrease in depression symptoms is associated with longer survival in patients with metastatic breast cancer: A secondary analysis. J Clin Oncol. 2011;29:413–420. doi: 10.1200/JCO.2010.28.4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cohen S, Janicki-Deverts D, Miller GE. Psychological stress and disease. JAMA. 2007;298:1685–1687. doi: 10.1001/jama.298.14.1685. [DOI] [PubMed] [Google Scholar]
- 44.Brown KW, Levy AR, Rosberger Z, et al. Psychological distress and cancer survival: A follow-up 10 years after diagnosis. Psychosom Med. 2003;65:636–643. doi: 10.1097/01.psy.0000077503.96903.a6. [DOI] [PubMed] [Google Scholar]
- 45.Lamont EB, Christakis NA. Prognostic disclosure to patients with cancer near the end of life. Ann Intern Med. 2001;134:1096–1105. doi: 10.7326/0003-4819-134-12-200106190-00009. [DOI] [PubMed] [Google Scholar]
- 46.Perni S, Rooney MK, Horowitz DP, et al. Assessment of use, specificity, and readability of written clinical informed consent forms for patients with cancer undergoing radiotherapy. JAMA Oncol. 2019;5:e190260. doi: 10.1001/jamaoncol.2019.0260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skidmore ZL, Wagner AH, Lesurf R, et al. GenVisR: Genomic Visualizations in R. Bioinformatics. 2016;32:3012–3014. doi: 10.1093/bioinformatics/btw325. [DOI] [PMC free article] [PubMed] [Google Scholar]