Abstract
PURPOSE
The US Food and Drug Administration recently announced reapproval of gemtuzumab ozogamicin (GO) for treatment of CD33-positive acute myeloid leukemia (AML), thus opening up opportunities to develop strategies for effective use of GO. In light of our recent report showing prognostic significance of CD33 splicing single nucleotide polymorphisms (SNPs), the objective of this study was to comprehensively evaluate CD33 SNPs for accurate prediction of patients with AML who are more or less likely to respond to GO.
PATIENTS AND METHODS
We investigated the five new CD33 SNPs (rs2455069, rs35112940, rs61736475, rs1803254, and rs201074739) for association with CD33 leukemic cell surface expression and clinical response in pediatric patients with AML enrolled in the Children’s Oncology Group AAML0531 trial. We further developed a composite CD33 pharmacogenetics (PGx) score using six CD33 SNPs (CD33_PGx6_score) for association with clinical outcome.
RESULTS
Four CD33 SNPs were associated with cell surface CD33 levels and clinical response in the GO versus no-GO arms. Therefore, the CD33_PGx6_score was built using directional genotype scores for the previously reported splicing SNP and five new SNPs. Patients with a CD33_PGx6_score of 0 or higher had higher CD33 expression levels compared with patients with a score of less than 0 (P < .001). In addition, patients with a score of 0 or higher demonstrated an improved disease-free survival in the GO versus no-GO arms (62.5% ± 7.8% v 46.8% ± 8.3%, respectively; P = .008) and a reduced risk of relapse (28.3% ± 7.2% v 49.9% ± 8.4%, respectively; P < .001). No improvement from GO was observed in patients with a CD33-PGx6_score of less than 0. Consistent results were observed across the risk groups.
CONCLUSION
In this study, we report a composite CD33_PGx6_score using directional genotype scores of CD33 SNPs. Once validated, our findings hold promise for use of the CD33_PGx6_score to guide efficient use of GO in patients with AML. In addition, because the CD33_PGx6_score considers SNPs with varying abundance in different ethnic groups, it has potential for global application.
INTRODUCTION
The recognition that anti-CD33 antibodies were internalized after binding to the target antigen1,2 led to the development of gemtuzumab ozogamicin (GO), an immunoconjugate between an anti-CD33 antibody (hP67.6) and a cytotoxin (calicheamicin).3 GO underwent a remarkable journey with accelerated approval in 20004 followed by withdrawal in 2010 as a result of increased deaths and no survival benefit observed in the S0106 postapproval phase III study.5 However, the withdrawal of GO was premature, because results from subsequent randomized studies (Medical Research Council [MRC] Acute Myeloid Leukemia [AML] 15, MRC/National Cancer Research Institute AML16, Groupe Ouest-Est des Leucémies et Autres Maladies du Sang AML 2006 IR, and ALFA-0701)6-14 demonstrated benefit from adding GO, which was confirmed in a meta-analysis using data from multiple clinical trials (total patients, N = 3,325).15 The Children’s Oncology Group (COG)–initiated AAML03P116 and AAML053117 clinical trials confirmed the tolerability and efficacy of GO in pediatric patients with AML. In light of these results, the US Food and Drug Administration announced reapproval of GO in September 2017 for treatment of newly diagnosed and relapsed or refractory CD33-positive AML.18,19
The most important factor with a critical impact on therapeutic efficacy of GO is the drug target CD33 itself. Interpatient variation in cell surface CD33 expression levels ranging up to 2 log-fold has been observed in patients with AML. In addition, CD33 expression levels have been correlated with disease characteristics such as presence of FLT3 internal tandem duplications (ITDs), NPM1 mutations, and high-risk group features.20,21 Recently, we reported a splicing single nucleotide polymorphism (SNP) in CD33 rs12459419 (C>T), resulting in a shorter isoform of CD33 that lacks exon 2 (CD33-D2).22 Lack of exon 2 results in loss of the IgV domain within the CD33 protein. This is clinically relevant, because the IgV domain is recognized by GO and other CD33 antibodies used in clinical immunophenotyping of AML specimens. Our results showed significant association of rs12459419 with diagnostic leukemic cell surface CD33 intensity (determined using IgV targeting p67.6 antibody), as well as differential response in GO versus no-GO treatment arms.22 Specifically, patients with at least one copy of the variant T allele (CT/TT genotypes) derived no benefit from addition of GO. In contrast, patients with homozygous CC genotype showed significantly better survival (event-free survival [EFS] and disease-free survival [DFS]) as well as lower risk of relapse with the addition of GO to standard chemotherapy.22 In contrast to these results, an investigation in the United Kingdom MRC adult AML cohorts showed consistent association of the splicing SNP with CD33 cell surface intensity but no association with clinical outcome.23 This was most likely a result of the inclusion of multiple GO random assignments and varying GO doses in these studies23 or the presence of other CD33 SNPs. These observations and an anticipated increase in the use of GO as a result of its recent approval prompted us to evaluate the prognostic role of other SNPs within CD33 in the context of the GO-based randomized COG AAML0531 trial. Given that our results identified multiple SNPs in CD33 that are associated with clinical outcome, we developed a composite CD33_PGx6_score derived from the six prognostically informative CD33 SNPs within the context of this GO randomized pediatric AML trial.
CONTEXT
Key Objective
The key objective of this study was to identify and establish the clinical impact of CD33 genetics on treatment response to gemtuzumab, a recently approved CD33-directed immunotherapeutic agent in patients with acute myeloid leukemia.
Knowledge Generated
Our results identified multiple CD33 single nucleotide polymorphisms (SNPs) of prognostic significance that were associated with gemtuzumab clinical response. Using the genotypes for six CD33 SNPs (CD33-PGx6), we generated a comprehensive CD33 genetic risk score for each patient. Patients with a CD33_PGx6_score of 0 or higher had higher CD33 expression levels compared with patients with a score of less than 0 and demonstrated improved disease-free survival and reduced risk of relapse when given gemtuzumab. No improvement from gemtuzumab was observed in patients with a CD33_PGx6_score of less than 0.
Relevance
Our results hold promise for developing strategies to personalize the use of CD33-directed agents such as gemtuzumab using CD33 genetics. Use of multiple SNPs in a CD33 comprehensive score allows for global application of the CD33 SNP score in different ethnic groups.
PATIENTS AND METHODS
Patients and Treatment
This study included specimens from pediatric patients enrolled in COG AAML0531. Details of the study design, treatment regimen, and clinical outcome have been reported previously.17 Overall, 1,022 patients with de novo AML (age, 0 to 29 years) were randomly assigned to either standard five-course chemotherapy (no-GO arm; n = 511) or the same chemotherapy plus two doses of GO 3 mg/m2 (GO arm; n = 511) during induction 1 and intensification 2. As described previously, low-risk features included t(8;21), inv(16), or t(16;16); high-risk features included monosomy 7, monosomy 5/5q deletion, or persistent disease at end of induction 1. All patients in the high-risk group received allogeneic stem-cell transplantation. Patients not classified as low or high risk were categorized as intermediate risk and received hematopoietic stem-cell transplantation if suitable donor was available.17,21 Specimens from patients who consented for biology studies were used in this study. The institutional review boards of all participating institutions approved the clinical protocol, and the COG Myeloid Disease Biology Committee approved this research study.
Genotyping of CD33 SNPs
On the basis of our previous findings,24,25 we selected CD33 SNPs with potential functional or clinical relevance that occurred with the minimum allele frequency of greater than 0.10 in either white or black patients. This included three coding SNPs (rs2455069-Arg60Gly, rs35112940-Arg304Gly, and rs61736475-Ser305Pro) and one 3′ untranslated region (UTR) SNP (rs1803254) that were genotyped using the Sequenom platform (Sequenom, San Diego, CA) at the Biomedical Genomics Center at the University of Minnesota (Minneapolis, MN). Genotype data on rs12459419 (Ala14 Val) splicing were already available in this cohort.22 All CD33 SNPs had a call rate of greater than 0.98, and all but rs1803254 were in accordance with the Hardy-Weinberg equilibrium. In addition, we genotyped a novel 4–base pair (bp) CCGG deletion variation (rs201074739) that creates a premature termination codon and thus loss of CD33 using a polymerase chain reaction–restriction fragment length polymorphism–based method (Data Supplement). Our recent data suggest a detrimental effect of this deletion on CD33 levels, with homozygous patients demonstrating complete loss of CD33 expression.26 Figure 1 shows the SNP map of CD33 SNPs included in this study as well as the splicing SNP recently reported.
FIG 1.
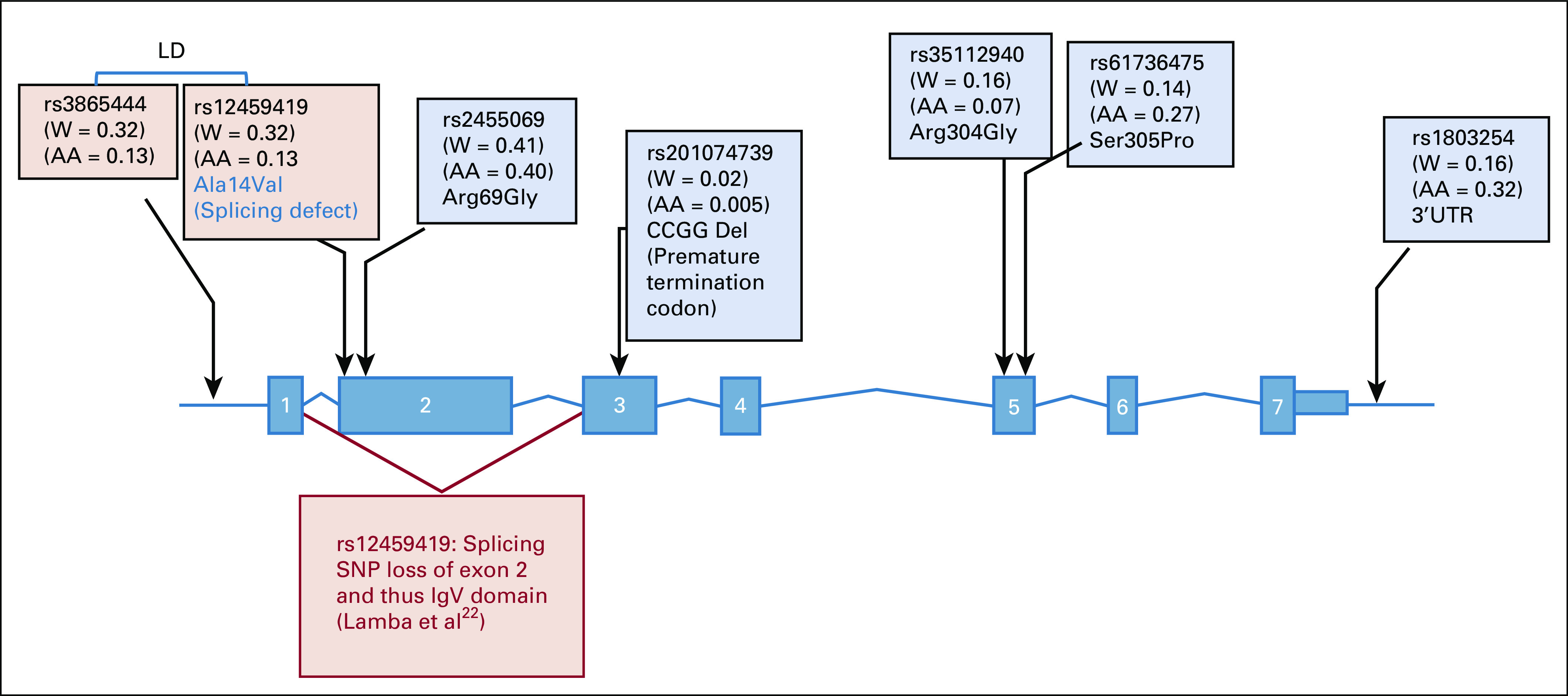
CD33 single nucleotide polymorphism (SNP) map. SNPs shaded in pink were reported recently (Lamba et al22); SNPs shaded in blue were evaluated in this study. For linked SNPs rs3865444 and rs12459419, only rs12459419 was used in creation of the CD33 composite SNP score. AA, African American; LD, linkage disequilibrium; UTR, untranslated region; W, white.
CD33 Expression Levels on Leukemic Cells
CD33 expression levels were determined by measuring CD33 mean fluorescence intensity (MFI) on AML blasts by flow cytometry using p67.6 antibody, as described previously.20,21,27
Development of Composite CD33_PGx6_Score
A composite CD33 SNP score was computed using genotype information on the six CD33 SNPs whose genotypes were significantly associated with cell surface CD33 levels. A directional genotype score was generated for each SNP by taking into the account the direction of association of the variant allele with CD33 cell surface levels.
Genotype score.
For CD33 SNPs rs12459419, rs1803254, rs35112940, rs6136475, and rs201074739, with variant alleles associated with lower leukemic cell surface CD33 intensity and inferior response by treatment arms, the genotype score was 0 for wild-type (wt)/wt genotype, −1 for wt/variant genotype, and −2 for variant/variant genotype (for rs201074739, none of the patients were homozygous for the variant allele). For CD33 SNP rs2455069, with variant allele associated with high CD33 cell surface intensity and superior response by treatment arms, the genotype score was 0 for wt/wt genotype, 1 for wt/variant genotype, and 2 for variant/variant genotype.
Composite CD33_PGx6_score = Σ (individual genotype scores of six SNPs [rs12459419, rs1803254, rs35112940, rs201074739, rs61736475, and rs2455069]).
Examples of composite scores follow. Patients with a CD33_PGx6_score of 2 could have a genotype score of 0 + 0 + 0 + 0 + 0 + 2 for rs12459419 + rs1803254 + rs35112940 + rs201074730 + rs61736475 + rs2455069, respectively. Patients with a CD33_PGx6_score of −4 could have any of the following possible combinations of genotype scores: −2 − 2 + 0 + 0 + 0 + 0 or −2 − 1 + 0 − 1 + 0 + 0 or −2 + 0 − 1 − 1 + 0 + 0 or −1 − 2 + 0 + 0 − 1 + 0 or −1 + 0 − 2 − 1 + 0 + 0 or −1 − 1 − 1 + 0 − 1 + 0 for rs12459419 + rs1803254 + rs35112940 + rs201074730 + rs61736475 + rs2455069, respectively.
Dichotomized CD33_PGx6_score.
Composite CD33 SNP score was further dichotomized to group specimens on the basis of their CD33 SNP score as CD33_PGx6_score of 0 or higher or CD33_PGx6_score of less than 0, representing high and low CD33 expression, respectively.
Statistical Analyses
Genotype and clinical data were available for 938 patients. Clinical outcome data for patients enrolled in COG AAML0531 were current as of September 30, 2014. The median follow-up time for eligible patients with de novo AML who were alive at last contact and included in our analysis was 1,856 days (range, 4 to 2,829 days). Patients were defined as being in complete remission (CR) if they had less than 5% blasts and absence of extramedullary disease after one course of induction chemotherapy. The Kaplan-Meier method was used to estimate overall survival (OS; defined as time from study entry to death), EFS (defined as time from study entry until failure to achieve CR during induction, relapse, or death), and DFS (defined as time from end of course 1 for patients in CR until relapse or death).28 Relapse risk (RR) was calculated by cumulative incidence methods defined as time from the end of induction 1 for patients in CR to relapse or death, where deaths without a relapse were considered competing events. The significance of predictor variables was tested using the log-rank statistic for OS, EFS, and DFS and using the Gray’s statistic for RR. All estimates are reported with two times the Greenwood SEs. Patients lost to follow-up were censored at their date of last known contact. Cox proportional hazards models were used to estimate the hazard ratios (HRs) for OS and EFS.29 The χ2 test was used to test the significance of observed differences in proportions, and Fisher’s exact test was used when data were sparse. The Mann-Whitney U or Wilcoxon signed rank test, as appropriate, compared differences in medians. P < .05 was considered statistically significant. No adjustments were made for multiple comparisons.
RESULTS
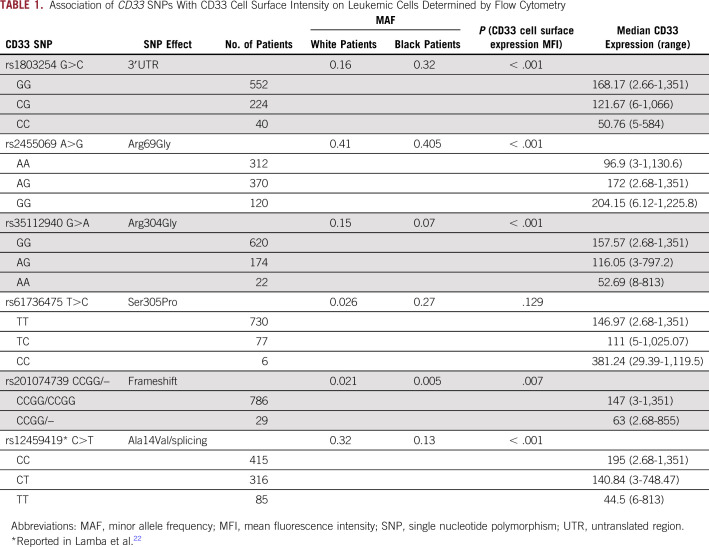
A total of 938 patients were included in our analyses. Patient- and disease-related characteristics with respect to CD33 SNP genotypes are listed in the Data Supplement. The allele frequencies of the rs1803254, rs2455069, rs35112940, and rs61736475 CD33 SNPs differed by race but not by treatment arm. Only the rs61736475 SNP differed significantly among cytogenetic risk groups, by FLT3-ITD status, and in patients who did or did not undergo hematopoietic stem-cell transplantation. As shown in Table 1, in contrast to the rs12459419 splicing SNP, which was more frequent in white patients (minor allele frequency [MAF], 0.32) compared with black patients (MAF, 0.13), the 3′UTR rs1803254 was more abundant in black patients (MAF, 0.32) compared with white patients (MAF, 0.16), and the coding SNP rs61736475 was also more common in black patients (MAF, 0.27) compared with white patients (MAF, 0.03).
TABLE 1.
Association of CD33 SNPs With CD33 Cell Surface Intensity on Leukemic Cells Determined by Flow Cytometry
Association of CD33 SNPs With CD33 Cell Surface Levels
Paired data on SNP genotype and CD33 cell surface intensity on AML blasts, expressed as MFI, were available from 817 patients (408 patients in the no-GO arm and 409 patients in the GO arm). Variant alleles for rs1803254 and rs35112940 demonstrated significantly low CD33 MFI (P < .001; Table 1 and Data Supplement). For the 4-bp deletion of CCGG (rs201074739) in exon 3, the presence of the 4-bp deletion resulted in lower CD33 MFI (P = .007). In contrast, for missense SNP rs2455069, the presence of the variant G allele was associated with higher CD33 levels (P < .001; Table 1 and Data Supplement). For completion and comparison sake, data on the previously reported SNP rs1245941922 were also included in Table 1. Given previous data reporting the rs12459419 SNP to be strongly associated with CD33 cell surface levels, we further screened for SNP-SNP interaction within rs12459419 genotypes for association with CD33 cell surface intensity. Although limited by sample size, in patients homozygous for the reference allele (rs12459419-CC genotype), we observed consistent and significant association between CD33 intensity and genotypes for rs2455069 (P = .032) and rs201074739 (P = .031).
The CD33 SNPs rs1803254, rs2455069, and rs35112940 demonstrated similar association with cell surface expression levels within each treatment arm, disease risk category, and FLT3-ITD status, consistent with overall results from the entire study cohort (Data Supplement). No difference in distribution within arms, in risk groups, or by FLT3-ITD status was observed (Data Supplement) for these SNPs, indicating the effect of these SNPs on CD33 cell surface intensity might not be associated with risk group characteristics. rs201074739 and rs61736475 occurred with a low frequency, and thus analysis within each group was limited by small sample size within genotype groups.
In a subset of patients (n = 496), we had mRNA levels of total CD33 available from the RNA sequencing data. As shown in the Data Supplement, variant alleles for 3′UTR rs1803254 (P = .010) and the rs201074739-CCGG deletion that results in a premature termination codon (P = .003) were associated with low CD33 mRNA expression.
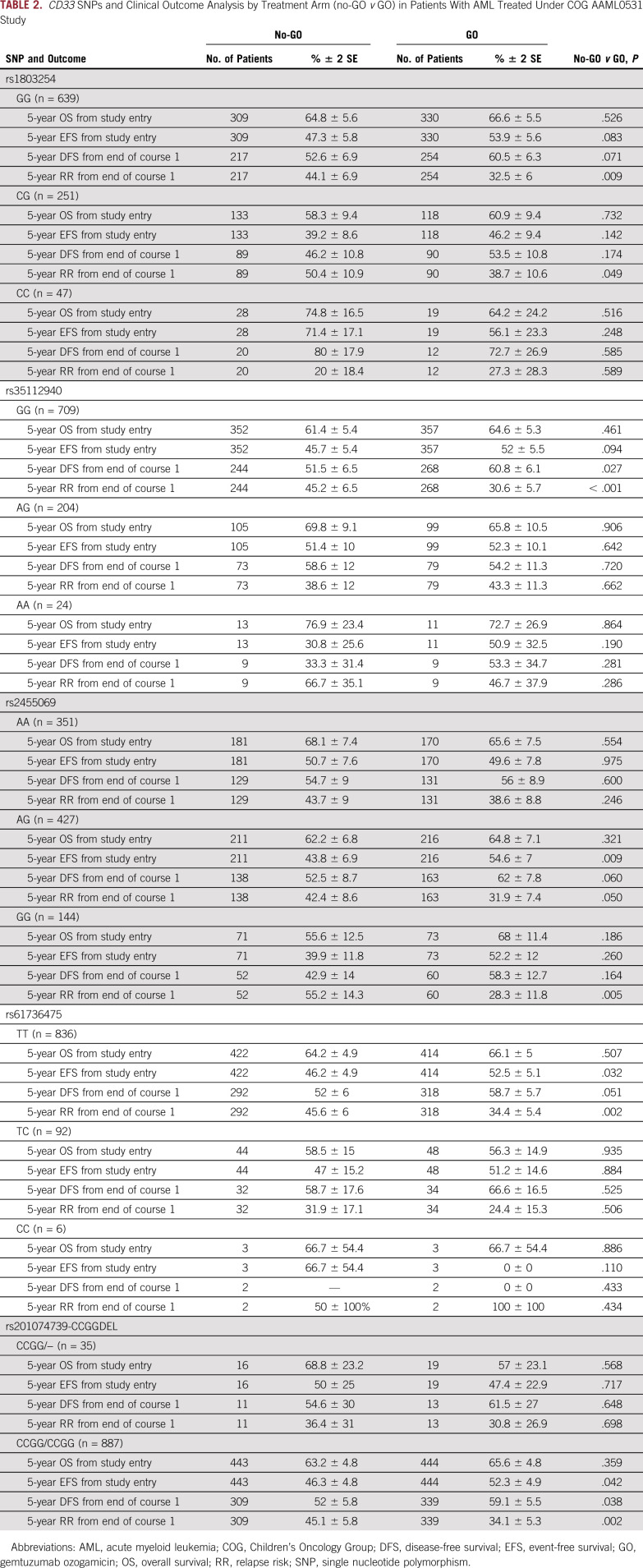
Association of CD33 SNPs With Clinical Outcome by Treatment Arm (No-GO v GO)
Survival estimates and RR indicated differences in treatment arm by CD33 SNPs (Table 2). Results for RR for selected SNPs are shown in Figure 2.
TABLE 2.
CD33 SNPs and Clinical Outcome Analysis by Treatment Arm (no-GO v GO) in Patients With AML Treated Under COG AAML0531 Study
FIG 2.
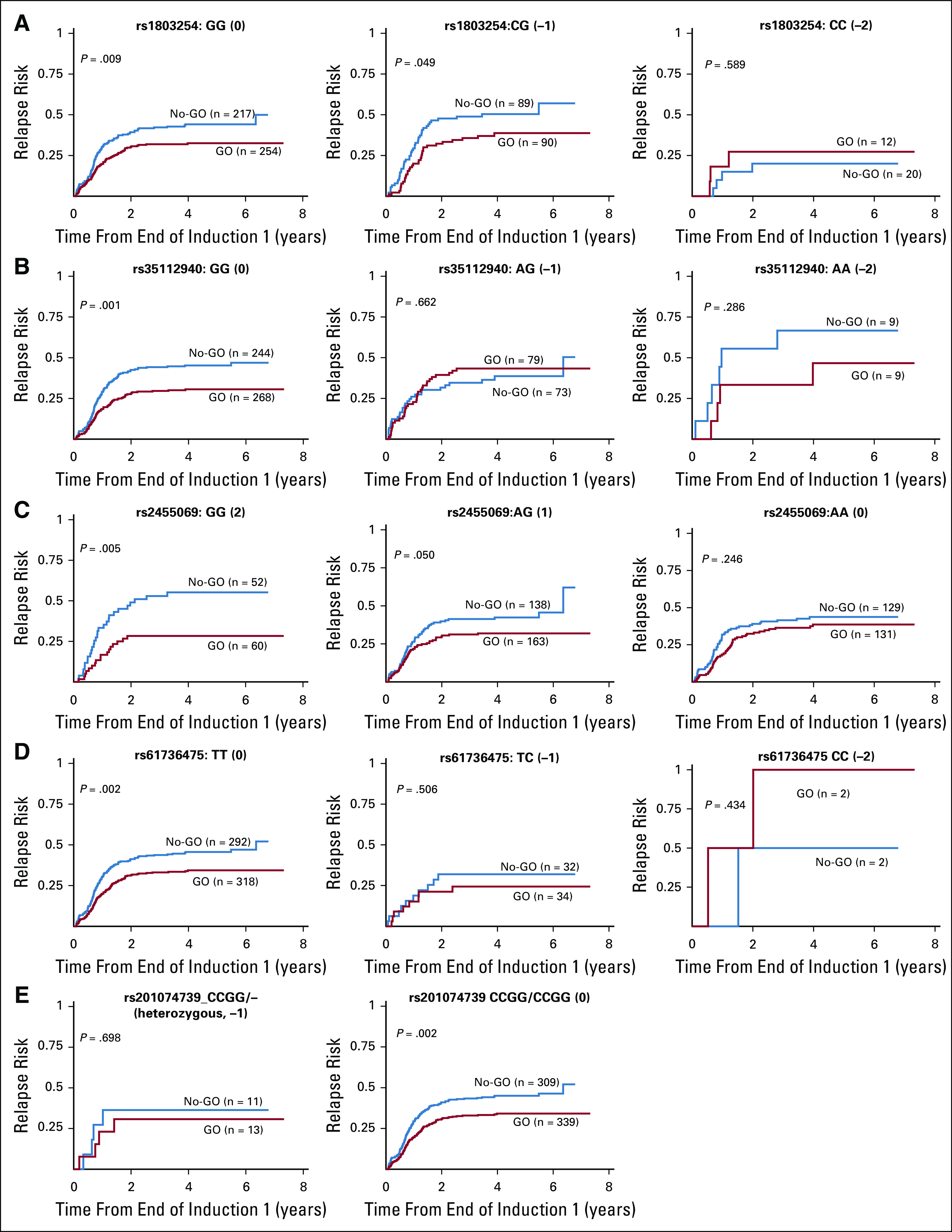
Risk of relapse (probability) from end of course 1 in the gemtuzumab ozogamicin (GO) arm versus the no-GO arm on the basis of different genotype groups for (A) rs1803254, (B) rs35112940, (C) rs2455069, (D) rs61736475, and (E) rs201074739 single nucleotide polymorphisms within CD33.
Specifically, for 3′UTR SNP rs1803254, patients with the GG or CG but not CC genotype demonstrated lower RR in the GO arm compared with the no-GO arm (GG: 32.5% ± 6% v 44.1% ± 7%, respectively; P = .009; CG: 38.7% ± 11% v 50.4% ± 11%, respectively; P = .049; CC: 27.3% ± 28.3% v 20% ± 18.4%, respectively; P = .59).
For coding SNP rs35112940 (Arg304Gly), in the GO versus the no-GO arm, only patients with GG genotype demonstrated better DFS (60.8% ± 6.1% v 51.5% ± 6.5%, respectively; P = .027) and lower RR (30.6% ± 5.7% v 45.2% ± 6.5%, respectively; P < .001). Patients with the AG or AA genotype for rs35112940 showed no difference in outcome by arm (P > .05).
For rs2455609 (Arg69Gly), patients homozygous for the variant allele (GG genotype) had higher CD33 intensity and better outcomes in the GO arm compared with the no-GO arm (RR, 28.3% ± 12% v 55.2% ± 14.3%, respectively; P = .005).
For rs201074739 (a frameshift SNP that causes premature termination codon), we did not identify any patient homozygous for CCGG deletion. However, in patients heterozygous for deletion, no benefit for adding GO was observed, whereas for patients without CCGG deletion, significantly lower RR (34.1% ± 5.3% in GO arm v 45.1% ± 6% in no-GO arm; P = .002) and improved DFS (59.1% ± 5.5% in GO arm v 52.1% ± 6% in no-GO arm; P = .038) were observed in the GO arm.
For rs61736475 (Ser305Pro), in the GO versus no-GO arms, patients with the TT genotype had lower RR (34.4% ± 5.4% v 45.6% ± 6%, respectively; P = .002) and superior EFS (52.5% ± 5.1% v 46.2% ± 5%, respectively; P = .032) and a trend toward better DFS (P = .051), whereas for patients with the TC or CC genotype, no difference was observed between treatment arms. It should be noted that the sample size for patients with the variant allele was small for SNPs rs61736475 and rs201074739.
The analysis of genotypes within each risk group, although limited by sample size, showed a consistent pattern of association of SNPs. In patients within the low-risk, intermediate-risk, and high-risk groups, rs1803254, rs35112940, and rs201074739; rs201074739; and rs2455069 and rs35112940, respectively, showed differences by treatment arms in RR after the end of course 1 by genotypes groups (Data Supplement).
Association of CD33 SNPs With Clinical Outcome by Treatment Arms (No-GO v GO) in Patients With CC Genotype for rs12459419 Splicing SNP
We recently reported significant impact of the rs12459419 C>T splicing SNP on clinical outcome by treatment arms. Our results showed no benefit of adding GO in patients with variant allele (CT and TT genotype); however, significant improvement in outcome with superior survival and lower incidence of RR was observed for patients with the CC genotype. In light of these results, we performed subgroup analysis of new CD33 SNPs within the rs12459419 CC genotype group. Although limited by numbers, we observed that presence of variant alleles for rs1803254 (C allele), rs35112940 (A allele), rs201074739 (CCGG deletion), and rs61736475 (C allele) within the 12459419 CC genotype resulted in no significant outcome benefit from adding GO to treatment (Data Supplement). Presence of the wt/reference genotype for these SNPs in conjunction with the wt/reference CC genotype for rs12459419 resulted in significant improvement in outcome with the addition of GO. For rs2455069, improvement with the addition of GO was seen across all genotype groups, which was expected given that the variant allele is associated with higher CD33 expression.
Development of Composite CD33_PGx6_Score and Its Association With CD33 Expression and Benefit From GO
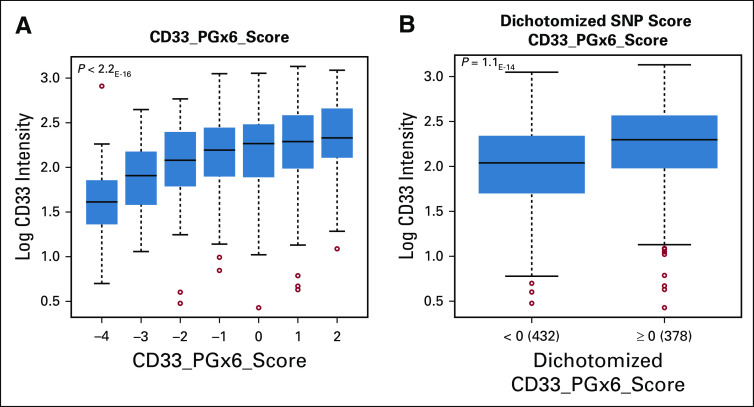
On the basis of the results for individual CD33 SNPs as well as our previous findings regarding rs12459419 splicing SNP,22 we developed a composite CD33_PGx6_score for each patient by taking into account the directional genotype scores of six SNPs (rs12459419, rs1803254, rs2455069, rs35112940, rs61736475, and rs201074739), as described in Patients and Methods. The composite CD33_PGx6_score ranged from −4 to +2. A higher SNP score was associated with higher CD33 cell surface expression, and as the score decreased, it was predictive of lower CD33 levels (P < .001; Fig 3A). We evaluated the incremental benefit of scores for the score categories of −3 to −4, −2 to −1, 0, and greater than 1. Significant benefit of adding GO, compared with the standard no-GO arm, was observed only for the score categories of 0 and greater than 1 (P < .05), and no difference in outcome was observed for the −3 to −4 and −2 to −1 categories. Given this observation and that a score of 0 indicated reference allele for each genotype, we dichotomized patients into those with a CD33_PGx6_score of 0 or higher and those with scores less than 0 (presence of at least one detrimental allele). As anticipated, CD33_PGx6_score was significantly associated with CD33 cell surface intensities (P < .001; Fig 3B).
FIG 3.
(A) Association of CD33_PGx6_score with CD33 leukemic cell surface intensity. The CD33_PGx6_score was created for each patient using genotype information for the following six single nucleotide polymorphisms (SNPs): rs12459419, rs1803254, rs35112940, rs2455069, rs61736475, and rs201074739. Genotype scores are as follows: wild type (wt)/wt = 0, wt/variant (Var) = −1, and Var/Var = −2 for rs12459419, rs1803254, rs35112940, rs61736475, and rs201074739 and wt/Var = 1 and Var/Var = 2 for rs2455069. CD33_PGx6_score was generated by adding the genotype score for each SNP. (B) Dichotomized CD33_PGx6_score (0 or higher v less than 0) and association with CD33 intensity.
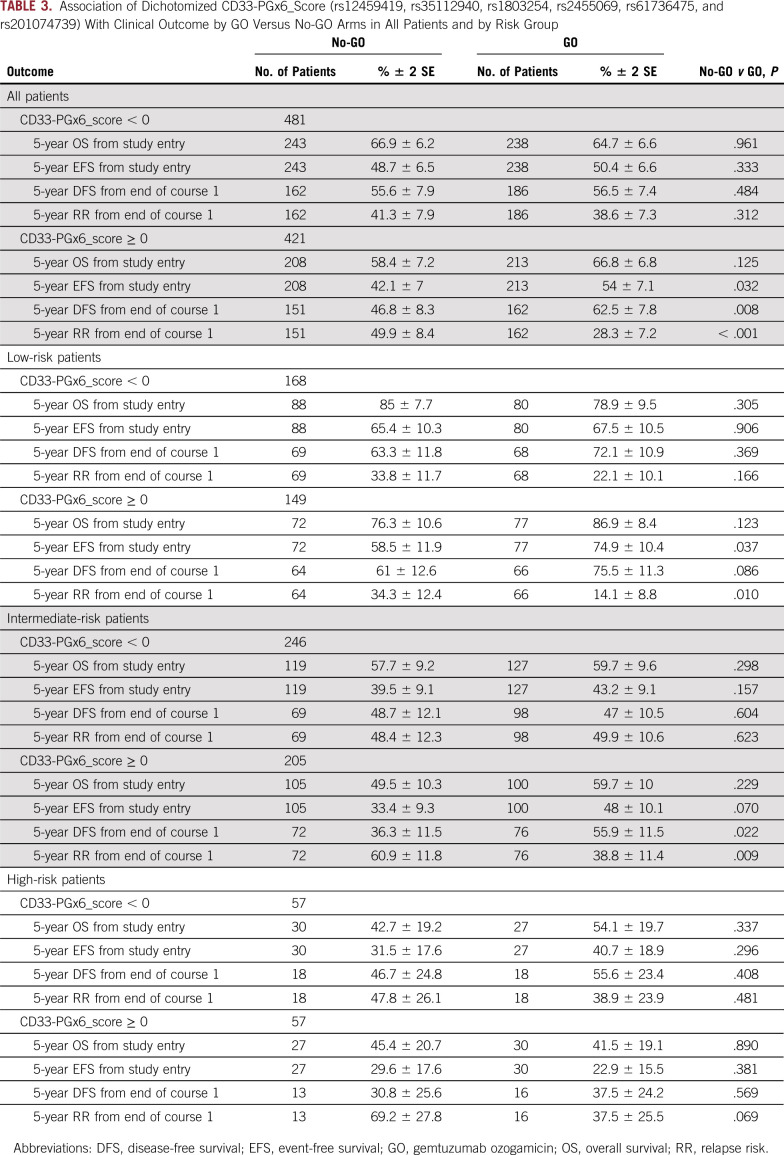
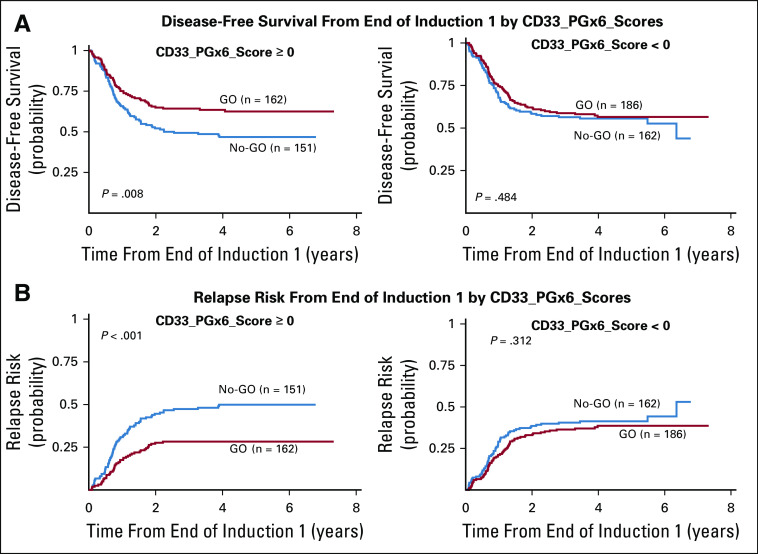
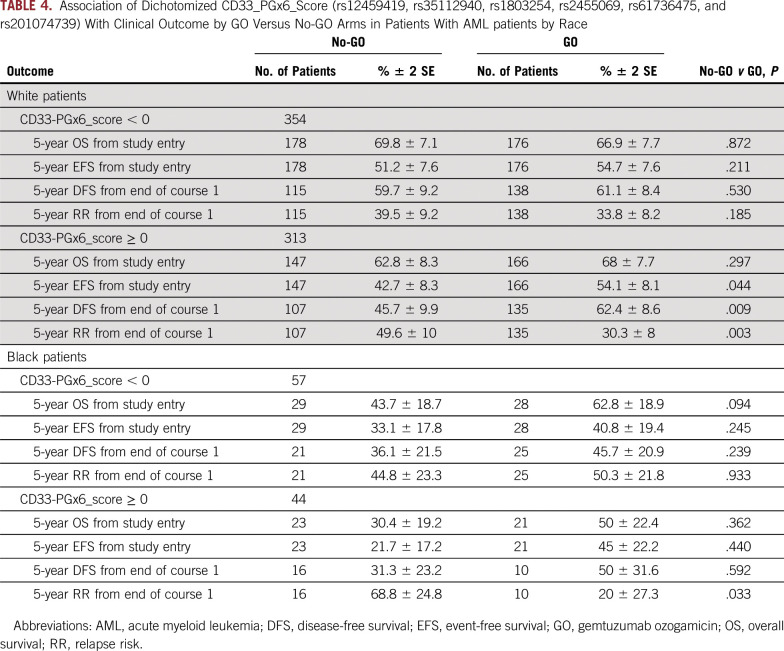
In the GO arm, compared with the no-GO arm, patients with a CD33_PGx6_score of 0 or higher, but not those with a score of less than 0, demonstrated improved DFS (62.5% ± 7.8% v 46.8 ± 8.3%, respectively; P = .008), improved EFS (54% ± 7.1% v 42.1 ± 7%, respectively; P = .032), and lower RR (28.3% ± 7.2% v 50 ± 8.4%, respectively; P < .001; Table 3 and Fig 4). Analysis by risk group demonstrated consistent results with significantly better outcome in low-risk group patients with a CD33_PGx6_score of 0 or higher when treatment included GO (RR, 14% ± 8.8% with GO v 34.3% ± 12.4% with no-GO; P = .010; Table 3). Similarly, within intermediate-risk group patients with a CD33_PGx6_score of 0 or higher, RR was significantly lower in the GO arm versus the no-GO arm (39% ± 11% v 61% ± 12%, respectively; P = .009; Table 3). Among high-risk group patients, we observed a trend toward lower RR in the GO arm versus the no-GO arm (37.5% ± 25.5% v 69% ± 28%, respectively; P = .069; Table 3). Because some of the CD33 SNPs occurred with varying frequency in patients with different race (Table 1 and Data Supplement; rs1803254 and rs61736475, in particular, occurred with higher frequency in black patients), we also investigated the association between the CD33_PGx6_score and GO separately within black and white patients. Although limited by sample size for black patients (n = 101), a CD33_PGx6_score of 0 or higher was associated with a lower RR for patients treated with GO versus no-GO (20% ± 27% v 68.8% ± 24.8%, respectively; P = .033). Similar results were observed for white patients with a CD33_PGx6_score of 0 or higher, with significant improvement in DFS (62.4% ± 8.6% in GO arm v 45.7% ± 10% in no-GO arm; P = .009) and lower RR (30.3% ± 8% in GO arm v 49.6% ± 10% in no-GO arm; P = .003) in the GO arm (Table 4 and Data Supplement). No difference in outcome was observed either in black or white patients with a CD33_PGx6_score of less than 0 (all P > .2). Patients in the GO arm with a CD33_PGx6_score of 0 or higher had a lower RR compared with patients with a score of less than 0 (28.3% ± 7% v 38.6% ± 7.3%, respectively; P = .073; Data Supplement), implying the clinical value of the score and the need to investigate it further.
TABLE 3.
Association of Dichotomized CD33-PGx6_Score (rs12459419, rs35112940, rs1803254, rs2455069, rs61736475, and rs201074739) With Clinical Outcome by GO Versus No-GO Arms in All Patients and by Risk Group
FIG 4.
Association of dichotomized CD33_PGx6_scores (score of 0 or higher v less than 0; generated using the following six CD33 SNPs: rs12459419, rs1803254, rs35112940, rs2455069, rs61736475, and rs201074739), with response by arm. (A) Differences in disease-free survival by arm in different CD33_PGx6_score groups. (B) Differences relapse risk from end of induction by arm in different CD33_PGx6_score groups.
TABLE 4.
Association of Dichotomized CD33_PGx6_Score (rs12459419, rs35112940, rs1803254, rs2455069, rs61736475, and rs201074739) With Clinical Outcome by GO Versus No-GO Arms in Patients With AML patients by Race
DISCUSSION
Encouraging results with and recent US Food and Drug Administration approval of GO, a CD33-directed agent, on the basis of several randomized trials constitute big steps in AML treatment. Success of GO has resulted in development of multiple new CD33-directed therapeutics that are currently at different stages of development and investigation, such as Fc-engineered unconjugated antibodies (BI 836858 [mAb 33.1]), antibody-drug conjugates (SGN-CD33A [vadastuximab talirine], IMGN779, and AVE9633), radioimmunoconjugates (actinium-225 lintuzumab), bi- and trispecific antibodies (AMG330, AMG673, AMV564, and 161533 TriKE fusion protein), and chimeric antigen receptor–modified immune effector cells.30 Thus, it is timely to define pharmacogenetic determinants of GO response.
We recently reported that the CD33 splicing polymorphism rs12459419 C>T regulates production of an alternatively spliced isoform of CD33 lacking exon 2 and, hence, the IgV domain of CD33.22 Our previous results demonstrated that rs12459419 was a significant predictor of cell surface CD33 intensities as well as clinical response. Most interestingly, we showed that genotype for rs12459419 is a significant predictor of whether pediatric patients enrolled in COG AAML0531 would (patients with CC genotype) or would not (patients with variant T allele that results in loss of exon 2) benefit from addition of GO to conventional chemotherapy.22 Despite the significant association of the splicing SNP with CD33 cell surface expression, there is still unexplained variability in CD33 levels within the rs12459419 CC genotype group (MFI ranging from 2.6 to 1,351 up to approximately 500-fold) as well as in clinical response within this group; thus, there is a need to further evaluate CD33 genetic variation.
In this study, we present a comprehensive evaluation of CD33 SNPs and their impact on cell surface CD33 intensities and association with clinical response in the GO and no-GO arms of the AAML0531 trial, a randomized pediatric AML clinical trial. Our results show that the rs1803254, rs2455069, rs201074739, and rs35112940 SNPs in CD33 are significantly associated with cell surface CD33 expression as well as outcome by treatment arm (GO v no-GO) within the context of the COG AAML0531 trial. rs1803254 is present in the 3′UTR of CD33, and our preliminary in vitro data suggest that it regulates CD33 transcript levels possibly through microRNA-mediated regulation (data not shown). The novel variant rs201074739 is a 4-bp deletion in exon 3 that changes the reading frame, creating a premature termination codon and thus nonfunctional truncated CD33 protein. The transcript with premature termination is degraded by the cellular nonsense-mediated decay pathway,26 consistent with the observed association of this variant with CD33 mRNA levels. rs35112940-Arg304Gly and rs61736475-Ser305Pro occur close to the ITIM motif of CD33 and thus might influence CD33 function. Our results demonstrate the clinical impact of these SNPs, warranting additional investigation in independent clinical cohorts as well as in-depth molecular characterization of these SNPs for impact on CD33 mRNA, protein stability, CD33 trafficking and signaling, and efficacy of other CD33-directed therapies.
Given that a patient with AML can inherit different genotype combinations for SNPs (as shown in CD33 onco-print; Data Supplement), it is important to evaluate the association of genotype combinations for the most influential SNPs in relation to CD33 cell surface levels and GO clinical response. Thus, to comprehensively evaluate the impact of multiple CD33 SNPs on outcome as well as to improve the clinical utility of CD33 SNP genotypes, we developed a composite CD33_PGx6_score and evaluated for association with outcome. A CD33_PGx6_score of 0 or higher was associated with significantly higher CD33 expression, better EFS and DFS, and lower RR in the GO arm compared with the no-GO arm. In patients with a score of less than 0, CD33 expression was low, and the addition of GO did not provide any clinical benefit. Consistent results were observed within each risk group category, with the greatest impact in the low-risk group followed by the intermediate- and high-risk groups. In addition, as shown in Figure 3B, there is overlap in the CD33 expression levels between the scores, indicating that the CD33 expression may not be the sole determinant of response. In fact, our previous data show that patients heterozygous (CT) for the splicing SNP rs12459419, despite having intermediate levels of CD33, did not benefit from the addition of GO, implying co-occurrence of the short isoform with the full-length isoform compromising the efficacy. Furthermore, as indicated earlier, the presence of the coding or splicing variations can affect CD33 intracellular trafficking, localization, internalization rates of the GO-CD33 complex, recycling, and signaling as the mechanisms underlying the observed impact on response. Pollard et al21 showed that patients classified as low CD33 expressers (quartile 1) on the basis of CD33 cell surface levels do not derive benefit from GO, whereas patients classified as high CD33 expressers (quartiles 2 to 4) show significant improvement with GO. We investigated the interaction of SNPs within high CD33 expressers (quartiles 2 to 4, as per Pollard et al21) and observed that, despite being in a high CD33 expression category, the presence of a detrimental variant allele for CD33 SNP compromises benefit from adding GO to chemotherapy (data not shown). These observations further strengthen the impact of CD33 SNPs on not only CD33 expression but also function and signaling as contributors to GO response.
We acknowledge the fact that the rs12459419 CC genotype was the major contributor to a CD33_PGx6_score of 0 or higher (among patients with a CD33_PGx6_score of 0 or higher, 94% had CC genotype and 6% had CT genotype for rs12459419, and among patients with a CD33_PGx6_score of less than 0, 7% had CC genotype, 73% had CT genotype, and 20% had TT genotype for rs12459419). However, this SNP is more abundant in the European population, occurring with an allele frequency of approximately 0.3, but is less frequent in other ethnic groups, ranging from approximately 0.10 to 0.15 in African Americans and Asians and being almost nonexistent in sub-Saharan Africans. In contrast, rs1803254, a 3′UTR SNP, has a completely flipped frequency of 0.32 in black patients and 0.16 in white patients, and rs61736475 is also more abundant in black patients (MAF, 0.27) compared with white patients (MAF, 0.03). Our results show that in black patients with a CD33_PGx6_score of less than 0, 54.5% of patients have the CC genotype, compared with only 6% of white patients with the CC genotype for the rs12459419 splicing group. This difference clearly demonstrates contribution of other SNPs to low CD33 score in black patients. Given that the CD33_PGx6_score considers multiple CD33 SNPs in addition to the splicing SNP, it is more likely to identify patients who are less likely to benefit from GO or other CD33-directed therapies across different ethnic groups. The novel SNP rs201074739, a 4-bp CCGG deletion that results in truncated protein, never occurs in patients in the rs12459419 TT genotype group (Data Supplement) but has been observed in patients with the CC and CT genotypes for rs12459419, indicating its contribution to lower CD33 levels and perhaps response within the rs12459419 CC and CT genotype groups.
Given the anticipated increase in GO use with its recent approval and several new CD33-directed therapies in the pipeline, development of the CD33_PGx6_score is timely and critical to establish its ability and clinical utility to personalize GO treatment in AML. Our future work is targeted toward validating the CD33 score in other clinical cohorts (United Kingdom trials) and an upcoming study from COG that plans to incorporate GO-containing regimens. Once validated, the CD33_PGx6_score holds promise in prospective assignment of GO and other CD33-directed therapies to patients who are more likely to benefit and thus may further improve clinical outcome in AML.
ACKNOWLEDGMENT
We thank the Children’s Oncology Group Acute Myeloid Leukemia Reference Laboratory for providing specimens for the study. Special thanks to Sommer Castro for providing specimens and Dr Roland Walter for providing critiques on the article.
Footnotes
Presented in part at the 59th Annual Meeting of the American Society of Hematology, Atlanta, GA, December 9-12, 2017.
Supported by National Institutes of Health Grants No. U10CA180899, U10CA180886, U10CA98413, U10CA098543, R21CA155524, and R01CA133881; the St Baldrick’s Foundation; the Children’s Oncology Group Foundation; and the Andrew McDonough B+ Foundation.
AUTHOR CONTRIBUTIONS
Conception and design: Michael Loken, Alan S. Gamis, Todd A. Alonzo, Soheil Meshinchi, Jatinder K. Lamba
Provision of study material or patients: Richard Aplenc
Collection and assembly of data: Lata Chauhan, Yi-Cheng Wang, Michael Loken, Jessica Pollard, Richard Aplenc, Betsy A. Hirsch, Susana Raimondi, Rhonda E. Ries, Alan S. Gamis, Todd A. Alonzo, Jatinder K. Lamba
Data analysis and interpretation: Lata Chauhan, Miyoung Shin, Yi-Cheng Wang, Michael Loken, Jessica Pollard, Richard Aplenc, Irwin D. Bernstein, Alan S. Gamis, Todd A. Alonzo, Soheil Meshinchi, Jatinder K. Lamba
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Michael Loken
Employment: Hematologics, Hematologics (I)
Leadership: Hematologics, Hematologics (I)
Stock and Other Ownership Interests: Hematologics, Hematologics (I)
Consulting or Advisory Role: Newlink Genetics
Richard Aplenc
Honoraria: Sigma-Tau
Expert Testimony: Wiggin and Dana
Travel, Accommodations, Expenses: Sigma-Tau
Irwin D. Bernstein
Stock and Other Ownership Interests: Nohla Therapeutics, Lyell
Consulting or Advisory Role: Nohla Therapeutics
Patents, Royalties, Other Intellectual Property: Multiple patents through Fred Hutchinson Cancer Research Center, share of royalties from Fred Hutchinson Cancer Research Center
Alan S. Gamis
Consulting or Advisory Role: Novartis
Jatinder K. Lamba
Patents. Royalties, Other Intellectual Property: Patent application No. 16/091,720 entitled ‘Biomarkers of Anti-Leukemic Therapy’
No other potential conflicts of interest were reported.
REFERENCES
- 1.Scheinberg DA, Lovett D, Divgi CR, et al. A phase I trial of monoclonal antibody M195 in acute myelogenous leukemia: Specific bone marrow targeting and internalization of radionuclide. J Clin Oncol. 1991;9:478–490. doi: 10.1200/JCO.1991.9.3.478. [DOI] [PubMed] [Google Scholar]
- 2.van der Jagt RH, Badger CC, Appelbaum FR, et al. Localization of radiolabeled antimyeloid antibodies in a human acute leukemia xenograft tumor model. Cancer Res. 1992;52:89–94. [PubMed] [Google Scholar]
- 3.Bernstein ID, Singer JW, Andrews RG, et al. Treatment of acute myeloid leukemia cells in vitro with a monoclonal antibody recognizing a myeloid differentiation antigen allows normal progenitor cells to be expressed. J Clin Invest. 1987;79:1153–1159. doi: 10.1172/JCI112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bross PF, Beitz J, Chen G, et al. Approval summary: Gemtuzumab ozogamicin in relapsed acute myeloid leukemia. Clin Cancer Res. 2001;7:1490–1496. [PubMed] [Google Scholar]
- 5.Petersdorf SH, Kopecky KJ, Slovak M, et al. A phase 3 study of gemtuzumab ozogamicin during induction and postconsolidation therapy in younger patients with acute myeloid leukemia. Blood. 2013;121:4854–4860. doi: 10.1182/blood-2013-01-466706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burnett A, Cavenagh J, Russell N, et al. Defining the dose of gemtuzumab ozogamicin in combination with induction chemotherapy in acute myeloid leukemia: A comparison of 3 mg/m2 with 6 mg/m2 in the NCRI AML17 trial. Haematologica. 2016;101:724–731. doi: 10.3324/haematol.2016.141937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Burnett AK, Hills RK. Who should be transplanted in first remission of acute myeloid leukaemia? Curr Treat Options Oncol. 2011;12:329–340. doi: 10.1007/s11864-011-0169-x. [DOI] [PubMed] [Google Scholar]
- 8.Burnett AK, Hills RK, Grimwade D, et al. Inclusion of chemotherapy in addition to anthracycline in the treatment of acute promyelocytic leukaemia does not improve outcomes: Results of the MRC AML15 trial. Leukemia. 2013;27:843–851. doi: 10.1038/leu.2012.360. [DOI] [PubMed] [Google Scholar]
- 9.Burnett AK, Hills RK, Hunter AE, et al. The addition of gemtuzumab ozogamicin to low-dose Ara-C improves remission rate but does not significantly prolong survival in older patients with acute myeloid leukaemia: Results from the LRF AML14 and NCRI AML16 pick-a-winner comparison. Leukemia. 2013;27:75–81. doi: 10.1038/leu.2012.229. [DOI] [PubMed] [Google Scholar]
- 10.Burnett AK, Hills RK, Milligan D, et al. Identification of patients with acute myeloblastic leukemia who benefit from the addition of gemtuzumab ozogamicin: Results of the MRC AML15 trial. J Clin Oncol. 2011;29:369–377. doi: 10.1200/JCO.2010.31.4310. [DOI] [PubMed] [Google Scholar]
- 11.Burnett AK, Russell NH, Hills RK, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy improves survival in older patients with acute myeloid leukemia. J Clin Oncol. 2012;30:3924–3931. doi: 10.1200/JCO.2012.42.2964. [DOI] [PubMed] [Google Scholar]
- 12.Castaigne S, Pautas C, Terré C, et al. Effect of gemtuzumab ozogamicin on survival of adult patients with de-novo acute myeloid leukaemia (ALFA-0701): A randomised, open-label, phase 3 study. Lancet. 2012;379:1508–1516. doi: 10.1016/S0140-6736(12)60485-1. [DOI] [PubMed] [Google Scholar]
- 13.Lambert J, Pautas C, Terre C, et al. Gemtuzumab ozogamicin for de novo acute myeloid leukemia: Final efficacy and safety updates from the open-label, phase III ALFA-0701 trial. Haematologica. 2018;104:113–119. doi: 10.3324/haematol.2018.188888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Addition of gemtuzumab ozogamycin to chemotherapy improves event-free survival but not overall survival of AML patients with intermediate cytogenetics not eligible for allogeneic transplantation: Results of the GOELAMS AML. Blood. 2011;118:79. Delaunay J, Recher C, Pigneux A, et al: (abstr) [Google Scholar]
- 15.Hills RK, Castaigne S, Appelbaum FR, et al. Addition of gemtuzumab ozogamicin to induction chemotherapy in adult patients with acute myeloid leukaemia: A meta-analysis of individual patient data from randomised controlled trials. Lancet Oncol. 2014;15:986–996. doi: 10.1016/S1470-2045(14)70281-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper TM, Franklin J, Gerbing RB, et al. AAML03P1, a pilot study of the safety of gemtuzumab ozogamicin in combination with chemotherapy for newly diagnosed childhood acute myeloid leukemia: A report from the Children’s Oncology Group. Cancer. 2012;118:761–769. doi: 10.1002/cncr.26190. [DOI] [PubMed] [Google Scholar]
- 17.Gamis AS, Alonzo TA, Meshinchi S, et al. Gemtuzumab ozogamicin in children and adolescents with de novo acute myeloid leukemia improves event-free survival by reducing relapse risk: Results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2014;32:3021–3032. doi: 10.1200/JCO.2014.55.3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jen EY, Ko CW, Lee JE, et al. FDA approval: Gemtuzumab ozogamicin for the treatment of adults with newly diagnosed CD33-positive acute myeloid leukemia. Clin Cancer Res. 2018;24:3242–3246. doi: 10.1158/1078-0432.CCR-17-3179. [DOI] [PubMed] [Google Scholar]
- 19.Norsworthy KJ, Ko CW, Lee JE, et al. FDA approval summary: Mylotarg for treatment of patients with relapsed or refractory CD33-positive acute myeloid leukemia. Oncologist. 2018;23:1103–1108. doi: 10.1634/theoncologist.2017-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pollard JA, Alonzo TA, Loken M, et al. Correlation of CD33 expression level with disease characteristics and response to gemtuzumab ozogamicin containing chemotherapy in childhood AML. Blood. 2012;119:3705–3711. doi: 10.1182/blood-2011-12-398370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pollard JA, Loken M, Gerbing RB, et al. CD33 expression and its association with gemtuzumab ozogamicin response: Results from the randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2016;34:747–755. doi: 10.1200/JCO.2015.62.6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamba JK, Chauhan L, Shin M, et al. CD33 splicing polymorphism determines gemtuzumab ozogamicin response in de novo acute myeloid leukemia: Report from randomized phase III Children’s Oncology Group trial AAML0531. J Clin Oncol. 2017;35:2674–2682. doi: 10.1200/JCO.2016.71.2513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gale RE, Popa T, Wright M, et al. No evidence that CD33 splicing SNP impacts the response to GO in younger adults with AML treated on UK MRC/NCRI trials. Blood. 2018;131:468–471. doi: 10.1182/blood-2017-08-802157. [DOI] [PubMed] [Google Scholar]
- 24.Lamba JK, Pounds S, Cao X, et al. Coding polymorphisms in CD33 and response to gemtuzumab ozogamicin in pediatric patients with AML: A pilot study. Leukemia. 2009;23:402–404. doi: 10.1038/leu.2008.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mortland L, Alonzo TA, Walter RB, et al. Clinical significance of CD33 nonsynonymous single-nucleotide polymorphisms in pediatric patients with acute myeloid leukemia treated with gemtuzumab-ozogamicin-containing chemotherapy. Clin Cancer Res. 2013;19:1620–1627. doi: 10.1158/1078-0432.CCR-12-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Papageorgiou I, Loken MR, Brodersen LE, et al. CCGG deletion (rs201074739) in CD33 results in premature termination codon and complete loss of CD33 expression: Another key variant with potential impact on response to CD33-directed agents. Leuk Lymphoma. 2019;5:1–4. doi: 10.1080/10428194.2019.1569232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. doi: 10.1016/j.leukres.2007.10.002. Wells DA, Loken MR: Flow cytometric mean fluorescence intensity: The biophysics behind the number. Leuk Res 32:845-846, 2008. [DOI] [PubMed] [Google Scholar]
- 28. Kaplan EL, Meier P: Nonparametric estimation from incomplete observations. J Am Stat Assoc 53:457-481, 1958. [Google Scholar]
- 29. Cox DR: Regression models and life tables. J R Stat Soc B 34:187-220, 1972. [Google Scholar]
- 30.Walter RB. Investigational CD33-targeted therapeutics for acute myeloid leukemia. Expert Opin Investig Drugs. 2018;27:339–348. doi: 10.1080/13543784.2018.1452911. [DOI] [PubMed] [Google Scholar]