Abstract
Although precision oncology is transforming clinical management of patients with cancer, many hospitals face challenges to effectively implement precision oncology. In addition, the cost and time exerted for genomic profiling needs to be balanced with expectations of benefit for each patient. This article summarizes the effort to implement precision oncology in Japan. The most promising development is that tests to profile the genomes of select cancers are now fully covered by the national health insurance system. In May 2019, two gene panels were approved with reimbursement: FoundationOne CDx Cancer Genomic Profile and OncoGuide NCC Oncopanel System, the latter of which was developed in Japan. To make better use of scarce resources, the reimbursement is restricted to patients with solid tumors that have progressed on standard chemotherapy, rare tumors, or tumors of unknown primary. To centralize Japanese precision oncology, the government designated approximately 170 hospitals and stratified them to three layers on the basis of their roles. In addition, Japan’s National Cancer Center launched a Center for Cancer Genomics and Advanced Therapeutics (C-CAT) that collects genomic information and clinical characteristics of patients who received genomic profiling tests. C-CAT is expected to be the central data repository, to match patients with clinical trials, and to assist translational research. The centralized system under the national health insurance system could be a double-edged sword. Although tight regulation may make it hard to keep up with the rapid development of precision oncology, a federated ecosystem for sharing clinical and genomic data will be a precious asset and allow for shared access to data. Access to unapproved drugs and administrative support from C-CAT will be keys for Japanese precision oncology to meet its full potential.
HEALTH CARE SYSTEM IN JAPAN
Japan’s health care system is characterized by universal health coverage.1 By law, all Japanese citizens and foreign nationals with a residence card must be enrolled in a health insurance program. There are two major types of insurance schemes in Japan: Employees’ Health Insurance and National Health Insurance. Employees’ health insurance covers public servants or employees in companies, and national health insurance covers the self-employed and unemployed. These health insurance policies cover 70%-90% of the cost for medical expenses, with the remainder left to the insured patient to pay, termed copayment. High-Cost Medical Expense Benefit also defines maximum out-of-pocket payment for the month according to household income of the patient. For example, the maximum is roughly $800 for a household income of approximately $34,000-$70,000. Although for-profit insurance companies have voluntary health insurance programs, holding this type of insurance does not exempt an individual from mandatory enrollment in the universal health coverage scheme. The role of voluntary health insurance is supplemental and complements social health insurance benefit packages.
Prices for all drugs and devices reimbursed by the universal health coverage system are the official price set by the government (Japanese Ministry of Health, Labor, and Welfare [MHLW]). In addition, the Japanese government does not allow treatments partially covered by insurance to control the whole medical expenses in the country. If a physician uses an unapproved device or off-label drug, any costs related to the patient, including but not limited to blood tests and physician fees, are not reimbursed. Therefore, any cancer genomic profiling tests must be examined and approved by the Pharmaceuticals and Medical Devices Agency (PMDA) and MHLW before insurance reimbursement is set by the government, unless all the cost is to be paid by the patients. Also, off-label drug use for nonapproved indications is prohibited except for clinical trials, even when gene profiling testing identifies actionable alterations.
CONTEXT
Key Objective
To summarize the framework of precision oncology recently arranged in Japan.
Knowledge Generated
The Japanese government designated core, hub, and liaison hospitals for cancer genomic medicine. A center was also established to coordinate an integrated network of these hospitals and function as the master database for cancer genomic medicine and research. After the establishment of a structured system, the government approved reimbursement of two cancer genome profiling systems under universal health coverage in May 2019. The biggest current challenge for Japanese precision oncology seems to be the accessibility to drugs, because off-label drug use for nonapproved indications is prohibited in Japan. Also, reimbursement applies only to patients who finished standard chemotherapy, as a way to restrict potentially unnecessary investigations.
Relevance
Each country must design a precision oncology initiative under their distinct social care system. Japan is an example of a centralized precision oncology under a universal health care coverage system.
The Japanese MHLW has a structured system to promote the development of drugs and devices under governmental regulations. First, in cases where MHLW anticipates the future full approval with reimbursement, MHLW grants the use of drug and devices under partial coverage. “The Advanced Medical Care System” is a governmental scheme introduced as a means to assess the efficacy and toxicity of new treatments/devices on a clinical trial basis. Although system A is designed for intervention with approved drugs/devices or minimally invasive intervention with unapproved drugs/devices, system B is designed for unapproved drugs or medical technologies. In addition, the MHLW-sanctioned Patient-Requested Therapy System covers unapproved medical care for patients not in clinical trials. The Patient-Requested Therapy System is also expected to be used as the Japanese version of what the United States has hailed as the compassionate use program. To start off-label drug treatment using Patient-Requested Therapy Systems, it is necessary to pass several review processes equivalent to clinical trials, and the cost of the unapproved medical care should be fully paid by the patients. Second, similar to the Breakthrough Therapy designation by the US Food and Drug Administration, MHLW has the SAKIGAKE program, an accelerated inspection scheme for rapid authorization of unapproved drugs and devices. Third, Japan’s Conditional Early Approval system was established for lethal and/or rare diseases where it is difficult to conduct verification studies because of limited patients. These schemes developed with the intention to promote the development of drugs and devices were used for the approval of genome profiling tests.
RESEARCH-BASED GENOME SCREENING IN JAPAN
Because of tightened regulations described above, genome profiling, like in most countries, began in the research setting in Japan. Beginning around 2010, pan-cancer genome screening started in Japan by using research-use only next-generation sequencing (NGS) panels.2 To promote genome-based clinical trials, the nationwide genome screening consortium for lung cancer was launched in February 2013. The group, LC-SCRUM-Japan, originally aimed to identify patients harboring ROS1 and RET fusions originally discovered in Japan.3 In February 2014, the GI-SCREEN-Japan multicenter screening project also started for GI cancer. The project began to screen for BRAF, NRAS, and PIK3CA mutations in patients with metastatic colorectal cancer, using multiplex polymerase chain reaction (PCR) and Luminex (xMAP) technology.4 In February 2015, these two groups merged into SCRUM-Japan and started to use Oncomine Comprehensive assays as a screening platform.5 With the advent of plasma-based genome screening, SCRUM-Japan subsequently started plasma-based NGS using Guardant 360. Currently, SCRUM-Japan has expanded their network to Asian countries, including Taiwan, enabling international clinical trials. The screening and associated clinical trials have been research based, funded by government agencies through grant mechanisms (The Japan Agency for Medical Research and Development or the National Cancer Center Research and Development Fund) as well as by pharmaceutical companies. Therefore, patients were not required to pay screening costs for any of these studies.
From February 2013 to December 2018, 6,860 and 6,391 patients were enrolled in LC-SCRUM-Japan and GI-SCREEN-Japan, respectively. On the basis of this screening platform, 28 umbrella and 20 basket-type genome-based studies have been conducted.6 One of the notable accomplishments was a clinical trial of vandetanib, for patients with RET-rearranged non–small-cell lung cancer (NSCLC) identified by the screening. Nine of 19 (47%) patients achieved an objective response, with a median progression-free survival of 4.7 months.7 Furthermore, results of clinical trials conducted by LC-SCRUM Japan led to the approval of crizotinib for the treatment of ROS1-translocated NSCLC and the combination of trametinib and dabrafenib for use in patients with BRAF V600E-mutant NSCLC. SCRUM-Japan also contributed to the approval of in vitro diagnostic testing by PMDA including simultaneous detection of RAS and BRAF mutations in colorectal cancer,8 Oncomine DX Target Test CDx NGS panel for EGFR and BRAF mutations and ROS1 and ALK translocations in NSCLC, PCR-based microsatellite instability testing for solid tumors,9 and plasma-based RAS mutation testing for colorectal cancer.10
Another notable genome screening is the TOP-GEAR (Trial of Oncopanel for Gene Profiling to Estimate both Adverse Events and Response) project, which is a prospective cohort study conducted by Japan’s National Cancer Center (NCC) to investigate the feasibility and utility of an NGS-based panel customized for Japanese patients with cancer (NCC Oncopanel).11 This work was carried out within the context of the SAKIGAKE program and clinical utility was investigated within the Advanced Medical Care B system. During the second stage of the TOP-GEAR project, 187 patients with advanced solid tumors obtained the gene profiling data, and 25 (13.3%) of them have received molecular-targeted therapy on the basis of their genome alterations.12 The achievements of this project led to governmental approval for the OncoGuide NCC Oncopanel System.
Some other medical university hospitals are also developing their own genome screening systems. Tokyo University Hospital and Osaka University Hospital have their own systems, called Todai OncoPanel and customized Oncomine Target Test, respectively. These tests are performed under the Advanced Medical Care B system. Kyoto University Hospital (OncoPrime)13,14 and Keio University Hospital (PleSSision-Rapid) offer their screening service outside of the national health care system. Recently, Keio University Hospital started a whole-exon sequencing service (PleSSision-Exome). Representative NGS-based panels available in Japan are listed in Table 1.
TABLE 1.
Next-Generation Sequencing–Based Tumor-Profiling Multiplex Gene Panels
ARRANGEMENTS OF INFRASTRUCTURE FOR PRECISION MEDICINE IN JAPAN
In Europe and the United States, government-funded initiatives were launched to implement precision medicine, including the 100,000 Genomes Project in the United Kingdom, Genomic Medicine 2025 in France, and the Precision Medicine Initiative in the United States.15-17 Partially in response to the development of precision medicine in other countries like these, the Japanese Headquarters for Healthcare Policy, under direction of the prime minister, started discussions regarding a national precision medicine program in 2015. In 2017, MHLW summoned a roundtable consortium on the promotion of cancer genomic medicine. The consortium required the development of an agile national cancer genome medicine system equivalent to the European and US models. In addition, they concluded there was a necessity to establish a nationwide cancer research framework and a corresponding ecosystem to move cancer genome medicine forward. Importantly, the report also concluded that cancer genome medicine should be accomplished under a universal health coverage system. After the report, the Japanese government designated 11 hospitals throughout Japan to serve as core hospitals for cancer genomic medicine. The requirements of designated core hospitals for cancer genome medicine are shown in Table 2. The government also appointed 156 facilities as liaison hospitals and, among these, designated 34 hub hospitals. The eligibility to be designed as a hub hospital is based on their ability to organize their resources and infrastructure akin to the core hospitals, which have their own molecular tumor board (MTB) and organic capabilities to run clinical trials. In addition, core hospitals have more responsibilities to train fellows and clinical coordinators and to be involved in translational research. Liaison hospitals, on the other hand, are dependent on core and hub hospitals for their sequencing, reports, and MTB (Fig 1).
TABLE 2.
Requirements for Core Hospitals

FIG 1.

The three-layer structure of designated institutes for precision oncology in Japan. Notably, core and hub hospitals are allowed to run genome panels in-house, whereas liaison hospitals refer these tests to core/hub hospitals. However, most hospitals outsource these genomic profiling tests to a clinical testing company in Japan.
From the academia side, three major Japanese cancer-related societies (the Japanese Society of Medical Oncology [JSMO], the Japanese Society of Clinical Oncology [JSCO], and the Japanese Cancer Association [JCA]) issued consensus clinical practice guidelines for NGS-based cancer tests in October 2017.18 The guideline defined the evidence level of each genomic alteration suitable for the Japanese medical care system in harmony with classifications of evidence levels set by regulators in the United States and European Union (EU).19
In June 2018, Japan’s National Cancer Center founded the Center for Cancer Genomics and Advanced Therapeutics (C-CAT) to coordinate an integrated network of core, hub, and liaison hospitals. C-CAT is expected to aggregate and deploy cancer genomic medicine information to advance the quality of health care offered under the universal health coverage system and to devise new modalities of health care (Fig 2). The center will function as the central repository—that is, the master database for cancer genomic medicine and research. First, the center has been constructing a cancer knowledge database (CKDB) optimized for Japan to assist in decision making by MTBs. C-CAT will collect and share updated information on clinical trials, promoting and improving matching between patients’ genomic data and clinical trials. Accumulated data will be shared for translational research, drug development matched to Japanese patients, and future administrative policy. Currently, C-CAT has been designing open and responsible data sharing. In principle, researchers and pharmaceutical companies will be able to access clinical and genomic data deposited in C-CAT after approval of their application by an independent access review committee. C-CAT has also discussed standard procedures for precision oncology, including preparation of a standard informed consent form, establishment of a flowchart for genetic counseling, and standardizing of MTB to promote and advance genome-based medicine in Japan.
FIG 2.
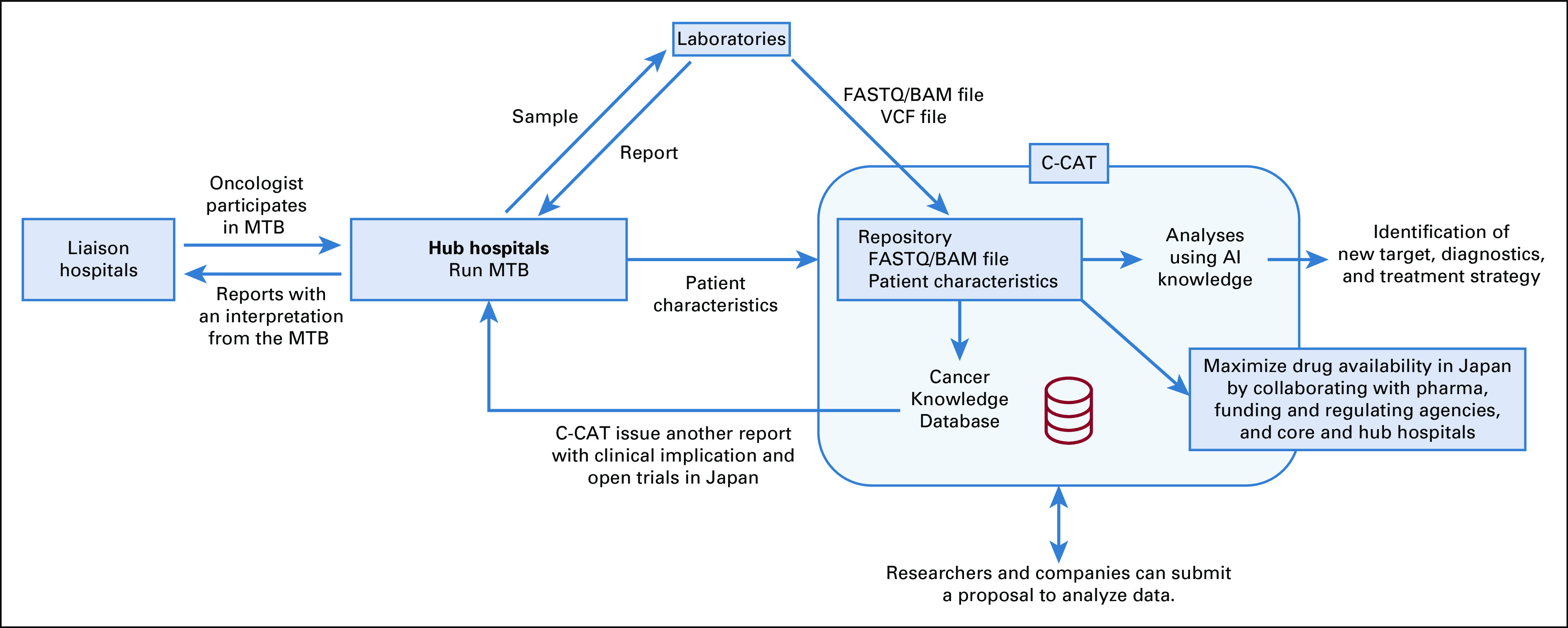
Expected roles of the Center for Cancer Genomics and Advanced Therapeutics (C-CAT). AI, artificial intelligence; MTB, molecular tumor board.
APPROVED CANCER GENOME PROFILING AND ITS CLINICAL PRACTICE
In December 2018, PMDA approved marketing of two cancer genome profiling systems. OncoGuide NCC Oncopanel System is a 114-gene NGS panel for tumor and germline analysis developed by Japan’s NCC and health instrument maker Sysmex Corp, and FoundationOne CDx Cancer Genomic Profile (Foundation Medicine) sequences 324 genes and also can detect microsatellite instability. OncoGuide NCC Oncopanel System was approved as a device combining the OncoGuide NCC Oncopanel kit and the OncoGuide NCC Oncopanel analyzing system. In contrast, although Foundation Medicine had to be reviewed their quality for sample analysis by PMDA for the approval, PMDA could not perform assessment as an in vitro diagnostic because the analysis was done in the United States. As a workaround, Foundation Medicine submitted and gained approval of an interpretation service. Therefore, although the sequencing process is not reimbursed, the annotation process and generation of the report are subject to reimbursement. The regulation stipulates a laborious process. First, physicians submit tumor samples for FoundationOne CDx analysis to Foundation Medicine, and Foundation Medicine performs sequencing and reports the variant calls in an XML file, uploaded to a portal site created by Chugai, a subsidiary of Roche. Because this step is not subject to reimbursement, and to set up a procedure suitable for reimbursement, physicians need to download the XML file from the portal site and then send it back to Foundation Medicine. Foundation Medicine annotates variants in the submitted XML file and returns to physicians a final report. All these steps are required for reimbursement. In addition, the Japanese Act on the Protection of Personal Information defines genomic sequence as personal information that was amended in response to the General Data Protection Regulation enacted by the EU. To comply with the law, written consent is required. On consent, samples are sent to third parties outside of Japan to analyze personal information and sequence data.
After approval by PMDA, the MHLW granted the use of these two gene panel analyses and set the official price of reimbursement at the end of May 2019. The reimbursement for the cost of these tests is ¥560,000 (approximately $5,185), and consists of two steps (Fig 3). The first reimbursement is ¥80,000, applied for the informed consent of genome profiling and preparation of tumor samples. The second reimbursement is ¥480,000, applied after the physician explains the results to patients following the assessment by MTB. The test is approved for patients with solid tumors that have progressed on standard chemotherapy, with rare tumors, or with cancers of unknown primary.
FIG 3.
Flow and reimbursements for cancer genomic profiling tests. C-CAT, Center for Cancer Genomics and Advanced Therapeutics; JPY, Japanese yen; NCC, National Cancer Center.
FoundationOne CDx is also approved for the use of companion diagnostic tests, such as those for the detection of EGFR, RAS, and BRAF mutations during standard care. However, the reimbursement for these companion diagnostic tests is much cheaper. Furthermore, the results obtained outside of companion diagnostics will not be taken into consideration for decision making about treatment even when the test analyzes a number of genes. If a physician uses a test result during standard care, the difference between the amount of reimbursement for companion diagnostics and the actual cost paid to Foundation Medicine will be a deficit for the hospital. If the physician wants to use the test results when the patient experiences progression while receiving standard therapy, additional reimbursement can be submitted as a fee for MTB (¥480,000). However, considering the possibilities that the patient dies during standard care or fails to follow-up, it is unlikely that genome profiling would be used during standard therapy in Japan under current reimbursement rules.
Although only a few hospitals run these genome panels in-house, most hospitals outsource these genomic profiling tests to a clinical testing company. Laboratories in core and hub hospitals are required to be certified by independent organizations such as ISO15189 to handle patient samples. Clinical testing companies also have Clinical Laboratory Improvement Amendments certification. Formalin-fixed and paraffin-embedded samples (and also blood samples in case of OncoGuide NCC Oncopanel System) are prepared in each hospital and sent to the company. The company analyzes the NCC Oncopanel by themselves or conveys samples to Foundation Medicine for FoundationOne CDx. Turnaround time in general is 16 to 22 days.
To fulfill its function as a data repository and to facilitate access to clinical data, a requirement of test granted by the government is that physicians need to submit detailed clinical data from patients with cancer to C-CAT, including diagnostics, treatment, and outcomes information, as well as the raw BAM or FASTQ and VCF or XML files (Fig 2; Appendix Table A1). In Japan, each hospital has an electronic platform that contains individual health records for patients. However, each platform is standalone and not shared with other hospitals. Test results are usually reported in PDF files that are incorporated into electronic medical records, making it difficult for physicians to match patients with ongoing clinical trials or to identify patients who are eligible for new drugs when they become available. Thus, a central data repository at C-CAT will make it easier to identify candidates for clinical trials in a timely manner.
Using the submitted data, C-CAT also issues their own report. Currently, CKDB consists of two databases. CKDB1 is further divided into four databases: marker database listing genomic abnormalities such as EGFR mutation or BRCA1 germline mutation; drug database listing approved drugs or drugs under clinical trials in and outside of Japan and their targets; evidence database curated from biologic, clinical, and therapeutic information in multiple public information resources, including CIViC (Clinical Interpretation of Variants in Cancer), BRCA Exchange, ClinVar, and COSMIC (Catalogue of Somatic Mutations in Cancer); and a clinical trial database generated from ClinicalTrials.gov as well as several Japanese clinical trial registries. CKDB2 uses the QIAGEN Clinical Insight interpretation platform. Through CKDBs, C-CAT can offer more attention to providing information customized for Japanese initiatives. For example, information of single nucleotide polymorphisms dominantly observed in Japanese or clinical trials run only in Japan can be included in the database. The evidence levels for therapeutic efficacy are categorized A to F, following the guideline from JSMO/JSCO/JCA (Table 3). MTBs in core and hub hospitals discuss reports from genomic tests (ie, NCC Oncopanel or FoundationOne CDx) as well as C-CAT and generate final reports for the patients.
TABLE 3.
Clinical and/or Experimental Evidence Levels Defined by C-CAT and Equivalent Evidence Levels in Other Guidelines
CURRENT ISSUES
Japan has made great strides in implementing a country-wide precision oncology system that has the ability to organize and harness the ever-increasing amount of genomic information to improve patient outcomes. The biggest current challenge for Japanese precision oncology seems to be the accessibility to drugs. For instance, a patient with a druggable mutation needs to be treated by the appropriate drug, even as an off-label indication in the absence of a clinical trial or current approval. However, off-label drug use is prohibited under Japanese regulations unless the patient pays the entire cost of care. Patient-Requested Therapy System, a Japanese compassionate use program, has to be initiated by the request from a patient to the government with required documentation from physician. Because the preparation of documents is a heavy burden for physicians, the system has not been widely adopted. Also, as mentioned above, the cost of the drug has to be fully paid by the patient. The flexible applications of this framework and the expansions of drug supply from pharmaceutical companies are expected. Furthermore, the drawback of drug access could be improved by launching large basket and umbrella trials. The consortium of core hospitals is expected to run clinical trials in collaboration with C-CAT. Patients at affiliated hospitals will be referred to core hospitals to enroll in clinical trials. To enable large basket/umbrella trials, a core facility supporting the protocol creation, drug distribution, monitoring, and audit will be needed, similar to the Cancer Therapy Evaluation Program in the United States.
The other big challenge facing Japanese precision oncology is the timing of reimbursements for genome profiling tests. According to the Japanese MHLW, genome profiling is currently applied only to patients who finished standard chemotherapy, to restrict unnecessary investigations and reduce the burden for MTBs. In addition, it was difficult to grant the use of cancer genomic profiling during standard care because clear evidence for the benefit of testing did not exist when consensus clinical practice guidelines for NGS-based cancer testing were published. For example, French clinical trials suggested the use of molecularly targeted agents outside established indications do not improve progression-free survival for heavily pretreated patients with cancer when compared with oncologists’ preferred treatment regimen.20 However, patients who experienced progression while receiving standard chemotherapy tend to have poor performance status and may not have enough time to wait for the results of genomic testing; even in the very best case scenarios, their tumors will continue to grow and their health worsen each day that they are required to wait. Therefore, tests before or during standard therapies are much more optimal for patients to promote their best care. Along these lines, the use of gene panel tests for companion diagnostic testing at the same time as genomic profiling during standard therapy could be considered by amending reimbursement rules. Currently, uncovered genome screening services are the only option for patients without rare tumors and before disease progression with standard therapy.
Quality and sustainability of MTBs are also challenging. The requirement of an MTB dictates it include medical oncologists, multiple geneticists, pathologists, and medical biologists. In addition, in the case of in-house genomic testing, a bioinformatician is also required. However, national guidelines for quality and procedure for MTBs do not exist. MTB meetings are also required to be held more than once a month. Ideally, MTB discussions are held once a week or every 2 weeks to shorten turnaround time. In addition, MTB meetings may routinely involve video conference system participation from liaison hospitals. However, a problem that arises is that it is challenging to demand this level of participation of physicians, given their limited availibility.21
Overall, the cost for precision oncology is a heavy burden for hospitals. Although the price for reimbursements has been defined, the net revenue is still unclear. Also, there is a need to hire technical assistants for administration. For example, the electronic data capture system in C-CAT requires submission of approximately 100 clinical characteristics, including prior treatments and toxicities worse than grade 3 (Appendix Table A1).
NEXT STEPS FOR JAPANESE PRECISION ONCOLOGY
It has been 4 years since the Japanese Headquarters for Healthcare Policy started discussing how to implement precision oncology in Japan. To put this into global perspective, Memorial Sloan Kettering Cancer Center has already achieved clinical sequencing of 10,000 patients in the United States.22 Furthermore, the United Kingdom finished sequencing for > 100,000 whole genomes, including 26,488 from patients with cancer. Although cancer genome profiling analysis is an efficient way to screen driver oncogenes under the national health care system, it cannot identify new targets and biomarkers. In September 2019, the MHLV unveiled a project with the goal of sequencing whole genomes from 100,000 patients with cancer over 3 years, a number chosen by referring to the United Kingdom’s sequencing achievements. Although details of the project have not emerged yet, it is expected to result in the analyses of fresh frozen samples in collaboration with biobanks at core and hub hospitals. In addition, these frozen samples may be used for multiomics analysis in the future. As the primary aim is to accelerate the development of new diagnostics and treatments, the project should be research based and funded by the Japanese government and private sources. In the case of the United Kingdom, the 100,000 Genomes Project was funded with £300 million over 5 years from various public and private parties. The MHLV also has a digital health initiative program, which began in 2017, in which precision oncology is one of the prioritized areas. Although the primary purpose of this initiative in precision oncology is to harness artificial intelligence (AI) for improved genomic analyses and drug target identification, the use of AI could connect genomic data with other prioritized areas in this initiative, such as pathology and radiology. The relatively homogenous genetic background of the Japanese population and the detailed clinical outcomes collected by C-CAT will be an advantage when harnessing the power of genomic data to develop new therapies. Also, the genomic and clinical data would be integrated with other Japanese databases, such as the Medical Genomics Japan Variant Database and the Japanese Multi Omics Reference Panel.23
CONCLUSION
In conclusion, precision oncology covered by the health insurance system has just begun in Japan. The total number of tests annually is estimated to be > 13,000, resulting in robust cancer genomic data storage of Asian patients in C-CAT. On one hand, harnessing the power of personalized genomics through this national system to better treat patients is an unprecedented opportunity. It could and should provide a significant advantage to establishing research databases including both genomic and clinical information and to conduct better and more successful and focused clinical trials. On the other hand, although the Japanese health care system has so far achieved excellent health outcomes with a relatively low cost,24 the centralized structure under the national health insurance system with its inherent tight regulation may cause difficulty in keeping up with the rapid development of precision oncology. This takes on even greater significance because of the aging population in Japan, with those ≥ 75 years of age making up > 30% of the population in 2025. In the United Kingdom, Genomics England was founded as a subsidiary limited company, as it was the most effective way to ensure the project running as quickly as possible. Even if concerns with keeping up with innovation are met, there is a more generalized skepticism that a centralized hub like C-CAT can truly work. In the United States, for instance, protocols and treatment decisions surrounding personalized medicine are largely decided by the institutions treating the patients, with governing bodies like the National Cancer Institute playing ancillary roles and the ability to pay for patient care largely dictated by individuals’ private insurance policies. Ultimately, like every great biomedical advance, there is a cost to harness its potential, and personalized cancer therapy is no different. As such, the success of personalized genomics in Japan may come down to how well the potential to improve cancer outcomes is balanced with a sensible and sustainable method to pay for it.
ACKNOWLEDGMENT
We thank Anthony Faber, MD (VCU Massey Cancer Center), for critical reading of the manuscript.
Appendix
TABLE A1.
Clinical Information Required to Submit to C-CAT
Footnotes
Supported by the Fund for the Promotion of Joint International Research Grant No. 15KK0303 from the Japan Society for the Promotion of Science (H.E.) and the Japan Agency for Medical Research and Development Grant No. 19lk0201065h0003 (H.E. and H.B.).
AUTHOR CONTRIBUTIONS
Conception and design: All authors
Collection and assembly of data: All authors
Data analysis and interpretation: All authors
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
Hiromichi Ebi
Honoraria: Taiho Pharmaceutical, AstraZeneca
Consulting or Advisory Role: Merck Serono, Eli Lilly, Astellas Pharma
Hideaki Bando
Honoraria: Taiho Pharmaceutical, Lilly Japan, Takeda, Chugai Pharma, Sanofi, Yakult Honsha
Research Funding: AstraZeneca, Sysmex
No other potential conflicts of interest were reported.
REFERENCES
- 1. Sakamoto H, Rahman M, Nomura S, et al: Japan Health System Review. World Health Organization. Regional Office for South-East Asia. https://apps.who.int/iris/handle/10665/259941.
- 2.Naito Y, Takahashi H, Shitara K, et al. Feasibility study of cancer genome alterations identified by next generation sequencing: ABC study. Jpn J Clin Oncol. 2018;48:559–564. doi: 10.1093/jjco/hyy052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kohno T, Ichikawa H, Totoki Y, et al. KIF5B-RET fusions in lung adenocarcinoma. Nat Med. 2012;18:375–377. doi: 10.1038/nm.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bando H, Yoshino T, Shinozaki E, et al. Simultaneous identification of 36 mutations in KRAS codons 61 and 146, BRAF, NRAS, and PIK3CA in a single reaction by multiplex assay kit. BMC Cancer. 2013;13:405. doi: 10.1186/1471-2407-13-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hovelson DH, McDaniel AS, Cani AK, et al. Development and validation of a scalable next-generation sequencing system for assessing relevant somatic variants in solid tumors. Neoplasia. 2015;17:385–399. doi: 10.1016/j.neo.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. National Cancer Center Hospital East: SCRUM-Japan. http://www.scrum-japan.ncc.go.jp/
- 7.Yoh K, Seto T, Satouchi M, et al. Vandetanib in patients with previously treated RET-rearranged advanced non-small-cell lung cancer (LURET): An open-label, multicentre phase 2 trial. Lancet Respir Med. 2017;5:42–50. doi: 10.1016/S2213-2600(16)30322-8. [DOI] [PubMed] [Google Scholar]
- 8.Taniguchi H, Okamoto W, Muro K, et al. Clinical validation of newly developed multiplex kit using Luminex xMAP technology for detecting simultaneous RAS and BRAF mutations in colorectal cancer: Results of the RASKET-B study. Neoplasia. 2018;20:1219–1226. doi: 10.1016/j.neo.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bando H, Okamoto W, Fukui T, et al. Utility of the quasi-monomorphic variation range in unresectable metastatic colorectal cancer patients. Cancer Sci. 2018;109:3411–3415. doi: 10.1111/cas.13774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bando H, Kagawa Y, Kato T, et al. A multicentre, prospective study of plasma circulating tumour DNA test for detecting RAS mutation in patients with metastatic colorectal cancer. Br J Cancer. 2019;120:982–986. doi: 10.1038/s41416-019-0457-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kohno T. Implementation of “clinical sequencing” in cancer genome medicine in Japan. Cancer Sci. 2018;109:507–512. doi: 10.1111/cas.13486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sunami K, Ichikawa H, Kubo T, et al. Feasibility and utility of a panel testing for 114 cancer-associated genes in a clinical setting: A hospital-based study. Cancer Sci. 2019;110:1480–1490. doi: 10.1111/cas.13969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kou T, Kanai M, Matsumoto S, et al. The possibility of clinical sequencing in the management of cancer. Jpn J Clin Oncol. 2016;46:399–406. doi: 10.1093/jjco/hyw018. [DOI] [PubMed] [Google Scholar]
- 14.Kou T, Kanai M, Yamamoto Y, et al. Clinical sequencing using a next-generation sequencing-based multiplex gene assay in patients with advanced solid tumors. Cancer Sci. 2017;108:1440–1446. doi: 10.1111/cas.13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. doi: 10.1136/bmj.k1687. Turnbull C, Scott RH, Thomas E, et al: The 100,000 Genomes project: Bringing whole genome sequencing to the NHS. BMJ 361:k1687, 2018 [Erratum BMJ 361:k1952, 2018] [DOI] [PubMed] [Google Scholar]
- 16.Lethimonnier F, Levy Y. Genomic medicine France 2025. Ann Oncol. 2018;29:783–784. doi: 10.1093/annonc/mdy027. [DOI] [PubMed] [Google Scholar]
- 17.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sunami K, Takahashi H, Tsuchihara K, et al: Clinical practice guidance for next-generation sequencing in cancer diagnosis and treatment (Edition 1.0). Cancer Sci 109:2980-2985, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li MM, Datto M, Duncavage EJ, et al. Standards and guidelines for the interpretation and reporting of sequence variants in cancer: A joint consensus recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J Mol Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Tourneau C, Delord JP, Gonçalves A, et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): A multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015;16:1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 21.Zuckerman DS. Cancer centers hitting the target: We are precise, but are we accurate? J Oncol Pract. 2019;15:305–306. doi: 10.1200/JOP.19.00117. [DOI] [PubMed] [Google Scholar]
- 22. doi: 10.1038/nm.4333. Zehir A, Benayed R, Shah RH, et al: Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med 23:703-713, 2017 [Erratum: Nat Med 23:1004, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tadaka S, Saigusa D, Motoike IN, et al. jMorp: Japanese multi omics reference panel. Nucleic Acids Res. 2018;46:D551–D557. doi: 10.1093/nar/gkx978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikegami N, Yoo BK, Hashimoto H, et al. Japanese universal health coverage: Evolution, achievements, and challenges. Lancet. 2011;378:1106–1115. doi: 10.1016/S0140-6736(11)60828-3. [DOI] [PubMed] [Google Scholar]






