Abstract
Purpose
The complexity of results generated from whole-genome sequencing (WGS) and whole-exome sequencing (WES) adds challenges to obtaining informed consent in pediatric oncology. Little is known about knowledge of WGS and WES in this population, and no validated tools exist in pediatric oncology.
Methods
We developed and psychometrically evaluated a novel WGS and WES knowledge questionnaire, the Precision in Pediatric Sequencing Knowledge Questionnaire (PIPseqKQ), to identify levels of understanding among parents and young adult cancer survivors (≥ 18 years old), off therapy for at least 1 year from a single-institution pediatric oncology outpatient clinic. Participants also completed health literacy and numeracy questionnaires. All participants provided written informed consent.
Results
One hundred eleven participants were enrolled: 76 were parents, and 35 were young adults. Of the total cohort, 77 (69%) were female, 63 (57%) self-identified as white, and 74 (67%) self-identified as non-Hispanic. Sixty-six (59%) had less than a college degree. Adequate health literacy (n = 87; 80%) and numeracy (n = 89; 80%) were demonstrated. Internal consistency was high (Cronbach’s α = .88), and test-retest reliability was greater than the 0.7 minimum requirement. Scores were highest for genetic concepts related to health and cancer and lowest for WGS and WES concepts. Health literacy and educational attainment were significantly associated with PIPseqKQ scores. Overall, participants felt the benefits of WGS and WES outweighed the potential risks.
Conclusion
Parents and young adult cancer survivors have some genetics knowledge, but they lack knowledge about WGS and WES. The PIPseqKQ is a reliable tool that can identify deficits in knowledge, identify perceptions of risks and benefits of WGS and WES, and help clinicians tailor their consent discussions to best fit families. The PIPseqKQ also may inform the development of educational tools to better facilitate the informed consent process in pediatric oncology.
INTRODUCTION
Advances in next-generation sequencing, particularly whole-genome sequencing (WGS) and whole-exome sequencing (WES) have led to a deeper understanding of the pathogenesis of cancer and offer promising avenues for clinical applications.1,2 In pediatric oncology, WGS and WES is being used to identify targetable mutations, clarify diagnosis, provide risk stratification, and identify pharmacogenomic modifiers.3-6 Although these genome-scale sequencing methods yield a wide spectrum of potential results, only a few are medically actionable; a large number are classified as variants of unknown significance (VUS). Furthermore, WGS and WES of matched tumor–normal tissue can identify pathogenic germline mutations not related to the indication for which testing was ordered, commonly referred to as secondary findings, and can identify carrier status for heritable conditions. The American College of Medical Genetics has defined a list of secondary findings that include noncancerous germline disease-causing mutations and cancer susceptibility mutations recommended for return by clinical laboratories.7,8 Such findings may be of clinical significance not only to the patient but also to family members not directly involved in testing.9-13 An understanding of these complexities is necessary for parents and patients to make informed decisions about consent to genomic sequencing.14,15
For most pediatric cancers, standard treatment protocols are highly effective, and WGS and WES results infrequently alter the initial treatment plan.16 However, among parents of pediatric patients with cancer, the hope that genomic sequencing will lead to a cure often exceeds the expected results of testing.17,18 Thus, parents may be more inclined to participate in WGS and WES without fully understanding the range of results that may be returned, including VUS and secondary findings.17,18 Even within the general public, including those who are well educated, there is a relatively low understanding of genetic concepts.19 Research to examine knowledge held by parents and young adult patients about the utility and limitations of genome sequencing is scarce, although there is evidence to suggest that participants with lower educational attainment or those from racial/ethnic minority groups may possess less knowledge.14 Within pediatric oncology, there are almost no data related to parent and young adult patient knowledge about genetics and WGS and WES, and there are no validated tools to assess these concepts in pediatric cancer.
As part of our clinical sequencing program at Columbia University Medical Center (CUMC), we developed the Precision in Pediatric Sequencing Knowledge Questionnaire (PIPseqKQ) to assess knowledge, attitudes, and expectations of parents and young adult patients about WGS and WES. The objectives of the study were to describe the development and psychometric properties of the PIPseqKQ and to identify preliminary levels of understanding within this population.
METHODS
Participants
Participants were recruited from the pediatric oncology outpatient clinic at CUMC between August 2015 and June 2016. Eligible participants included parents of pediatric patients with cancer and young adult cancer survivors age 18 years or older who were off therapy for at least 1 year. WGS and WES in pediatric oncology at CUMC commenced in January 2014, and 1 year off therapy was chosen as a cutpoint to minimize the likelihood that participants had undergone or had been exposed to the concepts of WGS and WES in the clinic setting. Study approval was received from the CUMC institutional review board.
Measures
PIPseqKQ development.
Initial items for the PIPseqKQ (Data Supplement) were selected or adapted from existing measures or measures in development that were identified in the literature.14,20-25 In addition, concepts presented in the CUMC WGS and WES clinical consent form were reviewed for additional item development. A total of 35 items and three knowledge domains were generated: general genetic concepts (n = 7 items), genetic concepts associated with general health and cancer (n = 12 items), and WGS and WES concepts (n = 16 items). A multidisciplinary team of experts from medical genetics, psychiatry, pediatric oncology, molecular pathology, nursing, and behavioral science reviewed the items and domains for content and clarity and to ensure good content validity across the three domains.26
Response categories included true, false, and not sure. Incorporation of the response of not sure minimized guessing and allowed uncertainty to be evaluated among participants. Correct responses were scored as one point; incorrect and not sure responses were scored as zero points. Total scores were generated for each domain and for the three domains combined. The maximum potential score was 35 for the total measure: one point per item. Scores also were converted to a percentage of correct responses. A fourth domain was developed to assess perceptions of the risks and benefits of sequencing; this domain consisted of nine subjective statements. Response categories included risk, benefit, or not sure for seven of nine items, and the remaining two items used true, false, or not sure. The final questionnaire was translated into Spanish by the Spanish Translation Center at CUMC.
Sociodemographic and medical characteristics.
Sociodemographic variables were self-reported on the intake questionnaire. Variables included sex, race/ethnicity, highest education level achieved, and household income. Patient diagnosis was abstracted from clinical records and was categorized into hematologic conditions, solid tumors, and brain tumors.
Health literacy and numeracy.
Health literacy was assessed with the Newest Vital Sign.27 The six-question validated tool was scored as follows: 4 or greater (adequate literacy), 2 to 3 (possibly limited literacy), and 0 to 1 (limited literacy). Numeracy was evaluated with an abbreviated (five-question) version of a validated objective numeracy scale,28 which assessed the ability to comprehend risks and probabilities of disease. Adequate numeracy was defined as correct answers to at least three questions. Both measures were available in English and Spanish.
Procedures
Electronic medical records were reviewed to identify potential participants. Eligible participants were approached by a treating physician and research coordinator during a routine clinic visit to discuss the study. After informed consent was obtained, participants completed a demographics questionnaire, the first PIPseqKQ, the Newest Vital Sign, and the numeracy questionnaire. Participants were contacted 2 weeks later by e-mail or standard post to complete the second PIPseqKQ (required to be completed within 4 weeks after the first administration). Participants who completed both surveys received a $15 gift card.
Data Analysis
PIPseqKQ responses were examined for high levels of nonresponse, which indicated an issue with an item. Descriptive statistics were examined for all items. Internal consistency reliability was evaluated by Cronbach’s α.29 Pearson’s correlation was used to assess test-retest reliability for participants who completed the PIPseqKQ twice. Minimum criteria for internal consistency and test-retest reliability was 0.7.30,31
Floor and ceiling effects were evaluated for item difficulty. A floor or ceiling effect was present if more than 10% of participants achieved the lowest or highest possible score, respectively. Item discrimination was measured by item-total correlations. Negative or low item-total correlations less than the standard 0.2 cutpoint indicated items to be reworded or discarded.32 To preserve content validity, items that tested an important aspect of knowledge not covered elsewhere in the questionnaire were retained.
Exploratory analyses were conducted by using two-tailed independent sample t tests to examine differences in PIPseqKQ scores for select dichotomized demographic variables: race/ethnicity (White non-Hispanic v others) and educational attainment (less than college/college v graduate/postgraduate). PIPseqKQ scores also were evaluated by literacy level (adequate v possibly limited/limited). All analyses were conducted with IBM SPSS Statistics for Windows, version 23 (IBM, Armonk, NY).
RESULTS
Participants
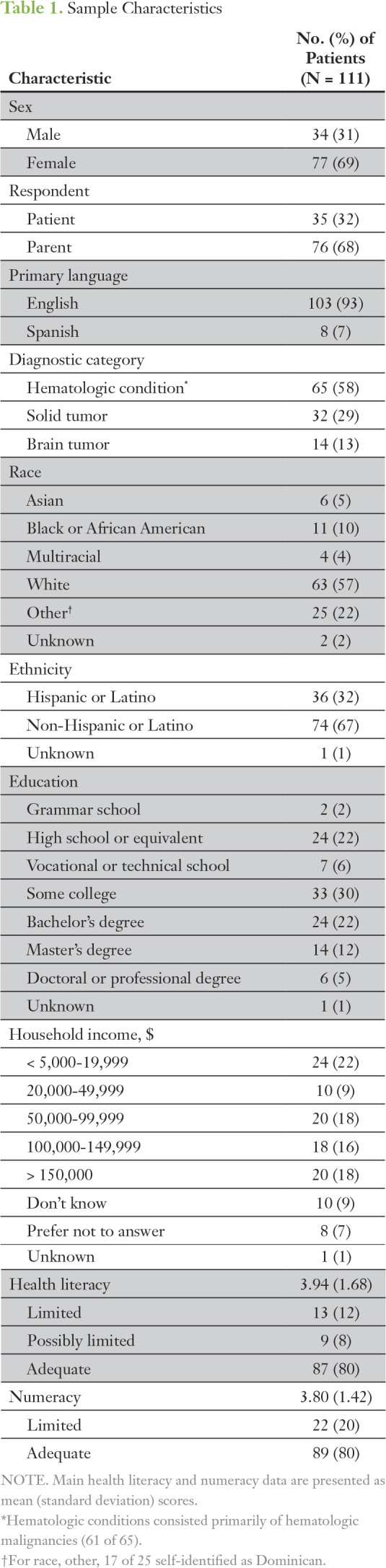
Of the 135 eligible participants, 11 (8%) declined participation and 13 (10%) were not approached because of timing conflicts (eg, rapid triage, radiology scans) during their clinic visits. One hundred eleven participants (82%) signed consent, and 110 participants completed the baseline questionnaires. None of the participants had WGS or WES or had received genetic counseling in relation to their cancer diagnosis. The final sample included 76 parents and 35 patients (Table 1). Just more than three quarters were women, and approximately 36% were racial/ethnic minorities. Eight participants (7%) spoke Spanish only. Thirty percent had not attended college, and nearly 40% had a college degree or higher. Household income less than $50,000 per year was reported by 31% of participants. The most common diagnoses were hematologic malignancies (55%). Approximately 20% had limited/possibly limited health literacy. Numeracy was high; the majority of participants answered more than three questions correctly. Seventy-five participants (68%) completed the PIPseqKQ twice. There was no difference in scores between those who completed the PIPseqKQ twice (responders) versus once (nonresponders) or by sex, diagnostic category, race/ethnicity, education, or income.
Table 1.
Sample Characteristics
PIPseqKQ Scale and Item Descriptive Statistics
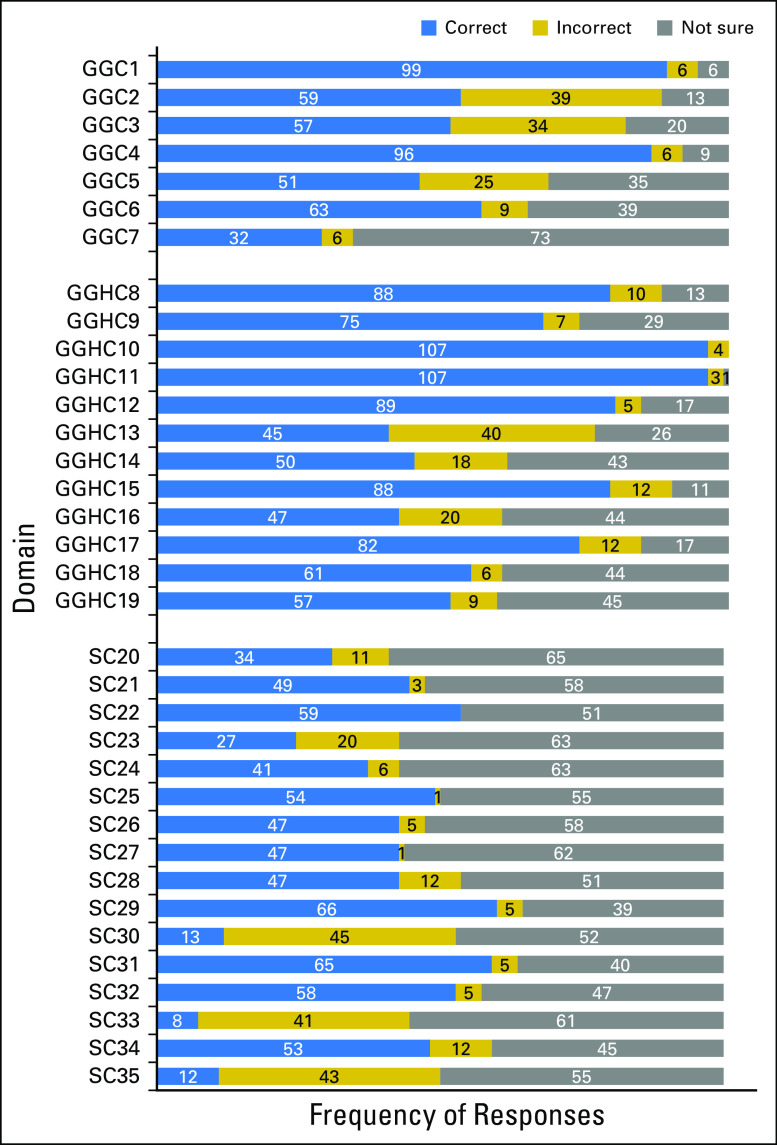
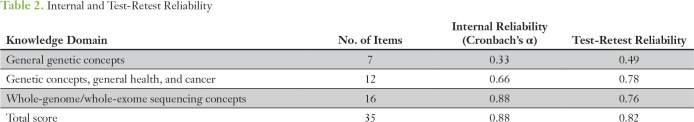
Item response distributions that showed items for which knowledge and uncertainty were highest and lowest are displayed in Figure 1. Items were well distributed across response choices. Only a ceiling effect was detected for three items, which were retained for their importance in evaluation of general genetic knowledge and genetics knowledge related to health and cancer. Correlations among all items were positive; the mean r was 0.52 (standard deviation [SD], 0.22; range, 0.07 to 0.96). All item-total correlations were positive; the mean r was 0.37 (SD, 0.20; range, 0.02 to 0.70), but 10 items were less than the 0.2 cutpoint and may require revision or removal (Data Supplement). Cronbach’s α was not affected by removal of any of the 10 items. Item rewording/removal will be considered for future versions of the PIPseqKQ. All 35 items were retained for analyses. Reliabilities are listed in Table 2. Internal consistency was high (Cronbach’s α, 0.88). Test-retest reliability from the 75 participants who completed the PIPseqKQ twice was 0.82.
Fig 1.
Precision in Pediatric Sequencing Knowledge Questionnaire (PIPseqKQ) item response distributions. GGC, general genetic concepts; GGHC, general genetic concepts related to health and cancer; SC, sequencing concepts.
Table 2.
Internal and Test-Retest Reliability
PIPseqKQ Scores
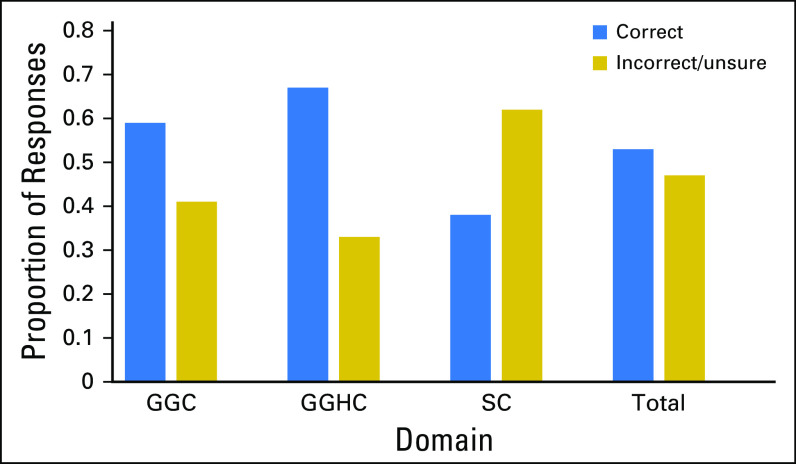
Figure 2 shows the proportion of correct and incorrect responses for the PIPseqKQ. Total scores ranged from 5 to 31 of 35, and the median score was 17.5 (mean, 18.39; SD, 6.87). The median proportion of patients who correctly answered each item was 0.50 (mean, 0.53; SD, 0.19; range, 0.14 to 0.89).
Fig 2.
Proportion of correct responses for the Precision in Pediatric Sequencing Knowledge Questionnaire and individual domains. GGC, general genetic concepts; GGHC, general genetic concepts related to health and cancer; SC, sequencing concepts.
Exploratory analyses revealed that higher health literacy and higher educational attainment were associated with a higher number of correct answers for the total measure and for genetic concepts associated with general health and cancer and with sequencing concepts. White non-Hispanic participants scored marginally higher for the total measure and for genetic concepts associated with general health and cancer (Fig 3).
Fig 3.
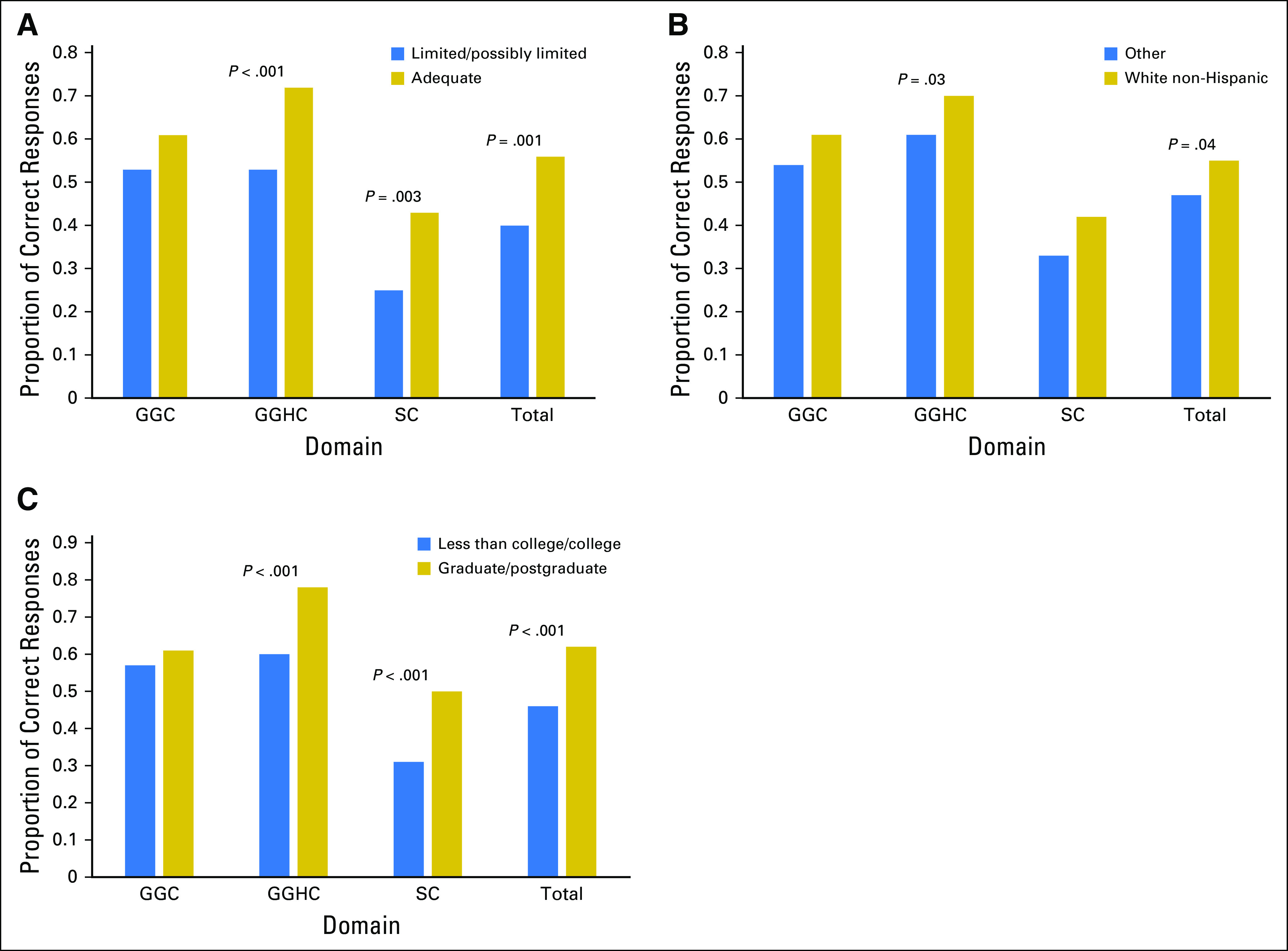
Proportion of items correct for select demographic characteristics: (A) literacy level, (B) race/ethnicity, and (C) educational attainment. GGC, general genetic concepts; GGHC, general genetic concepts related to health and cancer; SC, sequencing concepts.
General genetic concepts.
The median domain score for general genetic concepts was 4.0 (mean, 4.11; SD, 1.41; range, 1 to 7 of 7). The median proportion of participants who correctly answered each domain item was 0.57 (mean, 0.59; SD, 0.20; range, 0.14 to 1.00). The two items with the most incorrect or not sure responses were the definition of an exome (n = 79; 71%) and that statement about blood containing a full copy of your genes (n = 60; 53%).
Genetic concepts related to general health and cancer.
The median domain score for genetic concepts related to general health and cancer was 8.0 (mean, 8.07; SD, 2.37; range, 1 to 12 of 12). The median proportion of participants who correctly answered each domain item was 0.66 (mean, 0.67; SD, 0.19; range, 0.08 to 1.00). The items answered incorrect or not sure most often were “If a parent has a change in his/her DNA that causes a health problem, they can pass this change to their child” (n = 66; 59%); “A person with a change in a colon cancer gene may never develop colon cancer” (n = 61; 55%); and “A person’s genes can influence if they will experience side effects (bad reactions) or benefits from a medication” (n = 64; 58%).
WGS and WES concepts.
The median sequencing concepts domain score was 6.0 (mean, 6.18; SD, 4.44; range, 0 to 14 of 16). The median proportion of participants who correctly answered each domain item was 0.37 (mean, 0.38; SD, 0.27; range, 0.00 to 0.88). The most frequent items answered incorrectly or not sure were “WGS/WES in children with cancer performed at diagnosis will commonly guide initial treatment” (n = 97; 88%); “There are laws that prevent the use of genetic information to determine eligibility for life insurance, disability insurance, and long-term care insurance” (n = 102; 93%); and “WGS/WES will have no impact on cancer care for most children with cancer” (n = 98; 89%). Greater than 50% of participants were uncertain about the number of genes that can be tested with WGS and WES (ie, a small subset v the complete set of genes), whether WGS and WES compares the DNA of cancer cells to the DNA of normal cells, whether WGS and WES can find changes in genes related to diseases other than cancer, and what VUS means. Last, although 60% understood that secondary findings also may provide information about present and future health risks of other family members, 36% were unsure of the correct answer.
Perceptions of risks and benefits of WGS and WES.
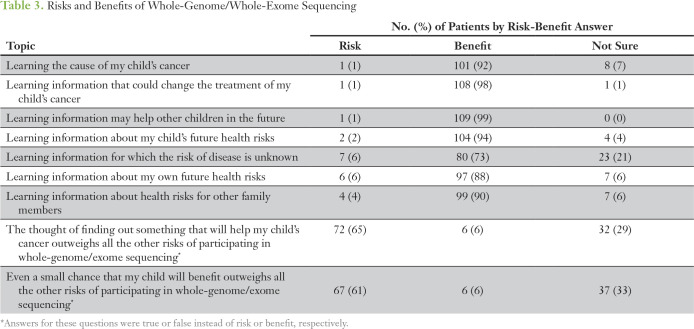
In general, participants view WGS and WES as beneficial (Table 3). Altruism was high; 99% of participants felt that learning information that may help other children in the future was a benefit. A majority felt that learning information about their own future health risks, future health risks for their child, and health risks for other family members was beneficial. Participants generally (73%) felt that learning information for which the risk of disease is unknown was beneficial. Sixty-one percent felt that even a small chance that their child would benefit from WGS and WES outweighed all other risks of WGS and WES.
Table 3.
Risks and Benefits of Whole-Genome/Whole-Exome Sequencing
DISCUSSION
This study used a newly developed questionnaire to examine genome-sequencing knowledge from parents and young adult pediatric cancer survivors to identify areas of comprehension that may require additional education before informed consent for WGS and WES is obtained. The PIPseqKQ performed better than the set minimum criteria for internal consistency and reliability. Furthermore, content validity was high, because item generation focused on areas of knowledge necessary to make informed decisions about WGS and WES.
Consistent with studies in other populations,14,24,33 participants had a basic understanding of genetics and heredity. Participants with limited literacy or with less than a college degree had lower knowledge. Limited literacy34,35 and educational attainment14,21,23,36-38 have been associated with genetic knowledge. Lower knowledge has been associated with racial and ethnic minority groups,14 and we observed this for knowledge related to genetic concepts associated with health and cancer.
The strengths and limitations of WGS and WES were not well understood. Deficits in knowledge were observed for items related to the likelihood of WGS and WES to guide initial treatment or to have an effect on cancer care for most children and whether laws existed to prevent discrimination for certain types of insurance on the basis of genetic information. Similar to Tolusso et al,39 we found that participants were uncertain about the ability of WGS and WES to examine the complete set of genes, compare tumor and normal DNA, find changes in genes related to diseases other than cancer, and identify VUS.
Although there is strong parent/patient interest in genomic sequencing,17,40-43 its potential benefit in cancer care was overestimated. In a recent study of adult patients with advanced cancer, 64% who underwent genomic testing had a strong belief that testing would significantly improve their cancer care.42 Similarly, results from the Dana Farber Cancer Institute44 revealed that parents and patients express significant hopes for molecular testing, particularly regarding increased number of treatment options (72%) and increased hope for a cure (59%).17 In our study, 61% felt that even a small chance that their child would benefit outweighed all other risks of WGS and WES. In addition, a majority thought WGS and WES performed at diagnosis would commonly guide initial treatment and would affect care for most children with cancer. Thus far, relatively few pediatric patients with cancer who undergo genomic testing are matched to a targeted therapy.3,4,6,44 These findings reflect two salient issues in pediatric oncology. The first reflects the reality that the majority of clinically significant findings from next-generation sequencing are not directly targetable by current anticancer therapies. The second relates to the ability to intervene with targeted therapies. Many of the newer molecularly targeted drugs lack efficacy data in pediatric diseases or safety data in children and, therefore, are not approved for administration in this population. Consequently, families should be provided with realistic information during consent discussions to manage expectations when they consider genomic testing.
Our previous work indicates that families strongly desire more education and improved modalities to communicate WGS and WES information before consent for genomic sequencing.18 Similarly, parents have reported that the standard consent process for genome-wide sequencing was not adequate and have requested availability of more resources to aid their understanding.45 The PIPseqKQ is a valid tool that can help identify knowledge deficits and guide the development of patient education materials to improve understanding of WGS and WES. Different educational resources, including Web-based technologies, written educational materials, and videos, should be developed. Our center is testing the effectiveness of a Web-based tool in English and Spanish as an educational resource to improve patient education and informed consent for clinical WGS and WES.
This study had a number of limitations to consider, which represent important areas for future research. First, this was a single-institution study, which affects the generalizability of results. Second, those surveyed had children who were off therapy for more than 1 year, and their prolonged exposure to treatment in a pediatric oncology center may have biased their knowledge of the concepts presented here, including their assessment of risk and benefit. Third, we did not have sufficient sample size to validate the PIPseqKQ Spanish version, evaluate cultural differences for non-English speaking participants, or conduct factor analyses or item-response theory analyses. Furthermore, for the 10 items that require revision or removal, scores may reflect misunderstanding of the statement rather than a lack of knowledge. Last, a factor not addressed by the PIPseqKQ that is being investigated through ancillary studies relates to evaluation of the potential cost to families for clinical WGS and WES. Insurance companies provide inconsistent coverage and variable payment for WGS and WES, and families will need to understand the financial responsibility they may incur from testing.
In conclusion, as WGS and WES become more ubiquitous in pediatric oncology, medical providers will need to help families understand the benefits and limitations of testing, including the potential financial cost to the family for testing and the possible identification of secondary findings. The PIPseqKQ can be administered by multidisciplinary specialists, including advanced genetics nurses or genetic counselors, to help address gaps in patient knowledge related to WGS and WES. This may be especially advantageous for providers who lack the expertise or time to fully engage with their patients about testing.
Our findings highlight that, although participants understand that there are risks and benefits of WGS and WES, many would accept any risk for the possibility of a small benefit, and their hope for a cure often exceeds the expected results from sequencing. The PIPseqKQ is a valid tool that can aid providers in precision oncology to identify levels of knowledge and tailor their informed consent discussions to best fit the patient and family. This will help foster more realistic expectations for testing and its potential impact on cancer care. Future research is needed to develop educational tools to facilitate informed consent discussions for WGS and WES in pediatric oncology.
Footnotes
Presented in part at the Annual Meeting of the American Society of Clinical Oncology, Chicago, IL, June 3-7, 2016.
Supported by Grants No. UL1 TR00004 (J.M.L., J.A.O., and T.A.-S.) and P50 HG007257 (P.S.A.) and by the Conquer Cancer Foundation of the American Society of Clinical Oncology Medical Student Rotation Award (J.R.).
AUTHOR CONTRIBUTIONS
Conception and design: Jennifer A. Oberg, Andrew L. Kung, Julia L. Glade Bender, Jennifer M. Levine
Collection and assembly of data: Jennifer A. Oberg, Jenny Ruiz, Trisha Ali-Shaw, Kathryn A. Schlechtweg, Wendy K. Chung, Jennifer M. Levine
Provision of study material or patients: Julia L. Glade Bender
Data analysis and interpretation: Jennifer A. Oberg, Angela Ricci, Wendy K. Chung, Paul S. Appelbaum, Jennifer M. Levine
Financial support: Andrew L. Kung
Administrative support: Trisha Ali-Shaw, Julia L. Glade Bender
Manuscript writing: All authors
Final approval of manuscript: All authors
Agree to be accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Jennifer A. Oberg
No relationship to disclose
Jenny Ruiz
No relationship to disclose
Trisha Ali-Shaw
No relationship to disclose
Kathryn A. Schlechtweg
No relationship to disclose
Angela Ricci
No relationship to disclose
Andrew L. Kung
Consulting or Advisory Role: Darwin Health, MI Bioresearch, Imago Bioscience
Patents, Royalties, Other Intellectual Property: Royalty from licensing agreements with MI Bioresearch
Wendy K. Chung
Consulting or Advisory Role: Regeneron Genetics Center
Research Funding: Biogen
Paul S. Appelbaum
No relationship to disclose
Julia L. Glade Bender
Research Funding: Bristol-Myers Squibb (Inst), Pfizer (Inst), Novartis (Inst), Ignyta (Inst), Amgen (Inst), Celgene (Inst), Eisai (Inst), Lilly (Inst), Merck (Inst)
Travel, Accommodations, Expenses: Novartis, Amgen, Merck
Jennifer M. Levine
No relationship to disclose
REFERENCES
- 1.Downing JR, Wilson RK, Zhang J, et al. : The Pediatric Cancer Genome Project. Nat Genet 44:619-622, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vogelstein B, Papadopoulos N, Velculescu VE, et al. : Cancer genome landscapes. Science 339:1546-1558, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mody RJ, Wu YM, Lonigro RJ, et al. : Integrative clinical sequencing in the management of refractory or relapsed cancer in youth. JAMA 314:913-925, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oberg JA, Glade Bender JL, Sulis ML, et al. : Implementation of next generation sequencing into pediatric hematology-oncology practice: Moving beyond actionable alterations. Genome Med 8:133, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsons DW, Roy A, Yang Y, et al. : Diagnostic yield of clinical tumor and germline whole-exome sequencing for children with solid tumors. JAMA Oncol 2:616-624, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Worst BC, van Tilburg CM, Balasubramanian GP, et al. : Next-generation personalised medicine for high-risk paediatric cancer patients: The INFORM pilot study. Eur J Cancer 65:91-101, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Green RC, Berg JS, Grody WW, et al. : ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 15:565-574, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kalia SS, Adelman K, Bale SJ, et al. : Recommendations for reporting of secondary findings in clinical exome and genome sequencing, 2016 update (ACMG SF v2.0): A policy statement of the American College of Medical Genetics and Genomics. Genet Med 19:249-255, 2017 [DOI] [PubMed] [Google Scholar]
- 9.Caulfield T, McGuire AL, Cho M, et al. : Research ethics recommendations for whole-genome research: Consensus statement. PLoS Biol 6:e73, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Facio FM, Eidem H, Fisher T, et al. : Intentions to receive individual results from whole-genome sequencing among participants in the ClinSeq study. Eur J Hum Genet 21:261-265, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cassa CA, Savage SK, Taylor PL, et al. : Disclosing pathogenic genetic variants to research participants: Quantifying an emerging ethical responsibility. Genome Res 22:421-428, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdul-Karim R, Berkman BE, Wendler D, et al. : Disclosure of incidental findings from next-generation sequencing in pediatric genomic research. Pediatrics 131:564-571, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGuire AL, Caulfield T, Cho MK: Research ethics and the challenge of whole-genome sequencing. Nat Rev Genet 9:152-156, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaphingst KA, Facio FM, Cheng MR, et al. : Effects of informed consent for individual genome sequencing on relevant knowledge. Clin Genet 82:408-415, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scollon S, Bergstrom K, Kerstein RA, et al. : Obtaining informed consent for clinical tumor and germline exome sequencing of newly diagnosed childhood cancer patients. Genome Med 6:69, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward E, DeSantis C, Robbins A, et al. : Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin 64:83-103, 2014 [DOI] [PubMed] [Google Scholar]
- 17.Marron JM, DuBois SG, Glade Bender J, et al. : Patient/parent perspectives on genomic tumor profiling of pediatric solid tumors: The Individualized Cancer Therapy (iCat) experience. Pediatr Blood Cancer 63:1974-1982, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Oberg JA, Glade Bender JL, Cohn EG, et al. : Overcoming challenges to meaningful informed consent for whole-genome sequencing in pediatric cancer research. Pediatr Blood Cancer 62:1374-1380, 2015 [DOI] [PubMed] [Google Scholar]
- 19.Bowling BV, Huether CA, Wang L, et al. : Genetic literacy of undergraduate non–science majors and the impact of introductory biology and genetics courses. Bioscience 58:654-660, 2008 [Google Scholar]
- 20.Furr LA, Kelly SE: The Genetic Knowledge Index: Developing a standard measure of genetic knowledge. Genet Test 3:193-199, 1999 [DOI] [PubMed] [Google Scholar]
- 21.Henneman L, Timmermans DRM, van der Wal G: Public experiences, knowledge, and expectations about medical genetics and the use of genetic information. Community Genet 7:33-43, 2004 [DOI] [PubMed] [Google Scholar]
- 22.Jallinoja P, Aro AR: Does knowledge make a difference? The association between knowledge about genes and attitudes toward gene tests. J Health Commun 5:29-39, 2000 [DOI] [PubMed] [Google Scholar]
- 23.Molster C, Charles T, Samanek A, et al. : Australian study on public knowledge of human genetics and health. Public Health Genomics 12:84-91, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Morren M, Rijken M, Baanders AN, et al. : Perceived genetic knowledge, attitudes towards genetic testing, and the relationship between these among patients with a chronic disease. Patient Educ Couns 65:197-204, 2007 [DOI] [PubMed] [Google Scholar]
- 25.Singer E, Antonucci T, Van Hoewyk J: Racial and ethnic variations in knowledge and attitudes about genetic testing. Genet Test 8:31-43, 2004 [DOI] [PubMed] [Google Scholar]
- 26.DeVellis R: Scale Development: Theory and Applications (ed 2). Newbury Park, CA, Sage, 2003 [Google Scholar]
- 27.Weiss BD, Mays MZ, Martz W, et al. : Quick assessment of literacy in primary care: The newest vital sign. Ann Fam Med 3:514-522, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lipkus IM, Samsa G, Rimer BK: General performance on a numeracy scale among highly educated samples. Med Decis Making 21:37-44, 2001 [DOI] [PubMed] [Google Scholar]
- 29.Cronbach L: Coefficient alpha and the internal structure of tests. Psychometrika 16:297-334, 1951 [Google Scholar]
- 30.Kline P: The Handbook of Psychological Testing. London, United Kingdom, Routledge,1993 [Google Scholar]
- 31.Streiner D, Norman G. Health Measurement Scales. A Practical Guide to Their Development and Use (ed 2). Oxford, England, Oxford University Press, 1995 [Google Scholar]
- 32.Kline P: A Handbook of Test Construction. London, United Kingdom, Methuen & Co, 1986 [Google Scholar]
- 33.Streicher SA, Sanderson SC, Jabs EW, et al. : Reasons for participating and genetic information needs among racially and ethnically diverse biobank participants: A focus group study. J Community Genet 2:153-163, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaphingst KA, Blanchard M, Milam L, et al. : Relationships between health literacy and genomics-related knowledge, self-efficacy, perceived importance, and communication in a medically underserved population. J Health Commun 21:58-68, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Langer MM, Roche MI, Brewer NT, et al. : Development and validation of a genomic knowledge scale to advance informed decision-making research in genomic sequencing. MDM Policy & Practice 2:1-13, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Calsbeek H, Morren M, Bensing J, et al. : Knowledge and attitudes towards genetic testing: A two year follow-up study in patients with asthma, diabetes mellitus and cardiovascular disease. J Genet Couns 16:493-504, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jallinoja P, Aro AR: Knowledge about genes and heredity among Finns. New Genet Soc 18:101-110, 1999 [Google Scholar]
- 38.Rose A, Peters N, Shea JA, et al. : The association between knowledge and attitudes about genetic testing for cancer risk in the United States. J Health Commun 10:309-321, 2005 [DOI] [PubMed] [Google Scholar]
- 39.Tolusso LK, Collins K, Zhang X, et al. : Pediatric whole exome sequencing: An assessment of parents’ perceived and actual understanding. J Genet Couns 26:792-805, 2017 [DOI] [PubMed] [Google Scholar]
- 40.Gray SW, Hicks-Courant K, Lathan CS, et al. : Attitudes of patients with cancer about personalized medicine and somatic genetic testing. J Oncol Pract 8:329-335, 2, 335, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller FA, Hayeems RZ, Bytautas JP, et al. : Testing personalized medicine: Patient and physician expectations of next-generation genomic sequencing in late-stage cancer care. Eur J Hum Genet 22:391-395, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Blanchette PS, Spreafico A, Miller FA, et al. : Genomic testing in cancer: Patient knowledge, attitudes, and expectations. Cancer 120:3066-3073, 2014 [DOI] [PubMed] [Google Scholar]
- 43.McCullough LB, Slashinski MJ, McGuire AL, et al. : Is whole-exome sequencing an ethically disruptive technology? Perspectives of pediatric oncologists and parents of pediatric patients with solid tumors. Pediatr Blood Cancer 63:511-515, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harris MH, DuBois SG, Glade Bender JL, et al. : Multicenter feasibility study of tumor molecular profiling to inform therapeutic decisions in advanced pediatric solid tumors: The individualized cancer therapy (iCat) study. JAMA Oncol 2:608-615, 2016 [DOI] [PubMed] [Google Scholar]
- 45.Li KC, Birch PH, Garrett BM, et al. : Parents’ perspectives on supporting their decision making in genome-wide sequencing. J Nurs Scholarsh 48:265-275, 2016 [DOI] [PubMed] [Google Scholar]