Abstract
PURPOSE
To assess the feasibility and utility of circulating tumor DNA (ctDNA) by amplicon-based next-generation sequencing (NGS) analysis in the daily clinical setting in a cohort of patients with advanced non–small-cell lung cancer (NSCLC), as an alternative approach to tissue molecular profiling.
PATIENTS AND METHODS
In this single-center prospective study, treatment-naïve and previously treated patients with advanced NSCLC were enrolled. Clinical validation of ctDNA using amplicon-based NGS analysis (with a 36-gene panel) was performed against standard-of-care tissue molecular analysis in treatment-naïve patients. The feasibility, utility, and prognostic value of ctDNA as a dynamic marker of treatment efficacy was evaluated. Results of tissue molecular profile were blinded during ctDNA analysis.
RESULTS
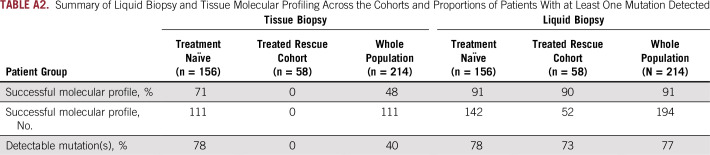
Of 214 patients with advanced NSCLC who were recruited, 156 were treatment-naïve patients and 58 were pretreated patients with unknown tissue molecular profile. ctDNA screening was successfully performed for 91% (n = 194) of all patients, and mutations were detected in 77% of these patients. Tissue molecular analysis was available for 111 patients (52%), and tissue somatic mutations were found for 78% (n = 87) of patients. For clinically relevant variants, concordance agreement between ctDNA and tumor tissue analysis was 95% among 94 treatment-naïve patients who had concurrent liquid and tumor biopsy molecular profiles. Sensitivity and specificity were 81% and 97%, respectively. Of the 103 patients with no tissue available, ctDNA detected potential actionable mutations in 17% of patients; of these, 10% received personalized treatment. ctDNA kinetics correlated with response rate and progression-free survival in 31 patients treated with first-line platinum-based chemotherapy.
CONCLUSION
These real-world data from a prospective study endorse ctDNA molecular profile by amplicon-based NGS as an accurate and reliable tool to detect and monitor clinically relevant molecular alterations in patients with advanced NSCLC.
INTRODUCTION
Tumor biomarker testing using clinical non–small-cell lung cancer (NSCLC) specimens in routine oncologic care evolved rapidly after approval of targeted therapies linked to diagnostic assays,1 and this therapeutic approach is considered standard in daily clinical practice2 because it has an impact on patient outcomes.3 This personalized therapeutic approach demands highly sensitive and effective technologies for molecular stratification.3 Both the European Society for Medical Oncology4 and ASCO guidelines5 make recommendations for next-generation sequencing (NGS) of molecular testing of key druggable alterations. However, real-world data show that less than 10% of patients with NSCLC are tested for the potential actionable (targetable) molecular alterations (EGFR mutations, ALK rearrangements, BRAF [p.v600E] mutations, ROS1 rearrangements, MET mutation, RET rearrangements, and HER2 mutations/amplifications) proposed by guidelines,6 and biopsy specimens can be inadequate for routine comprehensive profiling in up to 30% of cases.7,8 Targeted NGS of circulating tumor DNA (ctDNA) as a liquid biopsy offers the ability to profile a broad scope of genetic alterations on a single platform, which would overcome the limitations of tissue biopsy.9 ctDNA is now entering into routine clinical practice, and its use for patients with NSCLC is recommended in recent NCCN guidelines when tissue biopsy is not available or feasible10; however, there have been variable degrees of accuracy and performance results published to date.7
CONTEXT
Key Objective
To assess the feasibility and utility of circulating tumor DNA (ctDNA) by amplicon-based next-generation sequencing (NGS) analysis in a daily clinical setting in a cohort of patients with advanced non–small-cell lung cancer (NSCLC) as an alternative approach to tissue molecular profiling.
Knowledge Generated
These real-world data from a prospective study endorse ctDNA molecular profile by amplicon-based NGS as an accurate and reliable tool to detect and monitor clinically relevant molecular alterations in patients with advanced NSCLC. Also, they endorse the theory that liquid biopsy may predict earlier than standard radiologic criteria the effectiveness of platinum-based chemotherapy.
Relevance
This large, real-world, prospective study in patients with advanced NSCLC provides additional validation about ctDNA for molecular profiling and monitoring of alterations with high sensitivity and specificity to detect clinically relevant and actionable mutations when tissue biopsy is unavailable or uninformative. This study also suggests that ctDNA offers a potential prognostic biomarker for treatment efficacy.
Here, we assess prospectively in a real-world clinical setting the feasibility and effectiveness of an amplicon-based NGS assay (InVisionSeq, Inivata, Research Triangle Park, NC, and Cambridge, United Kingdom) with ctDNA analysis for routine molecular profiling in the largest cohort reported to date, to our knowledge, of patients with advanced NSCLC to identify clinically relevant mutations and evaluate those for whom tissue sequencing could not be conducted or was not performed.
PATIENTS AND METHODS
This single-center, prospective observational study was conducted at Gustave Roussy. Patients with advanced NSCLC were eligible for the study if they were treatment naïve for advanced disease and expected to receive first-line platinum-based chemotherapy (treatment-naïve cohort) or if the tissue-based molecular profile failed or was not performed on the primary tissue sample (treated rescue cohort; Appendix Fig A1). All patients provided written informed consent for biomedical research approved by the institutional ethics committee under one of the two studies (ClinicalTrials.gov identifier: NCT02105168; or ClinicalTrials.gov identifier: NCT02666612) to perform molecular analysis in tissue and plasma collections. In a subset of treatment-naïve patients (with or without tissue molecular profiles) who received standard platinum-based chemotherapy, additional blood specimens were collected at baseline and within 6 weeks of treatment initiation to match to the radiologic evaluation of the disease for clearance of baseline genomic alterations or emergence of new mutations after treatment.
The study aimed to correlate detection of molecular abnormalities in tissue versus blood in treatment-naïve patients, to evaluate the prognostic value of ctDNA as a dynamic biomarker of treatment efficacy, and to assess the feasibility and utility of liquid biopsy in patients for whom tissue analysis failed.
Blood Samples and ctDNA Isolation and Sequencing
Blood for plasma preparation was collected into a single 10-mL K2 EDTA tube before the start of platinum-based chemotherapy treatment or at the time of study enrollment for patients enrolled in the rescue cohort.
In this study, plasma analysis was performed using InVisionSeq Lung with an earlier 35-gene panel8 (n = 164) and a revised 36-gene panel9 (n = 50, treatment-naïve only; Appendix Fig A2). This tagged amplicon-based comprehensive genomic profiling assay identifies single nucleotide variants (SNVs), insertions, deletions, and amplifications. InVisionSeq Lung has recently undergone extensive analytic validation and demonstrates unprecedented sensitivity for identification of mutations by NGS across a gene panel. It detected 100% of SNVs with an expected mutation allele fraction (MAF) of 0.5% and greater; 99.48% of SNVs at an MAF in the range of 0.25% to 0.33%; 88.93% of SNVs at the MAF range of 0.13% to 0.16%; and more than 50% of SNVs at an MAF range of 0.06% to 0.08%.11 Methods were previously described .11,12 Results of tissue molecular profile were blinded during ctDNA analysis.
Tumor Tissue DNA Sequencing
The molecular analyses of EGFR (exons 18 to 21), HER2 (exon 20), KRAS (exon 2), BRAF (exon 15), and PIK3CA (exons 9 and 20) mutations were performed using either the Sanger sequencing method or a more sensitive validated allele-specific technique.3 Additional genes were analyzed on broader NGS panels in a subset of patients (n = 53).
Statistical Analyses: Clinical Validation, Utility, and Longitudinal Analyses
A review of the clinical history of eligible patients was done, and a clinical database was designed to collect patient demographics; diagnosis and histopathology details; and disease history, including tissue biopsy and prior cancer therapy. Descriptive summaries of liquid biopsy (ctDNA) and tissue molecular profiles were reported for all patients.
Clinical validation: concordance, sensitivity, and specificity analyses.
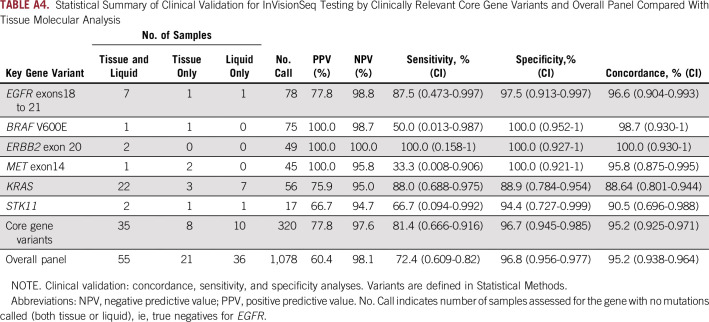
In this study, before analysis, we defined a core gene variant panel for clinically relevant gene mutation hotspots as follows: EGFR exons 18 to 21, BRAF V600, MET exon 14, ERBB2 ins 20, and KRAS; panel selections were based on recent recommendations of ASCO and International Association for the Study of Lung Cancer biomarker guidelines for NGS panels used in molecular profiling.5,13 On the basis of emerging clinical interest, STK1114 also was included in the core gene panel. Clinical validation (concordance, sensitivity, specificity) of InVisionSeq Lung for SNVs and indels was evaluated in patients with concurrent matched tissue analysis for these select gene variants. Overall performance of the assay also was reported for the broader gene panel, for which matched tissue and plasma testing was completed. Structural rearrangements (ALK, ROS1) were not evaluated in this study, because this testing was unavailable until the end of the study (as part of a revised plasma-based assay) when insufficient plasma volume remained from the initial collection.
All statistical analyses were performed using R version 3.2.5. In the concordance analysis, the data can be summarized as a two-by-two table, in which tissue data are the standard reference material. Described CIs are exact Clopper-Pearson CIs. Statistical methods are described in the Data Supplement. All patients with complete and partial tissue testing for the core genes and broader panel were included in the analysis, but they were only assessed for genes when both liquid biopsy and tissue molecular profiling were complete.
Reasons for exclusion from clinical validation analysis were as follows: tissue biopsy or molecular analysis failed or was not performed; ctDNA analysis failed; or the elapsed time between tissue biopsy and liquid biopsy specimen collection was longer than 100 days.
Clinical utility analysis.
The utility of an amplicon-based NGS liquid biopsy assay was performed by correlating the detection of clinically relevant molecular abnormalities in tissue versus blood in treatment-naïve patients and by assessing the feasibility and utility of liquid biopsy for reports of molecular profiles in patients whose tissue analysis failed or was not performed, specifically for actionable mutations that conferred sensitivity to approved or experimental therapies.
Longitudinal analysis.
To evaluate the prognostic value of ctDNA as a dynamic biomarker of treatment efficacy, all patients underwent tumor imaging, including computed tomography of the chest and abdomen and/or positron emission tomography scan at baseline. Brain imaging was performed in cases of symptoms. Restaging scans were obtained within 6 weeks after treatment initiation. The senior radiologist (C.C.) centrally reviewed the response rate (RR) and determined the best response according to RECIST version 1.1.15 The objective RR was defined as the percentage of patients with response (complete or partial) at first restaging after chemotherapy initiation. Progression-free survival (PFS) was calculated from the date of chemotherapy initiation until the date of progression by RECIST version 1.1 or death (that occurred without recorded progression), and censoring was the date of last follow-up if the patient had not experienced progression. Statistical methods for correlative and trend analyses are described in the Data Supplement.
RESULTS
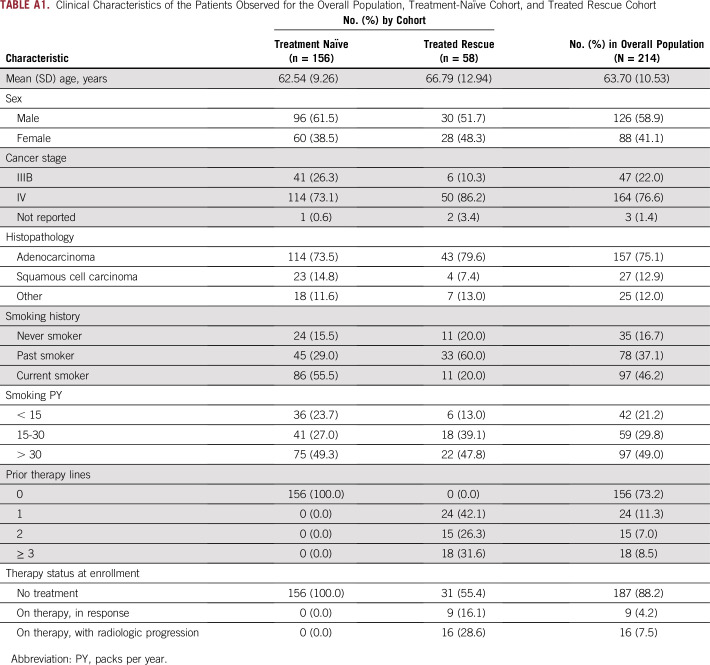
From June 2015 through April 2017, 214 patients with advanced NSCLC were recruited; 156 treatment-naïve patients were included (treatment-naïve cohort), and patients with unknown molecular status were also enrolled (n = 58; treated rescue cohort). Characteristics of the patients at enrollment are provided in Appendix Table A1. In the whole population, 41% were women and 17% were never smokers; the population had predominantly stage IV disease (77%) and the adenocarcinoma subtype (75%). Molecular profile was unknown at the time of enrollment for 29% of treatment-naïve patients and for 48% of the overall study population.
Tissue Molecular Profiling
Among the 156 treatment-naïve patients, 111 (71%) had successful tissue molecular profile analyses. Of these, 41% of patients had complete tissue molecular testing according to current guidelines. In these patients, somatic mutations were found in 78% of patients (n = 87); the highest frequencies were in KRAS (27%), EGFR (6%), and BRAF (5%). Of the remaining 103 patients (n = 45, treatment-naïve; n = 58 treated rescue cohort) without prior tissue molecular analyses, either molecular testing was not performed (44%) or tissue biopsies had insufficient quality or cellularity for testing (51%) or results were not reported in patient referral records.
ctDNA Molecular Profiling
Liquid biopsy ctDNA profiling was successfully performed for 91% of all patients (n = 194; Appendix Table A2), from a median plasma volume of 3.7 mL for DNA extraction (range, 2.4 to 5 mL). Because of the low volumes of plasma collected in this study, it was not feasible to repeat analysis in the case of technical fails. Mutations were detected in 77% of patients (149 of 194 patients). The time to complete testing for this research study was 10 days from receipt of sample.
Treatment-naïve cohort.
In the treatment-naïve cohort (n = 156; Appendix Table A2), liquid biopsy analysis was successful for 142 patients (91%) and, of these, mutations were detected in 111 patients (78%); the most frequent were TP53 (52%), KRAS (28%), STK11 (16%), EGFR (9%), and BRAF (6%; Fig 1). Of mutations detected, mutation allelic fraction was within a range of 0.038% to 63.5%, and 21% of mutation variants had an allelic fraction (AF) less than 0.5%. Of treatment-naïve patients without tissue molecular testing (n = 45; Appendix Fig A1), mutations were detected in 37 patients, of which clinically relevant mutations were detected in 16 patients (n = 1, ERBB2 exon 20; n = 1, EGFR exons 18 to 21; n = 11, KRAS with and without STK11; and n = 3, STK11).
FIG 1.
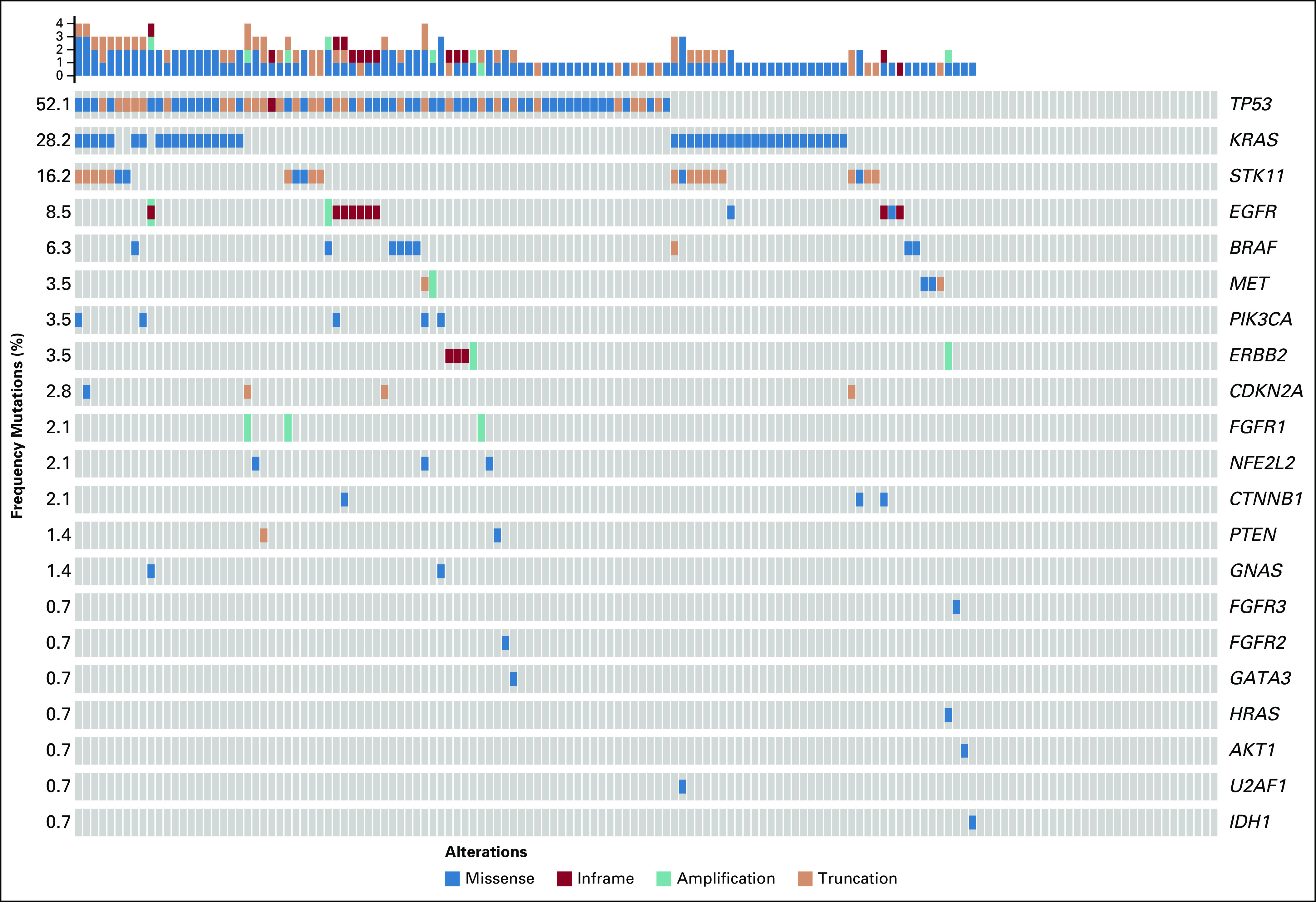
Liquid biopsy circulating tumor DNA (ctDNA) molecular profiles of treatment-naïve cohort with successful plasma-based testing by InVisionSeq (n = 142).
Treated rescue cohort.
In the rescue cohort (n = 58; Appendix Table A2), liquid biopsy analysis was successful in 52 patients (90%), of whom mutations were detected in 38 patients (73%; Fig 2). Mutations included EGFR exon 20/21 mutation (n = 3 patients), ERBB2 exon 20 in-frame insertion (n = 1 patient) and amplification (n = 1 patient), MET amplification (n = 1 patient); BRAF p.V600E (n = 1 patient), PIK3CA (n = 3 patients), FGFR1 amplification (n = 3 patients), and IDH1 (n = 1 patient); potentially actionable molecular alterations were identified in 27% of this cohort. KRAS was detected in an additional 10 patients (19% of the cohort, including one case with concurrent STK11), and just the STK11 mutation was detected in one additional patient. Overall, 48% of the rescue cohort had clinically relevant mutations detected with liquid biopsy, and, among those patients with mutations detected in the liquid biopsy, 10% received personalized treatment according to these results.
FIG 2.
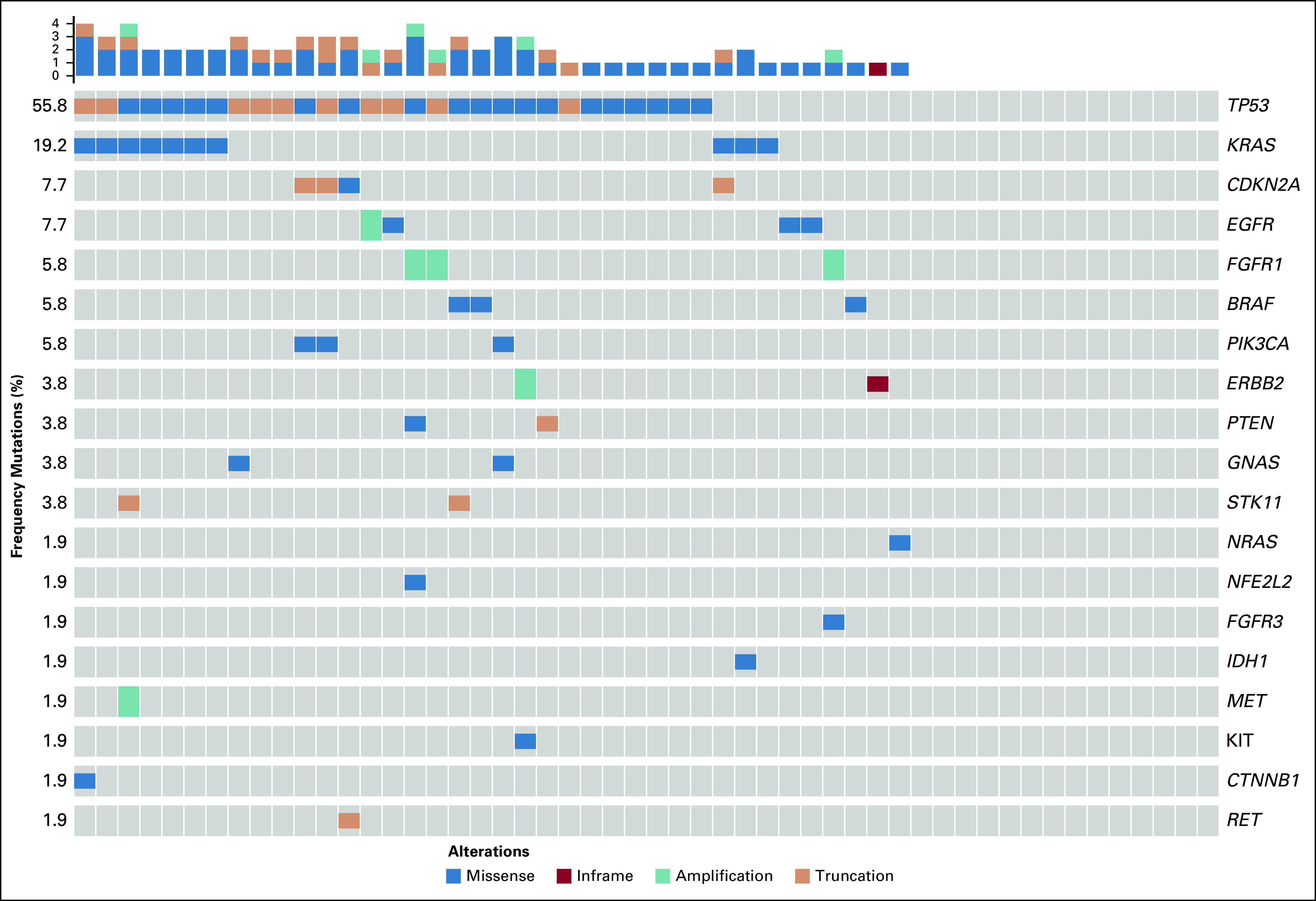
Liquid biopsy circulating tumor DNA (ctDNA) molecular profiles of the previously treated rescue cohort that had unknown tissue molecular profiles from primary tissue, with successful plasma-based testing by InVisionSeq (n = 52).
The feasibility of the liquid biopsy in the pooled analysis for all patients with NSCLC without tissue molecular profile regardless of treatment line and with a successful liquid biopsy (n = 41 treatment-naïve patients and n = 52 previously treated patients) was 90%, and 73% of samples had a mutation detected. In this population, utility of liquid biopsy was 17% (n = 16 patients with potentially actionable alterations: n = 2 treatment-naïve patients and n = 14 pretreated patients), and an additional 25 patients (27%) had clinically relevant molecular information, mainly KRAS mutation with or without STK11 mutation (Appendix Table A3). In addition, plasma provided broader molecular testing information than tissue, because not all patients with tissue were tested for recommended core gene variants (Fig 3).
FIG 3.
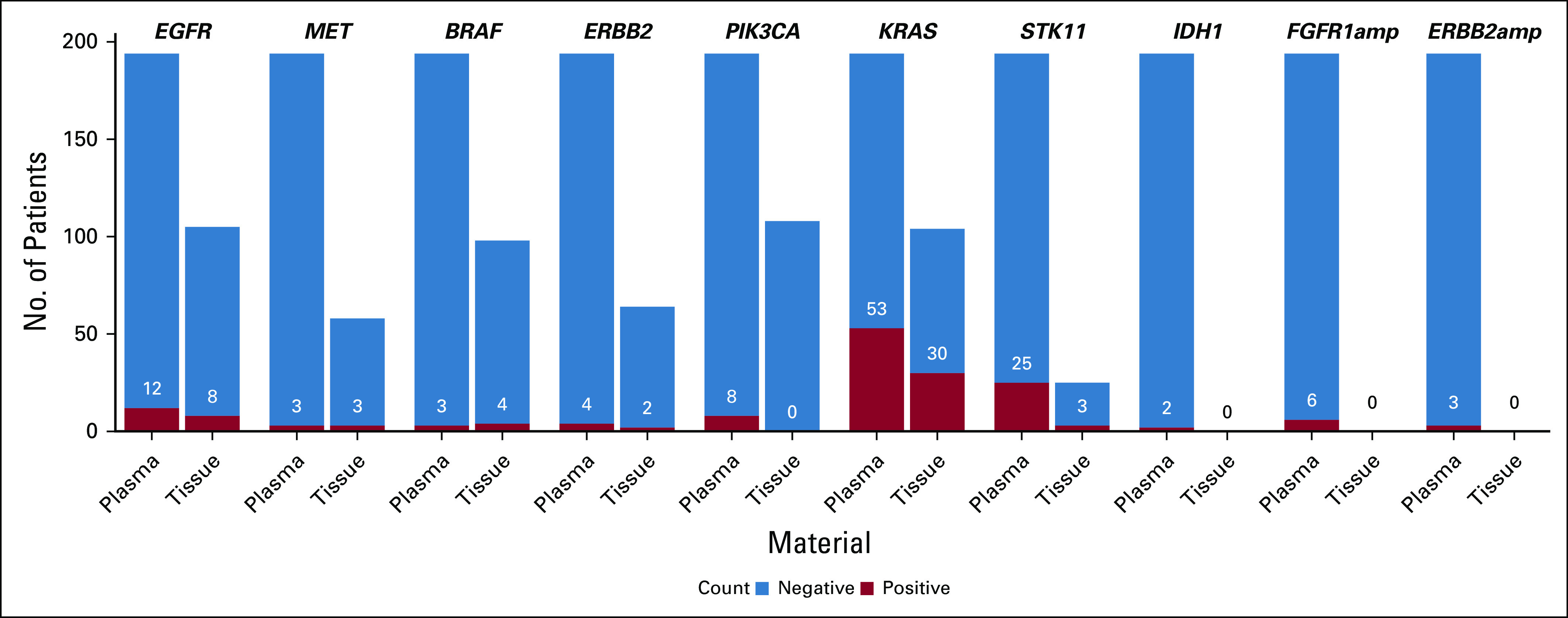
No. of patients with advanced NSCLC in whom molecular analysis was performed for tissue and liquid biopsy, which shows positive mutations detected. This analysis demonstrates the utility of plasma to support broader testing in patients with no or limited tissue analysis for actionable and clinically relevant mutations.
Tissue and Liquid Biopsy Performance: Concordance, Sensitivity, and Specificity
Of the treatment-naïve cohort, 94 patients had successful and concurrent tissue and liquid biopsy molecular profiles; the median time between tissue biopsy and liquid biopsy collection was 34 days. For the core gene variants, there was 95% (CI, 92.5% to 97.1%) concordance agreement. Sensitivity and specificity were determined to be 81% (CI, 66.6% to 91.6%) and 97% (CI, 94.5% to 98.5%), respectively Fig 4; Appendix Table A4). Overall, concordance for the broader panel in which concurrent tissue testing was performed was 95%; sensitivity and specificity were 72% and 97%, respectively (Appendix Table A4). There was an increase in detection when plasma testing was used in addition to tissue testing (Appendix Fig A3).
FIG 4.
Concordance of amplicon-based assay (InVisionSeq) for liquid biopsy compared with tissue biopsy.
Longitudinal Analysis of Liquid Biopsy Only: Unselected Chemotherapy First-Line Treatment
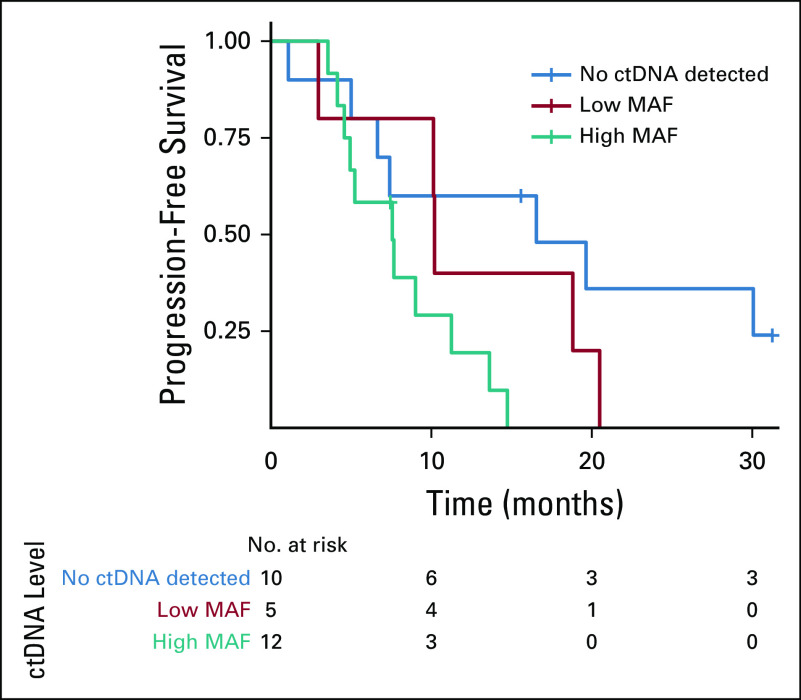
Within the treatment-naïve cohort, 31 patients had successful dynamic collection of ctDNA at baseline and at day 42 after treatment initiation (first radiologic evaluation). Serial liquid biopsies among patients with positive liquid biopsy at baseline who had at least one somatic mutation to track ctDNA (n = 20) demonstrated that the mutation burden ratio was significantly correlated with change in response per RECIST version 1.1 at clinical assessment on day 42 (P = .008; Fig 5). The median PFS for the cohort was 9.0 months. When patients were divided between no ctDNA detected, intermediate ctDNA load (AF, 0% to 2%), and high ctDNA load (AF > 2%), the PFS showed a consistent trend across these three groups; (16.1 months v 10.2 months v 7.5 months, respectively; P = .016; Fig 6). Moreover, there was a significant inverse correlation between high allele fraction at baseline and PFS (correlation P = .009).
FIG 5.
Liquid biopsy mutation correlation to response rate as calculated by change in radiologic response (RECIST 1.1) versus change in mutation molecules, representative of tumor mutation burden, after first-line platinum-based chemotherapy in an unselected cohort with advanced non–small-cell lung cancer (n = 31). Data from 20 patients are shown; others lacked baseline genomic alterations or RECIST data. (*) Two data points, with overlapping data. ctDNA, circulating tumor DNA. D42, day 42.
FIG 6.
Progression-free survival plot in unselected patients with advanced non–small-cell lung cancer treated with first-line platinum-based chemotherapy; plot compares circulating tumor DNA (ctDNA) mutations at baseline. Negative, those with mutations of low (< 2% mutation allele fraction [MAF]) and high (> 2% MAF) allele fractions, as defined by median allele fraction of baseline mutations across the cohort (n = 31).
DISCUSSION
Here, we present evidence for the feasibility and clinical utility of liquid biopsies for the management of advanced NSCLC. In our population, 29% of treatment-naïve patients had an unknown tissue molecular profile at the time of enrollment, similar to previous series8; however, in other series, the rate of failure had reached up to 46% of patients.16 By contrast, we have shown the successful feasibility of amplicon-based NGS ctDNA plasma analysis from patients with lung cancer: only 9% of patients did not achieve a ctDNA profile because of insufficient sequencing depth. The proportion of patients with detectable mutations was similar regardless of whether tissue or liquid biopsy testing was performed (78% v 77%), but the success for obtaining a molecular profile was higher in liquid than in tissue tests (91% v 52%). This detection rate of ctDNA is similar to recently published data in a population of patients with cancer, which reported genomic alterations in 80% of patients with NSCLC.17
Our data show overall concordance of 95% for mutation detection from blood-based ctDNA analysis compared with tissue; the sensitivity was 72% and increased to 81% for the defined core gene variant panel of gene hotspots within EGFR, MET, ERBB2, BRAF, STK11, and KRAS. True comparative analysis studies (with both analyses centralized) in an NSCLC population have reported similar rates of concordance.16,18 Other series in patients with NSCLC have reported concordance rates that range from 60% to 90% for specific mutations (EGFR mutations).19 However, some recent data in the cancer population have reported significant discordance between tissue- and plasma-based NGS sequencing tests20 and between different liquid biopsy tests,21 which highlights the importance of robust analytic and clinical validation data for the choice of tests used in clinical practice.
As the numbers of targeted therapies available and approved in lung cancer increase over time, a need exists for companion diagnostics for real-time detection of therapeutically targetable genetic lesions. Among those 103 patients without tissue molecular profiles regardless of treatment line, the amplicon-based ctDNA analysis was the only means for molecular profile in 90% of cases; it identified potential actionable molecular alterations in 17% of cases, which may have allowed an increased percentage of patients to get the benefit of personalized treatment. Although we did not perform tests for rearrangements in this study, because plasma volume collected was insufficient after the assay was available, the percentage of potential actionable alterations reported is similar to that of previous series3 and within the range of expected frequency of these mutations. High accuracy of rearrangements detection by the InVision amplicon-based platform has been reported.22,23 Prospective data endorse ctDNA-guided molecular testing treatment in advanced solid tumors and in patients with NSCLC when tissue is insufficient or unobtainable, and RRs and PFS are similar to those in tissue-based targeted therapy studies.24,25 These data suggest that ctDNA has the potential to provide clinically relevant information with a noninvasive procedure and can be more readily repeated in the case of technical failure. In our cohort, 44% of patients without tissue biopsy had ctDNA-detected clinically relevant mutations for personalized treatment as well as putative negative predictive markers of immunotherapy efficacy, such as STK11 mutation.14,26 Also, ctDNA testing demonstrated feasibility of use as complementary to tissue testing because detection of clinically relevant mutations increased when plasma testing was used. In our study, 10% of patients who had mutations detected with ctDNA received personalized treatment according to these results. Finally, we reported the dynamic evolution of ctDNA as a prognostic marker of RR and PFS for those patients who received first-line platinum-based chemotherapy. ctDNA testing may have clinical implications for monitoring treatment efficacy, especially in patients with high AFs or with decreased ctDNA and stable radiologic disease by RECIST, as a potential earlier marker of response. All of these observations merit additional evaluation.
It remains unknown what the appropriate number of genes to be screened in tumor type-specific gene panels or as universal tests irrespective of the tumor type will be. In practice, it is not really proven that larger gene panels may improve the chances of finding clinically relevant targetable alterations.27 In our cohort, use of the 36-gene panel highlights an appropriate balance of coverage between clinically relevant (rule-out/prognostic) and actionable genes with high sensitivity and specificity.
This study has some limitations. Plasma ctDNA release is affected by many factors25; previous reports have identified tumor burden and metastatic sites as factors associated with ctDNA release.26 These factors were not fully evaluated in this study but could explain lack of detected driver mutations in some patients. Another limitation of this study is that tissue analysis was not performed centrally, and performances may differ in terms of coverage and variant detection. Finally, another potential limitation was plasma availability for the validation of structural rearrangements using liquid biopsies compared with tissue. However, the ctDNA blood analysis was centralized and, in most cases, provided expanded gene coverage for molecular profiling.
In conclusion, our data provide additional validation that ctDNA with InVisionSeq Lung, an amplicon-based technology, can be used for molecular profiling and to monitor disease in patients with advanced NSCLC with high sensitivity and specificity to detect clinically relevant and actionable mutations when tissue biopsy is unavailable or uninformative. This study also suggests that ctDNA offers a potential prognostic biomarker for treatment efficacy.
ACKNOWLEDGMENT
We thank the patients, their families, the investigators, and the study teams for their contributions to this work. We thank Ivan Bièche and Fabrice André (SAFIR/UNICANCER) for tissue mutation data and panel information; Miki Mikidache and Natalie Jowett for operational support of the study, analytic testing, and data quality control; and Femke de Snoo of Medex15 for medical writing assistance.
APPENDIX
Blood Samples and Circulating Tumor DNA Isolation and Sequencing
Samples were considered acceptable for analysis if the DNA extraction procedure could recover at least 2,000 input copies of the genome, as measured by droplet digital polymerase chain reaction. Low input copies (< 2,000) were acceptable if sequencing read depth met additional quality-control criteria, defined by the panel version. After sequencing, for the earlier 35-gene panel, each sample required an average sequencing depth greater than ×5,000 across the overall panel. The revised 36-gene panel required an average depth of ×10,000, and a minimum locus-specific read-depth requirement of ×3,000 was applied for lung cancer stratification hotspots (including EGFR exon 19 deletions, L858R, T790M, BRAF V600E, and common KRAS activating mutations). Individual variants were called using the Inivata in-house analytic pipeline for somatic mutation detection.
Statistical Methods
All statistical analyses were performed using R version 3.2.5.
Clinical Validation: Concordance, Sensitivity, Specificity
For clinical validation analysis, the data can be summarized using a two-by-two table. Tissue data are the standard variables. Liquid positive and tissue positive are considered true positive (TP); liquid positive and tissue negative, false positive (FP); liquid negative and tissue positive, false negative (FN); and liquid negative and tissue negative, true negative (TN).
The following definitions were used: Concordance = (TP + TN)/(TP + TN + FP + FN). Sensitivity = TP/(TP + FN). Specificity = TN/(TN + FP). Positive predictive value = TP/(TP + FP). Negative predictive value = TN/(TN + FN).
Longitudinal Analysis
The correlation of circulating tumor DNA (ctDNA) mutation burden at day 42 with clinical response (assessed by RECIST version 1.1) was estimated using Pearson's correlation test between RECIST score and ctDNA ratio with respect to day1 (log scale; Fig 5).
Progression free-survival (PFS) in days according to by no, low (0% to 2%), or high (> 2%) mutation allele fraction (MAF) was as follows: 16.1, 10.2, and 7.5 days.
Patients with no ctDNA mutations at baseline showed significant advantages in progression-free survival compared with patients whose results were positive for ctDNA mutations (Cox proportional hazards model P = .022). Moreover, the maximum MAF at baseline was significantly inversely correlated with PFS (correlation P = .008967).
When patients were divided into groups of no ctDNA detected, intermediate ctDNA load (MAF, 0% to 2%), and high ctDNA load (MAF > 2%), the PFS showed a consistent trend across these three groups (Cox PH P = .016; Fig 6).
FIG A1.
Flowchart of enrollment and cohorts for analysis: patients with advanced non–small-cell lung cancer (NSCLC) were eligible for the study if they were treatment naïve and expected to receive first-line platinum-based chemotherapy (treatment-naïve cohort) or were previously treated but did not have tissue-based molecular profile of a primary tissue sample available. Patients with concurrent tissue and blood-based testing comprised the validation cohort. Patients without tissue molecular profiles comprised the utility cohort. QC, quality control.
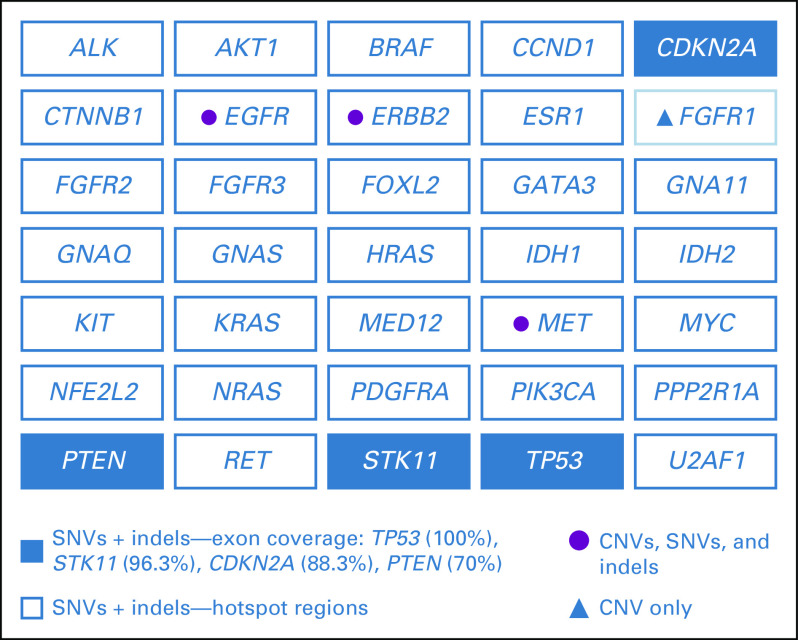
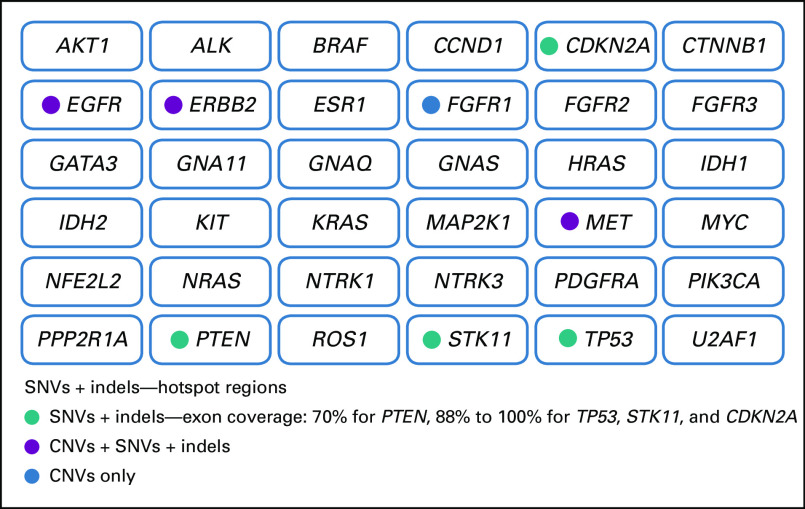
FIG A2.
InVision (Inivata, Research Triangle Park, NC, and Cambridge, United Kingdom) gene panels: Indicated is the coverage per gene, including hotspots, comprehensive or full coverage of coding regions (70% to 100% tiling coverage), and copy number variants (CNVs). In this study, two versions of the InVision gene panel were used (InvCore versions 1.4 and 1.5). (A) InVision liquid biopsy tumor profiling panel (InvCore version 1.4). (B) InVisionSeq Lung liquid biopsy tumor profiling panel (InvCore version 1.5). indels, short insertions or deletions; SNVs, single nucleotide variants.
FIG A3.
Agreement and complementary testing of liquid biopsy to tissue molecular profiling in the validation cohort (concurrent tissue and liquid biopsy) for clinically relevant core gene variants. CNVs, copy number variants; indels, short insertions or deletions; SNVs, single nucleotide variants.
TABLE A1.
Clinical Characteristics of the Patients Observed for the Overall Population, Treatment-Naïve Cohort, and Treated Rescue Cohort
TABLE A2.
Summary of Liquid Biopsy and Tissue Molecular Profiling Across the Cohorts and Proportions of Patients With at Least One Mutation Detected
TABLE A3.
Summary of Liquid Biopsy and Tissue Molecular Profiling to Detect Clinically Relevant Mutations
TABLE A4.
Statistical Summary of Clinical Validation for InVisionSeq Testing by Clinically Relevant Core Gene Variants and Overall Panel Compared With Tissue Molecular Analysis
Footnotes
Supported by Inivata funding to Gustave Roussy for sample acquisition.
AUTHOR CONTRIBUTIONS
Conception and design: Jordi Remon, Ludovic Lacroix, Caroline Caramella, Nitzan Rosenfeld, Clive Morris, Cecile Le Pechoux, Gilles Vassal, Emma Green, Jean-Charles Soria, Benjamin Besse
Collection and assembly of data: Jordi Remon, Ludovic Lacroix, Cecile Jovelet, Caroline Caramella, Karen Howarth, Laura Mezquita, Chloe Pannet, Maud Ngocamus, Julien Adam, Alina-Miruna Grecea, David Planchard, Jose Carlos Benitez, Anas Gazzah, Benjamin Besse
Provision of study material or patients: Ludovic Lacroix, Cecile Jovelet
Data analysis and interpretation: Jordi Remon, Ludovic Lacroix, Cecile Jovelet, Caroline Caramella, Vincent Plagnol, Clive Morris, Emma Green, Benjamin Besse
Administrative support: Emma Green
Manuscript writing: All authors
Final approval of manuscript: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org or ascopubs.org/po/author-center.
Jordi Remon
Consulting or Advisory Role: Pfizer, Bristol-Myers Squibb, Boehringer Ingelheim, MSD Oncology
Travel, Accommodations, Expenses: Roche, Genentech, Inivata, OSE Immunotherapeutics
Caroline Caramella
Consulting or Advisory Role: Bristol-Myers Squibb, Pfizer
Karen Howarth
Employment: Inivata
Stock and Other Ownership Interests: Inivata
Research Funding: Inivata
Patents, Royalties, Other Intellectual Property: Patents and patent applications relating to cancer classifications, detection or analysis of microRNA and circulating tumor DNA, detection of rare sequence variants, applications in molecular diagnostics
Vincent Plagnol
Employment: Inivata
Stock and Other Ownership Interests: Inivata
Patents, Royalties, Other Intellectual Property: Inivata patents
Nitzan Rosenfeld
Employment: Storm Therapeutics (I), Inivata
Leadership: Inivata
Stock and Other Ownership Interests: Inivata, Mission Therapeutics (I)
Research Funding: AstraZeneca (Inst)
Patents, Royalties, Other Intellectual Property: Patents and patent applications relating to cancer classifications, detection or analysis of microRNA and circulating tumor DNA, detection of rare sequence variants, applications in molecular diagnostics
Clive Morris
Employment: Inivata
Leadership: Inivata
Stock and Other Ownership Interests: Inivata
Cecile Le Pechoux
Consulting or Advisory Role: AstraZeneca, Lilly, Nanobiotix, Amgen
Julien Adam
Consulting or Advisory Role: Roche, Bristol-Myers Squibb, AstraZeneca, Merck Sharp & Dohme
Research Funding: Sanofi (Inst), Pierre Fabre (Inst), Merck Sharp & Dohme (Inst)
David Planchard
Consulting or Advisory Role: AstraZeneca, Boehringer Ingelheim, Bristol-Myers Squibb, Novartis, Roche, Pfizer, MSD Oncology, Celgene
Gilles Vassal
Consulting or Advisory Role: Bayer, Roche, Genentech, AstraZeneca, Bristol-Myers Squibb, Celgene, Lilly, Servier, Takeda, Incyte, Ipsen, Novartis, Merck Serono
Travel, Accommodations, Expenses: Bristol-Myers Squibb, Roche
Emma Green
Employment: Inivata
Stock and Other Ownership Interests: Inivata
Travel, Accommodations, Expenses: Inivata
Jean-Charles Soria
Employment: MedImmune
Stock and Other Ownership Interests: AstraZeneca, Gritstone Oncology
Honoraria: Roche, AstraZeneca, Sanofi, Servier, Pierre Fabre, Abbvie, Pharmamar-Zeltia
Benjamin Besse
Research Funding: AstraZeneca (Inst), Roche (Inst), Genentech (Inst), Pfizer (Inst), Boehringer Ingelheim (Inst), Lilly (Inst), Servier (Inst), Onxeo (Inst), Bristol-Myers Squibb (Inst), Ose Pharma (Inst), Inivata (Inst), Novartis (Ins), OncoMed (Inst), Loxo (Inst)
Travel, Accommodations, Expenses: Roche, Pfizer, Bristol-Myers Squibb, Medarex, Novartis, Pierre Fabre
No other potential conflicts of interest were reported.
REFERENCES
- 1.VanderLaan PA, Rangachari D, Majid A, et al. Tumor biomarker testing in non–small-cell lung cancer: A decade of change. Lung Cancer. 2018;116:90–95. doi: 10.1016/j.lungcan.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reck M, Rabe KF. Precision diagnosis and treatment for advanced non–small-cell lung cancer. N Engl J Med. 2017;377:849–861. doi: 10.1056/NEJMra1703413. [DOI] [PubMed] [Google Scholar]
- 3.Barlesi F, Mazieres J, Merlio J-P, et al. Routine molecular profiling of patients with advanced non–small-cell lung cancer: Results of a 1-year nationwide programme of the French Cooperative Thoracic Intergroup (IFCT) Lancet. 2016;387:1415–1426. doi: 10.1016/S0140-6736(16)00004-0. [DOI] [PubMed] [Google Scholar]
- 4.Novello S, Barlesi F, Califano R, et al. Metastatic non–small-cell lung cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2016;27:v1–v27. doi: 10.1093/annonc/mdw326. [DOI] [PubMed] [Google Scholar]
- 5.Kalemkerian GP, Narula N, Kennedy EB, et al. Molecular testing guideline for the selection of patients with lung cancer for treatment with targeted tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the Study of Lung Cancer/Association for Molecular Pathology clinical practice guideline update. J Clin Oncol. 2018;36:911–919. doi: 10.1200/JCO.2017.76.7293. [DOI] [PubMed] [Google Scholar]
- 6.Gutierrez ME, Choi K, Lanman RB, et al. Genomic profiling of advanced non–small cell lung cancer in community settings: Gaps and opportunities. Clin Lung Cancer. 2017;18:651–659. doi: 10.1016/j.cllc.2017.04.004. [DOI] [PubMed] [Google Scholar]
- 7.Tredan O, Corset V, Wang Q, et al. Routine molecular screening of advanced refractory cancer patients: An analysis of the first 2,490 patients of the ProfilER study. J Clin Oncol. 2017;35 suppl; abstr LBA100. [Google Scholar]
- 8.Hagemann IS, Devarakonda S, Lockwood CM, et al. Clinical next-generation sequencing in patients with non–small-cell lung cancer. Cancer. 2015;121:631–639. doi: 10.1002/cncr.29089. [DOI] [PubMed] [Google Scholar]
- 9.Couraud S, Vaca-Paniagua F, Villar S, et al. Noninvasive diagnosis of actionable mutations by deep sequencing of circulating free DNA in lung cancer from never-smokers: A proof-of-concept study from BioCAST/IFCT-1002. Clin Cancer Res. 2014;20:4613–4624. doi: 10.1158/1078-0432.CCR-13-3063. [DOI] [PubMed] [Google Scholar]
- 10.National Comprehensive Cancer Network https://www.nccn.org
- 11.Plagnol V, Woodhouse S, Howarth K, et al. Analytical validation of a next-generation sequencing liquid biopsy assay for high-sensitivity broad molecular profiling. PLoS One. 2018;13:e0193802. doi: 10.1371/journal.pone.0193802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gale D, Lawson ARJ, Howarth K, et al. Development of a highly sensitive liquid biopsy platform to detect clinically relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630. doi: 10.1371/journal.pone.0194630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lindeman NI, Cagle PT, Aisner DL, et al. Updated molecular testing guideline for the selection of lung cancer patients for treatment with targeted tyrosine kinase inhibitors: Guideline From the College of American Pathologists, the International Association for the Study of Lung Cancer, and the Association for Molecular Pathology. J Thorac Oncol. 2018;13:323–358. doi: 10.1016/j.jtho.2017.12.001. [DOI] [PubMed] [Google Scholar]
- 14.Skoulidis F, Goldberg ME, Greenawalt DM, et al. STK11/LKB1 mutations and PD-1 inhibitor resistance in KRAS-mutant lung adenocarcinoma. Cancer Discov. 2018;8:822–835. doi: 10.1158/2159-8290.CD-18-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eisenhauer EA, Therasse P, Bogaerts J, et al. New response evaluation criteria in solid tumours: Revised RECIST guideline (version 1.1) Eur J Cancer. 2009;45:228–247. doi: 10.1016/j.ejca.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 16.Thompson JC, Yee SS, Troxel AB, et al. Detection of therapeutically targetable driver and resistance mutations in lung cancer patients by next-generation sequencing of cell-free circulating tumor DNA. Clin Cancer Res. 2016;22:5772–5782. doi: 10.1158/1078-0432.CCR-16-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zill OA, Banks KC, Fairclough SR, et al. Clin Cancer Res. Vol. 24. 2018. The landscape of actionable genomic alterations in cell-free circulating tumor DNA from 21,807 advanced cancer patients; pp. 3528–3538. [DOI] [PubMed] [Google Scholar]
- 18.Müller JN, Falk M, Talwar J, et al. Concordance between comprehensive cancer genome profiling in plasma and tumor specimens. J Thorac Oncol. 2017;12:1503–1511. doi: 10.1016/j.jtho.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 19.Kwapisz D. The first liquid biopsy test approved: Is it a new era of mutation testing for non–small-cell lung cancer? Ann Transl Med. 2017;5:46. doi: 10.21037/atm.2017.01.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuderer NM, Burton KA, Blau S, et al. Comparison of 2 commercially available next-generation sequencing platforms in oncology. JAMA Oncol. 2017;3:996–998. doi: 10.1001/jamaoncol.2016.4983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torga G, Pienta KJ. Patient-paired sample congruence between 2 commercial liquid biopsy tests. JAMA Oncol. 2017;4:868–870. doi: 10.1001/jamaoncol.2017.4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guibert N, Hu Y, Feeney N, et al. Amplicon-based next-generation sequencing of plasma cell-free DNA for detection of driver and resistance mutations in advanced non–small-cell lung cancer. Ann Oncol. 2018;29:1049–1055. doi: 10.1093/annonc/mdy005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mezquita L, Jovelet L, Lacroix L, et al. An amplicon-based liquid biopsy for detecting ALK and ROS1 fusions and resistance mutations in advanced non–small-cell lung cancer (NSCLC) patients. J Clin Oncol. 2018;36 doi: 10.1200/PO.19.00281. suppl; abstr 9095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim ST, Banks KC, Lee S-H, et al. Prospective feasibility study for using cell-free circulating tumor DNA–guided therapy in refractory metastatic solid cancers: An interim analysis. JCO Precis Oncol. doi: 10.1200/PO.16.00059. doi: 10.1200/PO.16.00059 [published June 26, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Remon J, Caramella C, Jovelet C, et al. Osimertinib benefit in EGFR-mutant NSCLC patients with T790M-mutation detected by circulating tumour DNA. Ann Oncol. 2017;28:784–790. doi: 10.1093/annonc/mdx017. [DOI] [PubMed] [Google Scholar]
- 26.Skoulidis F, Carter BW, Zhang J, et al. Association of STK11/LKB1 mutations with primary resistance to PD-1/PD-L1 axis blockade in PD-L1–positive non-squamous NSCLC. J Clin Oncol. 2018;36 suppl; abstr 9028. [Google Scholar]
- 27.Jordan EJ, Kim HR, Arcila ME, et al. Prospective comprehensive molecular characterization of lung adenocarcinomas for efficient patient matching to approved and emerging therapies. Cancer Discov. 2017;7:596–609. doi: 10.1158/2159-8290.CD-16-1337. [DOI] [PMC free article] [PubMed] [Google Scholar]