Abstract
Purpose
To determine whether oncologists intended to change treatment as a result of tumor sequencing, and subsequently, whether patients experienced an alteration of clinical management or derived clinical benefit.
Patients and Methods
A prospective survey of oncologists referring adult patients with rare, advanced, or refractory cancer to the Michigan Oncology Sequencing program was conducted from June 2014 to March 2015 to assess the use of and intent to disclose sequencing findings. Oncologists’ responses were compared with the referred patients’ self-reported survey responses, and a content analysis of disclosure documented in the medical record was performed. Medical records were reviewed retrospectively to determine if clinical management was informed or changed by sequencing results.
Results
Oncologists (response rate, 93%) referring 112 consecutive patients were surveyed. Medical records of patients were reviewed for changes in clinical management on the basis of sequencing findings. Oncologists intended to change the treatment of 22% of patients (n = 24) on the basis of sequencing findings. Of these patients, 37.5% (n = 9) had an actual change in clinical management. Thirty-four patients with postsequencing survey data reported that a results disclosure discussion did not occur, despite documentation of disclosure by the physician in the medical record.
Conclusions
Findings demonstrate that many oncologists view next-generation sequencing results to be potentially valuable in directing subsequent therapy for their patients; however, barriers in communicating results to patients and implementing them in clinical management remain.
INTRODUCTION
Clinical implementation of tumor genomic sequencing is emerging as a promising strategy to improve patient outcomes in oncology.1 Integrating an individual’s clinical history with genomic data can inform both diagnostic and treatment strategies, potentially suggesting therapies that are tailored to the patient’s tumor mutational landscape.2
There are several ongoing national efforts to evaluate the effectiveness of treating cancers with targeted therapies informed by alterations in patients’ tumors. The National Cancer Institute's Molecular Analysis for Therapy Choice (MATCH) is a basket study matching patients with metastatic solid tumor malignancies to Food and Drug Administration–approved or experimental drugs on the basis of specific tumor alterations.3 The Tumor Agent and Profiling Utilization Registry study (TAPUR), sponsored by ASCO, matches patients to commercially available targeted antineoplastic agents on the basis of potentially actionable tumor events.4 The Initiative for Molecular Profiling in Advanced Cancer Therapy (IMPACT) program in the phase I clinic at MD Anderson Cancer Center matches targeted agents with tumor molecular alterations in patients with advanced cancer.5
Evaluating the evidence for improved outcomes for patients with cancer matched to targeted agents is an essential first step. However, many other barriers, including the lack of a standard framework for the classification of genomic alterations, complicate the integration of genomic information into clinical management. This is problematic because many alterations have not been validated as biomarkers to predict therapeutic response to drugs. Furthermore, there is debate among medical professionals about what should be considered clinically actionable and what types of findings from clinical sequencing should be disclosed to patients.6-12 Still, it is common for studies that use sequencing results to match patients with targeted therapy to report the frequency of actionable results.13 However, few report what percentages of these actionable results were eventually acted on in clinical practice.
Currently, most commercially available tumor-testing platforms focus on targetable alterations in a limited set of genes. In contrast, the Michigan Oncology Sequencing (MI-ONCOSEQ) program has adopted a more comprehensive approach by sequencing the genome, exome, and transcriptome of tumors, as well as a matched normal sample, in an effort to characterize novel variants that may increase cancer risk. This has led to the identification of alterations with important clinical implications, such as the activation of ESR1 mutations in patients with estrogen receptor-α–positive metastatic breast cancer refractory to hormonal therapies, implicating these mutations as a likely mechanism of resistance to endocrine therapy.3 Furthermore, novel fibroblast growth factor receptor fusions, potentially targetable with fibroblast growth factor receptor inhibitors currently being studied in clinical trials, have been identified across a diverse range of cancer types.4 These discoveries highlight how taking a comprehensive approach to identifying therapeutic targets for individual patients with cancer can expand the molecular taxonomy of cancer. To address whether oncologists are using sequencing findings to change a patient’s clinical management, and to what extent potentially actionable findings are being acted on, we surveyed oncologists referring patients with rare, advanced, or refractory cancer to the MI-ONCOSEQ program, a precision oncology research study at University of Michigan Comprehensive Cancer Center.14 We also examined whether there was concordance between an oncologist’s intention to disclose the finding and the referred patient’s perception of a disclosure of results. Finally, the clinical course of referred patients was reviewed retrospectively to establish how many patients actually had changes made to their clinical management, as well as how many derived clinical benefit from the sequencing results.
PATIENTS AND METHODS
Integrative Clinical Sequencing
Oncologists referring consecutive patients with advanced-stage solid-tumor malignancies to the MI-ONCOSEQ program were surveyed between June 2014 and March 2015. Detailed information on the program, including patient eligibility criteria, have been reported previously.14 Results were generated within approximately 6 to 8 weeks, and potentially actionable findings were discussed at an institutional precision medicine tumor board (PMTB) composed of members with expertise in medical oncology, hematology, clinical pathology, cancer genetics and genetic counseling, bioinformatics, and bioethics. A summary of the patient’s PMTB findings, and a brief online survey, were e-mailed to each referring oncologist approximately 1 week after PMTB. The patient was sent a survey approximately 2 weeks after the oncologist received the results. The MI-ONCOSEQ program received institutional review board approval from the University of Michigan.
Framework for Classification of Genomic Alterations
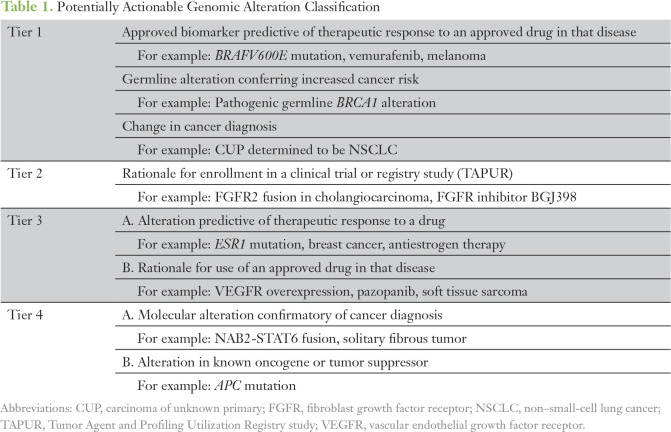
To determine the clinical relevance or potential actionability of comprehensive sequencing results, the study team developed a post hoc clinical tiering system to allow for a uniform and scalable approach for classifying alterations for research purposes (Table 1).
Table 1.
Potentially Actionable Genomic Alteration Classification
Tier 1 alterations represent the highest level of clinical evidence and are generally incorporated into National Comprehensive Cancer Network guideline–based recommendations for the treatment of malignancies. This includes Food and Drug Administration–approved therapies in the context of a molecular biomarker that predicts therapeutic response to a drug, pathogenic germline alterations conferring increased cancer risk, or sequencing information that contributed to a change in cancer diagnosis for tumors of unknown primary origin. Tier 2 alterations represent the identification of molecular alterations that meet the enrollment criteria for a clinical trial or registry study of a targeted therapeutic agent. Tier 3 alterations suggest a scientific rationale or preclinical data predictive of therapeutic response to a particular drug. Tier 4 alterations suggest a biologic relevance for a particular molecular alteration with regard to tumor pathogenesis, but are not considered to be clinically actionable.
Oncologist Survey
After receiving the PMTB report, oncologists were surveyed about their intended use of sequencing information. Each oncologist received a survey for every patient he or she referred during the 9-month period, and therefore, the same oncologist may have completed multiple surveys. Survey items assessed whether and how oncologists planned to change the patient’s treatment in light of findings from sequencing. For example, oncologists were asked, “Will you make any changes to this patient’s cancer treatment based on PMTB and/or the MI-ONCOSEQ report?” (Response options were “yes,” “no,” “not sure.”) If the oncologist selected “yes,” he or she was asked, “What changes will you make?” If the oncologist endorsed “no” or “not sure,” he or she was asked, “What is your reasoning for not making any changes?” Additional items asked about intention to disclose the findings to patients as well as how and when this communication would occur.
Patient Surveys
Patients enrolled in the MI-ONCOSEQ program were administered surveys after they consented to participate in the study and after the referring oncologist received the PMTB report. At baseline, a 23-item survey assessed patients’ knowledge and expectations regarding genomic sequencing, including beliefs about incidental findings, and preference for the return of results. A follow-up survey mailed approximately 2 weeks after the referring oncologist received the results assessed patients’ fulfillment of expectations, decisional regret, satisfaction with results, and behavioral changes.
Concordance Between Oncologist and Patient Responses
To assess gaps in the communication process, we compared items about an oncologist’s intention to disclose the sequencing results with the patient’s recollection of a results disclosure discussion. Specifically, “Will you share the genetic sequencing results with this patient?” (Response options were “yes,” “no,” “not sure.”) Similarly, in their follow-up survey, patients were asked, “Since you began participating in this research study, has your doctor(s) discussed one or more of your genome sequencing test results with you?” (Response options were “yes,” “no,” “not sure.”)
Next, we conducted a content analysis of patients’ medical records to find documentation of a disclosure discussion. Medical records were reviewed using search terms such as “MIONCOSEQ” and “results” in clinical visit notes, telephone calls, or e-mails preceding the PMTB presentation or when the oncologist received the results report. Two team members independently conducted the medical record review. Coding discrepancies were discussed and resolved by team members (N.B., L.Q.L., M.C.G., and J.S.R.).
Patient Outcome Information
A medical oncologist on the study team (E.C.) retrospectively reviewed the clinical course of all referred patients to determine the number of patients whose subsequent clinical management was informed or changed by sequencing results. Outcomes assessed included whether treatment was altered on the basis of results, the type of treatment or change in management, and the identification of a germline alteration with implications for management of patient or family member care (eg, cascade testing).
Data Analysis
Descriptive data, including frequencies and percentages, were calculated for demographic variables and responses to the survey questions for both the referring oncologists and the patients. Whether survey participants’ characteristics and responses predicted intention to change clinical management and report of a results discussion were compared using Fisher’s exact tests for categorical variables for low cell counts. All analyses were conducted using SPSS version 22 (SPSS, Chicago, IL).
RESULTS
Oncologist and Patient Surveys
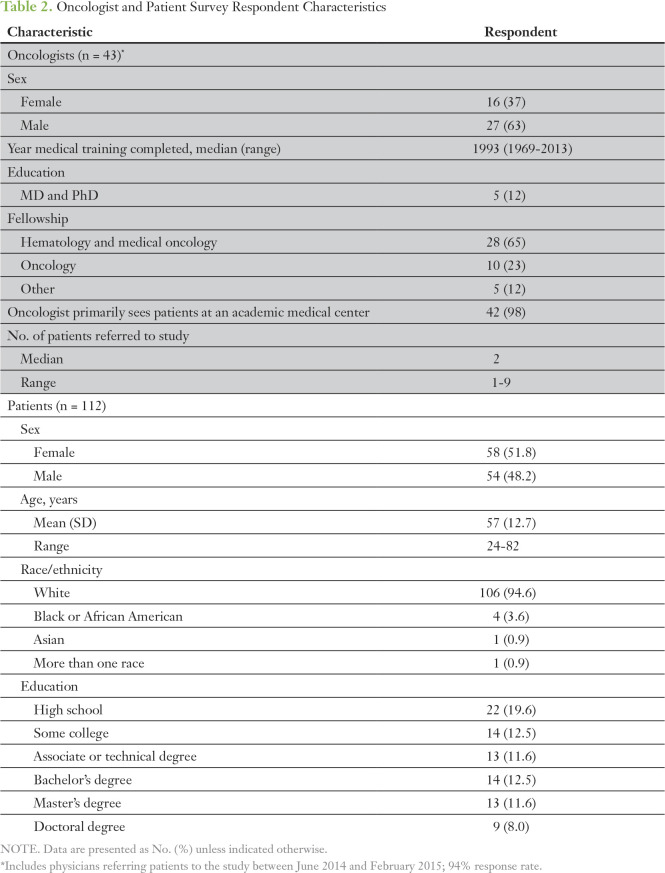
Forty-three oncologists referring 112 patients to the MI-ONCOSEQ study between June 2014 and March 2015 completed the survey (response rate, 93%). Twelve percent were MDs and PhDs. Median year of medical school graduation was 1993 (SD, 11.6). Approximately one half (52%) of the oncologists surveyed referred three or more patients (median, two patients; range, one to 12 patients).
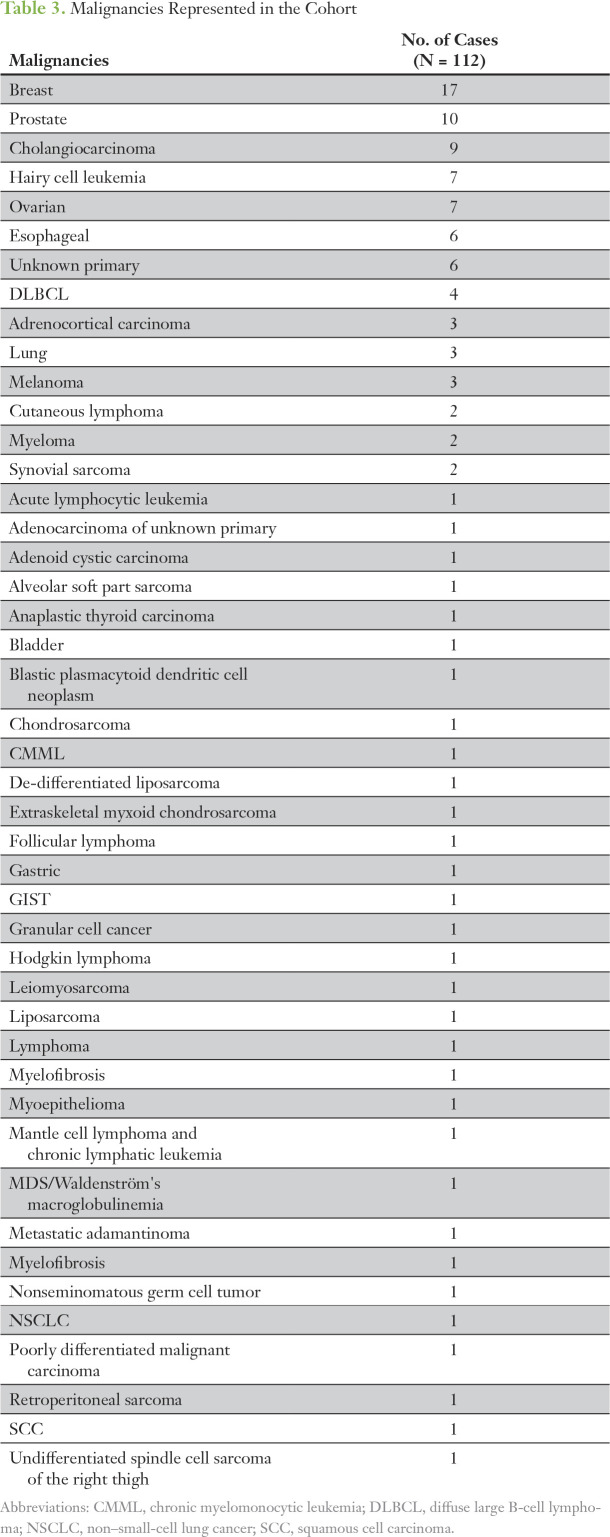
Of the 112 patients, the mean age was 57 years (SD, 12.7 years), 51.8% were female, 94.6% self-reported as being white, and 32.1% were college graduates (Table 2). Given that MI-ONCOSEQ recruitment centered on advanced stage solid tumor malignancies (including sarcomas and other rare cancers), a wide range of malignancies was represented in the cohort (Table 3).
Table 2.
Oncologist and Patient Survey Respondent Characteristics
Table 3.
Malignancies Represented in the Cohort
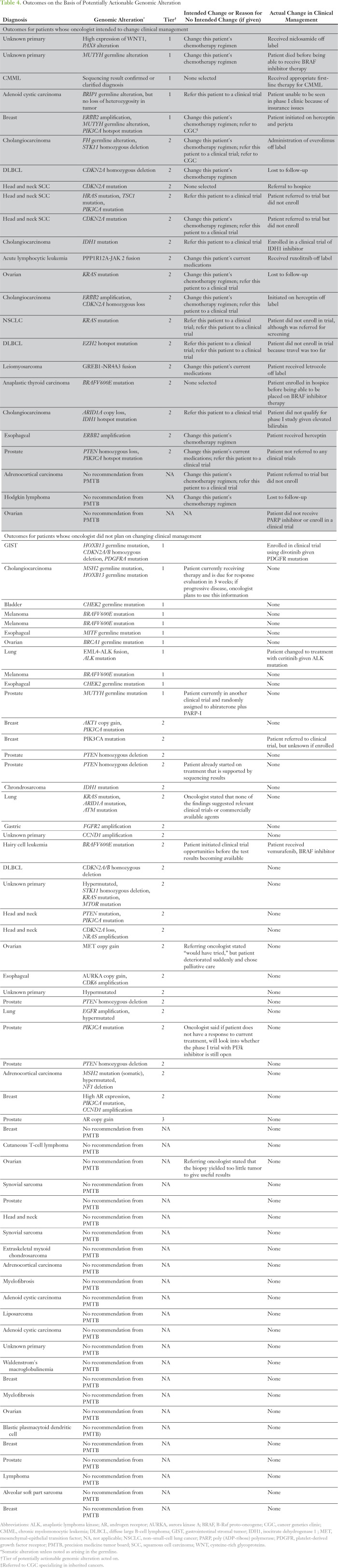
Surveyed oncologists reported that they planned to make changes to the treatment of 22% of patients (n = 24) on the basis of sequencing results. The oncologists had no plans to change the treatment of 51% of patients (n = 60), or were not sure if they were going to make any treatment changes for 28 patients. Specific reasons for treatment-related decisions are provided (Table 4). The most frequently endorsed reasons for not changing clinical management were a lack of locally available trials offering therapies relevant to the findings (59%), findings not actionable (ie, not enough clinical evidence or results not clinically significant; 27.4%), and the effectiveness of a patient’s current treatment (12%).
Table 4.
Outcomes on the Basis of Potentially Actionable Genomic Alteration
Regardless of an intention to change treatment, oncologists intended to share sequencing results with 94 of the 112 patients (84%). Surveyed oncologists planned to communicate the results to the majority of patients (68%) in person at a clinic visit. Several indicated that they would disclose results by telephone (20%) or before the patient’s next visit (24%).
Intention to change treatment and the mode of communication of findings were stratified by the median number of patients referred and the patient diagnosis. No variables significantly predicted intention to change treatment (Fig 1).
Fig 1.
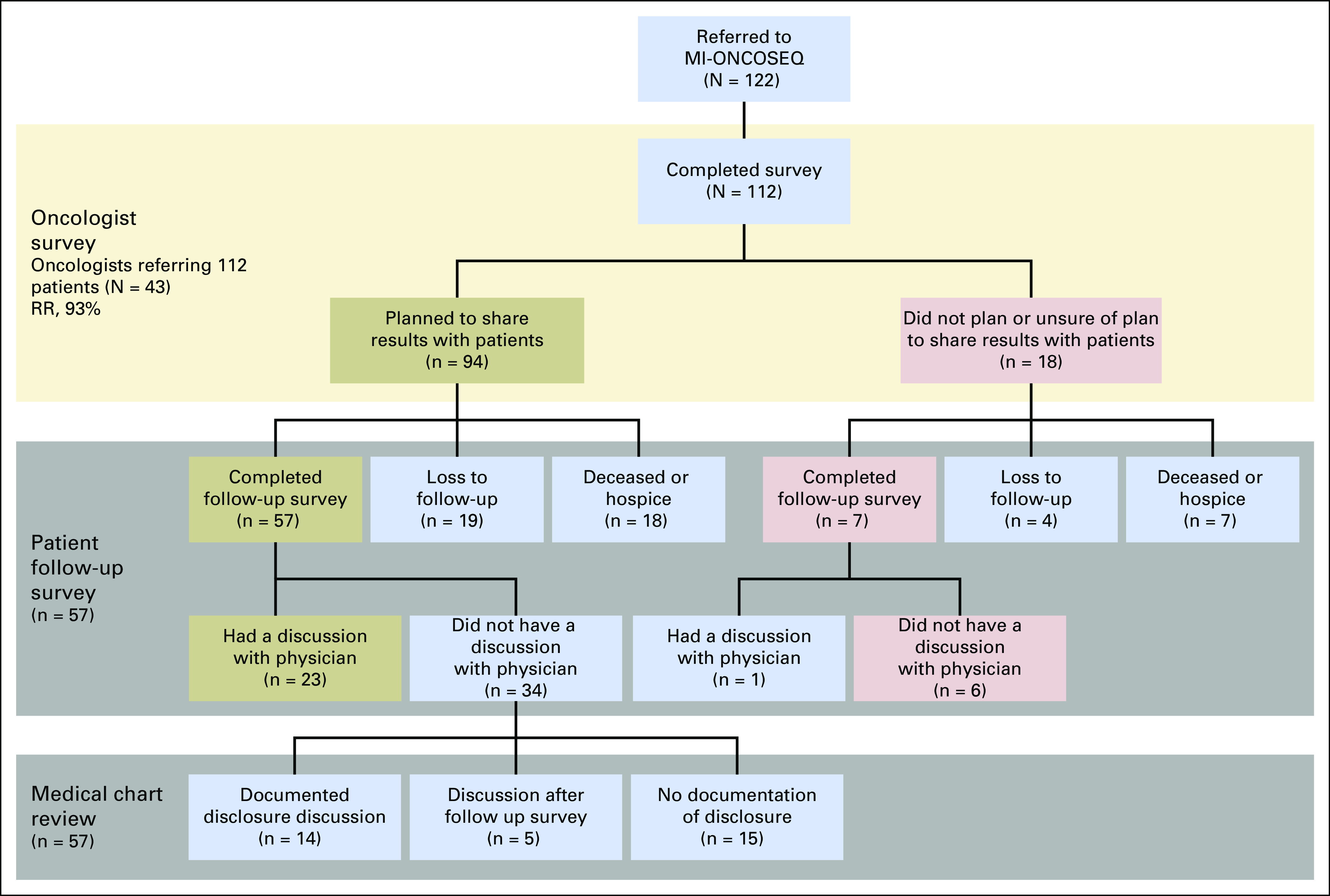
Study flowchart. MI-ONCOSEQ, Michigan Oncology Sequencing program; RR, response rate.
Concordance Between Oncologist and Patient Responses
Of the 94 patients for whom an oncologist intended to share results, 57 completed the follow-up survey. More than one half (n = 34) of the surveyed patients reported that a results disclosure discussion did not take place. However, in 14 of these patient records (representing 24.6% of respondents overall), there was documentation of results disclosure despite patients reporting otherwise. In addition, five medical records contained specific documentation that a discussion of results took place after the patient completed the follow-up survey. No evidence of a results disclosure discussion was found in the medical records of 15 patients. Twenty-three of the surveyed patients indicated that their oncologist discussed the sequencing results with them, and a documented note in their medical record supported their response. Controlling for referring oncologist or cancer type did not significantly predict report of a results discussion.
Actionability Classification of Tumor Genomic Alterations, and Patient Outcomes
On the basis of the retrospective chart review, nine of 24 patients had actual changes in clinical management informed by sequencing results. Three of the detected alterations were Tier 1 or were in accordance with guideline-based recommendations for treatment of that malignancy. The majority of alterations (n = 6) were Tier 2 and were relevant to enrollment in a clinical trial. Of note, three of the Tier 1- or 2-classified alterations identified are used widely in standard clinical practice and were known about before comprehensive sequencing was performed (eg, cholangiocarcinoma diagnosed before enrollment with FH germline mutation that had been clinically validated previously; Table 4).
Reasons varied greatly as to why results did not inform subsequent clinical management despite the identification of a clinically actionable result. On the basis of a review of medical records, these reasons included patient barriers (eg, unable to travel to referred trial), insurance or cost (eg, phase I clinical trial not covered), ineligibility for an identified study (eg, impaired liver or renal function), and loss to follow-up or patient deceased.
DISCUSSION
Comprehensive genomic sequencing offers the opportunity to characterize somatic and germline alterations for patients who have exhausted standard therapies and to potentially inform patients of additional treatment options. In this study, we found that oncologists reported intentions to make changes to treatment on the basis of genomic findings for 24 of 112 patients with rare or refractory cancers. Furthermore, oncologists planned to disclose findings to the majority of patients, regardless of whether test results were likely to inform the patient’s treatment plan. Of the 24 patients whose oncologists indicated an intention to change clinical management, fewer than one half actually had subsequent changes to clinical management informed by sequencing results. These findings demonstrate both the potential benefit that can result from taking a comprehensive approach to identify targeted therapies tailored to a patient’s mutational landscape and the barriers to implementation of tumor-related genomic results into clinical management. Significant obstacles include, but are not limited to, the need for additional educational support for both clinicians and patients, the need to define what constitutes an actionable finding, lack of access to clinical trials and off-label therapies, and the need for future research to develop improved patient-provider communication mechanisms.
Our findings are consistent with those of other studies that have reported notable barriers to the integration of sequencing results into clinical practice.15,16 We found several instances that suggested a need for educational resources to support both the patient and the clinician. Excessive, dense, or overly technical information can lead to misinterpretation, underscoring the need for improved and better-designed test reports.
Discordance between an oncologist and a patient’s perceptions of a disclosure of results suggests the need for improvements in patient-provider communication as a point of intervention. We also found notable gaps in the communication process. Determining the reasons for this discordance may inform strategies for improving communication in future studies. For example, patients with advanced or rare disease, such as the patients in this study, often join multiple research studies, go through multiple treatments regimens in a brief timeframe, and meet with several providers. It is reasonable that patients might not remember the specific study being discussed, be fatigued from treatments and appointments, or be confused by complex medical terminology.
Defining what constitutes an actionable finding is often challenging when multiple stakeholders are involved. Members of the PMTB bring expertise from a wide range of perspectives. Pathologists, for example, might consider a finding to be actionable if it informs diagnosis. Bioinformaticists or other basic science researchers could view as actionable information that expands the current understanding of the genetic mechanisms behind human cancers. Clinicians might define as actionable only findings with a therapeutic implication. In this study, all results given to the oncologist after deliberation at the PMTB were categorized in the summary as potentially actionable findings. However, several oncologists reported on the survey that they did not plan to make treatment changes because of a perceived lack of clinical significance of the sequencing results. In addition, we encountered unexpected examples of how clinicians defined an actionable finding. In one case, genomic sequencing of a tumor of a patient diagnosed with head and neck squamous cell carcinoma revealed a highly mutated phenotype. In the oncologist’s view, this confirmed that there was nothing that could be done. On the survey, the oncologist endorsed that this patient had an actionable finding because it aided in the decision to forego additional treatments, which in the end resulted in a change in clinical management (ie, the decision to refer to hospice). Actionable can mean different things to different individuals on the care team. Therefore, establishing a clear framework for defining actionable findings is critical to the implementation of comprehensive genomic sequencing information in clinical management. Our attempt to address this barrier was the tiering scalable framework for classification of alterations to be implemented in future test reports.
Other prominent barriers patients and oncologists faced included uncertainty about locally available clinical trials and the lack of access to off-label therapies. These barriers include a lack of information about existing trials and therapies, a lack of resources to identify trial eligibility, inability to obtain approval for compassionate use for off-label drugs, and logistical challenges for the patient (travel, lack of insurance coverage, or financial constraints).
Our study sample is not representative of oncologists in general given its over-representation of practitioners in an academic medical center. Patient outcomes in this study may not be reflective of those of common types of cancer. Enrollment eligibility criteria included metastatic or refractory cancer, where standard of care is often ineffective, or rare cancers for which no standard of care therapy exists. Furthermore, the sample size was limited by the rare nature of the cancer types and the survey time interval. For example, there was a higher frequency of patients with hematologic malignancies being referred during this 9-month interval than across the entire study timeframe. In addition, because medical record information is not always complete and may not accurately capture the existence or nature of discussions with patients, the verification of disclosures and outcomes was limited to a subset of individuals. It is possible that the views and experiences of these patients do not reflect the broader group of individuals in the research study.
Footnotes
The study funders had no role in the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
Supported by 5UM1HG006508 03 and 2UL1TR000433-06.
AUTHOR CONTRIBUTIONS
Conception and design: Michele C. Gornick, Lan Q. Le, Natalie Bartnik, Elena Stoffel, Scott Schuetze, Moshe Talpaz, Arul Chinnaiyan, J. Scott Roberts
Financial support: Arul Chinnaiyan, J. Scott Roberts
Administrative support: Arul Chinnaiyan
Provision of study material or patients: Scott Schuetze, Arul Chinnaiyan
Collection and assembly of data: Michele C. Gornick, Erin Cobain, Lan Q. Le, Natalie Bartnik, Elena Stoffel, Scott Schuetze
Data analysis and interpretation: Michele C. Gornick, Erin Cobain, Lan Q. Le, Natalie Bartnik, Elena Stoffel, Scott Schuetze, Moshe Talpaz
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/po/author-center.
Michele C. Gornick
No relationship to disclose
Erin Cobain
Consulting or Advisory Role: AstraZeneca
Lan Q. Le
No relationship to disclose
Natalie Bartnik
No relationship to disclose
Elena Stoffel
Research Funding: Cancer Prevention Pharmaceuticals (Inst)
Scott Schuetze
Consulting or Advisory Role: EMD Serono, Janssen Pharmaceuticals, Daiichi Sankyo
Research Funding: AB Science (Inst), Janssen Pharmaceuticals (Inst), Amgen (Inst), BioMed Valley Discoveries (Inst), CytRx (Inst), Plexxikon (Inst), Eli Lilly (Inst), Karyopharm Therapeutics (Inst), Adaptimmune (Inst)
Moshe Talpaz
Consulting or Advisory Role: Gilead Sciences, CTI BioPharma, Nynex
Research Funding: Gilead Sciences, Incyte, CTI BioPharma, ARIAD Pharmaceuticals, Novartis, Aptose Biosciences, Pfizer, Sanofi
Expert Testimony: Bristol-Myers Squibb Canada
Arul Chinnaiyan
Consulting or Advisory Role: Tempus
J. Scott Roberts
No relationship to disclose
REFERENCES
- 1.Collins FS, Varmus H. A new initiative on precision medicine. N Engl J Med. 2015;372:793–795. doi: 10.1056/NEJMp1500523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garraway LA. Genomics-driven oncology: Framework for an emerging paradigm. J Clin Oncol. 2013;31:1806–1814. doi: 10.1200/JCO.2012.46.8934. [DOI] [PubMed] [Google Scholar]
- 3. National Cancer Institute: NCI-MATCH Trial (Molecular Analysis for Therapy Choice (NCI-MATCH). https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/nci-match.
- 4. American Society of Clinical Oncology: Targeted Agent and Profiling Utilization Registry (TAPUR) study. http://www.tapur.org/
- 5.Tsimberidou AM, Wen S, Hong DS, et al. Personalized medicine for patients with advanced cancer in the phase I program at MD Anderson: Validation and landmark analyses. Clin Cancer Res. 2014;20:4827–4836. doi: 10.1158/1078-0432.CCR-14-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheuner MT, Peredo J, Benkendorf J, et al. Reporting genomic secondary findings: ACMG members weigh in. Genet Med. 2015;17:27–35. doi: 10.1038/gim.2014.165. [DOI] [PubMed] [Google Scholar]
- 7.Brandt DS, Shinkunas L, Hillis SL, et al. A closer look at the recommended criteria for disclosing genetic results: Perspectives of medical genetic specialists, genomic researchers, and institutional review board chairs. J Genet Couns. 2013;22:544–553. doi: 10.1007/s10897-013-9583-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hassler JR, Scheuner DL, Wang S, et al. The IRE1α/XBP1s pathway is essential for the glucose response and protection of β cells. PLoS Biol. 2015;13:e1002277. doi: 10.1371/journal.pbio.1002277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Townsend A, Adam S, Birch PH, et al. “I want to know what’s in Pandora’s Box”: Comparing stakeholder perspectives on incidental findings in clinical whole genomic sequencing. Am J Med Genet A. 2012;158A:2519–2525. doi: 10.1002/ajmg.a.35554. [DOI] [PubMed] [Google Scholar]
- 10.Yu JH, Harrell TM, Jamal SM, et al. Attitudes of genetics professionals toward the return of incidental results from exome and whole-genome sequencing. Am J Hum Genet. 2014;95:77–84. doi: 10.1016/j.ajhg.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Middleton A, Morley KI, Bragin E, et al. Attitudes of nearly 7000 health professionals, genomic researchers and publics toward the return of incidental results from sequencing research. Eur J Hum Genet. 2016;24:21–29. doi: 10.1038/ejhg.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green RC, Berg JS, Berry GT, et al. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet Med. 2012;14:405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meric-Bernstam F, Johnson A, Holla V, et al. A decision support framework for genomically informed investigational cancer therapy. J Natl Cancer Inst. 2015;107:djv098. doi: 10.1093/jnci/djv098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roychowdhury S, Iyer MK, Robinson DR, et al. Personalized oncology through integrative high-throughput sequencing: A pilot study. Sci Transl Med. 2011;3:111ra121. doi: 10.1126/scitranslmed.3003161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simon R, Roychowdhury S. Implementing personalized cancer genomics in clinical trials. Nat Rev Drug Discov. 2013;12:358–369. doi: 10.1038/nrd3979. [DOI] [PubMed] [Google Scholar]
- 16.Schilsky RL. Implementing personalized cancer care. Nat Rev Clin Oncol. 2014;11:432–438. doi: 10.1038/nrclinonc.2014.54. [DOI] [PubMed] [Google Scholar]