Abstract
Corynebacterium bovis is the causative agent of Corynebacterium-associated hyperkeratosis in immunocompromised mice. The resulting skin pathology can be profound and can be associated with severe wasting, making the animals unsuitable for research. Although the administration of antibiotics is effective in resolving clinical symptoms, antibiotics do not eradicate the offending bacterium. Furthermore, antibiotic use may be contraindicated as it can affect tumor growth and is associated with Clostridioides difficile enterotoxemia in highly immunocompromised murine strains. Lysins, which are lytic enzymes obtained from bacteriophages, are novel antimicrobial agents for treating bacterial diseases. The advantage of lysins are its target specificity, with minimal off-target complications that could affect the host or the biology of the engrafted tumor. The aim of this study was to identify lysins active against C. bovis. Chemical activation of latent prophages by using mitomycin C in 3 C. bovis isolates did not cause bacteriophage induction as determined through plaque assays and transmission electron microscopy. As an alternative approach, 8 lysins associated with other bacterial species, including those from the closely related species C. falsenii, were tested for their lytic action against C. bovis but were unsuccessful. These findings were congruent with the previously reported genomic analysis of 21 C. bovis isolates, which failed to reveal bacteriophage sequences by using the PHAST and PHASTER web server tools. From these results, we suggest C. bovis is among those rare bacterial species devoid of lysogenic bacteriophages, thus making the identification of C. bovis-specific lysins more challenging. However, C. bovis may be a useful model organism for studying the effects of antiphage systems.
Abbreviation: TEM, transmission electron microscopy
In 1998, the causative agent of Corynebacterium-associated hyperkeratosis, commonly known as ‘scaly skin disease,’ was identified as Corynebacterium bovis via 16S rRNA gene sequence analysis.12,34 C. bovis is a small gram-positive rod belonging to the family Corynebacteriaceae that colonizes the superficial layers of the epidermis of immunocompromised mice (for example, athymic nude, NOD.CB17-Prkdcscid/J [NOD SCID], and NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ [NSG] mice) and is highly contagious.5,9,11,12,31,41,52,63 The use of immunocompromised mouse models has increased exponentially over the past decade, given that they serve as valuable tools in oncology, immunology and other scientific disciplines because they support the growth of xenografts and allografts.29,50 Due to the prevalent use of these mouse models, infection with C. bovis has become increasingly problematic. The resulting skin pathology can be profound and, on occasion, can be associated with severe wasting, making the animals unsuitable for research.16 A recent study demonstrated the effect of C. bovis in a patient-derived chronic myelomonocytic leukemia model, where C. bovis infection decreased tumor engraftment in the immunocompromised murine host (NSGS mice).60 Recently, oral administration of amoxicillin (the therapy of choice for Corynebacterium-associated hyperkeratosis) to SCID and NSG mouse strains presumptively led to microbial dysbiosis, Clostridioides difficile colonization, enterotoxemia, diarrhea, and death.40 Furthermore, although the administration of antibiotics is effective in resolving disease, it does not eradicate the causative C. bovis organism.6
Currently, alternative antimicrobial therapies are being developed in response to both an increase in antibiotic-resistant bacteria and the decrease in antibiotic development.23 One such therapy involves the use of bacteriophage (that is, phage) lysins. At the end of phage replication and assembly inside a host bacterium, the progeny phage must escape.18,23,48 To this end, phage-encoded peptidoglycan hydrolases, termed lysins, degrade the bacterial cell wall, resulting in hypotonic lysis of the bacterium and release of phage progeny. When applied exogenously, purified forms of these enzymes are likewise able to access the peptidoglycan layer of the gram-positive cell envelope to produce the same lytic effect. Most lysins demonstrate high specificity, with lethal activity directed against the species that the lysin-encoding phage infects.23 However, some lysins exhibit broad-acting lytic activity.20 For example, PlySs2 displays strong lytic activity toward numerous Staphylococcus and Streptococcus species.23 Unlike broad-spectrum antibiotics, lysins do not disrupt the normal microbiota of the host.49 These enzymes have been shown to be highly effective in preclinical models of sepsis, endocarditis, pharyngitis, pneumonia, meningitis, and mucosal and skin decolonization, and a lysin against S. aureus has recently successfully completed phase 2 clinical trials for use in human medicine.18,48,49
With regard to using lysins as antimicrobial agents, several features distinguish lysins from traditional antibiotics for use in the animal research setting: (1) rapid bactericidal activity against both stationary and exponentially growing bacteria; (2) lack of observed resistance; (3) no loss of efficacy or adverse effects after repeated administration; and (4) minimal disruption of the resident microbiota—unlike antibiotics, which can be toxic, influence physiologic responses of the host (for example, immune function, gastrointestinal dysbiosis), and affect the use of animals as models. Lysins, due to their targeted mode of action, would not be expected to have the same negative effects.2,18,49 Using whole-genome sequencing and comparative genomic analyses, we recently reported on the genetic diversity of 21 C. bovis isolates obtained from various hosts, geographical locations, and time points.3,4,8,21,36,37,57,61 Genotypic differences between isolates, including the number of associated virulence factors, reflected the host from which they were obtaineds.8,19 Analyses using the PHAST (Phage Search Tool) and PHASTER (Phage Search Tool Enhanced Release) algorithms failed to identify the presence of intact or true (completeness score greater than 90) or questionable (completeness score, 70 to 90) phage in any of the isolates.1,64 Both PHAST and PHASTER are used to identify, annotate, and graphically display prophage sequences within bacterial genomes or plasmids. However, these prophage prediction methods, which are based on databases of previously identified prophage sequences, are limited with regard to determine the probabilities of putative phage sequences in submitted genomes.1 Notably, the prophage sequence databases used by the aforementioned algorithms are largely incomplete due to the ubiquity of phages. This understanding, along with their genetic diversity, allows PHAST and PHASTER to miss prophage. When phages are labeled as incomplete by these programs, excessive distance between possible prophage genes would prevent their combination into a single predicted region, and the completeness score would decrease. This limitation allows a prophage sequence to be classified as questionable or incomplete even though it represents an intact or true prophage. Finally, previous work demonstrated a sensitivity (no. of true positives / [no. of true positives + no. of false negatives]) of 85% for PHASTER (79% for PHAST), implying that about 15% of prophages would be missed.1 Furthermore, although no specific lysin sequences were found in our previous study, these methods did identify several incomplete phages in each isolate (completeness score of less than 70).8 For these reasons, additional in vitro studies are necessary to definitively conclude that C. bovis lacks phages.
The primary aim of the current study was to identify a phage lysin to use as a potential therapeutic for Corynebacterium-associated hyperkeratosis. Given both the ubiquity and abundance of phages and the known analytical limitations of PHAST and PHASTER, we hypothesized that C. bovis phage and their corresponding lysins potentially could be identified by using traditional phage induction methods.1,13 To this end, multiple C. bovis isolates were treated with mitomycin C to stimulate potential prophage to enter into a lytic cycle. The supernatant of the induced bacterial cultures was then visually inspected for phage by using transmission electron microcopy (TEM) and subsequently assayed for its ability to generate plaques when applied to multiple C. bovis strains. In addition, with the understanding that some lysins exhibit broad lytic activity, we analyzed the susceptibility of C. bovis to a diverse library of lysins.22,49,54 In total, 8 lysins were screened against C. bovis, including PlyCf, which is a lysin derived from a prophage from the closely related Corynebacterium falsenii.8 We hypothesized that lysins originating from non-C. bovis phage might exhibit nonspecific lytic activity against C. bovis. For example, the lysin from corynephage BFK20 obtained from Brevibacterium flavum has shown effective lytic activity against other bacterial species, including Corynebacterium glutamicum and Brevibacterium lactofermentum.20
Materials and Methods
Bacterial strains and culture conditions.
Strain information relating to C. bovis clinical isolates 7894, CUAMC1, 4826, 4828, MI 82-1021, F6900, and WCM1 was previously described.8 Each strain, as well as the C. falsenii strain ATCC BAA-596, was initially plated on Columbia colistin and nalidixic acid agar with 5% (v/v) sheep blood (Becton Dickinson, Franklin Lakes, NJ) and then routinely grown in brain–heart infusion medium (Becton Dickinson) supplemented with 0.1% (v/v) Tween 80 (Fisher Scientific, Hampton, NH). Streptococcus pyogenes strain D471 and S. pneumoniae strain DCC1490 were grown in Todd–Hewitt broth (Becton Dickinson) with 1% (w/v) yeast extract (Fisher Scientific). Clostridium difficile strain ATCC 43255 was grown anaerobically in brain–heart infusion medium supplemented with 0.5% (w/v) yeast extract and 0.1% (w/v) l-cysteine. Bacillus anthracis strain Sterne and Staphylococcus aureus strain RN4220 were grown in tryptic soy broth (Becton Dickinson).
Determining C. bovis growth kinetics.
A growth curve for C. bovis was generated by using previously described methods.14,28,43 Briefly, a turbid culture of C. bovis strain 7894 was diluted 1:100 in fresh growth medium and subsequently incubated at 37 °C with aeration for a total of 48 h. Every 6 h, an aliquot of bacteria was quantitated by either (1) measuring the optical density at 600 nm or (2) serially diluting and plating the bacteria to calculate the number of cfu per milliliter. The resulting growth curves were fit with a Boltzmann sigmoidal curve by using GraphPad Prism (GraphPad, San Diego, CA). All error bars correspond to the SEM of duplicate experiments.
Bacteriophage induction and TEM.
We attempted to chemically induce phage from C. bovis isolates WCM1, 7894, and CUAMC1 by using mitomycin C according to previously described methods, with modifications.15,39 In the initial set of experiments, each strain was grown to an OD600nm of 0.5. The culture was then divided, with one half being treated with 1 µg/mL mitomycin C and the other half serving as an untreated control. After 30 min, the bacteria were pelleted, resuspended in fresh growth medium, and grown for an additional 6 h. Growth curves for each strain were monitored by measuring the OD600nm at 1-h increments. In the second set of experiments, C. bovis strain 7894 was grown to an OD600nm of 0.5 and then divided into 5 flasks. Each flask was treated with either 0, 0.01, 0.1, 0.5 or 1 µg/mL mitomycin C. The subsequent steps were the same as described for the first set of experiments.
At the culmination of each induction experiment, the cells were pelleted by centrifugation. The supernatant was filtered and incubated with polyethylene glycol to precipitate any induced phage. Each sample was then centrifuged at 16,000 × g for 15 min at 4 °C. The supernatant was removed, and the pellet containing phage, if present, was resuspended in SM buffer (50 mM Tris-HCl, pH 7.5; 100 mM NaCl; 8 mM MgSO4). The samples were negatively stained with 3% (w/v) uranyl acetate and imaged by using TEM at the Core Laboratories Center Imaging Core Facility of Weill Cornell Medicine. The images were visually inspected for structures resembling phage components according to previous published TEM images of phages associated with other Corynebacterium spp.58
Plaque assay.
C. bovis strains 7894, CUAMC1, 4826, 4828, MI 82-1021, F6900, and WCM1 were each embedded at an initial concentration of approximately 106 cfu/mL in growth medium supplemented with 0.75% (w/v) agarose. Next, the filtered supernatant from C. bovis strain 7894 treated with 0.1 or 0.5 µg/mL mitomycin C was spotted onto the bacteria-containing plates. The filtered supernatant from bacteria absent mitomycin C treatment was spotted as a negative control. The plates were incubated at 37 °C for a maximum of 72 h to determine whether plaques formed.
Molecular cloning.
The plyCf gene encoding the translated C. falsenii lysin PlyCf (GenBank AHI04498) was synthesized and codon-optimized for protein expression in E. coli (GeneWiz, South Plainfield, NJ). The plyCf gene was PCR-amplified and then cloned into the NcoI and BamHI sites of the E. coli expression vector pET28a by using the NEBuilder HiFi DNA Assembly method (New England Biolabs, Ipswich, MA). After sequence confirmation, the pET28a::plyCf construct was transformed into E. coli BL21(DE3) for protein expression.
Lysin expression and purification.
The lysins PlyC, PlyCD, Cpl-1, ClyS, PlySs2, LysK, and PlyG were expressed and purified to homogeneity as previously described.10,22,38,45,46,53,62 The PlyCf lysin was expressed in Luria–Bertani medium (Fisher Scientific) supplemented with kanamycin (50 µg/mL; Fisher Scientific) for 16 h at 18 °C. Protein expression was induced at midlog phase by using 1 mM isopropyl β-d-1-thiogalactopyranoside (Biosynth Chemistry and Biology, Staad, Switzerland). Cells were washed and resuspended in 50 mM Tris-HCl (pH 7.5), 200 mM NaCl supplemented with 1 mM phenylmethanesulfonyl fluoride (Fisher Scientific). The bacteria were lysed by using a homogenizer (Emulsiflex-C5, Avestin, Ottawa, Ontario). The lysate was cleared by centrifugation at 27,000 × g for 1 h at 4 °C. The soluble lysate fraction was dialyzed against 20 mM Tris-HCl (pH 8.0) and then applied to a 5-mL HiTrap Q FF column (GE Healthcare Life Sciences, Pittsburgh, PA) in 20 mM Tris-HCl (pH 8.0). PlyCf was eluted from the column using a linear salt gradient from 0 to 500 mM NaCl. After removal of salt via dialysis, the protein sample was applied to a 5-mL HiTrap DEAE FF column (GE Healthcare Life Sciences) in 20 mM Tris-HCl (pH 8.0). The flow-through was collected and dialyzed against PBS (pH 7.4). Finally, the PlyCf sample was concentrated by using an Amicon Ultra Ultracel-10K filter (EMD Millipore, Burlington, MA) and applied to a HiLoad 16/60 Superdex 200 Prep Grade column (GE Healthcare Life Sciences) in PBS. Highly pure PlyCf elution fractions were combined and stored at –80 °C until use. All chromatography experiments were performed by using an AKTA fast protein liquid chromatography system (GE Healthcare Life Sciences, Pittsburgh, PA).
Turbidity reduction and ATP release assays.
Seven non-Corynebacterium lysins (PlyC, PlyCD, Cpl-1, ClyS, PlySs2, LysK, and PlyG) were initially assayed for bacteriolytic activity toward susceptible bacterial species by using the turbidity reduction assay to confirm that each lysin was enzymatically active. In brief, midlog S. pyogenes (PlyC), C. difficile (PlyCD), S. pneumoniae (Cpl-1), S. aureus (ClyS and PlySs2), and B. anthracis (PlyG) were incubated with their specific lysin(s) at 25 µg/mL (final concentration) in 50 mM sodium phosphate (pH 7.0) in 96-well microtiter plates. By using a SpectraMax M5 microplate reader (Molecular Devices, San Jose, CA), bacteriolytic activity was quantitated by measuring the OD600 of each sample every 15 s for 1 h at 37 °C. After confirmation of enzymatic activity, each lysin was then incubated with midlog C. bovis strain 7894 by using the aforementioned turbidity reduction assay protocol. All resulting data points were fit by using a 1- or 2-phase exponential decay curve (Prism, GraphPad Software). Each sample set was analyzed in duplicate and included untreated negative controls.
To further determine C. bovis susceptibility to each of the previously mentioned non-Corynebacterium lysins, ATP release assays were performed (ENLITEN, Promega, Madison, WI). Briefly, at the conclusion of the previously mentioned C. bovis turbidity reduction assays, the contents from each sample well were transferred to polypropylene microcentrifuge tubes. Intact cells and insoluble cellular debris were removed by centrifugation at 14,500 × g for 10 min. By using new microcentrifuge tubes, the sample supernatant was added to an equal volume of 1 M Tris-HCl (pH 7.75). After gentle mixing, 100 µL from each sample was transferred to a white, opaque 96-well microtiter plate and mixed with an equal volume of rL/L reagent (ENLITEN, Promega). The luminescence of each sample well was measured at 560 nm in a microplate reader by using a 1.5-s integration time. S. pyogenes treated with PlyC was used as positive control for ATP release, and ATP-free water, buffer, and untreated bacteria were used as negative controls. All error bars correspond to the SEM of duplicate samples.
A modified version of the turbidity reduction assay was used for evaluating the lytic activity of the C. falsenii lysin PlyCf toward C. bovis. Briefly, midlog C. bovis strain 7894 was incubated with 125 µg/mL PlyCf (final concentration) in PBS (pH 7.4). OD600 was measured every 30 s for 1 h at room temperature by using the microplate reader. As a positive control, PlyCf was incubated at 125 µg/mL with midlog C. falsenii strain ATCC BAA-596. Untreated bacteria were used as a negative control for lytic activity. The resulting data points were fit to a 1-phase exponential decay curve by using Prism (GraphPad Software). All samples were assayed in duplicate.
Results
C. bovis growth kinetics.
Generally, C. bovis are slow-growing bacteria, but a C. bovis-specific growth curve has yet to be published. In order to identify a lysin with lytic activity toward C. bovis, we must first understand when the bacterium has entered various stages of growth, given that logarithmic-phase bacteria are generally more susceptible than stationary phase cells to lysins.
To that end, C. bovis strain 7894 was grown in brain–heart infusion medium supplemented with Tween 80 at 37 °C for a total of 48 h, with the bacterial concentration calculated according to OD600 (Figure 1 A) or number of cfu per milliliter (Figure 1 B) measured every 6 h. The bacteria were in lag phase during the initial 6 to 12 h and then transitioned into the exponential phase of growth from 12 to 30 h. Early-log phase occurred at around 12 h, midlog phase was observed at 18 h, and late-log phase developed between 24 and 30 h. C. bovis began to enter stationary phase at approximately 30 h after inoculation.
Figure 1.
Growth curve of C. bovis. A turbid culture of C. bovis strain 7894 was diluted 1:100 in fresh BHI medium supplemented with Tween 80 and subsequently incubated at 37 °C with aeration for a total of 48 h. The bacterial concentration was quantitated every 6 h by either (A) measuring the OD600 or (B) serially diluting and plating to determine the number of cfu per mL. Error bars, SEM of duplicate experiments.
Bacteriophage induction, electron microscopy, and plaque assays.
Three C. bovis clinical isolates (WCM1, 7894, CUAMC1) were treated with 1 µg/mL mitomycin C for 30 min. After removal of mitomycin C, the OD600 of each sample was monitored for a total of 6 h at 37 °C. If lytic phage were induced successfully, a rapid decrease in the OD of the bacterial culture would be expected. Compared with untreated controls, the growth rate of each C. bovis strain treated with mitomycin C was reduced. However, the OD of each sample slowly increased during the 6-h incubation, suggesting either that lytic phage were not induced or only partially induced in the 3 C. bovis strains.
Given the potential that this finding could be an artifact resulting from the mitomycin C concentration used,15 additional induction experiments using various mitomycin C concentrations were performed. C. bovis strain 7894 was treated with mitomycin C concentrations of 0, 0.1, 0.5 and 1.0 µg/mL for a total of 30 min. After removal of mitomycin C, bacterial growth at 37 °C was determined for a total of 6 h (data not shown). The growth of the culture treated with 0.01 µg/mL mitomycin C was similar to that of the untreated control. Although the growth rates of C. bovis were significantly altered at mitomycin C concentrations of 0.1 µg/mL or greater, none of these concentrations rapidly decreased the OD of the culture over the duration of the experiment, indicating that phages likely were not induced. The absence of phage-like particles in the supernatant of mitomycin C-treated C. bovis was further confirmed through visual inspection by using TEM (data not shown). Furthermore, the same supernatants lacked the ability to generate plaques after spotting on soft agar overlays embedded with various strains of C. bovis (data not shown).
Evaluation of C. bovis susceptibility to various lysins.
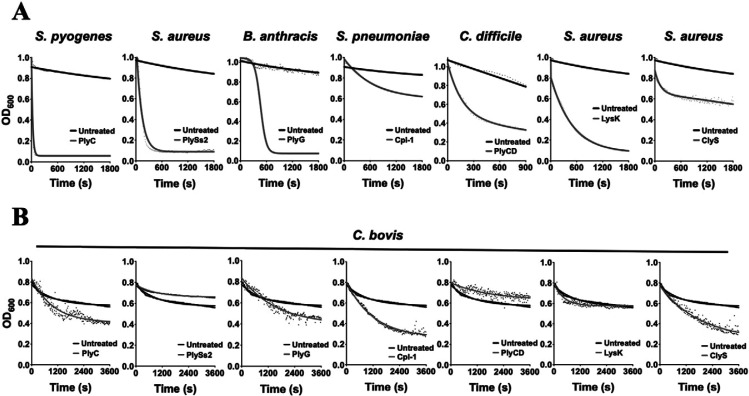
Several previously studied non-Corynebacterium lysins were assayed for lytic activity toward C. bovis. The lysins tested included PlyC, PlySs2, PlyG, Cpl-1, PlyCD, LysK, and ClyS. Because bacteria lyse on treatment with lysin, the turbidity reduction assay has become a standard method to determine the lytic activity of lysins.20,22,23 To confirm that each was enzymatically functional, all 7 lysins initially were incubated at a concentration of 25 µg/mL with a bacterial species known to be sensitive to the particular lysin10,22,38,45,46,53,62 (Figure 2 A). A rapid and significant reduction in turbidity was observed for PlyC (compared with S. pyogenes), PlySs2 (compared with S. aureus), PlyG (compared with B. anthracis), Cpl-1 (compared with S. pneumoniae), PlyCD (compared with C. difficile), LysK (compared with S. aureus), and ClyS (compared with S. aureus), thus confirming that each lysin was functional. Next, each lysin (concentration, 25 µg/mL) was incubated with C. bovis strain 7894 for a total of 1 h at 37 °C. Compared with the untreated control, a reduction in turbidity was observed when C. bovis was treated with PlyC, PlyG, Cpl-1, and ClyS (Figure 2 B). However, according to visual inspection of these samples, the decline in turbidity did not appear to be due to bacteriolytic activity of the lysins. Instead, the decrease in OD resulted from the formation of bacterial aggregates and subsequent settling of the sample, seen as white clumps at the bottom of the wells.
Figure 2.
Analyzing the lytic activity of several non-Corynebacterium lysins against C. bovis by means of the turbidity reduction assay. (A) To confirm each lysin was enzymatically functional, PlyC (S. pyogenes), PlySs2 (S. aureus), PlyG (B. anthracis), Cpl-1 (S. pneumoniae), PlyCD (C. difficile), LysK (S. aureus) and ClyS (S. aureus) were incubated at 25 µg/mL with a susceptible bacterial species. (B) The bacteriolytic activity of each lysin at 25 µg/mL was determined against C. bovis strain 7894. For each turbidity reduction assay, the OD600nm was measured every 15 s for 1 h at 37 °C. A decrease in turbidity is indicative of bacteriolytic activity. Bacteria absent lysin treatment were used as a negative control for lytic activity. Each sample was analyzed in duplicate.
Luminescence associated with ATP release is an assay used to confirm that the bacterial cell is disrupted as a result of lytic activity. To help verify the previous observation, ATP release was measured for each of the lysin-treated C. bovis samples (Figure 3). If the bacterial cells are indeed being lysed, then intracellular ATP would be released, with the amount of ATP observed in the supernatant directly correlating with the total number of cells lysed. As a positive control, S. pyogenes was treated with the streptococcal-specific lysin PlyC. The negative controls consisted of ATP-free water, buffer only, and untreated bacterial cells. The results show that all of the C. bovis samples treated with lysin had extracellular ATP concentrations comparable to that of the untreated control. This finding indicates that none of the tested lysins was capable of lysing C. bovis, thereby further confirming that the reduction in OD observed at the conclusion of the turbidity reduction assays was indeed a direct result of bacterial aggregation and settling and not due to lysis.
Figure 3.
Measuring the concentration of ATP released by C. bovis when treated with various non-Corynebacterium lysins. ATP assays were used to measure the amount of intracellular ATP released, as a function of luminescence, by C. bovis strain 7894 after incubation with PlyC, PlySs2, PlyG, Cpl-1, PlyCD, LysK, and ClyS (final concentration, 25 µg/mL) for 1 h at 37 °C. S. pyogenes treated with PlyC was used as a positive control; ATP-free water, buffer only, and untreated bacteria were used as negative controls. Error bar, SEM of duplicate experiments. RLU, relative light units.
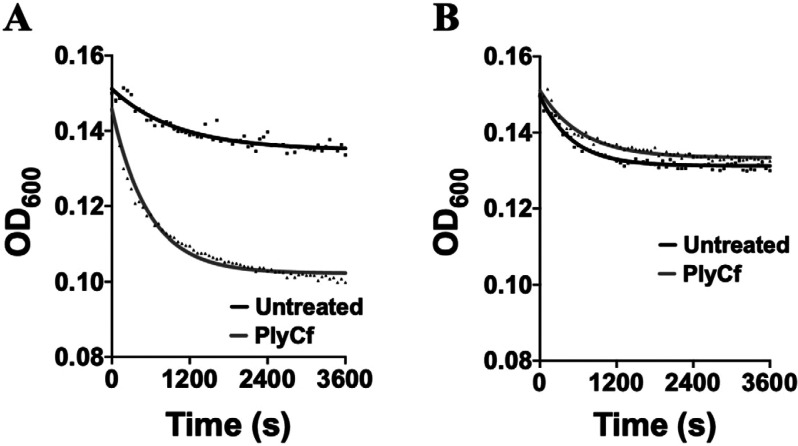
A lysin derived from a phage that infects a close relative of C. bovis, such as another Corynebacterium species, would be expected to have a greater probability of displaying nonspecific bacteriolytic activity toward the bacterium. Therefore, we tested the sensitivity of C. bovis against PlyCf, a lysin derived from a prophage encoded by C. falsenii strain DSM 44353. First, to establish that PlyCf was lytically active, it was incubated at 125 µg/mL with C. falsenii (its native target bacterial species) for 1 h (Figure 4 A). Results from the corresponding turbidity reduction assay confirmed that the lysin was enzymatically functional. PlyCf was subsequently incubated at 125 µg/mL with C. bovis for 1 h (Figure 4 B). Similar to the untreated negative control, PlyCf did not significantly reduce the turbidity of the bacterial sample, indicating that PlyCf lacks bacteriolytic activity toward C. bovis.
Figure 4.
Evaluating the bacteriolytic activity of the Corynebacterium lysin PlyCf against C. bovis. In a turbidity reduction assay, the C. falsenii lysin PlyCf (final concentration, 125 µg/mL) with either (A) C. falsenii strain ATCC BAA-596 (positive control) or (B) C. bovis strain 7894. OD600 was measured every 30 s for 1 h at room temperature. A decrease in turbidity is indicative of bacteriolytic activity. Bacteria absent lysin treatment were used as a negative control for lytic activity. Each sample was analyzed in duplicate.
Discussion
The eradication of C. bovis from vivaria that house immunodeficient mouse strains is particularly challenging because the organism can be found in drinking water, tumor and cell lines, within biosafety cabinets, and on fomites, including cages, bedding, and instruments.7,41 Although adherence to aseptic handling and cage-changing techniques can decrease the spread of C. bovis, studies have concluded that once C. bovis becomes enzootic, it is extremely difficult or even impossible to eradicate.6,41 Variable success in the management and eradication of C. bovis has been achieved through antibiotic therapy, rederivation, colony depopulation, and disinfection.6 However, control and elimination in academic programs is often unsuccessful because depopulation and restricted colony access usually are not options. Further evidence of the significance of the problem is demonstrated by the National Cancer Institute’s Requests for Applications for Patient-derived Xenograft (PDX) Development and Trial Centers (PDTC), which now requires that applicants indicate whether C. bovis-free animals can be maintained.44
The principal aim of this study was to identify a phage lysin for use as a potential therapeutic against Corynebacterium-associated hyperkeratosis. We attempted to use mitomycin C to chemically activate potential prophages latent within several C. bovis isolates. According to plaque assays and transmission electron microscopy, no phages were induced. We then analyzed the susceptibility of C. bovis to 9 lysins, including PlyCf, a lysin derived from a prophage of the closely related C. falsenii. None of the lysins tested showed lytic activity against C. bovis (Figures 2 B, 3, and 4 B).
Although treatment with mitomycin C at concentrations of 0.1 µg/mL or greater markedly decreased the growth rate of C. bovis, a sharp decline in optical density was never observed during the 6-h incubation. If lytic phage were induced, bacterial lysis—indicated as a rapid decrease in turbidity—would be expected 2 to 3 h after exposure.26,56 Therefore, when compared with untreated controls, the reduced growth rate of the mitomycin C–treated bacteria was not due to phage induction but instead to the toxic effects of the antibiotic. Visual inspection using TEM coupled with plaque assays further confirmed the absence of phage in the supernatant of bacteria treated with mitomycin C. Our inability to detect phage in C. bovis is consistent with recently reported findings, in which no prophage DNA was found in any of the 21 C. bovis isolates sequenced and analyzed with PHAST and PHASTER.8
Although C. bovis does not appear to have any chromosomally-integrated prophage, phages can be present extrachromosomally as lysogenic linear and circular plasmidial elements in the cytoplasm of the bacteria. These phages may switch between extrachromosomal and chromosomally integrated states.59 Although genomic sequencing might be biased toward bacterial chromosomes and thus ‘overlook’ the smaller, low-copy extrachromosomal elements unless they are isolated specifically, the methods used in the recently published C. bovis sequencing study would be expected to detect extrachromosomal elements.8 Therefore, the absence of phage in C. bovis is surprising, given the abundance and ubiquity of phages in bacterial populations.13 In addition, close relatives of C. bovis have associated phages. For example, C. falsenii contains the corynephage ɸCFAL8171I in both circular (extrachromosomal) and linear (chromosomally integrated) states.24
Evidence is available—albeit sparse—to show that not all bacteria can be infected by phages.32 Several antiphage systems found in bacteria have been acquired through evolution or horizontal transfer.51,55 These systems include surface alterations to block phage adsorption, inhibition of phage DNA penetration, DNA restriction or modification systems, phage-specific immunity (clustered regularly interspaced short palindromic repeats [CRISPR]–Cas systems), and abortive infection.13,17,26,27,30,51 Some of these bacterial defense mechanisms that have evolved against phage predation can help to explain the unusual lack of phages in C. bovis. Two likely phage defense mechanisms used by C. bovis were identified in our previous comparative genomic analyses: (1) CRISPR–Cas systems, which function as a form of adaptive immunity that targets foreign nucleic acids, and (2) phage-abortive infection proteins (toxin–antitoxin systems) that are activated by phages and elicit cellular inhibition by interfering with essential metabolic processes.8,13,25,27,33,35,42,47 Both of these defense mechanisms were detected in most of the C. bovis isolates analyzed. Whether these or other methods are used by C. bovis to resist phage attack remains unknown. As such, C. bovis may serve as a useful model for studies of phage resistance.
Acknowledgments
We thank Dr Christopher Manuel (University of Colorado–Denver) for providing the C. bovis isolate CUAMC1 and Dr David Bemis (University of Tennessee) for providing the bovine C. bovis isolate MI 82-1021. This research was funded in part through the NIH/NCI Cancer Center Support Grant P30 CA008748.
References
- 1.Arndt D, Grant J, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44 W1:W16–W21. 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker DG, Lipman NS. 2015. Factors that can influence animal research, p 1461. In: Fox JG, Anderson LC, Otto G, Pritchett-Corning KR, Whary MT, editors. Laboratory animal medicine, 3rd ed. San Diego (CA): Academic Press. [Google Scholar]
- 3.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brooks BW, Barnum DA. 1984. Characterization of strains of Corynebacterium bovis. Can J Comp Med 48:230–232. [PMC free article] [PubMed] [Google Scholar]
- 6.Burr HN, Lipman NS, White JR, Zheng J, Wolf FR. 2011. Strategies to prevent, treat, and provoke Corynebacterium-associated hyperkeratosis in athymic nude mice. J Am Assoc Lab Anim Sci 50:378–388. [PMC free article] [PubMed] [Google Scholar]
- 7.Burr HN, Wolf FR, Lipman NS. 2012. Corynebacterium bovis: Epizootiologic features and environmental contamination in an enzootically infected rodent room. J Am Assoc Lab Anim Sci 51:189–198. [PMC free article] [PubMed] [Google Scholar]
- 8.Cheleuitte-Nieves C, Gulvik CA, McQuiston JR, Humrighouse BW, Bell ME, Villarma A, Fischetti VA, Westblade LF, Lipman NS. 2018. Genotypic differences between strains of the opportunistic pathogen Corynebacterium bovis isolated from humans, cows, and rodents. PLoS One 13:1–30. 10.1371/journal.pone.0209231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chow SK, Bui U, Clarridge JE. 2015. Corynebacterium bovis eye infections, Washington, USA, 2013. Emerg Infect Dis 21:1687–1689. 10.3201/eid2109.150520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Daniel A, Euler C, Collin M, Chahales P, Gorelick KJ, Fischetti VA. 2010. Synergism between a novel chimeric lysin and oxacillin protects against infection by methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 54:1603–1612. 10.1128/AAC.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dole VS, Henderson KS, Fister RD, Pietrowski MT, Maldonado G, Clifford CB. 2013. Pathogenicity and genetic variation of 3 strains of Corynebacterium bovis in immunodeficient mice. J Am Assoc Lab Anim Sci 52:458–466. [PMC free article] [PubMed] [Google Scholar]
- 12.Duga S, Gobbi A, Asselta R, Crippa L, Tenchini ML, Simonic T, Scanziani E. 1998. Analysis of the 16S rRNA gene sequence of the coryneform bacterium associated with hyperkeratotic dermatitis of athymic nude mice and development of a PCR-based detection assay. Mol Cell Probes 12:191–199. 10.1006/mcpr.1998.0168. [DOI] [PubMed] [Google Scholar]
- 13.Dy RL, Richter C, Salomond GPC, Fineran PC. 2014. Remarkable mechanisms in microbes to resist phage infections. Annu Rev Virol 1:307–331. 10.1146/annurev-virology-031413-085500. [DOI] [PubMed] [Google Scholar]
- 14.Edwards DC. 1960. The growth and toxin production of Corynebacterium diphtheriae in submerged culture. J Gen Microbiol 22:698–704. 10.1099/00221287-22-3-698. [DOI] [PubMed] [Google Scholar]
- 15.Euler CW, Juncosa B, Ryan PA, Deutsch DR, McShan WM, Fischetti VA. 2016. Targeted curing of all lysogenic bacteriophage from Streptococcus pyogenes using a novel counterselection technique. PLoS One 11:1–22. 10.1371/journal.pone.0146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Field G. [Internet]. 2006. An update on scaly skin disease. ACLAM Newsletter. [Cited 15 September 2018]. Available at: https://www.aclam.org/newsletter.
- 17.Fineran PC, Blower TR, Foulds IJ, Humphreys DP, Lilley KS, Salomond GPC. 2009. The phage abortive infection system, ToxIN, functions as a protein-RNA toxin-antitoxin pair. Proc Natl Acad Sci USA 106:894–899. 10.1073/pnas.0808832106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fischetti VA. 2003. Novel method to control pathogenic bacteria on human mucous membranes. Ann N Y Acad Sci 987:207–214. 10.1111/j.1749-6632.2003.tb06050.x. [DOI] [PubMed] [Google Scholar]
- 19.Garg A, Gupta D. 2008. VirulentPred: a SVM based prediction method for virulent proteins in bacterial pathogens. Bioinformatics 9:62. 10.1186/1471-2105-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gerova M, Halgasova N, Ugorcakova J, Bukovska G. 2011. Endolysin of bacteriophage BFK20: evidence of a catalytic and a cell wall binding domain. FEMS Microbiol Lett 321:83–91. 10.1111/j.1574-6968.2011.02312.x. [DOI] [PubMed] [Google Scholar]
- 21.Ghyselinck J, Pfeiffer S, Heylen K, Sessitsch A, De Vos P. 2013. The effect of primer choice and short read sequences on the outcome of 16S rRNA gene based diversity studies. PLoS One 8:1–14. 10.1371/journal.pone.0071360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilmer DB, Schmitz JE, Euler CW, Fischetti VA. 2013. Novel bacteriophage lysin with broad lytic activity protects against mixed infection by Streptococcus pyogenes and methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother 57:2743–2750. 10.1128/AAC.02526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gilmer DB, Schmitz JE, Thandar M, Euler CW, Fischetti VA. 2017. The phage lysin PlySs2 decolonizes Streptococcus suis from murine intranasal mucosa. PLoS One 12:1–13. 10.1371/journal.pone.0169180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Glaub A, Bomholt C, Gravermann K, Brinkrolf K, Albersmeier A, Rückert C, Tauch A. 2014. Complete genome sequence of Corynebacterium falsenii DSM 44353 to study the evolution of Corynebacterium cluster 3 species. Genome Announc 2:1–2. 10.1128/genomeA.00158-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grissa I, Vergnaud G, Pourcel C. 2007. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 35 Web Server:W52–W57. 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Halgasová N, Majtán T, Ugorčáková J, Timko J, Bukovská G. 2005. Resistance of corynebacterial strains to infection and lysis by corynephage BFK 20. J Appl Microbiol 98:184–192. 10.1111/j.1365-2672.2004.02448.x. [DOI] [PubMed] [Google Scholar]
- 27.Hyman P, Abedon ST. 2010. Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70:217–248. 10.1016/S0065-2164(10)70007-1. [DOI] [PubMed] [Google Scholar]
- 28.Ikner LA, Schmitz B, Gerba C, Pepper I. 2014. Bacterial growth curve analysis and its environmental applications. JOVE [Internet]. Available from: https://www.jove.com/science-education/10100/bacterial-growth-curve-analysis-and-its-environmental-applications. 10.3791/10100. [DOI] [Google Scholar]
- 29.Ito R, Takahashi T, Katano I, Ito M. 2012. Current advances in humanized mouse models. Cell Mol Immunol 9:208–214. 10.1038/cmi.2012.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jansen R, Embden JD, Gaastra W, Schouls LM. 2002. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol 43:1565–1575. 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 31.Kim TH, Kim D, Han J, Chang S, Kim S, Seok S, Kim D, Park J, Park J. 2014. Detection of Corynebacterium bovis infection in athymic nude mice from a research animal facility in Korea. J Vet Sci 15:583–586. 10.4142/jvs.2014.15.4.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koskella B, Meaden S. 2013. Understanding bacteriophage specificity in natural microbial communities. Viruses 5:806–823. 10.3390/v5030806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Labrie SJ, Samson JE, Moineau S. 2010. Bacteriophage resistance mechanisms. Nat Rev Microbiol 8:317–327. 10.1038/nrmicro2315. [DOI] [PubMed] [Google Scholar]
- 34.Lane DJ. 1991.16S/23S rRNA sequencing, p 115–116. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York (NY): John Wiley and Sons. [Google Scholar]
- 35.Leplae R, Geeraerts D, Hallez R, Guglielmini J, Drèze P, Van Melderen L. 2011. Diversity of bacterial type II toxin-antitoxin systems: a comprehensive search and functional analysis of novel families. Nucleic Acids Res 39:5513–5525. 10.1093/nar/gkr131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li H. [Internet]. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA MEM; Database: arXiv. [Cited 10 October 2017]. Available from: http://arxiv.org/pdf/1303.3997.
- 37.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R, 1000 Genome Project Data Processing Subgroup. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loeffler JM, Nelson D, Fischetti VA. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170–2172. 10.1126/science.1066869. [DOI] [PubMed] [Google Scholar]
- 39.Lood R, Winer BY, Pelzek AJ, Diez-Martinez R, Thandar M, Euler CW, Schuch R, Fischetti VA. 2015. Novel phage lysin capable of killing the multidrugresistant Gram-negative bacterium Acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother 59:1983–1991. 10.1128/AAC.04641-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ma K, Lertpiriyapong K, Piersigilli A, Arbona RJ Ricart, Dobtsis I, Lipman NS. 2018. Clostridiodes difficile typhlocolitis resulting from amoxicillin treatment in highly immunocompromised mice. Abstract presented at the American Association for Laboratory Animal Science 69th National Meeting, Baltimore, Maryland, 28 October – 1 November 2018. [Google Scholar]
- 41.Manuel CA, Pugazhenthi U, Leszczyniski JK. 2016. Surveillance of a ventilated rack system for Corynebacterium bovis by sampling exhaust-air manifolds. J Am Assoc Lab Anim Sci 55:58–65. [PMC free article] [PubMed] [Google Scholar]
- 42.Modell JW, Jiang W, Marraffini LA. 2017. CRISPR-Cas systems exploit viral DNA injection to establish and maintain adaptive immunity. Nature 544:101–104. 10.1038/nature21719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mounier J, Rea MC, O’Connor PM, Fitzgerald GF, Cogan TM. 2007. Growth characteristics of Brevibacterium, Corynebacterium, Microbacterium, and Staphylococcus spp. isolated from surface-ripped cheese. Appl Environ Microbiol 73:7732–7739. 10.1128/AEM.01260-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.National Institutes of Health. [Internet]. 2016. PDX development and trial centers (PDTCs)(U54). [Cited 9 September 2019]. Available at: https://grants.nih.gov/grants/guide/rfa-files/RFA-CA-17-003.html
- 45.Nelson D, Loomis L, Fischetti VA. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc Natl Acad Sci USA 98:4107–4112. 10.1073/pnas.061038398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Flaherty S, Coffey A, Meaney W, Fitzgerald GF, Ross RP. 2005. The recombinant phage lysin LysK has a broad spectrum of lytic activity against clinically relevant staphylococci, including methicillin-resistant Staphylococcus aureus. J Bacteriol 187:7161–7164. 10.1128/JB.187.20.7161-7164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Örmälä A-M, Jalasvuori M. 2013. Phage therapy: Should bacterial resistance to phages be a concern, even in the long run? Bacteriophage 3:e24219. 10.4161/bact.24219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pastagia M, Euler C, Chahales P, Fuentes-Duculan J, Krueger JG, Fischetti VA. 2011. A novel chimeric lysin shows superiority to mupirocin for skin decolonization of methicillin-resistant and –sensitive Staphylococcus aureus strains. Antimicrob Agents Chemother 55:738–744. 10.1128/AAC.00890-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pastagia M, Schuch R, Fischetti VA, Huang DB. 2013. Lysins: the arrival of pathogen-directed anti-infectives. J Med Microbiol 62:1506–1516. 10.1099/jmm.0.061028-0. [DOI] [PubMed] [Google Scholar]
- 50.Puchalapalli M, Zeng X, Mu L, Anderson A, Hix Glickman L, Zhang M, Sayyad MR, Wangensteen SM, Clevenger CV, Koblinski JE. 2016. NSG mice provide a better spontaneous model of breast cancer metastasis than athymic (nude) mice. PLoS One 11:1–15. 10.1371/journal.pone.0163521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Samson JE, Magadán AH, Sabri M, Moineau S. 2013. Revenge of the phages: defeating bacterial defenses. Nat Rev Microbiol 11:675–687. 10.1038/nrmicro3096. [DOI] [PubMed] [Google Scholar]
- 52.Schröder J, Glaub A, Schneider J, Trost E, Tauch A. 2012. Draft genome sequence of Corynebacterium bovis DSM 20582, which causes clinical mastitis in dairy cows. J Bacteriol 194:4437. 10.1128/JB.00839-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schuch R, Nelson D, Fischetti VA. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884–889. 10.1038/nature01026. [DOI] [PubMed] [Google Scholar]
- 54.Schuch R, Pelzek AJ, Raz A, Euler CW, Ryan PA, Winer BJ, Farnsworth A, Bhaskaran SS, Stebbins CE, Xu Y, Cliffors A, Bearss DJ, Vankayalapati H, Goldberg AR, Fischetti VA. 2013. Use of a bacteriophage lysin to identify a novel target for antimicrobial development. PLoS One 8:1–9. 10.1371/journal.pone.0060754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seed KD. 2015. Battling phages: How bacteria defend against viral attack. PLoS Pathog 11:e1004847. 10.1371/journal.ppat.1004847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sonnen H, Schneider J, Kutzner HJ. 1990. Characterization of ɸGA1, an inducible phage particle from Brevibacterium flavum. J Gen Microbiol 136:567–571. 10.1099/00221287-136-3-567. [DOI] [PubMed] [Google Scholar]
- 57.Tatusova T, DiCuccio M, Badretdin A, Chetvernin V, Nawrocki EP, Zaslavsky L, Lomsadze A, Pruitt KD, Borodovsky M, Ostell J. 2016. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res 44:6614–6624. 10.1093/nar/gkw569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Uthman S, Liu S, Giorgini F, Stark MJR, Costanzo M, Schaffrath R. 2012. Diphtheria disease and genes involved in formation of diphthamide, key effector of the diphtheria toxin, p 340. Chapter 17. In: Priti R, editor. Insight and control of infectious disease in global scenario. London (United Kingdom): IntechOpen. 10.5772/31680. [DOI] [Google Scholar]
- 59.Utter B, Deutsch DR, Schuch R, Winer BY, Verratti K, Bishop-Lilly K, Sozhamannan S, Fischetti VA. 2014. Beyond the chromosome: The prevalence of unique extra-chromosomal bacteriophages with integrated virulence genes in pathogenic Staphylococcus aureus. PLoS One 9:1–11. Available at https://doi.org/10.1371. 10.1371/journal.pone.0100502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vedder AR, Miedel EL, Ragland NH, Balasis ME, Letson CT, Engelman RW, Padron E. 2019. Effects of Corynebacterium bovis on engraftment of patient-derived chronic myelomonocytic leukemia cells in NSGS mice. Comp Med 69:276–282. 10.30802/AALAS-CM-18-000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:1–14. https://doi:10.1371/journal.pone.0112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Q, Euler CW, Delaune A, Fischetti VA. 2015. Using a novel lysin to help control Clostridium difficile infections. Antimicrob Agents Chemother 59:7447–7457. 10.1128/AAC.01357-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whary MT, Baumgarth N, Fox JG, Barthold SW. 2015. Biology and diseases of mice, p 112–113. In: Fox JG, Anderson LC, Otto G, Pritchett-Corning KR, Whary MT, editors. Laboratory Animal Medicine, 3rd ed. San Diego (CA): Academic Press. [Google Scholar]
- 64.Zhou Y, Liang Y, Lynch K, Dennis JJ, Wishart DS. 2011. PHAST: A fast phage search tool. Nucleic Acids Res 39 suppl:W347–W352. 10.1093/nar/gkr485. [DOI] [PMC free article] [PubMed] [Google Scholar]