Abstract
The unexpected seroconversion of sentinel mice in our facility to murine T lymphotrophic virus (MTLV) positivity led to our identification of a novel murine astrovirus that we designated murine astrovirus 2 (MuAstV-2). During our investigation, MuAstV-2 was found to be a contaminant of the T helper cell line (D10.G4.1) that was used to generate the MTLV antigen that we included in the multiplex fluorometric immunoassay (MFIA) that we used for sentinel screening. We eventually determined that cross-reactivity with the astrovirus generated a positive result in the MTLV assay. A confirmatory immunofluorometric assay (IFA) using the same MTLV-infected cell line yielded a similar result. However, the use of antigen prepared from MTLV-infected neonatal mouse thymus did not reproduce a positive result, leading us to suspect that the seroreactivity we had observed was not due to infection with MTLV. A mouse antibody production test showed that mice inoculated with naïve D10.G4.1 cells and their contact sentinels tested positive for MTLV using cell-line generated antigen, but tested negative in assays using MTLV antigen produced in mice. Metagenomic analysis was subsequently used to identify MuAstV-2 in feces from 2 sentinel mice that had recently seroconverted to MTLV. Two closely related astrovirus sequences (99.6% capsid identity) were obtained and shared 95% capsid amino acid identity with the MuAstV-2 virus sequenced from the D10.G4.1 cell line. These viruses are highly divergent from previously identified murine astroviruses, displaying <30% capsid identity, yet were closely related to murine astrovirus 2 (85% capsid identity), which had recently been isolated from feral mice in New York City. A MuAstV-2 specific PCR assay was developed and used to eradicate MuAstV-2 from the infected colony using a test and cull strategy. The newly identified MuAstV2 readily transmits to immunocompetent mouse strains by fecal-oral exposure, but fails to infect NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl (NCG) mice, which have significantly impaired adaptive and innate immune systems. Neither immunocompetent nor immunodeficient mice showed any astrovirus-associated pathology. MuAstV-2 may provide a valuable model for the study of specific aspects of astrovirus pathogenesis and virus-host interactions.
Abbreviations: IFA, immunofluorescent assay; Lab 1, 2: Laboratory 1, 2; MFIA, multiplexed fluorometric immunoassay; MTLV, murine T lymphotrophic virus; MuAstV, murine astrovirus; MuAstV-2, murine astrovirus 2; MuLV, murine leukemia virus; NCG, NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl; NSG, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ; V1, V2, V3: vivarium 1, 2, 3
Astroviruses are nonenveloped, single stranded, positive sense RNA viruses, named for the stellate shape of their capsid. The family Astroviridae currently includes the genera Avastrovirus and Mamastrovirus, with 3 and 19 designated species respectively, although many additional species have been recently proposed.10 The first astrovirus was identified in 1975 from human stool, and astrovirus infection has since been implicated as a common cause of diarrhea in children worldwide.3 More recently, novel astrovirus variants have been associated with fatal encephalitis in immunocompromised human patients, as well as in cows, pigs, mink and sheep.14,24,31,38 Identification of these variants, and the numerous novel astroviruses found in other species, was made possible using metagenomic analysis.30
Despite the significant impact of astrovirus infection worldwide, until recently scientists placed relatively little effort on the search for treatments or prevention, perhaps due to the infection’s self-limiting course. As new variants have been identified as the cause of a fatal disease, this group of viruses has become more widely investigated.3 However, our understanding of astrovirus biology and pathogenesis has been confounded by the lack of a suitable laboratory animal model. Historically, investigations of human astrovirus were conducted in vitro, with in vivo studies limited to using turkeys infected with turkey astrovirus 1 and 2.8,17,25,34 The discovery of a murine astrovirus allows analyses of basic astrovirus biology, pathogenesis, and immunology using genetically engineered mouse models.6,7,40
In 1985, astrovirus particles were detected by electron microscopy in fecal samples from mice with diarrhea.20 The next report of astrovirus in rodents described 2 distinct, but related astroviruses identified in samples collected in 2007 from feral Rattus norvegicus in Hong Kong.4 The first astrovirus from a wild house mouse (Mus musculus) was less than 50% identical to the previously described rat astrovirus.29 Soon thereafter, additional astroviruses from wild mice and more significantly, laboratory mice, were identified and characterized. In 2012, a clade of astroviruses that was distinct from the previously identified rat and wild house mouse astroviruses was described.12 Furthermore, murine astrovirus was found in both wild type and highly immunocompromised laboratory mice without apparent clinical signs of disease.12 Also in that year, naïve healthy immunocompromised mice from 2 vendors were confirmed to be infected with an astrovirus that was almost identical to the one described earlier.40 Two other groups have also confirmed murine astrovirus infection in mice from commercial vendors.12,27 Murine astrovirus was found to be enzootic in a significant number of laboratory mouse colonies, including those in research universities and industry worldwide.27,32 In 2017, a high prevalence of astroviruses in multiple rodent species was described in China.36 Most recently, a novel astrovirus was identified in wild mice (Mus musculus) in New York City (NYC) using metagenomics.37 The sequence of this virus is significantly divergent from previously described wild and laboratory-derived murine astroviruses.9,12,27,29,40 However, based on pairwise alignments of the capsid protein, it shares greatest identity (76%) with the R. andamanensis astroviruses described earlier.36
Here we describe the serendipitous discovery of an astrovirus that is distinct from the astroviruses previously identified in laboratory-maintained Mus musculus. Infection was initially identified by serologic methods after soiled bedding sentinel mice tested positive to murine T lymphotropic virus (MTLV) using a multiplexed fluorescent immunoassay (MFIA). A series of investigations, described in this report, revealed that the MTLV antigen preparation had been obtained from a murine cell line that contained viral antigen from an astrovirus related to the one that had infected our mouse colony and the one detected in wild M. musculus from NYC.37 However, this new virus did not infect a strain of highly immunocompromised mice (NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl). This distinguishing feature, if confirmed, could be exploited as an additional important model for the study of specific aspects of astrovirus pathogenesis and virus-host interactions during infection and replication.
Materials and Methods
History.
In January 2016, 2 sentinel Swiss Webster mice (Tac:SW) housed in separate cages, each having received soiled bedding from 140 cages on 2 different racks in a single mouse holding room, tested positive for murine T lymphotropic virus (MTLV) during routine testing by multiplexed fluorometric immunoassay (MFIA). MFIA is routinely performed inhouse with commercially purchased reagents (Lab A; Charles River Laboratories, Wilmington, MA). Screening for MTLV antibodies is performed annually because of the low probability of infection, while antibody screening to a panel of other murine infectious agents is performed bimonthly, semiannually, or annually, based on the global prevalence of each specific agent. The holding room was 1 of 4 in a vivarium (V1) housing only mice. At the time of antibody detection, the vivarium had an average daily census of 815 mouse cages (95% capacity), and mice were routinely transferred between all 4 rooms. MFIA and indirect fluorescent antibody (IFA) confirmatory testing were conducted at Lab A. MFIA results were confirmed and an additional confirmatory test performed by Lab A was, at that time, interpreted as positive. The 2 MTLV MFIA and IFA positive sentinels were euthanized, exsanguinated, and necropsied. Their tissues were processed for histologic examination and frozen for potential further analysis. Immediately thereafter, 3 cage mates housed in the cages containing the 2 index cases and all sentinels (16 mice in 6 cages) in the 3 other holding rooms were tested for MTLV. Eleven were confirmed positive by MFIA and IFA.
After confirming that sentinels in all 4 holding rooms were MTLV positive, the movement of animals from V1 was stopped and the animals were quarantined in situ. Because animals and staff had historically moved between this vivarium and 4 other rooms in 2 additional vivaria (V2 and V3), all sentinels (31 mice) in these 4 rooms were tested for MTLV by MFIA and IFA. Two sentinel mice housed in 2 of 3 sentinel cages in a single holding room in a different vivarium (V2) were also found to be seropositive by MFIA and IFA. These sentinels were associated with 3 cage racks housing mice belonging to an investigator whose primary colony was maintained in V1. These 3 racks were moved to a separate room and quarantined, with no new mouse entries.
Sera from 2 MTLV positive sentinel mice were submitted to a second diagnostic laboratory (Lab B; IDEXX BioAnalytics, Columbia, MO) for MTLV testing by MFIA, IFA and Western blot. Both samples were negative by all methods. We subsequently discovered, after discussion with Lab A, that they had altered the methodology by which the MTLV antigen in the MFIA was produced. In the traditional method, the antigens for MFIA were prepared from suckling mouse thymus after MTLV infection. In early 2014, this method was replaced with an in vitro method in which the viral antigen was prepared by infecting an MTLV-permissive murine T helper cell line originating from an AKR/J mouse’s lymph node (D10.G4.1 [ATCC TIB224]; American Type Culture Collection, Manassas, VA). Approximately 1.5 yr later, this cell line was also used for the confirmatory IFA after the existing inventory of slides made with sucking mouse thymocytes was depleted. Lab B continued to perform the MTLV tests using antigen/slides prepared traditionally from suckling mice.
After being notified of these disparate results, Lab A performed a modified IFA panel that included comparing fluorescence in naive D10.G4.1 cells and the traditionally generated MTLV antigen using MTLV-infected suckling mouse thymocytes. The results confirmed that naïve D10.G4.1 cells were reacting to the MTLV positive sera from our sentinels. These findings, along with the apparent transmissibility of the agent through soiled bedding and epidemiology pointing to involvement of a single research group, suggested that an unusual infectious agent, most likely a virus, had been inadvertently introduced into the colony, and the introduced virus was reacting to antigens from the contaminating virus in the naïve D10.G4.1 cells.
We then undertook a series of investigations first to exclude MTLV and subsequently to understand and identify the causative contaminating agent. In parallel, we implemented procedures to contain the agent and reestablish agent-free colonies using a newly developed diagnostic qRT-PCR for the novel virus.
Animal housing.
All experimental and colony mice were housed in solid-bottom polysulfone microisolator cages maintained in an individually ventilated caging system (Maxi-Miser, Thoren Caging Systems, Hazelton, PA) on autoclaved aspen chip bedding (PWI Industries Canada, Quebec, Canada). Flash-autoclaved, γ-irradiated feed (LabDiet 5053, PMI, St Louis, MO) and acidified water (pH 2.5 to 2.8) were provided ad libitum in a polysulfone bottle with a neoprene stopper (Thoren Caging Systems).35 Two pieces of steam-sterilized, compressed, cotton nesting material (0.5 in2; Cotton squares, Ancare, Bellmore, NY) were supplied in each cage. Cages were changed weekly in a horizontal-flow, HEPA-filtered, mass air-displacement unit (NU301, Nuaire, Plymouth, MN). V1 and V2 operate with a single corridor traffic flow that provides direct access to the holding rooms, clean storage room, and clean and dirty cage washrooms. All rooms receive filtered (95% ASHRAE efficient), 100% outside air at 10 to 15 air changes per hour. All animal holding rooms and the dirty cage washroom are maintained at negative pressure relative to the corridor. The clean cage washroom and storage room are maintained at positive pressure relative to the corridor. Light:dark photoperiod cycle was maintained at 12:12 h intervals. Room temperature was maintained at 72 ± 2°F and relative humidity at 30% to 70%. Staff entering each room were required to don a disposable gown, hair bonnet and gloves. PPE was removed upon exiting each room. Investigators placed their mouse cages into bags to transport them to their laboratories and procedure rooms for experimental manipulation and returned to the housing rooms within 12 h. All mice in the V1 are used for neuroscience protocols, most involving neurosurgical procedures and/or behavioral testing. Investigators share several behavior and procedure rooms.
The animal care and use program at Weill Cornell Medicine (WCM) is accredited by AAALAC, and all animals are maintained in accordance to the recommendations provided in the Guide for the Use and Care of Laboratory Animals 8th Edition. All animal use described in this investigation was approved by WCM’s Institutional Animal Care and Use Committee.
Colony health monitoring.
The soiled bedding sentinel program has been previously described in detail.23 Briefly, 4 to 6 wk old female Tac:SW mice (Taconic Biosciences, Germantown, NY) are obtained for use as soiled bedding sentinels. On arrival, animals are free of antibodies to mouse hepatitis virus, lymphocytic choriomeningitis virus, Ectromelia virus, mouse parvovirus, minute virus of mice, murine norovirus, pneumonia virus of mice, Reovirus, Sendai virus, mouse rotavirus, Theiler meningoencephalitis virus, mouse adenovirus, K-Virus, murine polyoma virus, mouse cytomegalovirus, mouse T lymphotropic virus, Hantavirus, lactate dehydrogenase-elevating virus, Filobacterium rodentium, and Mycoplasma pulmonis, Helicobacter spp., Salmonella spp., Clostridium piliforme, Corynebacterium kutscheri, Citrobacter rodentium, and endoparasites and ectoparasites. Each sentinel cage serves a maximum of 4, single-sided, 70 cage racks and receives approximately 15 mL of dirty bedding from 40 different colony cages (1 column per rack) weekly. Soiled bedding is collected and provided to each soiled bedding sentinel cage from 280 colony cages over a 7 wk period. One sentinel mouse from each cage is identified every 8 wks and its’ blood collected for serologic testing and fecal samples and pelt swabs collected for PCR testing. At months 6 and 12 postplacement, one sentinel mouse from each cage is euthanized for blood collection, pelt and large intestinal content examination for ecto- and endoparasites, and a gross necropsy is performed with histologic examination if gross lesions are found, or if necessary to confirm positive parasitology or serologic tests. Survival blood collection (approximately 20 uL) is performed by tail vein nick, using a sterile 25G needle, collecting blood into a microsampler (HemaTIP; CRL, Wilmington, MA), or by cardiac puncture after euthanasia by CO2 inhalation. Select tissues are collected (heart, lungs, thymus, mediastinal lymph nodes, kidneys, liver, spleen, stomach, salivary glands, mandibular lymph node, mesenteric lymph node, uterus, cervix, skin, urinary bladder, adrenals, ovaries, oviducts, thyroid, trachea, esophagus, hind limb [femur, tibia, bone marrow, stifle joint, skeletal muscle, and peripheral nerves], vertebral column with spinal cord, sternum, head, coronal sections [including brain, eye, ears, nasal and oral cavities, teeth]. Tissues are preserved in 10% formalin and, if deemed necessary, are processed into paraffin blocks, cut into 5 micron-thick sections, and stained with hematoxylin and eosin for microscopic evaluation.
A more intensive soiled bedding sentinel protocol was implemented in V1 shortly after the initial MTLV-seropositive sentinel mice were identified. Briefly, 2 sentinel cages, each cage containing 2 Tac:SW and one NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG; Jackson Laboratories, Bar Harbor ME) mice were placed on each rack. Each sentinel cage received dirty bedding weekly from 34 different cages. Both Tac:SW mice from each sentinel cage were tested monthly by serology for MTLV using the MTLV MFIA assay that detected the initial seroconversion. Fecal samples from both Tac:SW mice in each cage were also harvested weekly and stored at -80°F until used for metagenomic analysis. Once any of the aforementioned Tac:SW sentinel mice tested positive, the NSG cage mate was removed and cohoused with 2 new Tac:SW sentinels which were tested by MFIA every 4 wk to confirm the NSG were infected and able to transmit infection to the new sentinels.
Serology assays.
Mouse sentinel testing by MFIA is routinely performed by our Laboratory of Comparative Pathology (LCP) using reagents obtained from Lab A. The MFIA platform is used to evaluate sera from the soiled bedding sentinel mice every 2 mo for antibody to mouse hepatitis virus, Sendai virus, mouse parvovirus, minute virus of mice, pneumonia virus of mice, Theiler meningoencephalitis virus, mouse rotavirus, murine norovirus, Reovirus, and Mycoplasma pulmonis; every 6 mo for Ectromelia virus, lymphocytic choriomeningitis virus, K virus, mouse adenovirus, and murine polyomavirus; and annually for mouse cytomegalovirus, mouse T lymphotropic virus, Hantavirus, Clostridium piliforme, and Filobacterium rodentium. Confirmatory testing (MFIA and IFA) is performed by Lab A. The MFIA platform has been previously validated and described.39 Two distinct IFA assays were used. In one assay, D10.G4.1 cells were cultured in Teflon delineated glass slide wells and subsequently infected with a low dose of MTLV that leaves most cells uninfected. Uninfected cells serve as an internal “tissue” control. In the second assay, suckling mouse thymocytes were prepared by inoculating a suspension of MTLV-infected thymus cells from suckling mice onto monolayers of immortalized thymus and spleen cells (JLS-V5[RRID:CVCL_2534]); ATCC, Manassas, VA) in slide wells. For testing, cells were incubated with test serum and then washed, followed by incubation with antimouse IgG FITC conjugated antibody. Slides were examined using a fluorescence microscope at 200× magnification.
MTLV qRT-PCR.
Buccal swabs were collected using cotton tipped applicators (Pigeon Corporation, Tokyo Japan) from 11 sentinel mice that had tested positive to MTLV by MFIA. Buccal swabs from 2 mice that had been used as sentinels for less than 1 mo were also tested. Filter tops and prefilters from the 2 cages in 2 racks housing the index cases were swabbed as previously described.13 Samples were submitted to Lab A for MTLV testing by qRT-PCR.
Pathologic Examination.
Four MTLV seropositive sentinel mice were euthanized and a complete necropsy performed. Tissue samples (heart, lung, thymus, mediastinal lymph nodes, kidneys, liver, pancreas, spleen, gallbladder, stomach, duodenum, jejunum, ileum, cecum, colon, salivary glands, mammary glands, mandibular and mesenteric lymph nodes, uterus, urinary bladder, adrenal glands, ovaries, oviduct, thyroid, trachea, esophagus, femur, tibia, bone marrow, stifle joint, skeletal muscle, peripheral nerves, vertebral column, spinal cord, sternum, brain, eyes, ears, nasal and oral cavities, and teeth) were harvested and preserved in 10% buffered formalin for at least 72 h. Samples were processed, embedded in paraffin and cut into 5-micron sections before staining with hematoxylin and eosin for microscopic evaluation.
Electron Microscopy.
Naïve and MTLV infected D10.G4.1 cells (approximately 3 × 107 cells) were fixed in 1 mL 4% paraformaldehyde, 2.5% glutaraldehyde in 0.1 M sodium cacodylate, pH 7.3 and kept on ice for 1 h. The fixative was decanted and 1 mL 0.1 M sodium cacodylate, pH 7.3 buffer was added. The sample was refrigerated at 4°C until evaluated by transmission electron microscopy.
Mouse antibody production (MAP) test.
The D10.G4.1 cell line (TIB-224; ATCC, Manassas, VA) was maintained following ATCC’s recommendations. Briefly, cells were cultured using RPMI-1640 Medium (ATCC) with 10% T-STIM with Con A (Beckton Dickinson, Franklin Lakes, NJ), 10% fetal bovine serum, 0.05mM 2-mercaptoethanol and 10 pg/mL recombinant mouse IL-1 α (R and D Systems, Minneapolis, MN). Two frozen vials containing MTLV infected or naïve cells (approximately 8 × 106 cells/vial) supplemented with an additional 10% fetal bovine serum and 5% DMSO were shipped to our laboratory for MAP testing and electron microscopy. Cells were allowed to thaw in a water bath at 20°C. Cells were centrifuged at 10,000 g for 20 min, supernatant removed and the cells washed twice by resuspending the pellet in 1 mL phosphate buffered saline (PBS) at 20°C.
A Tac:SW mouse was inoculated intraperitoneally (IP) with approximately 5 × 106 naïve cells in 0.5 mL PBS. A second mouse was inoculated IP with approximately 5 × 106 MTLV infected D10.G4.1 cells in 0.5 mL PBS solution. The 2 inoculated mice were housed in separate cages, each cohoused with a naïve Tac:SW contact sentinel. Twenty microliters of blood were collected from each mouse at 10, 35 and 48 d after inoculation. Blood samples were tested by MFIA and IFA at Lab A.
Metagenomic analysis of fecal samples.
Fecal samples collected from soiled bedding sentinels (intensified program) were selected for analysis based on positive serologic results. The samples used were those collected from the 2 sentinels in a single cage at 17 d before they tested positive for MTLV by MFIA. Fecal pellets were diluted with 750 µl Dulbecco phosphate-buffered saline, pooled, homogenized with a mortar and pestle, clarified by 15,000 g centrifugation for 5 min, and supernatantfiltered using a 0.45 µm filter (Millipore). Free nucleic acids in the filtrates (not protected in viral capsids) were digested using DNAse and RNAse to enrich for viral nucleic acids. Nucleic acids were then extracted (MagMAX Viral RNA Isolation Kit, Ambion, Austin, Tx) and amplified by random qRT-PCR followed by use of the Nextera XT Sample Preparation Kit (Illumina) to generate a library for Illumina MiSeq (2 × 250 bases) with dual barcoding as previously described.22 After de novo assembly using the Ensembl program,11 the contigs and singlets were analyzed using translated protein sequence similarity search (BLASTx v.2.2.7) to all annotated viral proteins available in GenBank. Geneious R1019 was then used to align reads and contigs to reference viral genomes in GenBank.
Whole virus genome sequencing (WGS) of virus from feces and D10.G4.1 cells.
WGS of the virus isolated from feces that had been used for metagenomic analysis (2 samples) and of the contaminated cell line was undertaken to obtain a more complete sequence. Four fecal pellets collected from sentinel mice were resuspended in PBS (Thermo Fisher Scientific, Waltham, MA) and homogenized (TissueLyser Cat. 85300; QIAGEN, Germantown, MD). RNA from the fecal pellets and naïve D10.G4.1 cells (approximately 5 × 106 cells) were extracted (RNeasy Isolation Kit Cat. 74104; QIAGEN) according to the manufacturer’s instructions with minor modifications. Specifically, extracted RNA was eluted into 60 µl elution buffer. Extracted nucleic acids were further treated with an endonuclease on an elution column (RNase-Free DNase Cat.79254; QIAGEN) for 5 min at ambient temperature before elution. Reverse transcription was performed on purified RNA using reverse transcriptase. Double-stranded cDNA was purified (AMPure XP Beads, Beckman Coulter Life Sciences, Indianapolis, IN) following the manufacturer’s instructions. The concentration of cDNA was determined (Qubit 3.0 Fluorometer and Qubit dsDNA HS Assay Kit, Thermo Fisher Scientific, Waltham, MA) and normalized to 0.2 ng/µl for sequencing library preparation. The sequencing library was prepared (Nextera XT library preparation kit, Illumina) with dual-indexing and sequenced on a sequencing instrument (Illumina MiSeq, Illumina) according to the manufacturer’s “MiSeq Sequencing System Guide” for 2 × 300 base pair reads. Adapters were removed and reads were de novo assembled with SPAdes (v 3.9.0).28
Phylogenetic analysis.
Capsid sequences from Mamastrovirus (n = 19) and Avastrovirus (n = 3) species were obtained from the National Center for Biotechnology Information reference sequence database. In total, 61 capsid protein sequences were aligned to the 4 MuAstV-2 sequences using ClustalW in Geneious 10.2.3.5 and exported to MEGA6 for model selection.19,33 The Le and Gascuel substitution model was employed to prepare a maximum likelihood tree with 500 bootstrap repetitions.21 A newick tree was exported to Figtree (http://tree.bio.ed.ac.uk/software/figtree/) for annotation.
MuAstV-2 qRT-PCR assay.
Contigs obtained from metagenomic analysis of fecal samples and cell line sequencing were subjected to similarity search against the nonredundant protein or nucleotide databases of GenBank. Based on comparison with related viruses, a conserved region was targeted for design of a real-time PCR TaqMan assay (proprietary) with 2 separate sets of primers based on methods previously described.15 Each primer set was used to test the naïve fecal pellets from both MFIA and IFA positive and negative sentinels, naïve D10.G4.1 cell derivatives (DNA and RNA pellets and supernatant), fresh cell culture media, and mouse thymocyte RNA.
MuAstV-2 Outbreak Eradication.
Investigators in V1 reduced the number of mouse cages to only those that were essential to maintain a specific strain, and/or complete ongoing investigations. The remaining cages were relocated into 2 rooms and quarantined in V2. V1 was emptied, all cage racks and caging equipment were sanitized in a rack washer; disposables, consumables, rack blower hoses, HEPA filters and prefilters were discarded and replaced. All surface areas in each holding room, including ventilated rack blower units and animal change stations/biologic safety cabinets were disinfected with chlorine dioxide solution (Clidox [1:4:1 dilution]; Pharmacal Research Labs, Waterbury, CT) providing at least 3 min contact. Surfaces were wiped dry, rinsed with water and allowed to air dry. All laboratories that used mice associated with V1 were also decontaminated using the same procedures. One week after depopulation and decontamination, MuAstV-2 free mice were purchased from approved vendors to repopulate the facility.
In parallel, the MuAstV-2 shedding status of mice in quarantine in V2 was evaluated. A fecal pellet was collected from each cage. Pellets were pooled (10 pellets/sample) and tested using the newly developed MuAstV-2 qRT-PCR assay. Individual cages (10 fecal pellets/cage) associated with the sentinel cage were tested after the detection of a positive pooled cage sample. If a cage was PCR positive, all cage occupants were euthanized. Immediately after the initial testing, these testing procedures were repeated. All remaining cages were then relocated to V1 and the intensive soiled bedding sentinel program previously described was implemented. If a sentinel tested positive, all cages on the associated cage rack were tested by PCR (10 cages/sample) and cages from each positive sample were then tested individually.
Infectivity experiment.
A virus stock was prepared from intestinal contents harvested from qRT-PCR positive mice. Intestinal contents (approximately 2.5 mL) were homogenized in sterile PBS (approximately 11.5 mL), clarified via centrifugation (5,000 RPM for 20 min), and passed through a sterile 0.22-µm filter (Nalgene 726 to 2520; Thermo Fisher Scientific, Waltham, MA). The stock was confirmed as MuAstV-2-positive using qRT-PCR (using the newly developed assay) and negative for murine astrovirus 1 using an established diagnostic assay performed by Lab A. Four NOD-Prkdcem26Cd52Il2rgem26Cd22/NjuCrl ([NCG], Charles River Laboratories) and 4 C57BL/6NCrl female mice and 3 female Crl:CD(SD) rats, all obtained from Charles River Laboratories, were each gavaged with 200 µL (mice) or 700 µL (rats) of the filtered virus stock solution. Animals were cohoused in separate cages by strain. Two additional C57BL/6NCrl were gavaged with 200 µL of PBS (controls) and housed together in a separate cage. At 3, 7, and 21 d after inoculation, all mice were separated and housed in clean individual cages until a fecal sample was obtained from each mouse; mice were then rehoused in their original cages. Fecal pellets were harvested in a similar fashion at 2, 3, and 6 d after inoculation from rats. All animals were euthanized for tissue harvest at the last time point. All fecal samples were maintained at -80°C until testing. Animals were observed daily for clinical signs of disease such as lethargy, hunched posture, unkempt hair coat, or diarrhea. Fecal samples from individual animals were assayed separately for MuAstV-2 by qRT-PCR.
Results
Outbreak with a Novel Infectious Agent.
After identification of the 2 index cases, an additional 13 of 21 soiled bedding sentinels in 7 of 8 sentinel cages in the 4 V1 animal holding rooms tested positive by MFIA and IFA using the D10.G4.1 cell-generated MTLV antigen and IFA slides, respectively. Two of 7 soiled bedding sentinel mice in 2 out of 3 cages in a single holding room in V2, having received mice from V1 on many occasions, were also seropositive. Soiled bedding sentinel mice in 7 cages, in 3 other holding rooms from V3 and receiving mice from V1, were all seronegative.
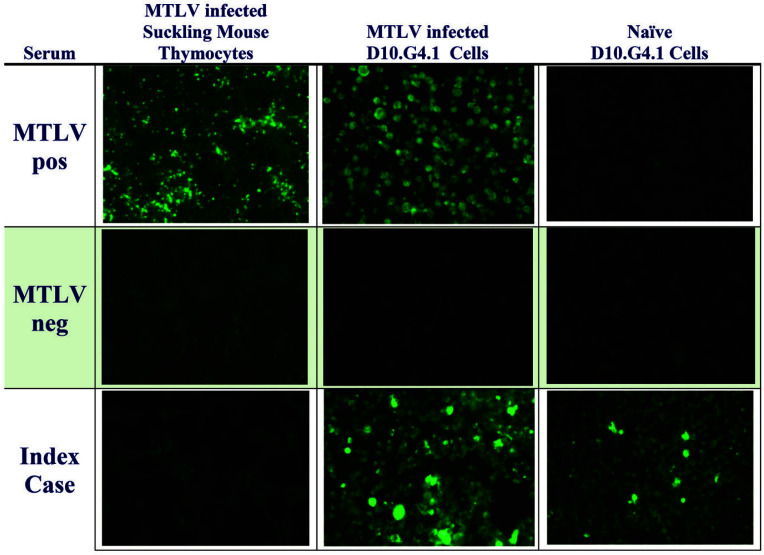
The MTLV-infected D10.G4.1 cell IFA slides reactive to the sera from the index cases were reevaluated due to the rarity of a true MTLV outbreak in laboratory mice. A modified IFA panel was performed by commercial Lab A, which demonstrated fluorescence on both MTLV infected and naïve D10.G4.1 cells, but not on MTLV infected suckling thymocytes. In addition, serum obtained from a negative control mouse showed no staining in either the IFA using MTLV infected sucking thymocytes or naïve (non MTLV infected) D10.G4.1 cells. Figure 1 shows a representative modified IFA panel. These findings strongly suggested that the sera from the index cases were cross-reacting with an unknown antigen in the naïve D10.G4.1 cells. Buccal swabs collected from 13 soiled bedding sentinels, filter tops from both index sentinel cages and prefilters from their respective racks all tested negative by qRT-PCR for MTLV, providing additional evidence that MTLV was not the offending infectious agent.
Figure 1.
Modified immunofluorescent assay (IFAs) panel conducted using MTLV-infected thymocytes, MTLV-infected D10.G4.1 cells, and naïve D10.G4.1 cells. Serum tested included sera from mice infected with MTLV (MTLV pos), sera from mice free of MTLV (MTLV neg), and sera from the index cases. Naive and MTLV infected D10.G.4 cells show strong fluorescence when exposed to serum from the index case. Naïve cells do not show fluorescence when exposed to serum from a mouse exposed to MTLV (positive control).
Pathology.
Gross or microscopic lesions attributable to MTLV or another infectious agent were not detected in the 4 MTLV seropositive sentinels examined by MFIA. Pathologic changes were limited to background lesions typically observed in mice of this age and stock.
Electron Microscopy.
Viral capsids with and without dense cores and prominent accumulations of filamentous structures, consistent with prior descriptions of MTLV ultrastructure, were commonly observed in the nucleus and less frequently in the cytoplasm (capsids only) of the MTLV experimentally infected D10.G4.1 cells used for antigen harvest.1 There was no evidence of viral particles or structural elements in the naïve D10.G4.1 cells.
MAP Test.
The mouse inoculated with MTLV infected cells was positive for MTLV by MFIA using D10.G4.1 cell-generated MTLV antigen at all 3 time points, but positive by IFA only at 48 d after inoculation. All other mice, including contact sentinels tested “positive” for MTLV by MFIA at days 35 and 48, but by IFA only at 48 d after inoculation.
Metagenomic Analysis of Fecal Samples from Seropositive Sentinels.
Two sequence reads, 238 and 212 base pairs long, were obtained from feces from the 2 soiled bedding sentinels that had subsequently seroconverted by MFIA at 17 d after collection of the feces. These reads were 89.9% and 91.3% identical at the nucleotide level with an astrovirus previously identified from feral mice in New York City (GenBank accession MF175073.1). Astrovirus from wild Rattus norvegicus (GenBank accession HM450380–HM450386) in Hong Kong were the next closest match at 80.8% and 73.8% nucleotide identity, respectively.4 Raw data is available in GenBank under accession PRJNA547824.
Whole Virus Genome Sequences.
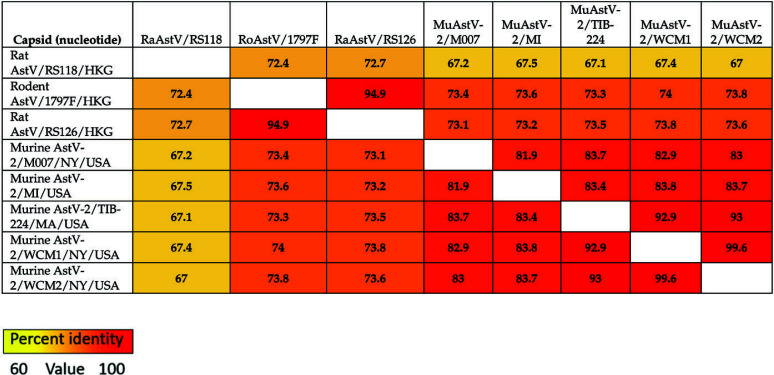
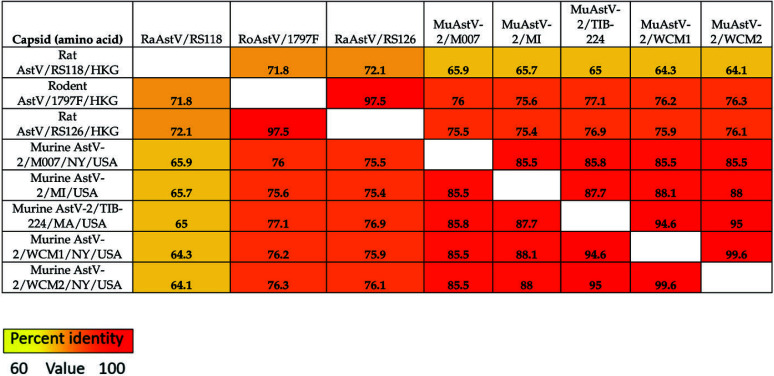
Astrovirus sequences were obtained from both fecal pellets and cell line samples. Full sequences are available in GenBank as accessions MN503236, MN503237, and MN503235, respectively. The virus identified as MuAstV-2, sequenced from feral mice in New York City, shares 89.9% and 89.2% and 89.3% nucleotide percent identity to the virus we sequenced from the cell line (MuAstV-2/TIB224) and the 2 fecal pellets (MuAstV-2/WCM1 and 2), respectively. MuAstV-2/TIB224 and MuAstV-2/WCM1 and 2 share 92.5% and 93.6% nucleotide identity, respectively, and share less than 50% identity with MuAstV commonly found in laboratory mice.27,40 Figures 2 and 3 summarize nucleotide and amino acid percent identities, respectively, of the ORF2 (capsid) sequence region of various astroviruses. The tables also provide a heat map in which increasing similarity between samples is shown by a color change, from yellow to orange to red. Figure 4 shows the phylogenetic relationship between each of the described MuAstV-2 capsid sequences and previously reported astroviruses. MuAstV-2 sequences form a unique clade (100% bootstrap support) separate from several astrovirus sequences identified in Rattus sp. from Hong Kong and China.
Figure 2.
Pairwise nucleotide identity matrix for the capsid protein of murine astrovirus 2 and related viruses
Figure 3.
Pairwise amino acid identity matrix for the capsid protein of murine astrovirus 2 and related viruses
Figure 4.
Phylogenetic analysis of murine astrovirus 2 sequences. The maximum likelihood tree was constructed using the capsid protein sequence. The scale bar represents the number of substitutions per site and bootstrap values are displayed when greater than 70%. Murine astrovirus 2 sequences are highlighted with blue shading.
MuAstV-2 qRT-PCR.
Naïve D10.G4.1 cells and fecal pellets from MTLV seropositive sentinels were tested using 2 primer sets. Only one set of primers detected viruses from both the mice and the D10.G4.1 cells and was therefore used in the infectivity experiments and outbreak eradication program. Results are summarized in Table 1.
Table 1.
MuAstV-2 qRT-PCR Primer Set Test Results
| Sample and template types | Copy no. primer set 1 | Copy no. primer set 2 |
|---|---|---|
| MTLV Seropositive Fecal Sample 1 RNA | 15,849 | 17,845 |
| MTLV Seropositive Fecal Sample 2 RNA | 2 | 2 |
| MTLV Seropositive Fecal Sample 3 RNA | 946 | 1,233 |
| MTLV Seropositive Fecal Sample 4 RNA | 2,712 | 3,845 |
| MTLV Seropositive Fecal Sample 5 RNA | 32,975 | 49,424 |
| D10.G4.1 cell pellet DNA | 0 | 0 |
| D10.G4.1 cell supernatant DNA | 0 | 0 |
| D10.G4.1 cell supernatant RNA | 0 | 107,227 |
| D10.G4.1 cell line pellet RNA | 0 | 1,995,262 |
| MTLV Seronegative Fecal Sample 1 RNA | 0 | 0 |
| Fresh cell culture media for D10.G4.1 cells | 0 | 0 |
| Naive Mouse Thymocyte RNA | 0 | 0 |
MuAstV-2 Outbreak Eradication.
Multiple pooled sentinel cage samples tested positive by PCR during the first round of testing in both quarantine rooms. Testing of individual cages associated with each positive pooled cage sentinel revealed at least one cage contributing to the pool of housed PCR positive mice; these animals were culled. MuAstV-2 positive samples were not detected by PCR during the second round of testing. Once all remaining cages were transferred back to V1, monthly testing of feces by PCR was conducted for 6 mo using the intensive soiled bedding sentinel protocol. The standard sentinel testing protocol was then restored, except that the sera samples were also tested every 2 mo using D10.G4.1 cell-generated MTLV serology assay. All serology results have been negative for more than 18 mo.
Infectivity Experiment.
All 4 B6 mice inoculated with MuAstV-2/WCM were PCR positive at the 3 time points tested. Both of the PBS-inoculated B6 mice remained negative throughout the experiment, confirming that cross-contamination did not occur. None of the mice had clinical signs of disease. All 4 NCG mice inoculated with MuAstV-2/WCM were negative at all 3 time points tested. Virus copy numbers for all mice are provided in Table 2. Individual fecal pellets, taken from a rat at each of the 3 time points, were all negative for MuAstV-2.
Table 2.
MuAstV-2 copy number at day 3, 7, and 21 post inoculation with MuAstV-2/WCM1
| Strain | Animal | Copy no. | ||
|---|---|---|---|---|
| Day 3 | Day 7 | Day 21 | ||
| C57BL/6NCrl | 1a | 0 | 0 | 0 |
| 2a | 0 | 0 | 0 | |
| 3 | 107227 | 432876 | 6579 | |
| 4 | 53367 | 432876 | 6579 | |
| 5 | 107227 | 215443 | 1630 | |
| 6 | 107227 | 869749 | 3275 | |
| NCG | 7 | 0 | 0 | 0 |
| 8 | 0 | 0 | 0 | |
| 9 | 0 | 0 | 0 | |
| 10 | 0 | 0 | 0 | |
Inoculated with sterile PBS
Discussion
We have shown the D10.G4.1 cell line (ATCC TIB224) was contaminated with a previously unknown astrovirus, MuAstV-2, and we present evidence that laboratory mice were infected with a similar virus strain. These discoveries would not have been possible had we not been testing mouse sentinels with an assay that, unbeknownst to us, included a serologic antigen preparation obtained from the contaminated cell line.
Initially, sentinels in V1 and a room in V2 tested positive for MTLV by MFIA using an inhouse, as well as a confirmatory assay conducted at a commercial diagnostic laboratory (Lab A) that also provided the reagents used in the inhouse MFIA. However, MTLV specific PCR results were negative, and the same sera samples were confirmed seronegative at a second diagnostic laboratory (Lab B). This suggested the initial results were false positives. The serology results were replicated by inoculating naïve mice with the uninfected (MTLV) cell line used by Lab A to obtain antigen for their MTLV serology assays. Metagenomic analysis confirmed that both our sentinels and the cell line were infected with similar strains of a novel murine astrovirus, explaining the cross reactivity with the MTLV serology assay.
It is unclear as to how and when the cell line became infected with MuAstV-2. The first report describing the establishment of the cell line describes the harvest of lymph nodes from AKR/J mice immunized with ovalbumin.18 These source animals could have been infected with the virus. However, cells from the harvested lymph nodes were cultured using several biologic products that also could have been contaminated. Notably, the culture medium included a T-cell culture supplement (T-STIM with Con A). This supplement, made from rat splenocytes exposed to the mitogen Concanavalin A, may also have been the source of contamination, as the virus isolated from the cell line showed up to 74% nucleotide identity to several astroviruses isolated from various rat species. As the cells were cocultured with splenocytes harvested from syngeneic mice, any of the various splenocyte donors used to maintain the cell line prior to providing it to the ATCC could also have served as the source of the virus. The current culture recommendations for the D10.G4.1 cell line use mouse interleukin 1 (IL1) rather than splenocytes. When this change occurred is not documented, but mouse IL1 harvested from activated murine cells, rather than recombinant IL1, may have been used. While we cannot speculate when the cell line became contaminated, our evidence shows that the current ATCC D10.G4.1 cell stock is infected with MuAstV-2. Furthermore, while we have not tested other commercially available cell lines for MuAstV-2 contamination, we have notified ATCC and suggest caution when interpreting experimental data obtained using D10.G4.1 cells.
We cannot connect the use of this cell line to the apparent MTLV outbreak identified in our vivarium. Furthermore, we could not find any publications describing the in vivo use of D10.G4.1 cells. We surveyed the scientists using animals in V1 and identified only a single biologic, a recombinant adeno-associated virus (AAV) vector, which had been administered to mice within the vivarium within the past several years. At the time of the survey, we had not identified MuAstV-2 and thus did not have a diagnostic PCR; however, the last available aliquot of the biologic had already been used experimentally. Sera from these mice were tested and found to be seronegative using the D10.G4.1 cell-generated MFIA. These results, and the fact that most AAV vectors are produced using human or insect cell platforms, make it highly unlikely the AAV was the source of the outbreak.26
AKR mice are carriers of an endogenous murine leukemia virus (eMuLV), AKV, which is associated with emergence of a unique MuLV called mink cell focus-forming (MCF) virus in the thymus at about 5 mo of age. MCF are formed by recombination of AKV with the replication-defective polytropic MuLV found in the mouse genome. The formation of MCF is promoted by the high level of AKV expression and leads to a 70% to 95% incidence of leukemia at approximately 6 mo of age.5 As the D10.G4.1 cells were of AKR origin and expressed high levels of ecotropic MuLV (data not shown), we speculated that the mice in our vivarium were potentially expressing and developing antibody to an endogenous MuLV or had become infected with a horizontally transmissible MuLV. We developed an AKV-specific IFA, but the results obtained were inconsistent.
Simultaneously, we confirmed that the MFIA and IFA results could be replicated in mice inoculated with MTLV-free D10.G4.1 cells, using their naïve cage mates as contact sentinels. Furthermore, sera obtained 48 d after inoculation with naïve or MTLV-infected cells were confirmed negative and positive for MTLV, respectively, by Lab B, using the in vivo-generated MTLV antigen. These findings, along with the modified IFA panel results, indicated that antibodies in mice from our vivarium were reacting with an unknown antigen(s) in the naïve D10.G4.1 cells. Although the antibodies may have been reacting to mouse antigens inherent to the D10.G4.1 cells, this was considered unlikely because the naïve SW contact sentinels also tested positive, suggesting transmission of an unknown agent. At the same time, SW sentinels from other vivaria did not have cross-reacting antibodies, nor did the thousands of samples tested by Lab A using this assay. We hypothesized, based on these results, the epidemiologic data, and the apparent fecal-oral mode of transmission, that an unidentified virus was infecting our mouse colony, and the cell line was infected with the same or an antigenically similar virus.
Lab A had screened the cell line by PCR for all known murine viruses as part of a quality assurance program prior to use for MTLV antigen production. The cell line was only positive for mouse parvovirus 1 (MPV-1); however, the virus appeared to be incomplete or nonreplicating because cells were MPV-negative by MAP test and by IFA with MPV-1 and other parvovirus antisera. Subsequent whole genome sequencing revealed integration of a partial MPV-1 sequence.
As we suspected a virus, and the D10.G4.1 cells were negative for replication competent mouse pathogens, we turned to metagenomics to identify the putative pathogen. Two contigs that most closely matched a novel astrovirus, identified from feral rats (Rattus norvegicus) in China, were assembled from fecal pellets collected from animals that subsequently seroconverted in the MTLV MFIA.36 Subsequently, an additional report identified a more closely related astrovirus from feral mice (M. musculus) in New York City.37
Whole virus genome sequencing of the cell line and the fecal pellets confirmed the presence of highly similar astroviruses (MuAstV-2/TIB-224 and MuAstV-2/WCM1 and 2). These viruses are most closely related to MuAstV-2, and the rodent astroviruses described previously36,37 These viruses are highly divergent from the first astrovirus identified from M. musculus and from the more common astroviruses of laboratory mice.9,12,27,29,40 This finding confirmed our speculation that MuAstV-2, or a closely related virus, had infected our laboratory mouse colony, as well as the cell line from which the antigen used for the MTLV screening assays was derived. This explained why the MTLV assay results were positive, despite the mice not having been exposed to MTLV.
With the virus sequences available, we developed a reliable PCR assay that detects MuAstV-2 and other closely related rodent astroviruses. This assay was used as the primary diagnostic tool to implement a test-and-cull eradication plan to repopulate V1 with mice free from MuAstV-2. Since completing that plan over 18 mo ago, we have continued to use the MTLV MFIA to test all sentinels every 2 mo as a surrogate serologic test for MuAstV-2. Results have consistently remained negative in all vivaria. We now also use PCR for MuAstV-2 to test feces from all mouse strains entering our facility from noncommercial sources. The prevalence of this virus appears to be very low. Over an 18-mo period we have tested over 350 groups of mice imported from other institutions; so far 2 groups of mice, also from academic institutions in NYC, have tested positive by PCR; these results have been confirmed with serologic assays. In addition, Lab A, which had been using the D10.G4.1 cells to generate MTLV antigen for approximately 18 mo prior to our first positive results, determined retrospectively that out of the thousands of samples tested using this assay, only one additional set of related samples, also collected from mice at a third academic institution in the NYC metropolitan area, had similar positive results. These findings indicate that this virus may be present in at least some other institutional colonies. The outbreak at our institution may have resulted from the importation of mice from another institution and passed through quarantine without the use of an appropriate diagnostic test. However, we consider this possibility less likely, because our quarantine records show imported mice that entered V1 in the 18 mo prior to the identification of the outbreak did not have questionable MTLV results consistent with MuAstV-2 contamination. However, we cannot be completely certain that the antigen used in all assays during this period was derived from the cell line.
In our opinion, the more likely root cause of the outbreak was the incursion of feral mice into laboratory areas where investigators work with research mice on the bench without the protection of cage filters, biologic safety cabinets, and/or animal change stations. While our pest control program had not identified feral mice in the vivarium or the adjacent laboratory spaces, feral mouse incursions could nonetheless go undetected. In fact, feral mice have been trapped in the building’s loading dock, and we confirmed that fecal pellets from a feral mouse trapped in this location tested MuAstV-2 positive by PCR.
Highly immunocompromised NSG mice were cohoused with immune competent SW mice during periods in which the latter were infected with and subsequently seroconverted to MuAstV-2. We expected that the NSG mice would become persistently infected, shed high levels of the virus for a protracted time, and potentially develop clinical disease, as occurs when immunocompromised mice are exposed to many murine viruses. However, this did not occur. To confirm our assumption that highly immunocompromised mice would not support MuAstV-2 infection, we inoculated both B6 and NCG mice with the virus. No clinical signs of disease were observed in either strain. Fecal PCR results were negative in NCG mice, whereas B6 mice has positive fecal PCR results. This finding suggests that infection did not occur in the NCG strain and supports the suspicions based on the NSG sentinels. Although additional confirmatory experiments are required, these results may highlight an unusual characteristic of this astrovirus in that perhaps at least one of the cell types or cytokine pathways absent or altered in NSG and NCG mice may be necessary to support virus entry and/or replication. A recent report found that enteric bacterial or viral species can prevent or shorten virus infection in highly immunocompromised mice. Therefore, our findings may indicate the presence of an unidentified infectious agent modulating the innate immune response in NCG mice and preventing MuAstV-2 infection.2,16 Our findings also suggest that MuAstV may serve as a valuable model to study virus (astrovirus)-host interactions and pathogenesis.
Because MuAstV-2 forms part of a phylogenetic clade shared with astroviruses identified from rats, we inoculated outbred rats with over a 3-fold greater infectious dose than was used to infect mice. Although we tested only one rat per time point, the results suggest that MuAstV-2 does not infect rats. Because astroviruses are considered to be species specific, this finding is not surprising, although some have suggested that cross-species infections may be responsible for the high number of astroviruses identified.10
In conclusion, the pursuit of false positive serologic results to MTLV led to the identification of a novel murine astrovirus, MuAstV-2, infecting both laboratory mice and the cell line in which the assay antigen was produced. MuAstV-2 appears to have the unusual feature of not replicating in highly immunocompromised mice. This finding is yet another example of how unexpected laboratory results can lead to discovery.
Acknowledgments
The authors would like to thank Vitalant Research Institute for their support with viral discovery, and Steve Jennings, Lisa Eldred, Antonio Bravo and the staff of the Laboratory of Comparative Pathology for their work with support, and Veterinary Services for their invaluable technical assistance with this project.
References
- 1.Athanassious R, Alain R, Lussier G. 1990. Electron microscopy of mouse thymic virus. Arch Virol 113:143–150. 10.1007/BF01316668. [DOI] [PubMed] [Google Scholar]
- 2.Baldridge MT, Nice TJ, McCune BT, Yokoyama CC, Kambal A, Wheadon M, Diamond MS, Ivanova Y, Artyomov M, Virgin HW. 2014. Commensal microbes and interferon- λ determine persistence of enteric murine norovirus infection. Science 347:266–269. 10.1126/science.1258025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bosch A, Pintó RM, Guix S. 2014. Human astroviruses. Clin Microbiol Rev 27:1048–1074. 10.1128/CMR.00013-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chu DK, Chin AW, Smith GJ, Chan KH, Guan Y, Peiris JS, Poon LL. 2010. Detection of novel astroviruses in urban brown rats and previously known astroviruses in humans. J Gen Virol 91:2457–2462. 10.1099/vir.0.022764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cloyd MW, Hartley JW, Rowe WP. 1980. Lymphomagenicity of recombinant mink cell focus-inducing murine leukemia viruses. J Exp Med 151:542–552. 10.1084/jem.151.3.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Compton SR, Booth CJ, Macy JD. 2017. Lack of effect of murine astrovirus infection on dextran sulfate-induced colitis in NLRP3-deficient mice. Comp Med 67:400–406. [PMC free article] [PubMed] [Google Scholar]
- 7.Compton SR, Booth CJ, Macy JD. 2017. Murine astrovirus infection and transmission in neonatal CD1 mice. J Am Assoc Lab Anim Sci 56:402–411. [PMC free article] [PubMed] [Google Scholar]
- 8.Cortez V, Meliopoulos VA, Karlsson EA, Hargest V, Johnson C, Schultz-Cherry S. 2017. Astrovirus biology and pathogenesis. Annu Rev Virol 4:327–348. 10.1146/annurev-virology-101416-041742. [DOI] [PubMed] [Google Scholar]
- 9.Cortez V, Sharp B, Yao J, Livingston B, Vogel P, Schultz-Cherry S. 2019. Characterizing a murine model for astrovirus using viral isolates from persistently infected immunocompromised mice. J Virol 93:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Benedictis P, Schultz-Cherry S, Burnham A, Cattoli G. 2011. Astrovirus infections in humans and animals—molecular biology, genetic diversity, and interspecies transmissions. Infect Genet Evol 11:1529–1544. 10.1016/j.meegid.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deng X, Naccache SN, Ng T, Federman S, Li L, Chiu CY, Delwart EL. 2015. An ensemble strategy that significantly improves de novo assembly of microbial genomes from metagenomic next-generation sequencing data. Nucleic Acids Res 43:1–11. 10.1093/nar/gkv002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Farkas T, Fey B, Keller G, Martella V, Egyed L. 2012. Molecular detection of novel astroviruses in wild and laboratory mice. Virus Genes 45:518–525. 10.1007/s11262-012-0803-0. [DOI] [PubMed] [Google Scholar]
- 13.Gerwin PM, Ricart Arbona RJ, Riedel ER, Henderson KS, Lipman NS. 2017. PCR testing of IVC filter tops as a method for detecting murine pinworms and fur mites. J Am Assoc Lab Anim Sci 56:752–761. [PMC free article] [PubMed] [Google Scholar]
- 14.Giannitti F, Caffarena RD, Pesavento P, Uzal FA, Maya L, Fraga M, Colina R, Castells M. 2019. The first case of bovine astrovirus-associated encephalitis in the southern hemisphere (Uruguay), uncovers evidence of viral introduction to the Americas from Europe. Front Microbiol 10:1–8. 10.3389/fmicb.2019.01240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henderson KS, Pritchett-Corning KR, Perkins CL, Banu LA, Jennings SM, Francis BC, Shek WR. 2015. A comparison of mouse parvovirus 1 infection in BALB/c and C57BL/6 mice: susceptibility, replication, shedding, and seroconversion. Comp Med 65:5–14. [PMC free article] [PubMed] [Google Scholar]
- 16.Ingle H, Lee S, Ai T, Orvedahl A, Rodgers R, Zhao G, Sullender M, Peterson ST, Locke M, Liu TC, Yokoyama CC, Sharp B, Schultz-Cherry S, Miner JJ, Baldridge MT. 2019. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat Microbiol 4:1120–1128. 10.1038/s41564-019-0416-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson C, Hargest V, Cortez V, Meliopoulos VA, Schultz-Cherry S. 2017. Astrovirus pathogenesis. Viruses 9:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaye J, Porcelli S, Tite J, Jones B, Janeway CA., Jr. 1983. Both a monoclonal antibody and antisera specific for determinants unique to individual cloned helper T cell lines can substitute for antigen and antigen-presenting cells in the activation of T cells. J Exp Med 158:836–856. 10.1084/jem.158.3.836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kjeldsberg E, Hem A. 1985. Detection of astroviruses in gut contents of nude and normal mice. Brief report. Arch Virol 84:135–140. 10.1007/BF01310560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 22.Li L, Deng X, Mee ET, Collot-Teixeira S, Anderson R, Schepelmann S, Minor PD, Delwart E. 2015. Comparing viral metagenomics methods using a highly multiplexed human viral pathogens reagent. J Virol Methods 213:139–146. 10.1016/j.jviromet.2014.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lipman NS, Homberger FR. 2003. Rodent quality assurance testing: use of sentinel animal systems. Lab Anim (NY) 32:36–43. 10.1038/laban0503-36. [DOI] [PubMed] [Google Scholar]
- 24.Lum SH, Turner A, Guiver M, Bonney D, Martland T, Davies E, Newbould M, Brown J, Morfopoulou S, Breuer J, Wynn R. 2016. An emerging opportunistic infection: fatal astrovirus (VA1/HMO-C) encephalitis in a pediatric stem cell transplant recipient. Transpl Infect Dis 18:960–964. 10.1111/tid.12607. [DOI] [PubMed] [Google Scholar]
- 25.Mor SK, Abin M, Costa G, Durrani A, Jindal N, Goyal SM, Patnayak DP. 2011. The role of type-2 turkey astrovirus in poult enteritis syndrome. Poult Sci 90:2747–2752. 10.3382/ps.2011-01617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naso MF, Tomkowicz B, Perry WL, 3rd, Strohl WR. 2017. Adeno-associated virus (AAV) as a vector for gene therapy. BioDrugs 31:317–334. 10.1007/s40259-017-0234-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng TF, Kondov NO, Hayashimoto N, Uchida R, Cha Y, Beyer AI, Wong W, Pesavento PA, Suemizu H, Muench MO, Delwart E. 2013. Identification of an astrovirus commonly infecting laboratory mice in the US and Japan. PLoS One 8:1–9. 10.1371/journal.pone.0066937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nurk S, Bankevich A, Antipov D, Gurevich A, Korobeynikov A, Lapidus A, Prjibelsky A, Pyshkin A, Sirotkin A, Sirotkin Y, Stepanauskas R, McLean J, Lasken R, Clingenpeel SR, Woyke T, Tesler G, Alekseyev MA, Pevzner PA. 2013. Assembling genomes and mini-metagenomes from highly chimeric reads, p 158–170. In: Deng M, Jiang R, Sun F, Zhang X, editors. Research in computational molecular biology. Lecture notes in computer science, vol 7821. Berlin (Heidelberg): Springer Berlin Heidelberg. [Google Scholar]
- 29.Phan TG, Kapusinszky B, Wang C, Rose RK, Lipton HL, Delwart EL. 2011. The fecal viral flora of wild rodents. PLoS Pathog 7:1–16. 10.1371/journal.ppat.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quan PL, Wagner TA, Briese T, Torgerson TR, Hornig M, Tashmukhamedova A, Firth C, Palacios G, Baisre-De-Leon A, Paddock CD, Hutchison SK, Egholm M, Zaki SR, Goldman JE, Ochs HD.2010. Astrovirus encephalitis in boy with X-linked agammaglobulinemia. Emerg Infect Dis 16:918–925. 10.3201/eid1606.091536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reuter G, Pankovics P, Boros A. 2018. Nonsuppurative (aseptic) meningoencephalomyelitis associated with neurovirulent astrovirus infections in humans and animals. Clin Microbiol Rev 31:1–23. doi: 10.1128/CMR.00040-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt K, Butt J, Mauter P, Vogel K, Erles-Kemna A, Pawlita M, Nicklas W. 2017. Development of a multiplex serological assay reveals a worldwide distribution of murine astrovirus infections in laboratory mice. PLoS One 12:1–22. 10.1371/journal.pone.0187174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Taylor MB, Grabow WO, Cubitt WD. 1997. Propagation of human astrovirus in the PLC/PRF/5 hepatoma cell line. J Virol Methods 67:13–18. 10.1016/S0166-0934(97)00071-2. [DOI] [PubMed] [Google Scholar]
- 35.Thurlow RW, Arriola R, Soll CE, Lipman NS. 2007. Evaluation of a flash disinfection process for surface decontamination of gamma-irradiated feed packaging. J Am Assoc Lab Anim Sci 46:46–49. [PubMed] [Google Scholar]
- 36.To KKW, Chan WM, Li KSM, Lam CSF, Chen Z, Tse H, Lau SKP, Woo PCY, Yuen KY. 2017. High prevalence of four novel astrovirus genotype species identified from rodents in China. J Gen Virol 98:1004–1015. 10.1099/jgv.0.000766. [DOI] [PubMed] [Google Scholar]
- 37.Williams SH, Che X, Garcia JA, Klena JD, Lee B, Muller D, Ulrich W, Corrigan RM, Nichol S, Jain K, Lipkin WI. 2018. Viral diversity of house mice in New York City. mBio 9:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wunderli W, Meerbach A, Gungor T, Berger C, Greiner O, Caduff R, Trkola A, Bossart W, Gerlach D, Schibler M, Cordey S, McKee TA, Van Belle S, Kaiser L, Tapparel C. 2011. Astrovirus infection in hospitalized infants with severe combined immunodeficiency after allogeneic hematopoietic stem cell transplantation. PLoS One 6:1–9. 10.1371/journal.pone.0027483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wunderlich ML, Dodge ME, Dhawan RK, Shek WR. 2011. Multiplexed fluorometric immunoassay testing methodology and troubleshooting. J Vis Exp 58:1–7. 10.3791/3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yokoyama CC, Loh J, Zhao G, Stappenbeck TS, Wang D, Huang HV, Virgin HW, Thackray LB. 2012. Adaptive immunity restricts replication of novel murine astroviruses. J Virol 86:12262–12270. 10.1128/JVI.02018-12. [DOI] [PMC free article] [PubMed] [Google Scholar]