Highlights
-
•
RT-qPCR and ddPCR were used for SARS-CoV-2 nucleic acids detection.
-
•
ddPCR shows higher sensitivity and lower limit of detection than RT-qPCR.
-
•
ddPCR successfully detected the dynamic changes in viral load while RT-qPCR failed to detect it.
-
•
Low-viral-load samples were not uncommon in clinical SARS-CoV-2 nucleic acids testing.
Keywords: Qualitative analysis, Quantitative analysis, COVID-19, Nucleic acid testing
Abstract
Background
Qualitative and quantitative detection of nucleic acids of SARS-CoV-2, the pathogen that causes coronavirus disease 2019 (COVID-19), plays a significant role in COVID-19 diagnosis, surveillance, prevention, and control.
Methods
A total of 117 samples from 30 patients with confirmed COVID-19 and 61 patients without COVID-19 were collected. Reverse transcriptase quantitative PCR (RT-qPCR) and droplet digital PCR (ddPCR) were used for qualitative and quantitative analyses of these samples to evaluate the diagnostic performance and applicability of the two methods.
Results
The positive detection rates of RT-qPCR and ddPCR were 93.3% and 100%, respectively. Among the 117 samples, 6 samples were tested single-gene positive by RT-qPCR but positive by ddPCR, and 3 samples were tested negative by RT-qPCR but positive by ddPCR. The viral load of samples with inconsistent results were relatively low (3.1–20.5 copies/test). There were 17 samples (37%) with a viral load below 20 copies/test among the 46 positive samples, and only 9 of them were successfully detected by RT-qPCR. A severe patient was dynamically monitored. All 6 samples from this patient were tested negative by RT-qPCR, but 4 samples were tested positive by ddPCR with a low viral load.
Conclusion
Qualitative analysis of COVID-19 samples can meet the needs of clinical screening and diagnosis, while quantitative analysis provides more information to the research community. Although both ddPCR and RT-qPCR can provide qualitative and quantitative results, ddPCR showed higher sensitivity and lower limit of detection than RT-qPCR, and it does not rely on the standard curve to quantify viral load. Therefore, ddPCR offers greater advantages than RT-qPCR.
1. Introduction
Coronavirus disease 2019 (COVID-19) is a new and emerging infectious disease that has spread rapidly across the globe. The disease is caused by a novel coronavirus, SARS-CoV-2. According to the data released by Johns Hopkins University, more than 10 million people have been diagnosed with COVID-19 across the world, with a death toll of more than half a million [1]. Due to the high infectious rate and the high percentage of severe outcomes of COVID-19, timely and effective diagnosis of COVID-19 is very important for the treatment of the disease, epidemic prevention, and epidemic control.
Viral nucleic acid testing is the primary method for diagnosing COVID-19. As the current gold standard of diagnosing COVID-19, reverse transcriptase quantitative polymerase chain reaction (RT-qPCR) uses specific primers and fluorescent probes to target specific regions of the SARS-CoV-2 genome. The fluorescent probes are used to monitor the progress of the amplification reaction, and the fluorescent intensity reflects the number of amplicons in the sample. A threshold cycle (Ct) value that can be directly correlated to the initial target concentration in the sample is used to determine whether a sample is positive or negative [2]. Although RT-qPCR can provide quantitative results using Ct values and a standard curve [3], it is mainly used for qualitative analysis in clinical COVID-19 detection.
In addition to qualitative diagnosis, quantitative detection also plays a significant role in COVID-19 diagnosis, surveillance, prevention, and control. As a technology for the absolute quantification of nucleic acids, digital polymerase chain reaction (dPCR) has also been applied to SARS-CoV-2 nucleic acid detection [4], [5], [6]. In a dPCR assay, the sample is first diluted and evenly partitioned into many independent reaction chambers. PCR amplification is then performed, where the SARS-CoV-2 nucleic acids are targeted by specific primers and fluorescent probes. Finally, PCR endpoint signals are detected and Poisson distribution statistical analysis is performed to determine the number of viral RNA copies [7]. In addition to providing qualitative results, dPCR can also provide the absolute number of SARS-CoV-2 without relying on the standard curve.
In this study, 117 clinical samples were analyzed qualitatively and quantitatively by RT-qPCR and droplet digital PCR (ddPCR), respectively. The purpose of this study was to investigate the diagnostic performance and applicability of the qualitative and quantitative analysis methods.
2. Material and methods
2.1. Participants
Forty pharyngeal swabs and 16 sputum specimens from 30 patients with confirmed COVID-19 and 51 pharyngeal swabs and 10 sputum specimens from 61 patients who were found to not have COVID-19 were collected in Beijing Youan Hospital affiliated to Capital Medical University between February and April 2020. Diagnostic criteria were the Diagnosis and Treatment Protocol for COVID-19 (trial version 7) established by the National Health Commission of the People’s Republic of China [8]. Patients’ laboratory test results and clinical data were also collected. Specimens that cannot be tested immediately were stored at −80 °C. This study was approved by the Beijing Youan Hospital Ethics Committee.
2.2. RNA extraction
Viral RNA was extracted using nucleic acids extraction kits (Lot. T124, Tianlong Science and Technology Co., Xi’an, China) on a nucleic acid extractor (GeneRotex, Tianlong Science and Technology) according to the manufacturer’s instructions. Sputum samples were treated with an equal volume of 7.5% Sputasol (Oxoid Ltd., Basingstoke, United Kingdom) until liquefaction was complete. The supernatant was drawn for nucleic acid extraction.
2.3. Qualitative analysis by RT-qPCR and quantitative analysis by ddPCR
RT-qPCR was conducted using the SARS-CoV-2 RNA detection kit (BioGerm Medical Biotechnology Co., Ltd., Shanghai, China) on the ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, Waltham, MA) following the manufacturer’s instructions. ORF1ab and N genes of SARS-CoV-2 were detected in this kit.
ddPCR was conducted via the SARS-CoV-2 Nucleic Acid Detection Kit (Digital PCR Method) (TargetingOne, Beijing, China) and the TD-1™ Droplet Digital™ PCR System (TargetingOne, licensed in China, registration number: 20170025; 20190065; 20192220517) following the manufacturer’s instructions. ORF1ab and N genes of SARS-CoV-2 were detected in this kit.
2.4. Positive standard of RT-qPCR and ddPCR detection
The results were considered valid when the reference gene was tested positive. According to the instructions of the RT-qPCR kit, a Ct value of ≤38 for both ORF1ab and N genes was defined as a positive test, while a Ct value of >38 for both genes was defined as a negative test. A Ct value of ≤38 for only one gene was defined as a single-positive test. In the ddPCR assay, if ORF1ab ≥ 3 copies/test and the sum of ORF1ab and N genes ≥ 5 copies/test or if N gene ≥ 5 copies/test, the result was considered positive; otherwise, the result was considered negative. The final viral RNA copy number was defined as the higher value between the copy numbers of the two genes.
3. Results
3.1. Information of the enrolled patients
A total of 30 patients with confirmed COVID-19 and 61 patients who did not have COVID-19 were included in this study, and the detailed information is shown in Table 1 . Fifteen out of the 30 COVID-19 patients were male and the median age was 62 years (IQR, 45–71 years). The main symptoms among these patients were fever and cough, reported in 83.3% and 63.3% of the patients, respectively. Fifteen of the patients had mild symptoms and were under 64 years of age, and the other 15 patients had severe symptoms and were over 60 years old. There were 38 male patients out of the 61 patients without COVID-19 and the median age was 37 years (IQR, 30–56 years). All 61 patients had fever.
Table 1.
Clinical features of the enrolled patients.
| Features | Value | ||
|---|---|---|---|
| Patients with confirmed COVID-19 | |||
| Age, median (IQR), years | 62 (45–71) | ||
| Male, n (%) | 15 (50) | ||
| Signs and symptoms at admission, n (%) | |||
| Fever | 25 (83.3) | ||
| Cough | 19 (63.3) | ||
| Fatigue | 9 (30) | ||
| Dyspnea | 8 (26.70) | ||
| Myalgia | 6 (20) | ||
| Anorexia | 5 (16.7) | ||
| Headache | 3 (10) | ||
| Pharyngodynia | 1 (3.3) | ||
| Nausea and vomiting | 1 (3.3) | ||
| Stomach ache | 1 (3.3) | ||
| No signs or symptoms | 1 (3.3) | ||
| Underlying chronic diseases, n (%) | |||
| Hypertension | 11 (36.7) | ||
| Mellitus | 4 (13.3) | ||
| Hyperlipemia | 2 (6.7) | ||
| Coronary heart disease | 2 (6.7) | ||
| Arrhythmia | 1 (3.3) | ||
| Bronchiectasis | 1 (3.3) | ||
| Chronic obstructive pulmonary disease | 1 (3.3) | ||
| Clinical classification, (%) | |||
| Mild type | 15 (50) | ||
| Severe type | 15 (50) | ||
| Clinical stage, n (%) | |||
| Early stage | 13 (43.3) | ||
| Progressive stage | 15 (50) | ||
| Recovery stage | 2 (6.7) | ||
| Patients without COVID-19 | |||
| Age, median (IQR), years | 37 (30–56) | ||
| Male, n (%) | 38 (62.3) | ||
3.2. Comparison of RT-qPCR results and ddPCR results with clinical diagnosis results
In this study, 117 samples were obtained via sampling once or repeatedly. After testing, 28 of the 30 COVID-19 patients were found to be positive by RT-qPCR, with a positive detection rate of 93.3%; and all patients were found to be positive by ddPCR, with a positive detection rate of 100% (Table 2 ).
Table 2.
Comparison of RT-qPCR and ddPCR with clinical diagnosis results.
| Analytical method | RT-qPCR |
ddPCR |
|||
|---|---|---|---|---|---|
| Positive | Negative | Positive | Negative | ||
| Clinical diagnosis results | Positive | 28 | 2 | 30 | 0 |
| Negative | 0 | 61 | 0 | 61 | |
| Positive detection rate | 93.3% | – | 100% | – | |
3.3. Comparison of RT-qPCR and ddPCR for all samples
The 117 samples tested included 91 pharyngeal swab samples and 26 sputum samples. As shown in Table 3 , 37 samples were tested positive by both RT-qPCR and ddPCR, while 71 samples were tested negative by both methods. Six samples were tested single-gene positive by RT-qPCR but positive by ddPCR, and 3 samples were tested negative by RT-qPCR but positive by ddPCR. The viral load of the 9 samples with inconsistent results between RT-qPCR and ddPCR was relatively low (ranging between 3.1 copies/test and 20.5 copies/test, Table 4 ). Five out of these 9 samples were collected each from a different COVID-19 patient. Three of these 5 patients had fever or respiratory symptoms, and the other 2 patients were re-admitted to the hospital after being tested positive in a follow-up examination and were asymptomatic during re-admission. The other 4 of the 9 samples were collected from a single patient at different times. This patient had a bacterial co-infection and was supported with extracorporeal membrane oxygenation (ECMO).
Table 3.
Comparison of the results of RT-qPCR and ddPCR.
| Sample type | RT-qPCR positive |
RT-qPCR single-gene positive |
RT-qPCR negative |
Sum | |||
|---|---|---|---|---|---|---|---|
| ddPCR Positive | ddPCR Negative | ddPCR positive | ddPCR negative | ddPCR positive | ddPCR negative | ||
| Pharyngeal swab | 27 | 0 | 4 | 0 | 1 | 59 | 91 |
| Sputum | 10 | 0 | 2 | 0 | 2 | 12 | 26 |
Table 4.
Detailed information of samples with inconsistent results between RT-qPCR and ddPCR.
| No. | Sample type | ddPCR (copies/test) |
RT-qPCR | ||
|---|---|---|---|---|---|
| ORF1ab | N gene | Viral load | |||
| 1 | Pharyngeal swab | 9.6 | 1.2 | 9.6 | ORF1ab (+) N (−) |
| 2 | Pharyngeal swab | 9.5 | 0 | 9.5 | ORF1ab (+) N (−) |
| 3 | Pharyngeal swab | 20.5 | 13.2 | 20.5 | ORF1ab (−) N (+) |
| 4 | Pharyngeal swab | 11.7 | 5.9 | 11.7 | ORF1ab (−) N (+) |
| 5 | Pharyngeal swab | 9.5 | 6.8 | 9.5 | ORF1ab (−) N (−) |
| 6 | Sputum | 13.8 | 15.2 | 15.2 | ORF1ab (−) N (+) |
| 7 | Sputum | 8.3 | 4.2 | 8.3 | ORF1ab (+) N (−) |
| 8 | Sputum | 5.9 | 4.4 | 5.9 | ORF1ab (−) N (−) |
| 9 | Sputum | 3.1 | 3.1 | 3.1 | ORF1ab (−) N (−) |
3.4. Analysis of the viral load of positive samples
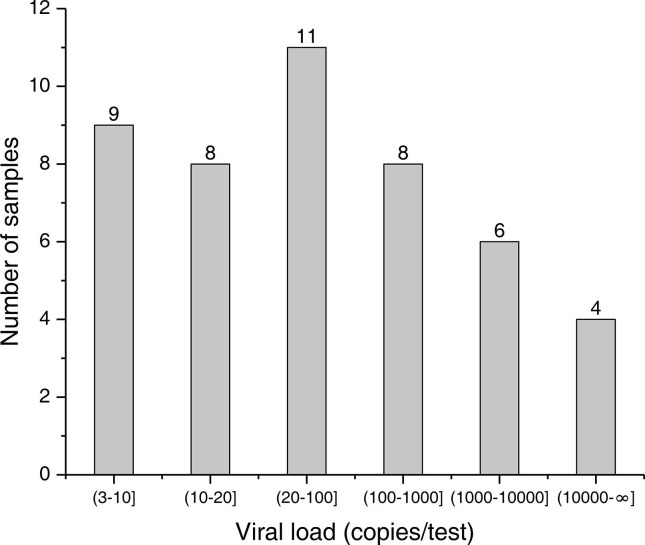
The viral load distribution of the 46 ddPCR positive samples was further analyzed, and the results are shown in Fig. 1 . There were only 4 samples with a viral load over 10,000 copies/test, while there were 17 samples (37%) with a viral load below 20 copies/test. Only 9 out of the 17 samples were successfully detected by RT-qPCR, and 6 samples and 2 samples were single-gene positive and negative, respectively.
Fig. 1.
Histogram showing distribution of viral load of the 46 positive samples. The viral load of the samples was quantified using ddPCR. The X-axis shows different ranges of viral load (copies/test) and the Y-axis shows the number of samples within each range of viral load.
3.5. Results of RT-qPCR and ddPCR for dynamic monitoring samples
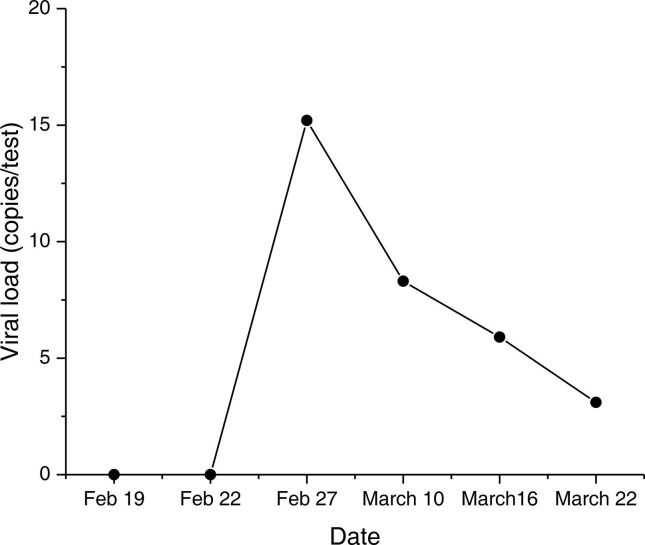
We used RT-qPCR and ddPCR to test the 6 sputum samples collected at different times from a severe patient. This patient was diagnosed with severe COVID-19 on February 5 and was diagnosed with a bacterial co-infection on February 9. All of the results of RT-qPCR were negative, but the results of ddPCR displayed a transition from negative to positive, with the first two tested negative and the last four positive. As shown in Fig. 2, the viral load of the positive samples was low, ranging from 3.1 to 15.2 copies/test. During the monitoring period (February 19–March 22), this patient developed symptoms including atrial fibrillation, hypoxemia, diarrhea, septic shock, respiratory failure, hematochezia, and pneumothorax. The ddPCR test results suggested that the incomplete clearance of the virus may continue to damage the host cells, leading to the above symptoms.
Fig. 2.
Results of ddPCR for dynamic monitoring. Six sputum samples from a severe patient were collected at different times. The X-axis shows the dates of sample collection and the Y-axis shows the viral load (copies/test) of each sample.
4. Discussion
Qualitative and quantitative detection of nucleic acids of SARS-CoV-2 plays a significant role in COVID-19 diagnosis, surveillance, prevention, and control. RT-qPCR and ddPCR are the primary methods for qualitative and quantitative detection of viral nucleic acids. In this study, we used RT-qPCR and ddPCR to perform qualitative and quantitative analyses of 117 samples from 30 patients with confirmed COVID-19 and 61 patients without COVID-19 and further evaluated the diagnostic performance and applicability of the two methods.
In this study, ddPCR was found to have a higher sensitivity and lower limit of detection (LoD), compared to RT-qPCR. The results showed that the positive detection rates of RT-qPCR and ddPCR were 93.3% and 100%, respectively. Nine samples (7.7%) from patients with confirmed COVID-19 were positively identified by ddPCR but found single-gene positive or negative by RT-qPCR. The viral load of these samples was relatively low (ranging between 3.1 copies/test and 20.5 copies/test). Furthermore, ddPCR successfully detected the dynamic changes in viral load in the samples from a severe COVID-19 patient while RT-qPCR failed to detect the presence of viral nucleic acids. This finding is consistent with previous reports [4], [5], [9], [10]. The sample partitioning step of ddPCR minimizes the effects of competition between targets and allows for higher tolerance to inhibitors [11], [12]. Therefore, ddPCR shows higher sensitivity than RT-qPCR.
Moreover, our results indicated that low-viral-load samples were not uncommon in clinical testing. Among the 46 positive samples, 17 (37%) were found to have a viral load of less than 20 copies/test. Thus, higher-sensitivity detection of SARS-CoV-2 is clinically valuable. RT-qPCR testing for SARS‐CoV‐2 has been reported to have a potentially high false-negative rate [13]; one of the possible reasons is that the LoD of RT-qPCR fails to meet the clinical requirement [14]. Furthermore, false-negative results may occur due to low viral productivity or improper sampling procedures.
Since ddPCR does not rely on calibration curves or Ct values for sample detection or quantification, the results are more objective than RT-qPCR results and it may avoid the gray zone of Ct values. Most of the commercial RT-qPCR diagnostic kits for COVID-19 include two or three targets, and a sample is considered positive if two or more targets are positive. Positive for only one target is defined as indeterminate and requires retesting. ddPCR results provide an absolute count of viral RNA, which prevents indefinite test results and repeated testing.
Furthermore, ddPCR can accurately detect the changes in viral load, which may provide clinicians with additional information that may be used to guide clinical judgment and treatment. During the monitoring period of the severe patient, we found that the patient developed many symptoms. The changes in viral load may be related to the patient’s symptoms. Further research to investigate the relationship between viral load and patient’s condition needs to be conducted.
5. Conclusions
Since COVID-19 is a new infectious disease, it is necessary to deepen our understanding of the disease to better prevent and control the outbreak. Quantitative analysis provides more information to the research community that may advance the research and treatment of COVID-19. Although both ddPCR and RT-qPCR can provide qualitative and quantitative results ddPCR showed higher sensitivity and lower LoD than RT-qPCR, and it does not rely on the standard curve to quantify viral load. Therefore, ddPCR offers greater advantages than RT-qPCR.
CRediT authorship contribution statement
Yan Dang: Data curation, Formal analysis, Writing - original draft. Ning Liu: Methodology, Validation. Chianru Tan: Formal analysis, Writing - original draft. Yingmei Feng: Investigation. Xingxing Yuan: Validation. Dongdong Fan: Visualization, Writing - original draft. Yanke Peng: Visualization. Ronghua Jin: Supervision. Yong Guo: Project administration, Writing - review & editing. Jinli Lou: Methodology, Project administration.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank the Department of Clinical Laboratory, the Department of Scientific Research and Bioinformatics Center of Beijing Youan Hospital for the timely help given in collecting the samples. The study has been financially supported by the National Science and Technology Major Project of China (2018ZX10305410-006 for Jinli Lou), the 2018 Beijing Youan Hospital Scientific Research Project for Young & Middle-Aged Talent’s Cultivation (YNKTQN20180204 for Yan Dang), Beijing Science and Technology Project (Z201100005420025 for Yong Guo), and Tsinghua University COVID-19 Project (for Yong Guo).
References
- 1.Johns Hopkins Coronavirus Resource Center, COVID-19 Map. https://coronavirus.jhu.edu/map.html, 2020 (accessed 8 July 2020).
- 2.Chu D.K.W., Pan Y., Cheng S.M.S., Hui K.P.Y., Krishnan P., Liu Y., Ng D.Y.M., Wan C.K.C., Yang P., Wang Q., Peiris M., Poon L.L.M. Molecular diagnosis of a novel coronavirus (2019-nCoV) causing an outbreak of pneumonia. Clin. Chem. 2020;66(4):549–555. doi: 10.1093/clinchem/hvaa029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/s1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yu F., Yan L., Wang N., Yang S., Wang L., Tang Y., Gao G., Wang S., Ma C., Xie R., Wang F., Tan C., Zhu L., Guo Y., Zhang F. Quantitative detection and viral load analysis of SARS-CoV-2 in infected patients. Clin. Infect. Dis. 2020 doi: 10.1093/cid/ciaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suo T., Liu X., Feng J., Guo M., Hu W., Guo D., Ullah H., Yang Y., Zhang Q., Wang X., Sajid M., Huang Z., Deng L., Chen T., Liu F., Xu K., Liu Y., Zhang Q., Liu Y., Xiong Y., Chen G., Lan K., Chen Y. ddPCR: a more accurate tool for SARS-CoV-2 detection in low viral load specimens. Emerg. Microbes. Infect. 2020;9(1):1259–1268. doi: 10.1080/22221751.2020.1772678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen H., Wu R., Xing Y., Du Q., Xue Z., Xi Y., Yang Y., Deng Y., Han Y., Li K., Luan Y., Zhang Y., Wei X., Yu T., Li H., Zhu L., Su S., Lian H., Lu L., Tan C., Zheng H., Chen B., Yu P., Guo Y., Ma C. Influence of different inactivation methods on severe acute respiratory syndrome coronavirus 2 RNA copy number. J. Clin. Microbiol. 2020 doi: 10.1128/JCM.00958-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salipante S.J., Jerome K.R. Digital PCR-An emerging technology with broad applications in microbiology. Clin. Chem. 2020;66(1):117–123. doi: 10.1373/clinchem.2019.304048. [DOI] [PubMed] [Google Scholar]
- 8.National Health Commission of the people’s republic of China, Diagnosis and Treatment Protocol for COVID-19 (Trial Version 7). http://en.nhc.gov.cn/2020-03/29/c_78469.htm, 2020 (accessed 8 July 2020).
- 9.Lu R., Wang J., Li M., Wang Y., Dong J., Cai W. SARS-CoV-2 detection using digital PCR for COVID-19 diagnosis treatment monitoring and criteria for discharge. medRxiv. 2020 doi: 10.1101/2020.03.24.20042689. [DOI] [Google Scholar]
- 10.Dong L., Zhou J., Niu C., Wang Q., Pan Y., Sheng S., Wang X., Zhang Y., Yang J., Liu M., Zhao Y., Zhang X., Zhu T., Peng T., Xie J., Gao Y., Wang D., Zhao Y., Dai X., Fang X. Highly accurate and sensitive diagnostic detection of SARS-CoV-2 by digital PCR. medRxiv. 2020 doi: 10.1101/2020.03.14.20036129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quan P.L., Sauzade M., Brouzes E. dPCR: A technology review. Sensors (Basel) 2018;18(4) doi: 10.3390/s18041271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basu A.S. Digital Assays Part I: Partitioning statistics and digital PCR. SLAS Technol. 2017;22(4):369–386. doi: 10.1177/2472630317705680. [DOI] [PubMed] [Google Scholar]
- 13.Li Y., Yao L., Li J., Chen L., Song Y., Cai Z., Yang C. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X., Yao H., Xu X., Zhang P., Zhang M., Shao J., Xiao Y., Wang H. Limits of detection of six approved RT-PCR kits for the novel SARS-coronavirus-2 (SARS-CoV-2) Clin. Chem. 2020 doi: 10.1093/clinchem/hvaa099. [DOI] [PMC free article] [PubMed] [Google Scholar]