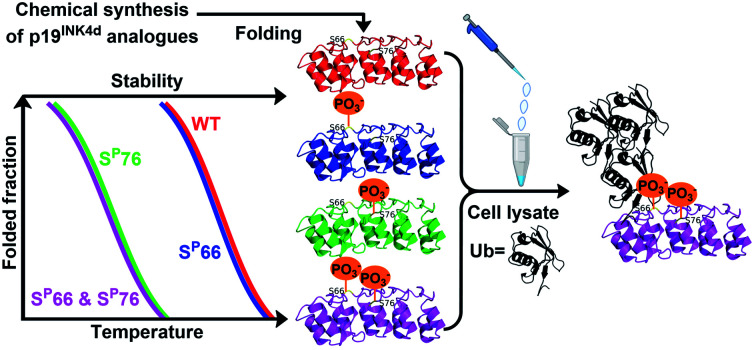
 This study describes the chemical synthesis of unmodified and phosphorylated p19INK4d that were characterized by circular dichroism and biochemical methods to examine the effect of phosphorylation on the thermal stability and ubiquitination.
This study describes the chemical synthesis of unmodified and phosphorylated p19INK4d that were characterized by circular dichroism and biochemical methods to examine the effect of phosphorylation on the thermal stability and ubiquitination.
Abstract
p19INK4d plays an important role in the regulation of the cell cycle by inhibiting the function of cyclin-dependent kinases 4/6 that is responsible for the phosphorylation and deactivation of the retinoblastoma protein (pRb) tumour suppressor. Recently, it was reported that phosphorylation of p19INK4d at Ser76 and Ser66 causes structural changes, which lead to its ubiquitination and degradation. Yet the exact contribution of each phosphorylation site remains unclear. To shed light on the role of these sites, we developed the chemical synthesis of unmodified, mono- and doubly phosphorylated p19INK4d using state of the art methods for chemical protein synthesis. The synthesized proteins were characterized by circular dichroism and biochemical methods to examine the effect of phosphorylation on the thermal stability and ubiquitination, respectively. Our results provide clear determination of p19INK4d stability upon phosphorylation at different sites and reveal that phosphorylation of both Ser residues might be necessary for promoting ubiquitination of p19INK4d.
Introduction
p19INK4d is a member of the INK4 family that includes p16INK4a, p15INK4b and p18INK4c proteins.1 The INK4 family shares a high degree of sequence and structural homology, which contributes to its function in regulating the cell cycle.2 Specifically, this family induces cell cycle arrest through the inhibition of cyclin-dependent kinases 4/6 (CDK4/6) and prevents the formation of the CDK4/6-Cyclin D complex that deactivates the pRb tumour suppressor via its phosphorylation.3,4 p19INK4d is a 166 amino acid protein made of five ankyrin repeats of helices and beta-turns,5,6 of which Ser66 and Ser76 residues undergo phosphorylation, while Lys62 was found to be ubiquitinated. Interestingly, among the INK4 family members, only p19INK4d showed a high level of phosphorylation and exhibited rapid proteasome mediated-degradation in vivo.7,8
Balbach and coworkers proposed a crosstalk between p19INK4d phosphorylation at Ser66 and Ser76 and its ubiquitination at Lys62.9,10 The group prepared different analogues of p19INK4d using a glutamic acid mimic of phosphoserine at Ser66 and Ser76 (S66E/S76E) and found that unlike S66E, S76E strongly destabilizes the protein; however for efficient ubiquitination both S66E and S76E are required.9 More recently, the group enzymatically prepared p19INK4d mono-phosphorylated at Ser66 and doubly phosphorylated at both Ser residues and studied the effect of phosphorylation on the p19INK4d structure and ubiquitination.10 Based on this study, the group proposed a mechanism where the stepwise phosphorylation, first at Ser66 using p38 kinase and then at Ser76 using CDK1 kinase, causes significant structural changes, leading to the dissociation of CDK4/6 and subsequent ubiquitination and degradation of p19INK4d. However, phosphorylation of Ser76 could not be introduced separately and was only detected after Ser66 phosphorylation. Therefore, the exact effect of mono-phosphorylation of either Ser66 or Ser76 on both the thermal stability and ubiquitination of p19INK4d is still not clear.
Chemical protein synthesis is a powerful approach to prepare homogeneous and workable amounts of proteins bearing post-translational modifications such as phosphorylation and ubiquitination, essentially, without restrictions on the number and sites of these modifications.11,12 In addition, incorporation of modified amino acids during the chemical synthesis bypasses the need for the involvement of enzymes such as kinases, which in many cases are unknown for the specific protein. Therefore, we reasoned that by using protein chemical synthesis one could prepare phosphorylated p19INK4d analogues, in particular phosphorylated p19INK4d at Ser76, with high homogeneity and in workable quantities to shed light on its exact role.
Here we report the chemical synthesis of four different analogues of p19INK4d including the non-phosphorylated p19INK4d(WT), the mono-phosphorylated at Ser66 or Ser76 p19INK4d(SP66)/p19INK4d(SP76) and the doubly phosphorylated p19INK4d(SP66 & SP76). The synthetic proteins were folded in vitro and characterized for their stabilities and ubiquitination levels. Our results clearly show that the stability of p19INK4d is affected by the position of phosphorylation; moreover, phosphorylation of both of Ser residues might be required for higher stimulation of ubiquitination of p19INK4d.
Results & discussion
Chemical synthesis of p19INK4d analogues
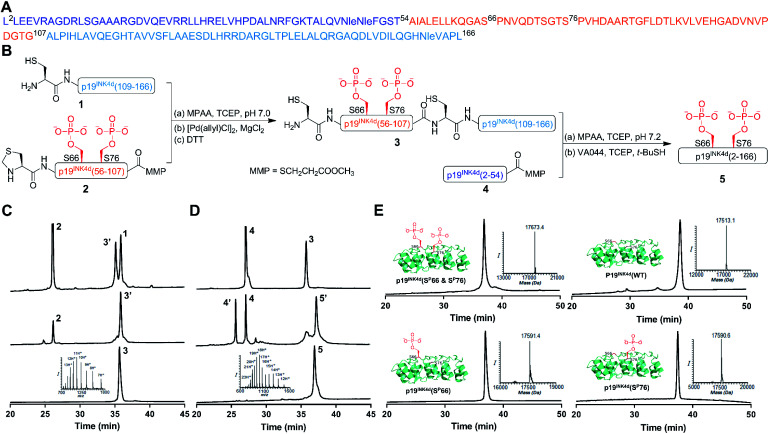
Our strategy for the synthesis of p19INK4d is outlined in Fig. 1, in which we divided the polypeptide sequence into three fragments (Fig. 1A); Cys-p19INK4d(109-166) (fragment 1), Thz-p19INK4d(SP66 & SP76)(56-107)-thioester (fragment 2), and p19INK4d(2-54)-thioester (fragment 4). All the fragments were prepared according to the Fmoc-SPPS strategy on a Rink-amide resin with the appropriate linker depending on the desired C-terminus functionality for each fragment. Fragment 2 was prepared using the 3-amino-4-(methylamino)benzoic acid linker (MeDbz) since it has a C-terminal Gly residue to afford the peptide with the N-acyl-N′-methylbenzimidazolinone (MeNbz) functionality, while fragment 4 which bears a C-terminal Thr residue was prepared using a Dbz linker to obtain the peptide with N-acylbenzimidazolinone (Nbz).13,14 The thioester forms of these fragments were obtained by substituting the MeNbz and Nbz units with methyl 3-mercaptoproprionate (MMP), to allow a longer storage time and reduce hydrolysis during ligation. The N-terminal Ala residue in fragments 1 and 2 was replaced with Cys and thiazolidine (Thz), respectively, to allow native chemical ligation (NCL).15,16
Fig. 1. General scheme for the synthesis of p19INK4d analogues. (A) Sequence of p19INK4d. Met49, Met50 and Met162 were replaced with Nle to avoid oxidation during the synthesis. (B) Synthetic strategy for obtaining p19INK4d(SP66 & SP76). (C) HPLC and mass analysis of ligation and Thz-opening to acquire 3. (D) HPLC and mass results for final ligation and desulfurization steps to afford 5, p19INK4d(SP66 & SP76). (E) HPLC and mass analysis of the purified analogues of p19INK4d: p19INK4d(SP66 & SP76) with an observed mass of 17 673.4 ± 2.1 Da (calculated 17 671.9 Da), p19INK4d(WT) with an observed mass of 17 513.1 ± 2.1 Da (calculated 17 513.9 Da), p19INK4d(SP66) with an observed mass of 17 591.4 ± 2.1 Da (calculated 17 592.9 Da), and p19INK4d(SP76) with an observed mass of 17 590.6 ± 2.1 Da (calculated 17 592.9 Da).
Once these fragments were prepared in high yield and excellent purity, we performed NCL between fragments 1 and 2 under typical NCL conditions of 4-mercaptophenylacetic acid (MPAA) and tris(2-carboxyethyl)phosphine hydrochloride (TCEP) in 6 M Gn·HCl at pH 7. The ligation step was fast and completed within 1 h at 37 °C. The reaction mixture was then treated with MgCl2 followed by [Pd(allyl)Cl]2 for Thz-opening in a one-pot manner.17 The reaction was completed within an hour (Fig. 1B), quenched with DTT and the crude mixture was purified by reversed-phase HPLC to obtain the ligated product in a 42% isolated yield (Fig. 1C). To obtain the full-length polypeptide, NCL between ligated peptide 3 and fragment 4 was executed, which required 24 h for completion because of the sterically hindered ligation site. Subsequently, the Cys residues were desulfurized by applying radical desulfurization conditions (TCEP, VA-044, and tBuSH) (Fig. 1D).18 The desulfurized product was purified and lyophilized to obtain the synthetic p19INK4d(SP66 & SP76) in ∼18% yield. Using this chemical strategy, we synthesized all the p19INK4d analogues in milligram quantities and excellent purity (Fig. 1E).
Folding of p19INK4d analogues
While attempting the in vitro folding of p19INK4d analogues, we encountered difficulties despite using different folding conditions since we often observed precipitation of the protein over time both while diluting and during the circular dichroism (CD) measurements. Eventually, we found that dissolving the analogues in 6 M Gn·HCl followed by adding 50 mM sodium phosphate in 25% glycerol (pH 7.4 and final 2% Gn·HCl), which has been shown to increase the folding yield and reduce aggregation in various systems,19,20 led to obtaining a clear solution of the protein ready for further characterization.
Secondary structure characterization and thermal stability analysis of p19INK4d analogues
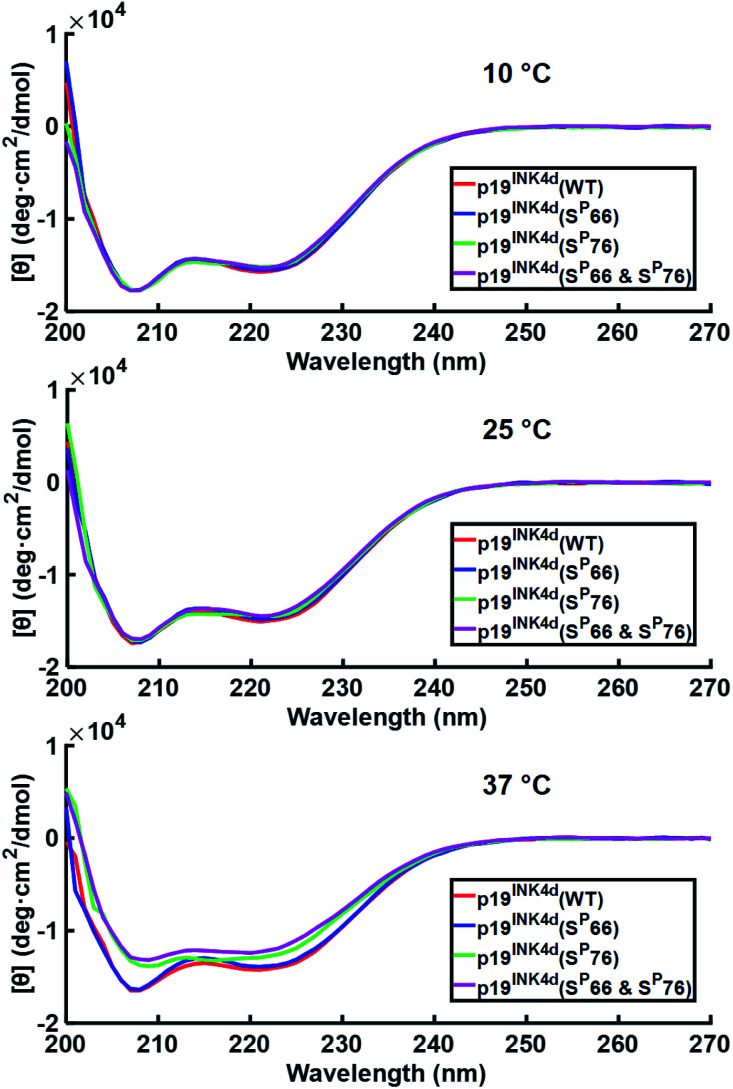
After successfully folding all the different synthetic analogues we attempted to determine the effect of phosphorylation on p19INK4d thermal stability by monitoring the changes in the CD of each analogue upon increasing the temperature of the protein solution. Specifically, we followed the changes in the ellipticity at 208 nm and 222 nm – the signature of the alpha-helical secondary structure, upon increasing the temperature.21,22
At 10 °C, we observed the expected characteristic peaks for all the p19INK4d analogues. Increasing the temperature to 25 °C did not induce any major changes in the CD signal. However, after a further increase in the temperature to 37 °C, we observed a significant decrease in the molar ellipticity curve for p19INK4d(SP76) and p19INK4d(SP66 & SP76) (Fig. 2). We quantified this change for each analogue by calculating the alpha-helical content at both 25 °C and 37 °C using the Greenfield and Fasman equation.23 The changes in the alpha-helical content from 25 °C to 37 °C for p19INK4d(WT) and p19INK4d(SP66) analogues were 2.8% and 3.7% respectively. These changes were negligible compared to those of p19INK4d(SP76) and p19INK4d(SP66 & SP76) analogues, which were 11.6% and 13.4% accordingly, thus indicating that the latter analogues are less stable than the former.
Fig. 2. CD analysis of p19INK4d synthetic analogues where the CD signal is monitored at different temperatures; 10 °C, 25 °C, and 37 °C. Concentrations determined for this experiment were 7 μM for p19INK4d(WT) and p19INK4d(SP66 & SP76), and 5 μM for p19INK4d(SP66) and p19INK4d(SP76).
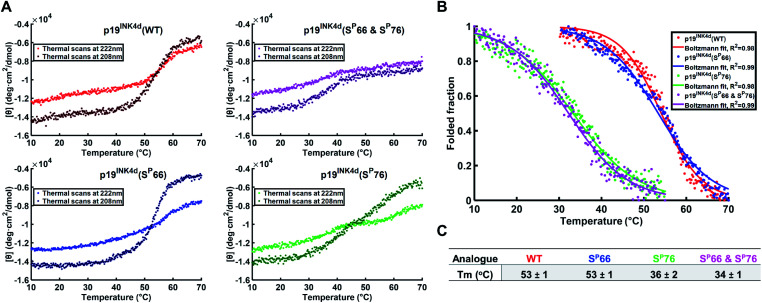
To further verify the thermal stability of the different analogues, thermal-denaturation scans were collected with a wider range of temperatures at both 222 nm and 208 nm. From the initial observation of the thermal-denaturation curves, we noticed similar thermal stability for p19INK4d(WT) and p19INK4d(SP66) analogues and correspondingly for p19INK4d(SP76) and p19INK4d(SP66 & SP76) (Fig. 3A). The melting temperature (Tm) of each analogue was determined, giving us direct evidence and accurate determination of their stabilities. The CD signal at 222 nm was converted to the folded fraction and the Tm at which the folded fraction was 0.5 was extrapolated using the Boltzmann fit (Fig. 3B).21,24 The Tm values reported in Fig. 3C support our alpha-helical content change results and clearly indicate that p19INK4d(WT) and p19INK4d(SP66) are much more stable analogues than p19INK4d(SP76) and p19INK4d(SP66 & SP76) with more than 10 °C differences. Complementary results were obtained when calculating the Tm according to the CD signal at 208 nm wavelength (ESI, Fig. S20†). Our findings do not only suggest that there is a remarkable effect of phosphorylation on the p19INK4d stability, but also uncover that phosphorylation of different Ser residues has a different effect on thermal stability, where phosphorylation at Ser76 significantly decreases p19INK4d stability compared with Ser66 phosphorylation.
Fig. 3. Stability analysis of p19INK4d analogues using CD. (A) Single measurement of thermal scans from 10 °C to 70 °C obtained for all the analogues of p19INK4d at 222 nm and 208 nm wavelength. (B) Folded fraction plotted against temperature calculated using the Boltzmann equation to fit the sigmoidal curves obtained from thermal scans at 222 nm in Fig. 3A. (C) Mean Tm values ± SD °C of the four analogues acquired from three independent measurements for each analogue (ESI, Fig. S21†).
Additional H-D exchange experiments were performed for the four analogues to check any tertiary structural differences at room temperature, wherein the CD spectra of these analogues did not exhibit any secondary structure differences (ESI, Fig. S22†). The results show a higher rate of exchange for p19INK4d(SP76) and p19INK4d(SP66 & SP76) analogues compared to p19INK4d(WT) and p19INK4d(SP66), reflecting tertiary structural changes even at room temperature. Since Ser76 phosphorylation seems to have an effect on the tertiary structure, these results agree with the CD analysis as well as support the previously reported 1H–15N 2D HSQC NMR study which has shown major structural changes upon the incorporation of the second phosphorylation at Ser76.10
Studying the crosstalk between phosphorylation and ubiquitination
Next, we wanted to study the crosstalk between phosphorylation and ubiquitination of our synthetic analogues. This would allow us to find out first, whether the ubiquitination level will change upon mono- or di-phosphorylation of p19INK4d and second, if there are different effects of the phosphorylation positions on ubiquitination and correlate these results with the protein stability. To enable additional verification of the ubiquitination level, HA-tagged analogues were prepared. The synthesis of these analogues was performed according to the synthesis of the previous analogues; however, fragment 4 was prepared with an HA tag (ESI, Scheme S7, S16–S19†).
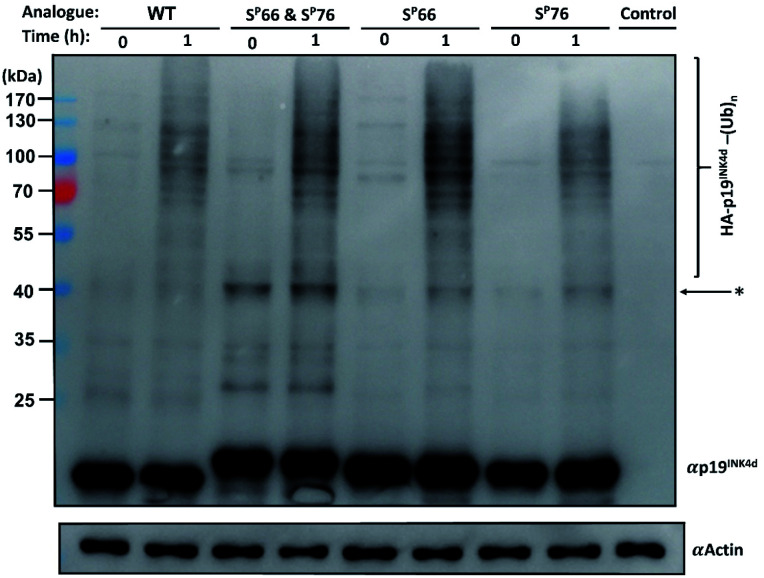
Since the E3 ligase of p19INK4d has yet to be discovered, we used S-phase synchronized HeLa or U2OS cells as a source for the E3 enzyme. The analogues were loaded separately in four in vitro ubiquitination reactions, reacted simultaneously, and incubated at 37 °C for 1 h. The analogues can be distinguished through western blot by a slight but clear shift in gel migration, according to the difference in the number of phosphorylated residues. The doubly phosphorylated HA-p19INK4d(SP66 & SP76) band appears slightly higher than the bands of mono-phosphorylated HA-p19INK4d(SP66)/HA-p19INK4d(SP76) analogues while the latter migrate higher than the band of HA-p19INK4d(WT). Phosphorylation or dephosphorylation of the different analogues can thus be monitored throughout the ubiquitination assay and since the dephosphorylation of HA-p19INK4d(SP66 & SP76) was observed, the phosphatase inhibitor was added to the lysis buffer to prevent it (Fig. S23†). The signal of HA-p19INK4d polyubiquitin conjugate bands for HeLa cells is reasonably low after 1 h (Fig. S24†) but can be seen for a similar assay performed using S-phase U2OS cells, with the incorporation of p38 and CDK1 kinase inhibitors (Fig. 4). Based on Fig. 4 and S26,† in comparison to HA-p19INK4d(WT), HA-p19INK4d(SP66 & SP76) and HA-p19INK4d(SP66) exhibited increased ubiquitination of 62 ± 22% and 14 ± 7% respectively, while HA-p19INK4d(SP76) ubiquitination was reduced by 8.9 ± 2%, indicating that phosphorylation at Ser76 is not enough for efficient ubiquitination.
Fig. 4. Ubiquitination study of HA-p19INK4d analogues. Immunoblot of each synthetic analogue of HA-p19INK4d with addition of S-phase U2OS lysate obtained before incubation in 37 °C (0 h) and after 1 h. The control contained assay compounds with cell lysate. Actin is shown below as a loading control. *Unidentified bands which can correspond to impurity and poly-ubiquitinated p19INK4d.
Based on these results, we suggest that phosphorylation at both Ser66 and Ser76 might potentially promote the ubiquitination of p19INK4d. We also suggest that other factors might be involved in the productive ubiquitination of p19INK4d. Thullberg et al. suggested, for instance, that the association with CDK4, even in its mutant form, determines the efficient ubiquitination of p19INK4d, by notably increasing the ubiquitination level.8
Phosphorylation among other effects can either inhibit or induce ubiquitination and stability is often referred to as a linking bridge towards understanding the effect of phosphorylation on ubiquitination.25 Phosphorylation that stabilizes the protein can lead to a decrease in ubiquitination such as in the case of MDM2 phosphorylation via PKB, or otherwise, cause destabilization and increase ubiquitination as in the case of p53 phosphorylation by aurora kinase A.26,27 In the case of p19INK4d, based on the results presented here, phosphorylation at different positions have an eminently different effect on stability, where phosphorylation at Ser76, unlike phosphorylation at Ser66, causes major destabilization of p19INK4d but its sole incorporation is not enough for inducing efficient ubiquitination. Phosphorylation at both Ser66 and Ser76 might be required for the recognition of the E3 ligase, which triggers a higher ubiquitination signal of p19INK4d.
Conclusions
In this work, we report the first total chemical synthesis of p19INK4d protein – p19INK4d(WT) along with its phosphorylated analogues; p19INK4d(SP66 & SP76), p19INK4d(SP66) and the p19INK4d(SP76) analogue that was not obtained using enzymatic approaches. We were able to determine by measuring the Tm for the different synthetic analogues, the changes in the thermal stability of p19INK4d upon phosphorylation of Ser66 or/and Ser76 residues, where we show that p19INK4d(SP76) and p19INK4d(SP66 & SP76) have similar low stability while p19INK4d(SP66) does not affect the stability as the calculated Tm is identical compared to p19INK4d(WT). While p19INK4d(SP76) causes major thermal destabilization, our in vitro ubiquitination study reveals that it is not sufficient for inducing a high polyubiquitination signal. However p19INK4d(SP66 & SP76) induces the highest ubiquitination signal supporting the mechanism suggested by the Balbach group.10,28 Since polyubiquitination was also detected for the other analogues, we suggest that other factors might affect ubiquitination and should be taken into consideration such as the direct interaction with CDK4/6. Also, since the phosphorylation of both Ser residues occurred at the Ser-Pro junction, S-phase lysate may contain enzymes such as peptidyl-prolyl cis–trans isomerases (PPIas) that assist the proper folding of p19INK4d. Although the use of prolyl isomerase was mentioned before for folding p19INK4d to its native-like order structure in vitro, its function was not reported in vivo.29,30 Apart from the possible involvement of these enzymes in folding p19INK4d, there are various enzymes essential for understanding the functional pathway of p19INK4d that have still not been identified, such as the ligase and the phosphatases. Finding the specific E3 ligase, for example, would aid in minimizing variables during in vitro ubiquitination assay performed with lysate. Our synthetic route for the different analogues opens the door for proteomic analysis to unravel these enzymes and other unknown aspects of p19INK4d biology.
Conflicts of interest
The authors declare no conflict of interest.
Supplementary Material
Acknowledgments
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation programme (grant agreement no. [831783]). A. B. holds The Jordan and Irene Tark Academic Chair. We thank Dr Sumeet K. Singh for his contribution in peptide synthesis.
Footnotes
†Electronic supplementary information (ESI) available. See DOI: 10.1039/c9sc06300e
References
- Chan F. K., Zhang J., Cheng L., Shapiro D. N., Winoto A. Mol. Cell. Biol. 1995;15:2682–2688. doi: 10.1128/mcb.15.5.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumgartner R., Fernandez-Catalan C., Winoto A., Huber R., Engh R. A., Holak T. A. Structure. 1998;6:1279–1290. doi: 10.1016/s0969-2126(98)00128-2. [DOI] [PubMed] [Google Scholar]
- Cánepa E. T., Scassa M. E., Ceruti J. M., Marazita M. C., Carcagno A. L., Sirkin P. F., Ogara M. F. IUBMB Life. 2007;59:419–426. doi: 10.1080/15216540701488358. [DOI] [PubMed] [Google Scholar]
- Roussel M. F. Oncogene. 1999;18:5311–5317. doi: 10.1038/sj.onc.1202998. [DOI] [PubMed] [Google Scholar]
- Luh F. Y., Archer S. J., Domaille P. J., Smith B. O., Owen D., Brotherton D. H., Raine A. R. C., Xu X., Brizuela L., Brenner S. L., Laue E. D. Nature. 1997;389:999–1003. doi: 10.1038/40202. [DOI] [PubMed] [Google Scholar]
- Kalus W., Baumgartner R., Renner C., Noegel A., Chan F. K. M., Winoto A., Holak T. A. FEBS Lett. 1997;401:127–132. doi: 10.1016/s0014-5793(96)01465-2. [DOI] [PubMed] [Google Scholar]
- Thullberg M., Bartkova J., Khan S., Hansen K., Rönnstrand L., Lukas J., Strauss M., Bartek J. FEBS Lett. 2000;470:161–166. doi: 10.1016/s0014-5793(00)01307-7. [DOI] [PubMed] [Google Scholar]
- Thullberg M., Bartek J., Lukas J. Oncogene. 2000;19:2870–2876. doi: 10.1038/sj.onc.1203579. [DOI] [PubMed] [Google Scholar]
- Löw C., Homeyer N., Weininger U., Sticht H., Balbach J. ACS Chem. Biol. 2009;4:53–63. doi: 10.1021/cb800219m. [DOI] [PubMed] [Google Scholar]
- Kumar A., Gopalswamy M., Wolf A., Brockwell D. J., Hatzfeld M., Balbach J. Proc. Natl. Acad. Sci. U. S. A. 2018;115:3344–3349. doi: 10.1073/pnas.1719774115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondalapati S., Jbara M., Brik A. Nat. Chem. 2016;8:407–418. doi: 10.1038/nchem.2476. [DOI] [PubMed] [Google Scholar]
- Kulkarni S. S., Sayers J., Premdjee B., Payne R. J. Nat. Rev. Chem. 2018;2:0122. [Google Scholar]
- Blanco-Canosa J. B., Dawson P. E. Angew. Chem., Int. Ed. 2008;47:6851–6855. doi: 10.1002/anie.200705471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Canosa J. B., Nardone B., Albericio F., Dawson P. E. J. Am. Chem. Soc. 2015;137:7197–7209. doi: 10.1021/jacs.5b03504. [DOI] [PubMed] [Google Scholar]
- Dawson P. E., Muir T. W., Kent S. B. H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Agouridas V., El Mahdi O., Diemer V., Cargoët M., Monbaliu J. C. M., Melnyk O. Chem. Rev. 2019:119. doi: 10.1021/acs.chemrev.8b00712. [DOI] [PubMed] [Google Scholar]
- Jbara M., Maity S. K., Seenaiah M., Brik A. J. Am. Chem. Soc. 2016;138:5069–5075. doi: 10.1021/jacs.5b13580. [DOI] [PubMed] [Google Scholar]
- Wan Q., Danishefsky S. J. Angew. Chem., Int. Ed. 2007;46:9248–9252. doi: 10.1002/anie.200704195. [DOI] [PubMed] [Google Scholar]
- V Rariy R., Klibanov A. M. Proc. Natl. Acad. Sci. U. S. A. 1997;94:13520–13523. doi: 10.1073/pnas.94.25.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa F., Farah C. S. Biophys. J. 2007;92:2463–2475. doi: 10.1529/biophysj.106.098541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N. J. Nat. Protoc. 2007;1:2527–2535. doi: 10.1038/nprot.2006.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N. J. Nat. Protoc. 2007;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Biochemistry. 1969;8:4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Brown A. M. Comput. Methods Progr. Biomed. 2001;65:191–200. doi: 10.1016/s0169-2607(00)00124-3. [DOI] [PubMed] [Google Scholar]
- Hunter T. Mol. Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Feng J., Tamaskovic R., Yang Z., Brazil D. P., Merlo A., Hess D., Hemmings B. A. J. Biol. Chem. 2004;279:35510–35517. doi: 10.1074/jbc.M404936200. [DOI] [PubMed] [Google Scholar]
- Katayama H., Sasai K., Kawai H., Yuan Z. M., Bondaruk J., Suzuki F., Fujii S., Arlinghaus R. B., Czerniak B. A., Sen S. Nat. Genet. 2004;36:55–62. doi: 10.1038/ng1279. [DOI] [PubMed] [Google Scholar]
- Phosphorylation using CDK1 kinase occurred in the p19INK4d(SP66) analogue only, confirming the stepwise phosphorylation mechanism suggested by the Balbach group, see Fig. S28, page 51
- Zeeb M., Rösner H., Zeslawski W., Canet D., Holak T. A., Balbach J. J. Mol. Biol. 2002;315:447–457. doi: 10.1006/jmbi.2001.5242. [DOI] [PubMed] [Google Scholar]
- Löw C., Weininger U., Zeeb M., Zhang W., Laue E. D., Schmid F. X., Balbach J. J. Mol. Biol. 2007;373:219–231. doi: 10.1016/j.jmb.2007.07.063. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.