Abstract
Background -
Left ventricular ejection fraction (EF) is an indicator of cardiac function, usually assessed in individuals with heart failure and other cardiac conditions. Although family studies indicate that EF has an important genetic component with heritability estimates up to 0.61, to date only six EF-associated loci have been reported.
Methods -
Here, we conducted a genome-wide association study (GWAS) of EF in 26,638 adults from the GERA and the UK Biobank cohorts.
Results -
A meta-analysis combining results from GERA and UK Biobank identified a novel locus: TMEM40 on chromosome 3p25 (rs11719526; β=0.47 and P=3.10x10−8) that replicated in Biobank Japan and confirmed recent findings implicating the BAG3 locus on chromosome 10q26 in EF variation, with the strongest association observed for rs17617337 (β=-0.83 and P=8.24x10−17). Although the minor allele frequencies (MAF) of TMEM40 rs11719526 were generally common (between 0.13 and 0.44) in different ethnic groups, BAG3 rs17617337 was rare (MAF<0.05) in Asian and African ancestry populations. These associations were slightly attenuated, after considering antecedent cardiac conditions (i.e., heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation, valvular disease, and revascularization procedures). This suggests that the effects of the lead variants at TMEM40 or BAG3 on EF are largely independent of these conditions.
Conclusions -
In this large and multiethnic study, we identified two loci, TMEM40 and BAG3, associated with EF at a genome-wide significance level. Identifying and understanding the genetic determinants of EF is important to better understand the pathophysiology of this strong correlate of cardiac outcomes and to help target the development of future therapies.
Keywords: Basic Science Research, Genetics, Genetic, Association Studies, Heart Failure, Echocardiography, human, Genome Wide Association Study, single nucleotide polymorphism, left ventricular ejection fraction, cardiac disease
Left ventricular ejection fraction (EF) is a reliable measurement reflecting left ventricular contractile structure and function, and the length of the sarcomere, which is the basic contractile unit of skeletal muscle1, 2. EF is defined as the stroke volume divided by the end-diastolic volume. This measurement enables the classification of cardiac phenotypes3, such as heart failure or cardiomyopathy, with low EF (≤40%) increasing the risk of poor clinical outcomes. Although the utility of EF measurement for distinguishing cardiac phenotypes has been recently questioned4, to date EF remains the best means for successfully stratifying cardiac phenotypes3. A better understanding of the genetic factors underlying EF could provide important insights into the pathophysiology underlying cardiac conditions and may suggest the basis for novel treatments.
Genetic factors have been suggested to be one of the determinants of the variation of left ventricular structural features5, 6. Twin and family studies indicate that EF has a moderate to important genetic component, with heritability estimates ranging from 0.27 to 0.617–10. However, these classical twin/family studies may be biased by the shared environment within families leading to overestimation of heritability estimates11–13. Importantly, no study has estimated the heritability of EF based on genome-wide array data of unrelated individuals.
Over the last decade, genome-wide association studies (GWAS) have accelerated the discovery of genomic regions contributing to a vast number of diseases and complex traits14, 15. Recently, a GWAS of 58 quantitative traits in Japanese individuals identified genome-wide significant associations (P<5x10−8) with EF at three genetic loci (SLC1A4, MTSS1, and DERL3)16. Another recent study conducted in the UK Biobank confirmed the implication of the MTSS1 locus in EF variation and identified an additional three genetic loci (CLCNKA, TTN, and BAG3)17. However, the mechanisms controlling EF are still poorly understood and the gene identification may help uncover the mechanisms underlying EF variation, and, consequently, related cardiac conditions.
Here, we undertake a multiethnic meta-analysis for EF combining GWAS data from the large and multiethnic Genetic Epidemiology Research in Adult Health and Aging (GERA) cohort18, 19, and the UK Biobank (UKB) cohort20–22. We then replicate the novel genome-wide significant association using summary statistics from the previously published GWAS of EF in Biobank Japan16. We also conducted sensitivity analyses adjusting for antecedent cardiac conditions, and estimated the array-heritability for EF and the variance explained by previous and newly identified loci. The different stages and datasets used for this study are summarized in an overview diagram (Figure 1).
Figure 1:

Flowchart of the Study Design
Methods
The complete methods used in this study are available in the Data Supplement. The meta-analysis GWAS summary statistics are available from the NHGRI-EBI GWAS Catalog (https://www.ebi.ac.uk/gwas/summary-statistics). In brief, 22,155 adults (18 years and older) of non-Hispanic white, Hispanic/Latino, Asian, or African-American ethnicity from the GERA cohort18, 19, who had EF measured were included in the study (Table 1). All study procedures were approved by the Institutional Review Board of the Kaiser Permanente Northern California Institutional Review Board. Written informed consent was obtained from all participants. The GERA genotype data are available upon application to the KP Research Bank (https://researchbank.kaiserpermanente.org/). The UK Biobank22 (UKB) European sample (Supplementary Table 1) included in the analysis consisted of 4,483 individuals with EF measured during a multi-modality imaging visit, which was part of the UK Biobank Cardiovascular magnetic resonance (CMR) protocol23.
Table 1.
Characteristics of the GERA subjects with EF measurements by sex and ethnic group
| Participants N (%) |
1st EF meas. (%) Mean ± SD |
Age at 1st EF (years) Mean ± SD |
||
|---|---|---|---|---|
| All | 22,155 (100.0) | 60.96 ± 10.97 | 72.61 ± 11.21 | |
| Sex | Female | 11,333 (51.2) | 63.67 ± 10.14 | 71.91 ± 11.83 |
| Male | 10,822 (48.8) | 58.11 ± 11.09 | 73.34 ± 10.48 | |
| Ethnicity | NHW | 18,670 (84.3) | 60.92 ± 10.88 | 73.26 ± 10.88 |
| H/L | 1,514 (6.8) | 60.73 ± 11.61 | 69.03 ± 12.50 | |
| EAS | 1,207 (5.5) | 62.24 ± 10.93 | 69.16 ± 12.37 | |
| AA | 764 (3.4) | 60.27 ± 11.75 | 69.33 ± 11.75 | |
Abbreviations: N, number; SD, standard deviation; meas, measurement; NHW: non-Hispanic whites; H/L: Hispanic/Latinos; EAS: East Asians; AA: African-Americans.
Results
GWAS of EF and Meta-Analysis (GERA+UKB)
In both GERA and UKB, we observed that, on average, males had lower EF values compared to females (Table 1 and Supplementary Table 1), as previously reported24; this was observed within each GERA ethnic group. Further, East Asians from GERA had higher EF values compared to the other groups. A multiethnic meta-analysis of EF, combining the GERA and UKB cohorts identified two loci that exceeded genome-wide significance: a novel locus, TMEM40 on chromosome 3 (lead intronic SNP rs11719526; β=0.47; P=3.10x10−8), and a previously reported locus, BAG3 on chromosome 10 (lead intronic SNP rs17617337; β=−0.83, P=8.24x10−17) (Figure 2; Supplementary Figure 1). Regional plots at the 2 identified EF-associated loci, TMEM40 and BAG3, are presented in Supplementary Figure 2 and include SNP association values from our meta-analysis results (GERA+UKB European samples) ± 1 Mb upstream and downstream of our lead significant SNPs.
Figure 2:
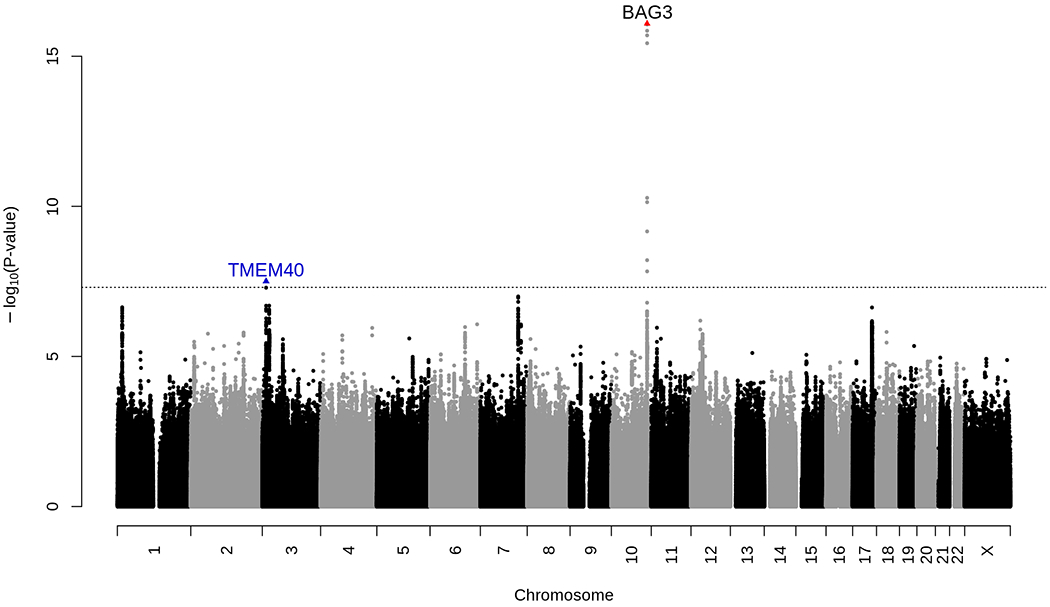
Manhattan plot of the combined (GERA+UKB European sample) meta-analysis of EF. This combined meta-analysis included 26,638 individuals from the GERA and UKB cohorts. Association results (−log10 P-values) are plotted for each chromosome.
Names of loci and lead variants that reach genome-wide level of significance are indicated as follows: previously identified BAG3 locus is in black font with red triangle indicating the lead SNP; a blue triangle indicates the lead variant at novel TMEM40 locus (in blue font) identified in our combined (GERA+UKB European sample) meta-analysis of EF.
Replication in Biobank Japan
We then tested the lead SNP rs11719526 at the novel TMEM40 locus for replication in an independent external cohort; GWAS summary statistics from the study of Kanai et al16, consisting of 19,516 Japanese participants from the Biobank Japan Project (BBJ), were publicly accessible. The association between TMEM40 rs11719526 and EF replicated at Bonferroni significance (β=0.032; P=0.016) with the same direction of effect in the replication sample, albeit with a much reduced magnitude of effect (Table 2).
Table 2.
Genome-wide significant loci identified in the GERA meta-analysis of EF or combined (GERA+UKB) meta-analysis and replication in UK Biobank (European sample) or BioBank Japan
|
BAG3 rs17617337 (chr10:121,426,884) Allele C |
TMEM40 rs11719526 (chr3:12,803,313) Allele C |
||||||
|---|---|---|---|---|---|---|---|
| Cohort | Freq | β (SE) | P | Freq | β (SE) | P | |
| GERA | NHW (n=18,670) | 0.79 | −0.82 (0.13) | 6.73x10−10 | 0.57 | 0.45 (0.12) | 1.10x10−4 |
| H/L (n=1,514) | 0.85 | −1.34 (0.56) | 0.016 | 0.67 | 0.27 (0.42) | 0.52 | |
| EAS (n=1,207) | 0.98 | −1.19 (1.58) | 0.45 | 0.81 | 0.47 (0.59) | 0.43 | |
| AA (n=764) | 0.95 | −0.11 (1.31) | 0.94 | 0.87 | −1.33 (0.92) | 0.15 | |
| Combined (n=22,155) | - | −0.84 (0.13) | 4.72x10−11 | - | 0.41 (0.11) | 1.58x10−4 | |
| UKB | EUR (n=4,483) | 0.78 | −0.81 (0.16) | 3.42x10−7 | 0.56 | 0.55 (0.13) | 3.79x10−5 |
| GERA+UKB (n=26,638) | - | −0.83 (0.10) | 8.24x10−17 | - | 0.47 (0.08) | 3.10x10−8 | |
| BioBank Japan (n=19,516) | NA | NA | NA | 0.83 | 0.032 (0.013) | 0.016 | |
Abbreviations: Freq, allele frequency; chr, chromosome; β, beta; P, P-value; NHW: non-Hispanic whites; H/L: Hispanic/Latinos; EAS: East Asians; AA: African-Americans; NA, not available as rs17617337 is rare (MAF<0.05) in Asian populations.
In GERA, linear regression of EF was adjusted for age at specimen collection, age at EF measurement, sex, data sources, and ancestry principal components (PCs)
Conditional Analysis
We next searched for additional genome-wide significant SNPs within a 2 Mb window (±1.0 Mb with respect to the lead SNP) including the 2 lead SNPs (BAG3 rs17617337 and TMEM40 rs11719526) identified in the multiethnic meta-analysis as covariates. We conducted this conditional analysis in the meta-analysis combining results from GERA and UKB European samples. We did not identify any additional genome-wide significant SNPs or SNPs with suggestive evidence of association with EF that appeared to be independent signals. Replication threshold was set up at 0.0001=0.05/500 (corresponding to an estimate of ~500 independent variants per locus for 2Mb interval surrounding each of our original signals), as previously used25.
Gene-Based Association Analysis
To identify additional genes associated with EF at a gene level we conducted a gene-based association analysis using the FUMA26 integrative tool using the GWAS meta-analysis (GERA + UKB European samples) results. FUMA implements MAGMA27 (Multi-marker Analysis of GenoMic Annotation) gene-based analysis, which employs a multiple linear regression approach to properly incorporate linkage disequilibrium (LD) between genetic variants and to detect multi-variant effects. As 19,420 genes were tested, the P-value adjusted for Bonferroni correction was set as P<2.57x10−6 (0.05/19,420). We found significant associations with EF for 8 genes, with the strongest association for BAG3, followed by CCDC136, BPTF, TTN, HSPB7, TLN1, TIAL1, and CSRP3 (Supplementary Table 2).
Replication of Previous EF GWAS Results
We also investigated in GERA the lead SNPs within 5 loci (in addition to BAG3) associated with EF at a genome-wide significance level in previous studies conducted in Biobank Japan or UKB16, 17. Here, we used a nominal significance level of 0.05, and a more stringent multiple testing correction accounting for the number of SNPs tested (Bonferroni-corrected alpha level of 0.01 (=0.05/5)). No evidence of heterogeneity was detected among GERA samples for any of the 5 SNPs tested. Three of the 5 replicated at Bonferroni significance (P<0.05/5=0.01) in our GERA multiethnic meta-analysis (i.e. rs945425 at CLCNKA, rs34866937 at MTSS1, and rs5760061 in DERL3) with consistent direction of effect (Supplementary Table 3 and Supplementary Figure 3). In addition, TNN rs2042995 which was reported as genome-wide significant in a previous GWAS of EF17, was nominally associated with EF in GERA (P=0.05) with concordant direction of effect. This is consistent with the previously reported nominal replication at this locus in the MESA cohort17. The previously reported16 rs6546120 in SLC1A4 did not replicate in our GERA meta-analysis, although it was nominally associated in the GERA East Asian sample but with inconsistent direction of effect compared to the original study.
Associations Between EF and Cardiac Conditions In GERA
Because left ventricular EF varies in association with a number of cardiac conditions3, we investigated whether certain antecedent cardiac conditions (i.e., heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation, valvular disease, or revascularization procedures) were associated with EF in GERA. To preserve statistical power, we conducted the analysis in the largest group from GERA, non-Hispanic whites. Among those with EF measurements, 25.03% had heart failure and/or cardiomyopathy, 75.06% had hypertension, 10.55% had a myocardial infarction recorded, 29.70% had atrial fibrillation, 15.5% had valvular disease (mitral or aortic) and 15.10% had had revascularization procedures (Table 3). All the cardiac antecedent conditions were significantly associated with lower EF (Table 4–Univariate Model).
Table 3.
Characteristics of the GERA non-Hispanic whites with EF measurements by antecedent cardiac conditions
| Participants N (%) |
1st EF meas. Mean ± SD |
Age at 1st EF (years) Mean ± SD |
||
|---|---|---|---|---|
| HF/cardiomyopathy | Cases | 4,674 (25.03%) | 54.42 ± 14.37 | 76.49 ± 9.83 |
| Hypertension | Cases | 14,013 (75.06%) | 60.81 ± 11.33 | 74.72 ± 10.05 |
| MI | Cases | 1,970 (10.55%) | 55.83 ± 13.10 | 74.81 ± 9.88 |
| Atrial fibrillation | Cases | 5,545 (29.70%) | 58.95 ± 11.74 | 76.40 ± 9.39 |
| Valvular disease | Cases | 2,896 (15.51%) | 58.95 ± 12.25 | 74.22 ± 10.33 |
| Revascularization procedures | Cases | 2,819 (15.10%) | 57.67 ± 12.15 | 73.00 ± 10.08 |
| None of the above | Controls | 2,834 (15.18%) | 62.96 ± 7.21 | 67.36 ± 12.19 |
| Any condition | Cases | 15,836 (84.82%) | 60.55 ± 11.38 | 74.32 ± 10.27 |
| Multiple cardiac conditions | *Cases | 8,846 (47.38%) | 58.20 ± 12.54 | 75.64 ± 9.75 |
| 2 conditions | 4,281 (48.4%) | 60.15 ± 11.43 | 74.94 ± 10.04 | |
| 3 conditions | 2,617 (29.6%) | 57.44 ± 12.78 | 76.29 ± 9.67 | |
| 4 conditions | 1,314 (14.9%) | 55.65 ± 13.59 | 76.26 ± 9.21 | |
| 5 conditions | 546 (6.2%) | 53.69 ± 13.50 | 76.36 ± 8.83 | |
| 6 conditions | 88 (0.90%) | 52.06 ± 16.23 | 75.93 ± 8.65 | |
Abbreviations: N, number; SD, standard deviation; meas, measurement; HF: heart failure; MI, myocardial infarction.
Patients having multiple cardiac conditions were defined as having at least 2 of the cardiac conditions listed in the above table.
Table 4.
Association of genetic variants and antecedent conditions with EF in GERA non-Hispanic whites
| Univariate Models‡ | BAG3 Multivariable Model§ | TMEM40 Multivariable Model‖ | ||||
|---|---|---|---|---|---|---|
| Risk Factor/condition | β (SE) | P | β (SE) | P | β (SE) | P |
| BAG3 rs17617337 | −0.82 (0.13) | 6.73x10−10 | −0.78 (0.13) | 1.53x10−9 | - | - |
| TMEM40 rs11719526 | 0.45 (0.12) | 1.10x10−4 | - | - | 0.45 (0.11) | 5.96x10−5 |
| HF/cardiomyopathy | −8.70 (0.33) | 3.88x10−150 | - | - | - | - |
| Hypertension | −1.36 (0.22) | 5.40x10−10 | - | - | - | - |
| MI | −6.43 (0.32) | 2.25x10−84 | - | - | - | - |
| Atrial fibrillation | −3.04 (0.26) | 1.45x10−30 | - | - | - | - |
| Valvular disease | −3.24 (0.29) | 2.57x10−28 | ||||
| Revascularization procedures | −3.78 (0.28) | 2.22x10−40 | - | - | - | - |
| *Any condition | −1.51(0.22) | 1.78x10−11 | - | - | - | - |
| Multiple conditions | −4.07(0.24) | 2.88x10−65 | - | - | - | - |
| None of the above conditions | 1.51(0.22) | 1.78x10−11 | - | - | - | - |
| †Total number of conditions | −2.03(0.063) | 4.21x10−222 | −2.03(0.063) | 9.76x10−222 | −2.03(0.063) | 2.51x10−222 |
Each analysis was adjusted for age at specimen collection, age at 1st EF measurement, sex, data sources, and genetic ancestry PCs.
‘Any condition’ collapses all cardiac conditions (i.e. HF/cardiomyopathy, hypertension, MI, atrial fibrillation, valvular disease, and revascularization procedures).
‘Total number of conditions’ is a variable which accounts for the number of conditions a patient had (i.e., 0 to 6 for the following conditions: heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation, valvular disease, or revascularization procedures).
The Univariate Model tests the association of each variable individually with EF;
Included BAG3 rs17617337 and total number of conditions;
Included TMEM40 rs11719526 and total number of conditions.
As these relationships may mediate the observed associations between our 2 lead SNPs (BAG3 rs17617337 and TMEM40 rs11719526) and EF, we conducted sensitivity analyses of the effects of these loci on EF adjusting for a covariate named “total number of conditions” which accounts for the number of conditions a patient had (i.e., 0 to 6 for the following conditions: heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation, valvular disease, or revascularization procedures). When we included “total number of conditions” as a covariate in the models, only the association between BAG3 rs17617337 with EF was slightly attenuated but remained significant (β=-0.78; P=1.53x10−9). The association between TMEM40 rs11719526 with EF was unchanged after including the ““total number of conditions” covariate in the model (β=0.45; P=5.96x10−5) (Table 4–Multivariable Models).
Association of the 2 EF-associated Loci with Cardiac Conditions
Because BAG3 is a well-known familial cardiomyopathy susceptibility locus28, 29, and polymorphisms at the TMEM40/CAND2 locus have been previously reported to be associated with atrial fibrillation30, 31, we investigated whether our 2 lead EF-associated SNPs (BAG3 rs17617337 and TMEM40 rs11719526) were associated with antecedent cardiac conditions. To do so, we conducted case–control analyses for heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation and revascularization procedures in GERA non-Hispanic whites (Table 3). We found a nominal association between BAG3 rs17617337 and heart failure/cardiomyopathy (OR=1.11; P=0.029); but no significant association was observed between BAG3 rs17617337 and hypertension, myocardial infarction, valvular disease or revascularization procedures. Further, TMEM40 rs11719526 was not associated with atrial fibrillation or any other antecedent cardiac conditions (Supplementary Table 4).
To confirm our findings, we then evaluated whether the 2 EF-associated SNPs identified in the current study were also associated with cardiac conditions in the UKB European sample using Gene ATLAS integrative tool32. Overall, the results were quite similar across the two cohorts with respect to associations with heart failure, myocardial infarction, and hypertension – the cardiac conditions available Gene ATLAS (Supplementary Figure 4 and Supplementary Table 4). The BAG3 rs17617337 was significantly associated with heart failure in UKB and nominally significantly associated with heart failure in GERA, with very similar effects in both cohorts and very comparable associations with EF in both cohorts.
We also examined linear associations of the 2 lead EF-associated SNPs (BAG3 rs17617337 and TMEM40 rs11719526) with EF within each cardiac condition (i.e. heart failure/cardiomyopathy, hypertension, myocardial infarction, atrial fibrillation, valvular disease, or revascularization procedures) and in the absence of these conditions in GERA non-Hispanic whites. While the association of BAG3 rs17617337 with EF showed different effect sizes according to the cardiac condition tested, and in comparison to the control group; the association of TMEM40 rs11719526 with EF showed very similar effect sizes across the different conditions (Supplementary Table 5).
Genetic correlations between EF and other phenotypes
To estimate the genetic correlation of EF phenotype with more than 700 diseases/traits, including heart attack, atrial fibrillation, and blood pressure, from different publicly available resources/consortia, we used the LD Hub web interface, which performs automated LD score regression. In the LD Score regressions, we included only HapMap3 SNPs with MAF>0.01. Genetic correlations were considered significant after Bonferroni adjustment for multiple testing (P<6.5x10−5 which corresponds to 0.05/771 phenotypes tested). We did not detect significant genetic correlations between EF and other diseases/traits after Bonferroni correction.
In Silico Analyses Provide Relevant Biological Insights
We examined associations with gene expression for each of the 2 identified lead SNPs (TMEM40 rs11719526 and BAG3 rs17617337) using the Genotype-Tissue Expression (GTEx) project33 tool (https://gtexportal.org/). Although no significant expression quantitative trait loci (eQTL) were detected for our lead BAG3 SNP or its proxies (R2>0.90), our lead TMEM40 SNP had 7 significant GTEx cis-eQTLs, which affect the expression of CAND2, KRT18P17, or TSEN2 in different tissues, including skeletal muscle (Supplementary Table 6).
FUMA26 gene-set enrichment analysis (using the MAGMA27 tool, which employs multiple regression to obtain gene-set P-values) highlighted many gene-sets involved in heart tissue development, including cardiac myofibril assembly (P=5.26x10−6), cardiac muscle cell differentiation (P=1.37x10−5), striated muscle cell differentiation (P=1.86x10−4), cardiac cell development (P=2.37x10−4), or cardiocyte differentiation (P=5.08x10−4) (Supplementary Table 7). For this analysis, the gene-set P-value is computed using the gene-based P-value for 4,728 curated gene sets (including canonical pathways) and 6,166 GO terms obtained from MsigDB v5.2, and a false discovery rate (FDR) was used to correct for multiple testing.
FUMA tissue eQTL specificity analysis highlighted the heart atrial appendage as the main tissue for which expression was affected by EF-associated variants (P <9.43x10−4; Bonferroni significance after correcting for 53 GTEx tissues tested - Supplementary Figure 5).
Heritability Estimate and Variance Explained
We estimated array heritability to be 11.12% (SE=3.53%) in the GERA non-Hispanic white sample using a LD Score regression method34. We then determined the proportion of variance explained by lead variants (2 identified in the current study and 5 previously reported) in the GERA non-Hispanic white sample and found an estimate of 0.46%. The limited proportion of variance explained by those loci suggests that additional loci influencing EF variation remain to be discovered, and future studies investigating the genetic complexity of EF would require extremely large sample sizes.
Discussion
In this study, a large meta-analysis combining GERA and UKB cohorts identified a novel locus, TMEM40, that was validated in an external replication cohort, Biobank Japan, albeit with a much more modest effect. This combined meta-analysis also confirmed the strong implication of BAG3 in EF variation, as recently reported17. Our results also confirmed the association of variants with EF in 3 loci (i.e. CLCNKA, MTSS1, and DERL3) previously reported to be associated with EF in recent studies conducted in Biobank Japan or UK Biobank16, 17. Finally, we provided for the first time a modest but statistically significant heritability estimate of EF based on genome-wide array data of unrelated individuals.
Our study led to the identification of a new region on chromosome 3 associated with EF at TMEM40 locus. TMEM40 encodes the transmembrane protein 40, a protein of 233 amino acids about which little is known35. Common variants in the CAND2 gene (within 1Mb of TMEM40 locus) have been previously reported to be associated with atrial fibrillation at a genome-wide level of significance30, 31, 36. The current study demonstrates that the association signal at TMEM40 was not attenuated after adjusting for antecedent cardiac conditions including atrial fibrillation, suggesting that variation at TMEM40 may influence EF levels through different mechanisms.
Our findings are consistent with a recent study17 reporting genome-wide association at the BAG3 locus with EF, although the strongest SNPs reported by Aung et al. differed from ours. However, the lead SNP (rs17617337) at BAG3 locus from the current study was relatively close to the lead SNP (rs72840788) from the recent Aung and colleagues’ study17 (11.2 kb apart) and were strongly correlated in European-ancestry populations (r2=0.99 and D’=1.0), suggesting that they represent the same signal. Our findings also demonstrate that common variation at the BAG3 locus remains significantly associated with EF after adjusting for cardiac conditions, including heart failure/cardiomyopathy. These findings extend a recent study of heart failure, which identified BAG3 as a genome-wide significant locus for nonischemic cardiomyopathy and reported a suggestive association between BAG3 rs2234962 and reduced left ventricular EF in UKB individuals without clinical heart failure29.
In addition to identifying 2 loci that exceeded genome-wide significance with EF (TMEM40 and BAG3), our findings also highlighted 7 additional genes (CCDC136, BPTF, TTN, HSPB7, TLN1, TIAL1, and CSRP3) associated with EF at a gene-level in European population samples from GERA and UKB. Early work demonstrated the contribution of some of these genes to cardiac conditions. TTN encodes a large abundant protein of striated muscle, and mutations in this gene are associated with familial hypertrophic cardiomyopathy37. Further, TTN locus has been recently reported to exceed genome-wide significance with EF as well as left ventricular end-diastolic volume17. HSPB7 encodes the heat shock protein family B (small) member 7 and common variants in this gene are associated with advanced heart failure38. TLN1 encodes a cytoskeletal protein named talin-1, and in mice models, Tln1 plays an important role in preserving heart function, and loss of Tln1 from the heart-muscle cell leads to myocyte instability and a dilated cardiomyopathy39. CSRP3 encodes the cysteine and glycine rich protein 3, and mutations in this gene can cause heritable forms of hypertrophic cardiomyopathy and dilated cardiomyopathy in humans40–42. The multiple associations of these genes with cardiomyopathies and advanced heart failure highlight the importance of understanding genetic influences on EF. Future investigations may provide insights into whether these genes influence EF in conditions other than cardiomyopathies and heart failure.
The main strengths of this study include the large samples with EF measures, enabled in the GERA cohort by extraction of routine clinical measurement of EF from EHRs. In addition, the EF measures are based on different methods in different cohorts and were obtained on unselected groups of patients with a variety of clinical indications and antecedent conditions. The combination of large, clinically diverse samples and different examination methods contribute to the robustness of the study findings, enabling the identification of risk loci contributing to the variation of EF across varying populations and antecedent conditions. We showed that combining GWAS data across large samples with a combination of methods for measuring EF increases the power of gene mapping for this strong correlate of cardiac outcomes. However, we found that previously and newly identified loci explained only 0.46% of the variance of this complex trait. Future GWAS with increasing sample sizes and including individuals from diverse ethnic backgrounds should enable the discovery of additional loci, explaining a much greater proportion of EF variance.
In conclusion, in this large and multiethnic study, we identified two loci, TMEM40 and BAG3, associated with EF at a genome-wide significance level. Moreover, our gene-based association analyses confirmed BAG3 and highlighted 3 other genes (CCDC136, TTN, and TLN1) associated with EF at a gene-level in European ancestry populations. Our results also extend previous findings implicating MTSS1 and DERL3 loci in EF variation. Because of its clinical value in diagnosis, prognosis, and prediction of clinical outcomes of cardiac conditions, identifying and understanding the genetic architecture of EF remains essential for understanding the pathophysiology underlying variation of this strong correlate of cardiac outcomes and the development of future therapies.
Supplementary Material
Acknowledgments:
We thank the Kaiser Permanente Northern California members who have generously agreed to participate in the Kaiser Permanente Research Program on Genes, Environment, and Health.
Sources of Funding: This study is based on data and samples collected as part of the Research Program on Genes, Environment and Health, supported by the Wayne and Gladys Valley Foundation, The Ellison Medical Foundation, Robert Wood Johnson Foundation, and Kaiser Permanente Community Benefit Programs. Genotyping of samples was supported by a grant from the National Institutes of Health (NIA, NIMH, and NIH Common Fund) RC2 AG036607. Analyses for this study were also supported by funding from GlaxoSmithKline.
Nonstandard Abbreviations and Acronyms
- BAG3
BAG cochaperone 3
- BBJ
Biobank Japan Project
- CMR
Cardiovascular magnetic resonance protocol
- EF
Ejection fraction
- EHR
Electronic health records
- eQTL
expression quantitative trait loci
- FUMA
Functional mapping and annotation of genetic associations
- GERA
Genetic Epidemiology Research on Adult Health and Aging cohort
- GTEx
Genotype-Tissue Expression project
- GWAS
Genome-wide association study
- KPNC
Kaiser Permanente Medical Care Plan, Northern California Region
- LD
Linkage disequilibrium
- MAF
Minor allele frequency
- MAGMA
Multi-marker Analysis of GenoMic Annotation
- SNP
Single nucleotide polymorphism
- TMEM40
Transmembrane protein 40
- UKB
UK Biobank cohort
Footnotes
Disclosures: None
References:
- 1.Kennedy JW, Baxley WA, Figley MM, Dodge HT, Blackmon JR. Quantitative angiocardiography. I. The normal left ventricle in man. Circulation. 1966;34:272–8. [DOI] [PubMed] [Google Scholar]
- 2.Sonnenblick EH. Correlation of myocardial ultrastructure and function. Circulation. 1968;38:29–44. [DOI] [PubMed] [Google Scholar]
- 3.Bristow MR, Kao DP, Breathett KK, Altman NL, Gorcsan J 3rd, Gill EA, Lowes BD, Gilbert EM, Quaife RA,Mann DL. Structural and Functional Phenotyping of the Failing Heart: Is the Left Ventricular Ejection Fraction Obsolete? JACC Heart Fail. 2017;5:772–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konstam MA, Abboud FM. Ejection Fraction: Misunderstood and Overrated (Changing the Paradigm in Categorizing Heart Failure). Circulation. 2017;135:717–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bielen E, Fagard R, Amery A. The inheritance of left ventricular structure and function assessed by imaging and Doppler echocardiography. Am Heart J. 1991;121:1743–9. [DOI] [PubMed] [Google Scholar]
- 6.Swan L, Birnie DH, Padmanabhan S, Inglis G, Connell JM, Hillis WS. The genetic determination of left ventricular mass in healthy adults. Eur Heart J. 2003;24:577–82. [DOI] [PubMed] [Google Scholar]
- 7.Noh HM, Lee SC, Park SW, Sung J, Song YM. Genetic influence on left ventricular structure and function: a Korean twin and family study. Twin Res Hum Genet. 2015;18:281–9. [DOI] [PubMed] [Google Scholar]
- 8.Jin Y, Kuznetsova T, Bochud M, Richart T, Thijs L, Cusi D, Fagard R, Staessen JA. Heritability of left ventricular structure and function in Caucasian families. Eur J Echocardiogr. 2011;12:326–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fox ER, Klos KL, Penman AD, Blair GJ, Blossom BD, Arnett D, Devereux RB, Samdarshi T, Boerwinkle E, Mosley TH Jr. Heritability and genetic linkage of left ventricular mass, systolic and diastolic function in hypertensive African Americans (From the GENOA Study). Am J Hypertens. 2010;23:870–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kovacs A, Molnar AA, Kolossvary M, Szilveszter B, Panajotu A, Lakatos BK, Littvay L, Tarnoki AD, Tarnoki DL, Voros S, et al. Genetically determined pattern of left ventricular function in normal and hypertensive hearts. J Clin Hypertens (Greenwich). 2018;20:949–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A. 2012;109:1193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zaitlen N, Pasaniuc B, Sankararaman S, Bhatia G, Zhang J, Gusev A, Young T, Tandon A, Pollack S, Vilhjalmsson BJ, et al. Leveraging population admixture to characterize the heritability of complex traits. Nat Genet. 2014;46:1356–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayhew AJ, Meyre D. Assessing the Heritability of Complex Traits in Humans: Methodological Challenges and Opportunities. Curr Genomics. 2017;18:332–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tam V, Patel N, Turcotte M, Bosse Y, Pare G, Meyre D. Benefits and limitations of genome-wide association studies. Nat Rev Genet. 2019. [DOI] [PubMed] [Google Scholar]
- 15.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 2017;101:5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kanai M, Akiyama M, Takahashi A, Matoba N, Momozawa Y, Ikeda M, Iwata N, Ikegawa S, Hirata M, Matsuda K, et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat Genet. 2018;50:390–400. [DOI] [PubMed] [Google Scholar]
- 17.Aung N, Vargas JD, Yang C, Cabrera CP, Warren HR, Fung K, Tzanis E, Barnes MR, Rotter JI, Taylor KD, et al. Genome-Wide Analysis of Left Ventricular Image-Derived Phenotypes Identifies Fourteen Loci Associated With Cardiac Morphogenesis and Heart Failure Development. Circulation. 2019;140:1318–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banda Y, Kvale MN, Hoffmann TJ, Hesselson SE, Ranatunga D, Tang H, Sabatti C, Croen LA, Dispensa BP, Henderson M, et al. Characterizing Race/Ethnicity and Genetic Ancestry for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kvale MN, Hesselson S, Hoffmann TJ, Cao Y, Chan D, Connell S, Croen LA, Dispensa BP, Eshragh J, Finn A, et al. Genotyping Informatics and Quality Control for 100,000 Subjects in the Genetic Epidemiology Research on Adult Health and Aging (GERA) Cohort. Genetics. 2015;200:1051–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Allen NE, Sudlow C, Peakman T, Collins R, Biobank UK. UK biobank data: come and get it. Sci Transl Med. 2014;6:224ed4. [DOI] [PubMed] [Google Scholar]
- 21.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, Motyer A, Vukcevic D, Delaneau O, O’Connell J, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:e1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Petersen SE, Matthews PM, Bamberg F, Bluemke DA, Francis JM, Friedrich MG, Leeson P, Nagel E, Plein S, Rademakers FE, et al. Imaging in population science: cardiovascular magnetic resonance in 100,000 participants of UK Biobank - rationale, challenges and approaches. J Cardiovasc Magn Reson. 2013;15:46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung AK, Das SR, Leonard D, Peshock RM, Kazi F, Abdullah SM, Canham RM, Levine BD, Drazner MH. Women have higher left ventricular ejection fractions than men independent of differences in left ventricular volume: the Dallas Heart Study. Circulation. 2006;113:1597–604. [DOI] [PubMed] [Google Scholar]
- 25.Fritsche LG, Chen W, Schu M, Yaspan BL, Yu Y, Thorleifsson G, Zack DJ, Arakawa S, Cipriani V, Ripke S, et al. Seven new loci associated with age-related macular degeneration. Nat Genet. 2013;45:433–9, 439e1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Leeuw CA, Mooij JM, Heskes T, Posthuma D. MAGMA: generalized gene-set analysis of GWAS data. PLoS Comput Biol. 2015;11:e1004219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Villard E, Perret C, Gary F, Proust C, Dilanian G, Hengstenberg C, Ruppert V, Arbustini E, Wichter T, Germain M, et al. A genome-wide association study identifies two loci associated with heart failure due to dilated cardiomyopathy. Eur Heart J. 2011;32:1065–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aragam KG, Chaffin M, Levinson RT, McDermott G, Choi SH, Shoemaker MB, Haas ME, Weng LC, Lindsay ME, Smith JG, et al. Phenotypic Refinement of Heart Failure in a National Biobank Facilitates Genetic Discovery. Circulation. 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, et al. Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet. 2018;50:1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sinner MF, Tucker NR, Lunetta KL, Ozaki K, Smith JG, Trompet S, Bis JC, Lin H, Chung MK, Nielsen JB, et al. Integrating genetic, transcriptional, and functional analyses to identify 5 novel genes for atrial fibrillation. Circulation. 2014;130:1225–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Consortium GT. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bulik-Sullivan BK, Loh PR, Finucane HK, Ripke S, Yang J, Schizophrenia Working Group of the Psychiatric Genomics C, Patterson N, Daly MJ, Price AL, Neale BM. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Q, Huang D, Zhang Z, Feng Y, Fu M, Wei M, Zhou J, Huang Y, Liu S, Shi R. High expression of TMEM40 contributes to progressive features of tongue squamous cell carcinoma. Oncol Rep. 2019;41:154–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, et al. Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet. 2018;50:1225–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andersen PS, Havndrup O, Hougs L, Sorensen KM, Jensen M, Larsen LA, Hedley P, Thomsen AR, Moolman-Smook J, Christiansen M, et al. Diagnostic yield, interpretation, and clinical utility of mutation screening of sarcomere encoding genes in Danish hypertrophic cardiomyopathy patients and relatives. Hum Mutat. 2009;30:363–70. [DOI] [PubMed] [Google Scholar]
- 38.Cappola TP, Li M, He J, Ky B, Gilmore J, Qu L, Keating B, Reilly M, Kim CE, Glessner J, et al. Common variants in HSPB7 and FRMD4B associated with advanced heart failure. Circ Cardiovasc Genet. 2010;3:147–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Manso AM, Okada H, Sakamoto FM, Moreno E, Monkley SJ, Li R, Critchley DR, Ross RS. Loss of mouse cardiomyocyte talin-1 and talin-2 leads to beta-1 integrin reduction, costameric instability, and dilated cardiomyopathy. Proc Natl Acad Sci U S A. 2017;114:E6250–E6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Janin A, Bessiere F, Chauveau S, Chevalier P, Millat G. First identification of homozygous truncating CSRP3 variants in two unrelated cases with hypertrophic cardiomyopathy. Gene. 2018;676:110–116. [DOI] [PubMed] [Google Scholar]
- 41.Hershberger RE, Parks SB, Kushner JD, Li D, Ludwigsen S, Jakobs P, Nauman D, Burgess D, Partain J, Litt M. Coding sequence mutations identified in MYH7, TNNT2, SCN5A, CSRP3, LBD3, and TCAP from 313 patients with familial or idiopathic dilated cardiomyopathy. Clin Transl Sci. 2008;1:21–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Geier C, Gehmlich K, Ehler E, Hassfeld S, Perrot A, Hayess K, Cardim N, Wenzel K, Erdmann B, Krackhardt F, et al. Beyond the sarcomere: CSRP3 mutations cause hypertrophic cardiomyopathy. Hum Mol Genet. 2008;17:2753–65. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


