Abstract
Nucleoside derivatives, in particular those featuring uridine, are familiar components of the nucleoside family of bioactive natural products. The structural complexity and biological activities of these compounds have inspired research from organic chemistry and chemical biology communities seeking to develop novel approaches to assemble the challenging molecular targets, to gain inspiration for enzyme inhibitor development and to fuel antibiotic discovery efforts. This review will present recent case studies describing the total synthesis and biosynthesis of uridine natural products, and de novo synthetic efforts exploiting features of the natural products to produce simplified scaffolds. This research has culminated in the development of complementary strategies that can lead to effective uridine-based inhibitors and antibiotics. The strengths and challenges of the juxtaposing methods will be illustrated by examining select uridine natural products. Moreover, structure-activity relationships (SAR) for each natural product-inspired scaffold will be discussed, highlighting the impact on inhibitor development, with the aim of future uridine-based small molecule expansion.
Keywords: nucleoside analog, uridine, natural product antibiotics, inhibitor discovery, structure-activity relationship
Graphical Abstract
1. Introduction
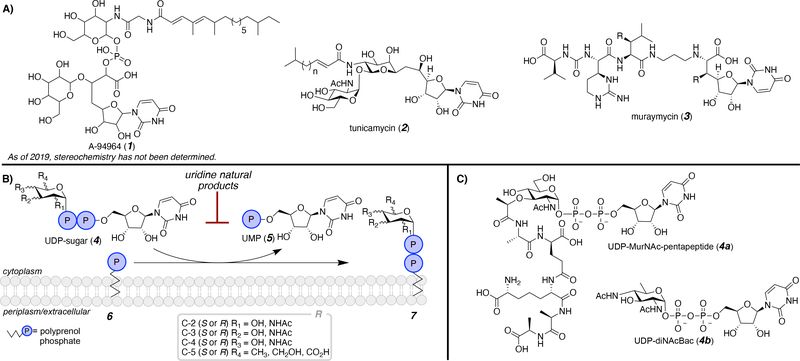
Uridine consists of uracil, a member of the pyrimidine family of nitrogenous bases, and a ribose. Of all the characterized and isolated pyrimidine natural products and natural product-derived compounds, uridine is the most abundant.1 Additionally, uridine appears to be a privileged scaffold for many purposes due to favorable cellular uptake. Specifically, uridine natural products have shown antibiotic activity by targeting intracellular processes, such as glycan assembly (Figure 1). These representative natural products have diverse structures with a common uridine core. A-94964 (1) is a natural product featuring a phosphodiester linking monosaccharides, a long chain acyl tail, and a uridinyl moiety.2,3 The stereochemistry has not yet been defined, making it an important target for future studies. In addition to a uridinyl moiety, tunicamycin (2) consists of a pseudodisaccharide with a long chain acyl tail.4 Tunicamycin has been the focus of many reviews and the scaffold has been extensively targeted as a model for inhibitor development. Recently, total chemical and biochemical syntheses of tunicamycin have been highlighted.5,6 Unlike A-94964 and tunicamycin, muraymycin (3) lacks a long chain acyl moiety and instead contains a pseudo peptide.7,8 Significantly, the aforementioned natural products are just a selection of scaffolds that are currently available for inhibitor development and new uridine-containing compounds are continually being isolated, producing new targets for future studies.9
Figure 1.
A) Representative uridine-based natural product, noting that the stereochemistry of A-94964 (1) has not yet been determined; B) the first membrane-committed step of complex glycan assembly resulting in transfer of a phospho-sugar from a UDP-sugar to a membrane-bound polyprenol phosphate; and C) select uridine diphosphate (UDP)-linked glycan donors.
Some uridine-based antibiotics show inhibition of bacterial cell well peptidoglycan biosynthesis.10 For example, muraymycin has been extensively studied due to inhibition of the E. coli phospho-MurNAc-peptapeptide translocase, an integral membrane protein, that catalyzes the first step in the peptidoglycan assembly or the peptidoglycan assembly pathway (Figure 1B).11, 12 It is not surprising that uridine-based natural products inhibit this process as uridine diphosphate (UDP)-linked substrates are involved in the construction of the cell wall. Two examples of UDP-sugar donors (4) involved in this type of transformation are illustrated in Figure 1C, UDP-MurNAc-pentapeptide (4a, Park’s nucleotide) and UDP-diNAc-bacillosamine (4b, UDP-diNAcBac). These UDP-sugars (4) donate a glycosyl-phosphate to a membrane-bound polyprenol phosphate (6), producing uridine monophosphate (UMP, 5) and the glycan-linked polyprenol diphosphate (7). In addition to uridine natural products, uridine nucleotide derivatives have therapeutic applications.13 Although nucleotides are not within the purview of this review, major synthetic efforts have focused on these compounds due to their prominence as antiviral agents.14
Isolation of these valuable uridine-linked natural products at native levels from natural sources can pose a challenge for production, and often only small quantities are isolated.15 Furthermore, spectroscopic methods do not always provide unambiguous structural characterization, which makes total chemical synthesis the only viable avenue.16–19 However, total synthesis of such complex natural products is labor-intensive and, similar to isolation from natural sources, typically only produces small quantities.
This review will discuss selected initiatives towards 1) total chemical synthesis, 2) biosynthesis, and 3) de novo synthesis, culminating in the development of complementary strategies that can lead to effective uridine-based inhibitors and antibiotics. The contrasting approaches will be compared to illustrate strengths and challenges of each approach through examination of select natural products. Furthermore, inhibitor development for each natural product will be discussed, probing structure-activity relationships (SAR), with the goal of inspiring future opportunities for uridine-based small molecule elaboration.
2. Total chemical synthesis
Total chemical synthesis, involving the chemical construction of natural products from simple building blocks, contributes to methodology development, positively impacting the field of synthetic organic chemistry as a whole.20 Additionally, total synthesis aids in the unambiguous determination of structure and absolute stereochemistry.15 A recent synthesis of a macrocyclic peptide exemplifies the role of total synthesis in determining natural product stereochemistry.21, 22 However, limited overall yields from multistep syntheses often only provides small quantities of the natural product,23 and the optimization of regio- and stereoselective transformations requires specialized expertise. Critically, extensive SAR studies are challenging to develop for natural products using total synthesis due to difficulty of modifying the scaffold.15 Furthermore, if a compound exhibits poor bioavailability, chemical modifications must be made to improve uptake and efficacy, which may ultimately be more readily achieved through de novo synthesis of natural product mimetics with simpler structures.24, 25 Here, a synthetic achievement providing access to a uridine natural product from the caprazamycin family, caprazamycin A, will be highlighted, followed by an illustration of the utilization of semisynthesis to access a panel of caprazamycin-based inhibitors.
2.1. Caprazamycin
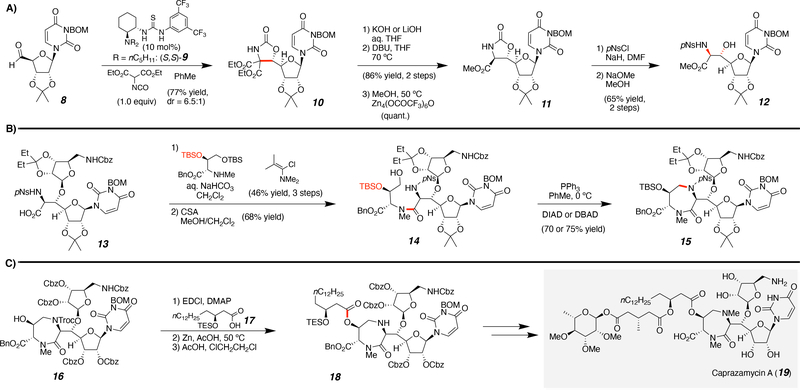
Caprazamycin was first isolated from fermentation of Streptomyces sp. MK730–6272 by Igarashi and co-workers in 2003.26 This natural product exhibits antibacterial activity against Mycobacterium tuberculosis by inhibition of MraY (translocase I), a key enzyme in peptidoglycan biosynthesis. The gene cluster encoding the enzymes for the biosynthesis of caprazamycin, represented the first gene cluster of a translocase I inhibitor to be identified and cloned. Prior to the synthesis presented, attempts had been made to access liponucleoside natural products and researchers successfully accessed the core of caprazamycin.27–29 However, until recently, the final complex compound of a caprazamycin family member, caprazamycin A (19, Scheme 1), including the unstable long chain acyl tail modification, had proven synthetically inaccessible. The first total synthesis of (−)-caprazamycin A was performed by Takemoto and co-workers in 2015,30 and the synthesis was further developed in 2019.31
Scheme 1.
Highlighting key transformations from the total synthesis of caprazamycin A. Specifically, the A) installation of the syn-b-hydroxyamino acid, B) formation of the 1,4-diazepanone core, and C) introduction of the fatty acid side chain.
2.1.1. A recent total chemical synthesis of a complex uridine natural product
Challenges from previous efforts towards caprazamycin natural products (19) involved construction of the syn-b-hydroxyamino acid moiety, formation of the 1,4-diazepanone core, and the introduction of the fatty acid side chain (Scheme 1). Therefore, these synthetic achievements within the total synthesis will be highlighted. Within this approach, the fatty acyl side chain was introduced later in the synthesis due to its instability under acidic and basic conditions; presumably due to the presence of the β-acyloxyester and acyloxy acetal functional groups. This structural feature emphasizes the downside of some natural products and is hypothesized to pose a metabolic liability,32, 33 which can be avoided through redesign to more structurally robust targets.34
Starting from the aldehyde-protected uridine (8), an organocatalyzed, diastereoselective aldol was performed with chiral-catalyst 9 to yield the oxooxazolidine 10 (Scheme 1). Then selective hydrolysis, decarboxylation, and transesterification produced the methyl ester 11. The oxazolidinone (11) underwent a ring opening via hydrolysis to produce the syn-β-hydroxyamino acid 12. The next challenge in the total synthesis was to form the 1,4-diazepanone core. After several steps, including a β-selective glycosylation, the carboxylic acid 13 was produced, which was coupled to the pseudo-amino acid. A selective tert-butyldimethylsilyl (TBS) deprotection poised 14 for cyclization via an intramolecular Mitsunobu reaction (15). The final obstacle was the introduction of the fatty acid side chain. After formation of the ring and protecting group manipulations, the long chain acyl tail 17 was attached to 16 via an esterification followed by trichloroethoxycarbonyl (Troc) removal to afford amine 18. Alkylation of the diazepanone secondary amine, Yamaguchi esterification, and global deprotection yielded the natural product caprazamycin A (19). This total synthesis highlights the complexity of these nucleoside natural products. Although these molecules can only be accessed through lengthy and challenging total chemical syntheses, the scaffolds can certainly be utilized as blueprints for future inhibitor design and development.
2.1.2. Semi-synthesis
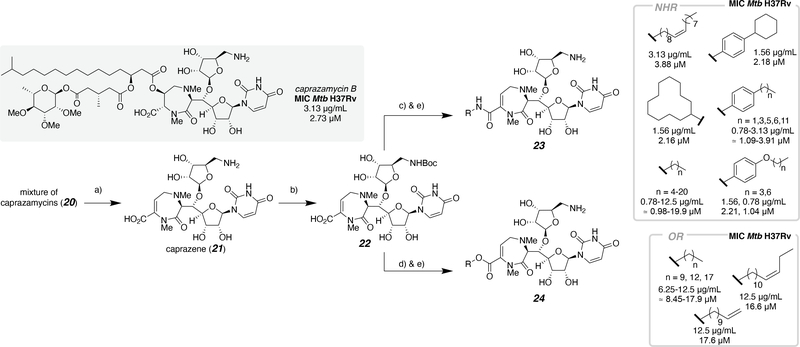
Harnessing a combination of bioprocesses and chemical syntheses to access complex small molecules is defined as semi-synthesis. Prior to the total chemical synthesis of caprazamycin A, a mixture of caprazamycins (Scheme 2, 20) from fermentation were exploited for inhibitor development.29,35 In these efforts, the caprazamycins were treated with aqueous acetic acid, to afford caprazene (21), a core scaffold amenable to diversification by chemical synthetic methods (Scheme 2).36 The free amine was Boc-protected (22) and the carboxylic acid was coupled with either an amine or alcohol to produce a series of amides (23) and esters (24), respectively. The compounds were tested for antibacterial activity after global deprotection with trifluoroacetic acid. The derivatives were classified as alkylamides, anilides, and esters, which allowed for SAR studies of caprazamycin-derived compounds. Only the M. tuberculosis (Mtb) H37Rv antibacterial activity is shown in Scheme 2, because the inhibitors exhibited the most promising antibacterial activity with this strain; however, one additional strain of Mtb and seven other bacterial species were treated with each inhibitor. For the alkylamides, a significant increase in antibacterial activity was observed when the length of the lipidic tail was larger than six carbons (MIC: 12.5 → 3.13 μg/mL; ≈ 19.9 → 4.89 μM) and further enhancement of antibacterial activity was observed when the length ranged between 9 and 21 carbons (MIC: 0.78–1.56 μg/mL; ≈ 0.98–2.03 μM), with C11–15 representing the optimal range. Overall anilides had excellent antibacterial activity, however activity decreased with the presence of an alkoxy substituent, comparing para-substituted anisole and phenyl inhibitors. Interestingly, derivatives 4-butylanilide, 4-cyclohexylanilide, and 4-butoxyanilide, all with MIC values of 1.56 μg/mL (≈ 2.27 μM) were more potent than caprazamycin B (3.13 μg/mL; ≈ 2.73 μM) with antimycobacterial activity in vitro against M. tuberculosis H37Rv. Lastly, carboxyl derivatives were tested and compared to the free carboxylic acid, which displayed no antibacterial activity (deprotected 22). When a lipophilic side chain was installed, recovery of antibacterial activity was observed. Overall, carboxyl derivatives demonstrated inferior activity compared to amine/amide derivatives. Given the results of the structure-based investigation, the authors hypothesize that the amide proton adjacent to the carbonyl group is required for antibacterial activity.
Scheme 2.
Structure-activity relationship of the caprazamycin core with selected antimicrobial activity against Mtb H37Rv. Conditions: a) 80% AcOH/H2O; b) Boc2O, triethylamine, 1,4-dioxane(aq); c) R-NH2, bis(2-oxo-3-oxazolidinyl)phosphinic chloride (BOP-Cl), triethylamine, THF, or R-NH2, 4-(4,6-dimethoxy-1,3,5-triazin-2-yl)-4-methylmorpholinium chloride (DMT-MM), 2-propanol(aq); d) R-OH, BOP-Cl, pyridine; e) trifluoroacetic acid, MeOH.
3. Biosynthesis
In a second strategic approach, uridine analogs can be accessed via biosynthesis using natural product biosynthetic enzymes. Biosynthetic pathways for uridine natural products have been extensively studied,1 and an in-depth review of biosynthetic strategies to access nucleoside antibiotics was recently published by Shiraishi and Kuzuyama.37 Enzymes for converting simple building blocks into complex natural products have evolved in nature, and some transformations that cannot be readily achieved via chemical synthesis can be accomplished efficiently by bioprocess steps.
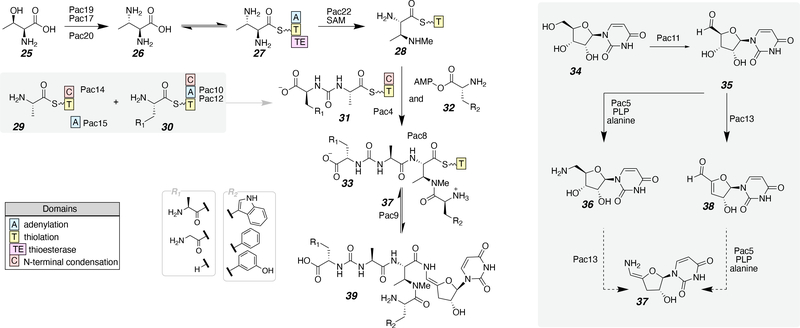
The identification of the biosynthetic gene clusters involved in the production of pacidamycin (39), a uridine-peptide natural product, was concurrently investigated by two research groups (Scheme 3).38–40 Pacidamycin and similar compounds include an N-methyl-diaminobutyric acid (DABA) within the peptide fragment. The N-terminal amine of DABA is connected to a pseudo dipeptide consisting of alanine and a urea-linked amino acid, a site of variability. The pathway starts with L-threonine (25), which is converted to (2S,3S)-2,3-diaminobutanoic acid (26) by three enzymes: Pac19, a bidomain protein including pyridoxal phosphate (PLP)-dependent DABA synthase and ATP-grasp domains, Pac17, a putative argininosuccinate lyase, and Pac20, a threonine aldolase (Scheme 3). The acid 26 is in equilibrium with the corresponding thioester 27 covalently attached to the thiolation domain (T domain) of Pac22, a putative S-adenosyl methionine (SAM)-dependent methyltransferase during non-ribosomal peptide synthesis (NRPS)-mediated assembly. This enzyme is responsible for N-methylation of the side chain of DABA (28). Next the pseudo dipeptide is made with L-Ala on the T domain of Pac14 (29) and an aromatic amino acid on the T domain of Pac12 (30). The condensation domain of Pac12 is hypothesized to be sufficient to catalyze the acetylation with sodium bicarbonate, the carbonyl source of the carbamide linkage, to produce 31.40, 41 Then transthioesterification occurs and Pac4 catalyzes the amide bond formation of 28 and 31 with 32 to produce the final peptide fragment 33. The uridine modifications are initiated by oxidation of uridine (34) with Pac11 to the aldehyde (35). There are two possible mechanistic pathways to the dehydrated enamine uridine derivative (37) from 35. The first mechanism involves a reductive amination to the amine (36) catalyzed by Pac5 in the presence of PLP and alanine followed by dehydration and tautomerization (37). The second possible mechanism consists of a dehydration of the ribose catalyzed by Pac13 (38) followed by Pac5 installing an amine and tautomerization to the enamine (37). After the modified uridine is formed (37), the thioester peptide undergoes a coupling catalyzed by Pac9 with 33. All analogs of pacidamycin are shown with R1 and R2 representing alkyl and aromatic modifications, respectively. For their work on precursor-directed biosynthesis, Goss and co-workers isolated pacidamycin tryptophan analogs from cultures of Streptomyces coeruleorubidus in yields ranging from 2–5 mg/L. Additional biosynthetic investigations of uridine natural products, caprazamycin (20) and A-94964 (1), have been recently studied.42, 43
Scheme 3.
The proposed biosynthetic pathway for pacidamycin.
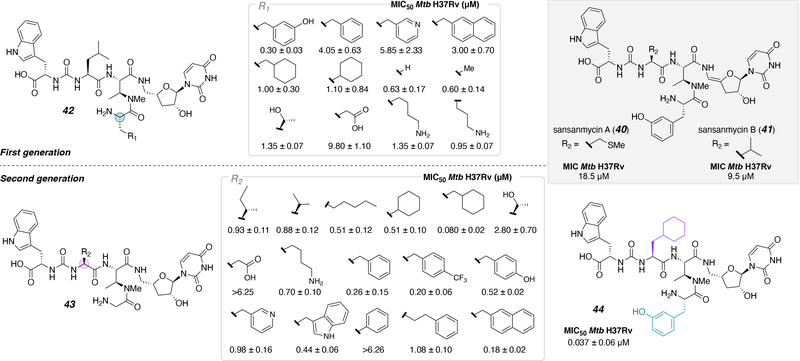
Both pacidamycin and sansanmycin, a structurally similar antibiotic to pacidamycin, are assembled by NRPS and a re-examination of the sansanmycin biosynthetic gene cluster also led to the discovery of another analog.44 Prior to the study, analogs had been made through biosynthesis, semi-synthesis, and chemical synthesis approaches.34, 45–48 However, to more effectively assess SAR Payne and co-workers synthesized a library of sansanmycin analogs (A, 40 and B, 41) with antibacterial activity against M. tuberculosis (Figure 2). Previous studies demonstrated that the enamide linkage was not necessary for activity and, due to its lability,48 was eliminated for this work. First (42) and second (43) generation libraries were synthesized using a partial solid phase synthetic approach. The peptide fragment was made on 2-chlorotrityl chloride (CTC) resin and, after cleavage from the resin, subsequently coupled to the 4’-deoxy-5’-aminouridine in solution. After global deprotection, the molecules were tested and the MIC50 against Mtb H37Rv for each modification was determined. The difference between the first and second sets are the substitutions at R1 and R2 of the peptide fragment. The two series of inhibitors established that 1) the substitution of meta-tyrosine, found naturally in sansanmycin, on DABA and 2) cyclohexyl-Ala of the linear pseudo peptide, were the most promising substitutions, displaying the most potent antibacterial activity. The culminating results of the aforementioned studies was implemented into the design of a new inhibitor to test additive effects. This strategy produced the most potent inhibitor (44), which displayed an MIC50 of 37 nM against Mtb.
Figure 2.
First and second-generation inhibitors based on the scaffold of sansanmycin.
4. De novo synthesis
Since 2010, over half of approved drugs have been inspired by natural products.49–52 Strategically, the de novo approach produces simplified structures, making the chemical synthesis of target small molecule inhibitors more straightforward. Additionally, some natural products have undesired side effects and it is easier to control structural modifications to minimize these properties through de novo synthesis. For example, salicin, a natural product used for pain treatment, exhibited undesired side effects, which led to the chemical synthesis of aspirin, an analog of salicin.20 Aspirin exhibited the same desired outcomes for pain treatment without the undesired side effects. This is an example of function-oriented synthesis (FOS), where comparable activity to a natural product can be achieved, further emphasizing that natural products can be utilized as a blueprint to probe SAR.
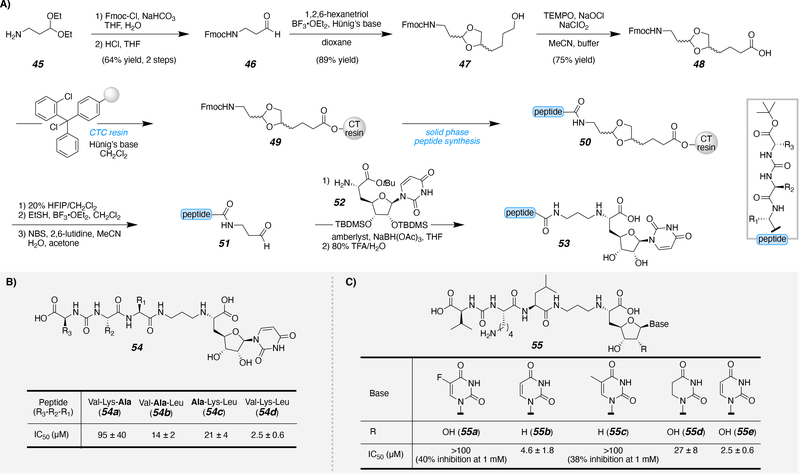
Solution-phase synthesis of uridine-based inhibitors was previously performed; however, the study was limited by technical issues with the synthesis and purification of the various inhibitors.53 In general, switching to a solid phase synthesis approach is advantageous for multiple reasons. The solid support allows for reagents to be easily rinsed away after each manipulation while the product of interest remains on the resin, reducing technical issues and improving yields. Meanwhile, it promotes a modular approach to analog or library development; one resin can act as the basis for an entire inhibitor library for SAR investigations. Attachments of uridine to solid supports have been previously explored. A series of linkages through the nitrogenous base have been studied. One approach attached a hydroxyalkyl linker to uracil of uridine via a Mitsunobu reaction.54 Specifically, the amide of uracil displaces the hydroxyl linkage of the solid support. An additional nitrogenous base-linked approach attached a thiol-tethered solid support to the C2-carbonyl of uracil through a carbamimidothioate linkage.55 In addition to base attachments, uridine can be attached to the solid support through the 2’ and 3’ hydroxyl groups of the ribose moiety via an acetal or hydroxysilane linkage.56–59 An additional advantage of switching to a solid phase system is that a key intermediate can be synthesized on resin in a single large batch and diversified with ease. Recently, Ducho and co-workers have been developing muraymycin (3)-based inhibitors for SAR investigation with MraY.60, 61 They developed a solid phase approach to access the peptidic component on resin (Scheme 4A). The key functionalized linker 48, ultimately serving to join the peptidic and nucleoside components, was obtained from the amine 45 after N-fluorenylmethyloxycarbonyl (N-Fmoc) protection, acetal deprotection (46), acetal protection (47), followed by TEMPO-catalyzed oxidation (48). The carboxylic acid linker 48 was attached to CTC resin (49), followed by solid phase peptide synthesis (SPPS) to produce the elongated tripeptide on resin (50). After SPPS, the peptide fragment was cleaved from the resin and the acetal was deprotected to generate the C-terminal aldehyde (51). The resulting aldehyde underwent a reductive amination with the modified uridine (52) to produce the final inhibitor scaffold (53). The scaffold that most closely mimics muraymycin 54d includes Val-Lys-Leu and exhibited the most potent inhibition of MraY. An alanine screen was then implemented with the peptide fragment to determine if a specific amino acid side chain is vital for bioactivity (Scheme 4B). When any of the positions in that fragment were replaced with alanine, loss of activity was observed (54a-c). For that reason, future modifications were made with the Val-Lys-Leu peptide fragment. The effects of the base and ribose substitutions were next tested (Scheme 4C). Substitution at the 5-position of the uracil caused a significant loss in inhibitory activity compared to the uracil with no substitution, whether unsaturated (55b,e) or not (55d).
Scheme 4.
A) Synthetic strategy to access muraymycin-inspired inhibitors and B)-C) biological evaluation of various modifications.
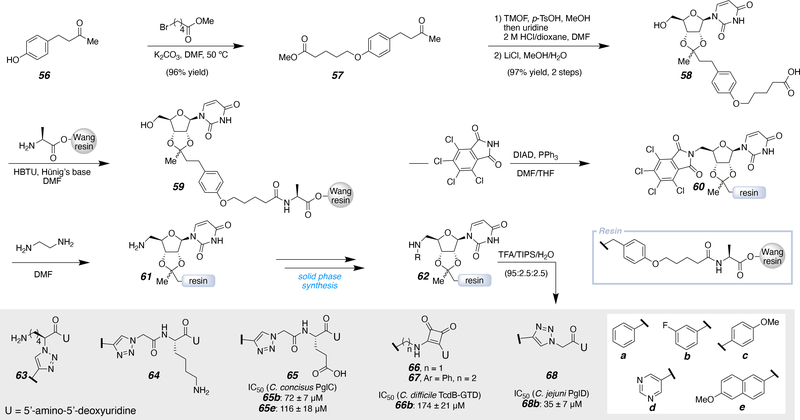
In parallel to the previously mentioned efforts, the Imperiali laboratory developed a solid phase approach to minimize in-solution manipulations of the uridine analogs (Scheme 5).62 This approach was inspired by a previously designed uridine linker, attaching uridinyl ribose through a similar acetal linkage.59 Starting from the anisole with a para-substituted ketone (56), the hydroxyl group was alkylated with methyl 5-bromovalerate. The resulting ketone (57) was converted to the uridine acetal and subsequent hydrolysis produced the free carboxylic acid 58. Then 58 was coupled to H-Ala-Wang resin, which anchored the uridine to the solid support (59). To install the C5’ amine on the ribose (61), a Mitsunobu reaction was performed with pentachlorophthalimide (60), followed by deprotection. Deprotection poised the compound (61) for further solid phase-based manipulations to access a uridine library on solid phase (62). The final compounds (63 to 68a-e) were isolated after resin cleavage, global deprotection, and HPLC purification. The inhibitors that were produced in this study were designed to mimic UDP-sugars, as these compounds are accepted by many bacterial enzymes of interest. Specifically, these inhibitors were tested with a phosphoglycosyl transferase (PGT) involved in N-linked protein glycosylation from Campylobacter concisus, PglC, which is responsible for catalyzing the transfer of phosphor-N,N'-diacetylbacillosamine onto polyprenol phosphate (Figure 1B). Additionally, TcdB-GTD, a glycosyltransferase (GT) from Clostridium difficile was tested, which is an enzyme that transfers glucose from UDP-glucose onto a glycosyl acceptor protein. Lastly, an acetyltransferase from Campylobacter jejuni, PglD, was tested and this specific enzyme is responsible for acetylating an amino group on the sugar portion of UDP-4-amino from an acetyl source, such as acetylcoenzyme A. All of which validated the selectivity of the small molecules towards specific enzymes. SAR of the inhibitor was investigated by installing a series of terminal aryl groups (a-e) to mimic the sugar portion of UDP-sugars. Specifically taking advantage of the target enzyme sugar-binding sites, as carbohydrate-binding sites often contain aromatic amino acids that engage in favorable CH-π interactions.63 The key results and lead inhibitors for future development are shown with the respective IC50 values, which were determined with various assays.64 Of the glutamic acid series, 65b and 65e exhibited the most promising activity against the phosphoglycosyl transferase PglC, with IC50 values of 72 ± 7 μM and 116 ± 18 μM, respectively. To mimic the diphosphate section of UDP-sugars, squaric acid was used as it is a convenient phosphomimetic building block.65 The squaramic acid series produced one inhibitor (66b) with an IC50 of 174 ± 21 μM against the glycosyl transferase TcdB-GTD.66, 67 The glycine inhibitor (68b) revealed an IC50 of 35 ± 7 units with the acetyltransferase PglD. Furthermore, pseudo uridinyl substitution of the uridinyl moiety for three inhibitors (68e, 65b, and 65e) exhibited comparable activity to the parent uracil derivatives. These inhibitors could in principle bind in a similar hydrogen-bonding pattern as uracil, allowing for minor modifications to uridine without sacrificing essential uridine binding pocket interactions.
Scheme 5.
Implementing a solid phase synthesis approach to access uridine-containing inhibitors.
5. Conclusions
Natural products provide a rich source of inspiration that promote method development for chemical synthesis, biosynthesis, and the design of novel bioactive compounds. Herein, we have discussed recent approaches to access select examples of uridine-containing small molecules. Uridine-based natural products and small molecules are intriguing targets due to their potential inhibitory activities against a plethora of uridine-nucleotide utilizing enzymes and as leads for antiviral and antibiotic agents. However, the chemical complexity of many uridine nucleoside natural products may render chemical synthesis and the application of biosynthetic approaches extremely challenging. Furthermore, this complexity may limit access to the quantities of products and analogs needed for detailed biochemical and biological studies. Nevertheless, both chemical and biosynthetic approaches produce natural product scaffolds that can be utilized for the construction of selective, potent molecules. More recently, de novo synthesis has been implemented to access small molecule nucleoside analogs. Inspired by nature, these compounds have simplified scaffolds relative to the respective natural product. In this case, the ability to tailor small molecule scaffolds rapidly facilitates investigation of structure-activity relationships (SAR) with targets of interest. Overall the synergistic nature of these approaches will continue to contribute to inhibitor development, which will ultimately aid in the production of effective antibiotics.
Acknowledgements
Financial support from the National Institutes of Health (GM131627 to B.I. and F32GM136023 to C.A.A.) is gratefully acknowledged. The authors would like to thank Hannah Bernstein for providing substantial edits to the manuscript.
This research was supported by the National Institutes of Health GM131627 and F32GM136023.
References
- [1].Michailidou F, Burnett D, Sharma SV, Van Lanen SG, Goss RJM, Natural Products Incorporating Pyrimidine Nucleosides, Reference Module in Chemistry, Molecular Sciences and Chemical Engineering, Elsevier; 2020. [Google Scholar]
- [2].Fujita Y, Murakami R, Muramatsu Y, Miyakoshi S, Takatsu T, A-94964, Novel Inhibitor of Bacterial Translocase I, Produced by Streptomyces sp. SANK 60404, J. Antibiot. 2008; 61: 545–549. [DOI] [PubMed] [Google Scholar]
- [3].Murakami R, Fujita Y, Kizuka M, Kagawa T, Muramatsu Y, Miyakoshi S, Takatsu T, Inukai M, A-94964, a Novel Inhibitor of Bacterial Translocase I, Produced by Streptomyces sp. SANK 60404, J. Antibiot. 2008; 61: 537–544. [DOI] [PubMed] [Google Scholar]
- [4].TAKATSUKI A ARIMA K, TAMURA G, Tunicamycin, a new antibiotic. I. Isolation and characterization of tunicamycin., J. Antibiot. 1971; 24: 215–223. [DOI] [PubMed] [Google Scholar]
- [5].Yamamoto K, Ichikawa S, Tunicamycin: chemical synthesis and biosynthesis, J. Antibiot. 2019; 72: 924–933. [DOI] [PubMed] [Google Scholar]
- [6].Yamamoto K, Sato T, Hikiji Y, Katsuyama A, Matsumaru T, Yakushiji F, Yokota S-I, Ichikawa S, Synthesis and biological evaluation of a MraY selective analogue of tunicamycins, Nucleos. Nucleot. Nucl 2019; 1–16. [DOI] [PubMed] [Google Scholar]
- [7].Wiegmann D, Koppermann S, Wirth M, Niro G, Leyerer K, Ducho C, Muraymycin nucleoside-peptide antibiotics: uridine-derived natural products as lead structures for the development of novel antibacterial agents, Beilstein J. Org. Chem. 2016; 12: 769–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].McDonald LA, Barbieri LR, Carter GT, Lenoy E, Lotvin J, Petersen PJ, Siegel MM, Singh G, Williamson RT, Structures of the Muraymycins, Novel Peptidoglycan Biosynthesis Inhibitors, J. Am. Chem. Soc. 2002; 124: 10260–10261. [DOI] [PubMed] [Google Scholar]
- [9].Fang Z, Chen S, Zhu Y, Li J, Khan I, Zhang Q, Zhang C, A new uridine derivative and a new indole derivative from the coral-associated actinomycete Pseudonocardia sp. SCSIO 11457, Nat. Prod. Res 2019; 1–7. [DOI] [PubMed] [Google Scholar]
- [10].Gentle AC, Harrison AS, Inukai M, Bugg DHT, Structure—function studies on nucleoside antibiotic mureidomycin A: synthesis of 5'-functionalised uridine models, J. Chem. Soc., Perkin Trans 1 1999; 1287–1294. [Google Scholar]
- [11].Brandish PE, Burnham MK, Lonsdale JT, Southgate R, Inukai M, Bugg TDH, Slow Binding Inhibition of Phospho-N-acetylmuramyl-pentapeptide-translocase (Escherichia coli) by Mureidomycin A, J. Biol. Chem. 1996; 271: 7609–7614. [DOI] [PubMed] [Google Scholar]
- [12].Bugg TDH, Walsh CT, Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance, Nat. Prod. Rep. 1992; 9: 199–215. [DOI] [PubMed] [Google Scholar]
- [13].Connolly GP, Duley JA, Uridine and its nucleotides: biological actions, therapeutic potentials, Trends Pharmacol. Sci. 1999; 20: 218–225. [DOI] [PubMed] [Google Scholar]
- [14].Roy B, Depaix A, Périgaud C, Peyrottes S, Recent Trends in Nucleotide Synthesis, Chem. Rev. 2016; 116: 7854–7897. [DOI] [PubMed] [Google Scholar]
- [15].Mahoney KPP, Smith DRM, Bogosyan EJA, Goss RJM, Access to High Value Natural and Unnatural Products through Hyphenating Chemical Synthesis and Biosynthesis, Synthesis 2014; 46: 2122–2132. [Google Scholar]
- [16].Nuhant P, Roush WR, Enantio- and Diastereoselective Synthesis of N-Acetyl Dihydrotetrafibricin Methyl Ester, J. Am. Chem. Soc. 2013; 135: 5340–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hu DX, Clift MD, Lazarski KE, Thomson RJ, Enantioselective Total Synthesis and Confirmation of the Absolute and Relative Stereochemistry of Streptorubin B, J. Am. Chem. Soc. 2011; 133: 1799–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Takada A, Hashimoto Y, Takikawa H, Hikita K, Suzuki K, Total Synthesis and Absolute Stereochemistry of Seragakinone A, Angew. Chem. Int. Ed. 2011; 50: 2297–2301. [DOI] [PubMed] [Google Scholar]
- [19].Custar DW, Zabawa TP, Hines J, Crews CM, Scheidt KA, Total Synthesis and Structure—Activity Investigation of the Marine Natural Product Neopeltolide, J. Am. Chem. Soc. 2009; 131: 12406–12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nicolaou KC, Sorensen EJ, Winssinger N, The Art and Science of Organic and Natural Products Synthesis, J. Chem. Educ. 1998; 75: 1225. [Google Scholar]
- [21].Mondal J, Sarkar R, Sen P, Goswami RK, Total Synthesis and Stereochemical Assignment of Sunshinamide and Its Anticancer Activity, Org. Lett. 2020; 22: 1188–1192. [DOI] [PubMed] [Google Scholar]
- [22].Srivastava N, Macha L, Ha H-J, Total Synthesis and Stereochemical Revision of Biemamides B and D, Org. Lett. 2019; 21: 8992–8996. [DOI] [PubMed] [Google Scholar]
- [23].Mulzer J, Trying to rationalize total synthesis, Nat. Prod. Rep. 2014; 31: 595–603. [DOI] [PubMed] [Google Scholar]
- [24].Mushtaq S, Abbasi BH, Uzair B, Abbasi R, Natural products as reservoirs of novel therapeutic agents, EXCLI journal 2018; 17: 420–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Gurnani N, Mehta D, Gupta M, Natural Products: Source of Potential Drugs, Afr. J. Basic Appl. Sci. 2014; 6: 171–186. [Google Scholar]
- [26].IGARASHI M, NAKAGAWA N, DOI N, HATTORI S, NAGANAWA H, HAMADA M, Caprazamycin B, a Novel Anti-tuberculosis Antibiotic, from Streptomyces sp., J. Antibiot. 2003; 56: 580–583. [DOI] [PubMed] [Google Scholar]
- [27].Hirano S, Ichikawa S, Matsuda A, Total Synthesis of Caprazol, a Core Structure of the Caprazamycin Antituberculosis Antibiotics, Angew. Chem. Int. Ed. 2005; 44: 1854–1856. [DOI] [PubMed] [Google Scholar]
- [28].Hirano S, Ichikawa S, Matsuda A, Development of a Highly β-Selective Ribosylation Reaction without Using Neighboring Group Participation: Total Synthesis of (+)-Caprazol, a Core Structure of Caprazamycins, J. Org. Chem. 2007; 72: 9936–9946. [DOI] [PubMed] [Google Scholar]
- [29].Hirano S, Ichikawa S, Matsuda A, Synthesis of Caprazamycin Analogues and Their Structure-Activity Relationship for Antibacterial Activity, J. Org. Chem. 2008; 73: 569–577. [DOI] [PubMed] [Google Scholar]
- [30].Nakamura H, Tsukano C, Yasui M, Yokouchi S, Igarashi M, Takemoto Y, Total Synthesis of (−)-Caprazamycin A, Angew. Chem. Int. Ed. 2015; 54: 3136–3139. [DOI] [PubMed] [Google Scholar]
- [31].Nakamura H, Tsukano C, Yoshida T, Yasui M, Yokouchi S, Kobayashi Y, Igarashi M, Takemoto Y, Total Synthesis of Caprazamycin A: Practical and Scalable Synthesis of syn-β-Hydroxyamino Acids and Introduction of a Fatty Acid Side Chain to 1,4-Diazepanone, J. Am. Chem. Soc. 2019; 141: 8527–8540. [DOI] [PubMed] [Google Scholar]
- [32].Tanino T, Ichikawa S, Al-Dabbagh B, Bouhss A, Oyama H, Matsuda A, Synthesis and Biological Evaluation of Muraymycin Analogues Active against Anti-Drug-Resistant Bacteria, ACS Med. Chem. Lett. 2010; 1: 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tanino T, Al-Dabbagh B, Mengin-Lecreulx D, Bouhss A, Oyama H, Ichikawa S, Matsuda A, Mechanistic Analysis of Muraymycin Analogues: A Guide to the Design of MraY Inhibitors, J. Med. Chem. 2011; 54: 8421–8439. [DOI] [PubMed] [Google Scholar]
- [34].Ichikawa S, Yamaguchi M, Hsuan LS, Kato Y, Matsuda A, Carbacaprazamycins: Chemically Stable Analogues of the Caprazamycin Nucleoside Antibiotics, ACS Infect. Dis. 2015; 1: 151–156. [DOI] [PubMed] [Google Scholar]
- [35].Patel B, Ryan P, Makwana V, Zunk M, Rudrawar S, Grant G, Caprazamycins: Promising lead structures acting on a novel antibacterial target MraY, Eur. J. Med. Chem. 2019; 171: 462–474. [DOI] [PubMed] [Google Scholar]
- [36].Takahashi Y, Igarashi M, Miyake T, Soutome H, Ishikawa K, Komatsuki Y, Koyama Y, Nakagawa N, Hattori S, Inoue K, Doi N, Akamatsu Y, Novel semisynthetic antibiotics from caprazamycins A—G: caprazene derivatives and their antibacterial activity, J. Antibiot. 2013; 66: 171–178. [DOI] [PubMed] [Google Scholar]
- [37].Shiraishi T, Kuzuyama T, Recent advances in the biosynthesis of nucleoside antibiotics, J. Antibiot. 2019; 72: 913–923. [DOI] [PubMed] [Google Scholar]
- [38].Rackham EJ, Grüschow S, Ragab AE, Dickens S, Goss RJM, Pacidamycin Biosynthesis: Identification and Heterologous Expression of the First Uridyl Peptide Antibiotic Gene Cluster, ChemBioChem 2010; 11: 1700–1709. [DOI] [PubMed] [Google Scholar]
- [39].Zhang W, Ntai I, Bolla ML, Malcolmson SJ, Kahne D, Kelleher NL, Walsh CT, Nine Enzymes Are Required for Assembly of the Pacidamycin Group of Peptidyl Nucleoside Antibiotics, J. Am. Chem. Soc. 2011; 133: 5240–5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Zhang W, Ostash B, Walsh CT, Identification of the biosynthetic gene cluster for the pacidamycin group of peptidyl nucleoside antibiotics, Proc. Natl. Acad. Sci. U. S. A. 2010; 107: 16828–16833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Imker HJ, Walsh CT, Wuest WM, SylC Catalyzes Ureido-Bond Formation During Biosynthesis of the Proteasome Inhibitor Syringolin A, J. Am. Chem. Soc. 2009; 131: 18263–18265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Shiraishi T, Nishiyama M, Kuzuyama T, Biosynthesis of the uridine-derived nucleoside antibiotic A-94964: identification and characterization of the biosynthetic gene cluster provide insight into the biosynthetic pathway, Org. Biomol. Chem. 2019; 17: 461–466. [DOI] [PubMed] [Google Scholar]
- [43].Kimura K i, Liposidomycin, the first reported nucleoside antibiotic inhibitor of peptidoglycan biosynthesis translocase I: The discovery of liposidomycin and related compounds with a perspective on their application to new antibiotics, J. Antibiot. 2019; 72: 877–889. [DOI] [PubMed] [Google Scholar]
- [44].Shi Y, Wang X, He N, Xie Y, Hong B, Rescrutiny of the sansanmycin biosynthetic gene cluster leads to the discovery of a novel sansanmycin analogue with more potency against Mycobacterium tuberculosis, J. Antibiot. 2019; 72: 769–774. [DOI] [PubMed] [Google Scholar]
- [45].Roy AD, Grüschow S, Cairns N, Goss RJM, Gene Expression Enabling Synthetic Diversification of Natural Products: Chemogenetic Generation of Pacidamycin Analogs, J. Am. Chem. Soc. 2010; 132: 12243–12245. [DOI] [PubMed] [Google Scholar]
- [46].Ragab AE, Grüschow S, Rackham EJ, Goss RJM, New pacidamycins biosynthetically: probing N- and C-terminal substrate specificity, Org. Biomol. Chem. 2010; 8: 3128–3129. [DOI] [PubMed] [Google Scholar]
- [47].Boojamra CG, Lemoine RC, Lee JC, Léger R, Stein KA, Vernier NG, Magon A, Lomovskaya O, Martin PK, Chamberland S, Lee MD, Hecker SJ, Lee VJ, Stereochemical Elucidation and Total Synthesis of Dihydropacidamycin D, a Semisynthetic Pacidamycin, J. Am. Chem. Soc. 2001; 123: 870–874. [DOI] [PubMed] [Google Scholar]
- [48].Tran AT, Watson EE, Pujari V, Conroy T, Dowman LJ, Giltrap AM, Pang A, Wong WR, Linington RG, Mahapatra S, Saunders J, Charman SA, West NP, Bugg TDH, Tod J, Dowson CG, Roper DI, Crick DC, Britton WJ, Payne RJ, Sansanmycin natural product analogues as potent and selective anti-mycobacterials that inhibit lipid I biosynthesis, Nat. Commun. 2017; 8: 14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Harvey AL, Natural products in drug discovery, Drug Discov. Today 2008; 13: 894–901. [DOI] [PubMed] [Google Scholar]
- [50].Newman DJ, Cragg GM, Natural Products as Sources of New Drugs over the Last 25 Years, J. Nat. Prod. 2007; 70: 461–477. [DOI] [PubMed] [Google Scholar]
- [51].Butler MS, Natural products to drugs: natural product-derived compounds in clinical trials, Nat. Prod. Rep. 2008; 25: 475–516. [DOI] [PubMed] [Google Scholar]
- [52].Newman DJ, Cragg GM, Natural Products As Sources of New Drugs over the 30 Years from 1981 to 2010, J. Nat. Prod. 2012; 75: 311–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Walvoort MTC, Lukose V, Imperiali B, A Modular Approach to Phosphoglycosyltransferase Inhibitors Inspired by Nucleoside Antibiotics, Chem. Eur. J. 2016; 22: 3856–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Champdoré Md, Napoli LD, Fabio GD, Messere A, Montesarchio D, Piccialli G, New Solid Supports Linking Nucleoside Scaffolds, Nucleos. Nucleot. Nucl 2003; 22: 695–697. [DOI] [PubMed] [Google Scholar]
- [55].Petricci E, Renzulli M, Radi M, Corelli F, Botta M, Solid-phase synthesis (SPS) of substituted uracils via Oxone® cleavage methodology, Tetrahedron Lett. 2002; 43: 9667–9670. [Google Scholar]
- [56].Sun D, Jones V, Carson EI, Lee REB, Scherman MS, McNeil MR, Lee RE, Solid-phase synthesis and biological evaluation of a uridinyl branched peptide urea library, Bioorg. Med. Chem. Lett. 2007; 17: 6899–6904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sun D, Lee RE, Solid-Phase Synthesis of a Thymidinyl Dipeptide Urea Library, J. Comb. Chem. 2007; 9: 370–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Epple R, Kudirka R, Greenberg WA, Solid-Phase Synthesis of Nucleoside Analogues, J. Comb. Chem. 2003; 5: 292–310. [DOI] [PubMed] [Google Scholar]
- [59].Bozzoli A, Kazmierski W, Kennedy G, Pasquarello A, Pecunioso A, A solid-phase approach to analogues of the antibiotic mureidomycin, Bioorg. Med. Chem. Lett. 2000; 10: 2759–2763. [DOI] [PubMed] [Google Scholar]
- [60].Leyerer K, Koppermann S, Ducho C, Solid Phase-Supported Synthesis of Muraymycin Analogues, Eur. J. Org. Chem. 2019; 2019: 7420–7431. [Google Scholar]
- [61].Heib A, Niro G, Weck SC, Koppermann S, Ducho C, Muraymycin Nucleoside Antibiotics: Structure-Activity Relationship for Variations in the Nucleoside Unit, Molecules 2020; 25: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Madec AGE, Schocker NS, Sanchini S, Myratgeldiyev G, Das D, Imperiali B, Facile Solid-Phase Synthesis and Assessment of Nucleoside Analogs as Inhibitors of Bacterial UDP-Sugar Processing Enzymes, ACS Chem. Biol. 2018; 13: 2542–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hudson KL, Bartlett GJ, Diehl RC, Agirre J, Gallagher T, Kiessling LL, Woolfson DN, Carbohydrate—Aromatic Interactions in Proteins, J. Am. Chem. Soc. 2015; 137: 15152–15160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Das D, Walvoort MTC, Lukose V, Imperiali B, A Rapid and Efficient Luminescence-based Method for Assaying Phosphoglycosyltransferase Enzymes, Sci. Rep 2016; 6: 33412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Niewiadomski S, Beebeejaun Z, Denton H, Smith TK, Morris RJ, Wagner GK, Rationally designed squaryldiamides — a novel class of sugar-nucleotide mimics?, Org. Biomol. Chem. 2010; 8: 3488–3499. [DOI] [PubMed] [Google Scholar]
- [66].Sato K, Seio K, Sekine M, Squaryl Group as a New Mimic of Phosphate Group in Modified Oligodeoxynucleotides: Synthesis and Properties of New Oligodeoxynucleotide Analogues Containing an Internucleotidic Squaryldiamide Linkage, J. Am. Chem. Soc. 2002; 124: 12715–12724. [DOI] [PubMed] [Google Scholar]
- [67].Seio K, Miyashita T, Sato K, Sekine M, Synthesis and Properties of New Nucleotide Analogues Possessing Squaramide Moieties as New Phosphate Isosters, Eur. J. Org. Chem. 2005; 2005: 5163–5170. [Google Scholar]