Abstract
Previous work demonstrates that the hearing loss in Alport mice is caused by defects in the stria vascularis. As the animals age, progressive thickening of strial capillary basement membranes (SCBMs) occurs associated with elevated levels of extracellular matrix expression and hypoxia-related gene and protein expression. These conditions render the animals susceptible to noise-induced hearing loss. In an effort to develop a more comprehensive understanding of how the underlying mutation in the COL4A3 gene influences homeostasis in the stria vascularis, we performed vascular permeability studies combined with RNA-seq analysis using isolated stria vascularis from 7-week old wild-type and Alport mice on the 129 Sv background. Alport SCBMs were found to be less permeable than wild-type littermates. RNA-seq and bioinformatics analysis revealed 68 genes were induced and 61 genes suppressed in the stria from Alport mice relative to wild-type using a cut-off of 2-fold. These included pathways involving transcription factors associated with the regulation of pro-inflammatory responses as well as cytokines, chemokines, and chemokine receptors that are up- or down-regulated. Canonical pathways included modulation of genes associated with glucose and glucose-1-PO4 degradation, NAD biosynthesis, histidine degradation, calcium signaling, and glutamate receptor signaling (among others). In all, the data point to the Alport stria being in an inflammatory state with disruption in numerous metabolic pathways indicative of metabolic stress, a likely cause for the susceptibility of Alport mice to noise-induced hearing loss under conditions that do not cause permanent hearing loss in age/strain-matched wild-type mice. The work lays the foundation for studies aimed at understanding the nature of strial pathology in Alport mice. The modulation of these genes under conditions of therapeutic intervention may provide important pre-clinical data to justify trials in humans afflicted with the disease.
Introduction
The stria vascularis is a highly specialized tissue lining the lateral wall comprised of marginal cells at the luminal surface that form basolateral infolds with intermediate cells and basal cells that are attached to the spiral ligament fibrocytes. Specialized pericytes and endothelial cells along with the strial capillary basement membranes maintain a tight fluid barrier of the strial capillaries [1]. The stria functions through various channels and transporters to maintain the endocochlear potential (EP) of the endolymph in the scala media of the cochlear duct. This EP is what drives the depolarization of cochlear hair cells when the mechanotransduction channels in the hair cell stereocilia open in response to movement of the basilar membrane. Thus, defects in strial function result in hearing loss. Alport mice are used as a model to study strial dysfunction.
Alport syndrome is characterized by delayed onset glomerular disease associated with progressive hearing loss [2]. The syndrome results from mutations in basement membrane type IV collagen genes COL4A3, COL4A4 (autosomal recessive Alport syndrome, [3, 4], and COL4A5 [X-linked Alport syndrome, [5]]. In the glomerulus of the kidney, the glomerular basement membranes, which are initially thinner than that of normal GBM owing to the lack of the type IV collagen α3/4/5 network, become progressively and irregularly thickened and thinned with multiple laminations of electron dense areas. This diagnostic phenotype, which is linked to the onset and progression of proteinuria, ultimately culminates in focal segmental glomerulosclerosis associated with renal failure.
In the inner ear we previously suggested basement membrane thickening occurs specifically in the strial capillary basement membranes (SCBMs) based on electron microscopy analysis of basement membrane thickening in 7–9 week-old autosomal Alport mice [6]. This was associated with the accumulation of basement membrane proteins, including laminins, entactin, and type IV collagen, and elevated expression of hypoxia-associated genes and matrix metalloproteinases [7, 8].
Alport mice showed permanent ABR threshold shifts after a noise exposure that was insufficient to cause permanent threshold shifts in age/strain matched wild-type mice [8]. These data strongly suggest that the stria vascularis, which shows a loss of metabolic homeostasis, is the source of pathologic hearing loss in the Alport mouse model.
In order to further our understanding of the nature of the changes that occur in the Alport stria vascularis compared to healthy wild-type stria, we verified that the thickening of the SCBM negatively influenced vascular permeability, which may account for the metabolic stress. We then performed RNA-seq analysis of strial RNA from wild-type and Alport mice at an age when SCBM thickening is apparent. The data confirm that the stria vascularis in the 7-week-old 129 Sv Alport mouse model is injured and dysfunctional. Moreover, the data provide new information informing whether current or planned pre-clinical trials of novel therapeutics can restore normal strial homeostasis.
Methods
Mice
129 Sv autosomal Alport mice were developed in the Cosgrove lab [9]. All mice were on a pure 129 Sv genetic background and maintained in house. Lab diet was Teklad Envigo diet # 7912.15. All procedures involving animals were conducted in accordance of an approved IACUC protocols at both sites (Boys Town National Research Hospital and Saint Louis University) and consistent with the NIH guide for the care and use of laboratory animals. Every effort was made to minimize usage as well as minimize any pain or distress. Both males and females were utilized. Animals were housed in groups with in rooms with a 14/10 hour light/dark cycle.
Strial microdissection
A detailed procedure for strial microdissection was described previously [7]. The temporal bones were harvested from non-transcardially perfused mice following cervical dislocation (within two minutes) and transferred to ice-cold HBSS buffer in a specialized petri dish. Both stria were microdissected within 10 minutes and transferred to TriZol for later RNA isolation. The stria from eight wild-type and eight Alport mice were combined for a single analysis and the experiment was performed two independent times.
RNA-seq analysis
Microdissected striae were lysed in Trizol® (Ambion®, Carlsbad, CA) and RNA isolated from the aqueous phase using PureLink® RNA Mini Kit (Ambion®). An RNA Quality Number (RQN) was determined for each sample using a Fragment Analyzer™ Automated CE System (Advanced Analytical Technologies, Inc. Ames, IA). Samples with RQN’s of ≥ 8 were used in SMART-Seq®v4 Ultra® (Takara Bio USA, Inc.). cDNA synthesis and libraries generated utilizing Nextera™ XT DNA Library Preparation Kit (Illumina® San Diego, CA). RNA seq analysis was performed using the Illumina® NextSeq™ 500 system (San Diego, CA). The data was analyzed using Ingenuity Pathway Analysis software (QIAGEN Bioinformatics) by the University of Nebraska Medical Center bioinformatics core facility. This is a classification of the data into categories and then sub-categories. For example, major categories include canonical pathways, transcriptional regulators, disease functions, and toxicity functions. These are further broken down to sub-categories based on disease functions, these include immunological disease, metabolic disease endocrine disorders and organismal injury. For each sub-category the molecules that are significantly modulated and the degree of that modulation are listed by the software based on a two-fold cut-off for up- or down-regulation. Because this categorization has considerable redundancy, given that the program was written as for broad-spectrum analysis, we present general categories that the literature suggests reflect the underlying causes of strial pathology, which is not a category probed by the software because it is too specific [1, 10, 11]. This is the data presented in Tables 1–4. The unbiased nature of this approach is reflected in the fact that >80% of these genes have never been ascribed as functionally important in the stria vascularis. Standard deviations are provided for two independent RNA-seq experiments. The complete set of raw data can be accessed at https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA602720.
Table 1. Dysregulation of genes involved in cell morphology.
| Gene Name | Protein | Function | Fold Change Alp/WT | |
|---|---|---|---|---|
| Average | Std Dev | |||
| Card11 | Caspase recruitment domain-containing protein 11 | NF-kappa-B activation | 20.00 | 11.31 |
| Gata1 | Erythroid transcription factor | transcription factor | 12.00 | 8.49 |
| Il17f | Interleukin-17F | pro-inflammatory cytokine | 9.50 | 5.66 |
| Klf1 | Krueppel-like factor 1 | transcription factor | 9.50 | 1.41 |
| Prss16 | Thymus-specific serine protease | protease | 8.83 | 1.18 |
| Lta | Lymphotoxin-alpha | TNF-B in cochlea | 7.00 | 2.36 |
| Dcdc2a | Doublecortin domain-containing protein 2 | tubulin polymerization | 6.75 | 3.18 |
| Tnf | Tumor necrosis factor | NF-kappa-B activation, cytokine | 6.60 | 3.11 |
| Hamp2 | Hepcidin-2 | iron storage/SMAD signaling | 6.43 | 1.82 |
| Ntrk1 | High affinity nerve growth factor receptor | MAPK activation | 5.00 | 1.41 |
| Dlx2 | Homeobox protein DLX-2 | transcriptional activator | 4.00 | 0.00 |
| Bdkrb2 | B2 bradykinin receptor | ERK 1/2 activation | 3.83 | 0.24 |
| Efna3 | Ephrin-A3 | tyrosine kinase | 3.59 | 1.22 |
| Cldn24 | Claudin 24 | cell adhesion | 3.57 | 1.5 |
| Pvalb | Parvalbumin | calcium signaling | 3.55 | 2.70 |
| Ly9 | T-lymphocyte surface antigen Ly-9 | Regulates IL-17 production | 3.50 | 2.50 |
| Nefh | Neurofilament heavy polypeptide | Neurofilament | 3.47 | 2.91 |
| Slc8a1 | Sodium/calcium exchanger 1 | calcium signaling | 3.33 | 1.41 |
| Tulp1 | Tubby-related protein 1 | Cell survival | 3.27 | 1.03 |
| Prkg2 | cGMP-dependent protein kinase 2 | Serine/threonine kinase | 3.00 | 2.36 |
| Pou2af1 | POU domain class 2-associating factor 1 | transcription factor | 3.00 | 0.61 |
| Spink5 | Serine protease inhibitor Kazal-type 5 | Serine protease inhibitor | 2.94 | 0.11 |
| Ccl7 | C-C motif chemokine 7 | chemokine | 2.79 | 0.34 |
| Cd79b | B-cell antigen receptor complex-associated protein beta chain | B lymphocyte antigen receptor | 2.72 | 1.02 |
| Ccr5 | C-C chemokine receptor type 5 | chemokine | 2.71 | 2.14 |
| Fgf2 | Fibroblast growth factor 2 | development | 2.69 | 0.11 |
| Col10a1 | Collagen alpha-1(X) chain | Cell matrix | 2.67 | 0.94 |
| Gpr3 | G-protein coupled receptor 3 | G-protein coupled receptor activity. | 2.63 | 0.53 |
| Nefm | Neurofilament medium polypeptide | Neurofilament | 2.62 | 0.87 |
| Bco1 | Beta, beta-carotene 15,15'-dioxygenase | metabolism | 2.45 | 1.16 |
| Prkcg | Protein kinase C gamma type | Serine/threonine kinase | 2.42 | 0.24 |
| Cd247 | T-cell surface glycoprotein CD3 zeta chain | signal transduction | 2.41 | 1.58 |
| Trpm2 | Transient receptor potential cation channel subfamily M member 2 | oxidative stress, activated calcium influx | 2.41 | 0.04 |
| Slc26a4 | Pendrin | Sodium-independent transporter | 2.25 | 0.06 |
| Grid2 | Glutamate receptor ionotropic, delta-2 | neurotransmitter | 2.19 | 0.39 |
| Fgf21 | Fibroblast growth factor 21 | development | 2.18 | 0.00 |
| Iqgap3 | IQ motif-containing GTPase-activating protein 3 | actin cytoskeleton/adhesion | 2.13 | 0.46 |
| Bach2 | Transcription regulator protein BACH2 | transcription factor, activates NFkB | 2.04 | 0.27 |
| Fgf22 | Fibroblast growth factor 22 | Development | 2.00 | 0.42 |
| Icam1 | Icam1 | Cell adhesion | 1.74 | 0.31 |
| Jph1 | Junctophilin-1 | cell surface ion channels | -1.30 | 0.51 |
| Clip3 | CAP-Gly domain-containing linker protein 3 | cytoplasmic linker protein | -2.08 | 0.48 |
| Vcan | Versican core protein | large ECM chondroitin sulfate proteoglycan | -2.27 | 2.43 |
| Elf4 | ETS-related transcription factor Elf-4 | transcription factor | -2.27 | 0.93 |
| Igf2 | Insulin-like growth factor II | WNT signaling | -2.27 | 1.50 |
| Ncam2 | Neural cell adhesion molecule 2 | cell adhesion | -2.50 | 1.44 |
| Grik2 | Glutamate receptor ionotropic, kainate 2 | neurotransmitter | -2.56 | 0.72 |
| Slc4a4 | Electrogenic sodium bicarbonate cotransporter 1 | anion exchange | -2.56 | 1.31 |
| Jph4 | Junctophilin-4 | cell surface ion channels | -2.63 | 2.42 |
| Kcna5 | Potassium voltage-gated channel subfamily A member 5 | mediates potassium transport | -2.86 | 3.02 |
| Slc17a8 | Vesicular glutamate transporter 3 | vesicular glutamate transporter | -3.03 | 2.20 |
| Astn1 | Astrotactin-1 | Cell migration | -3.03 | 0.00 |
| Pla2g10 | Group 10 secretory phospholipase A2 | Phospholipase | -3.23 | 1.56 |
| Sez6 | Seizure protein 6 | neuronal membrane signaling | -3.33 | 1.89 |
| Islr2 | Immunoglobulin superfamily containing leucine-rich repeat protein 2 | -3.45 | 0.29 | |
| Spock1 | Testican-1 | Protease inhibitor | -3.52 | 0.34 |
| Fshb | Follitropin subunit beta | hormone | -3.57 | 0.38 |
| Pou3f2 | POU domain, class 3, transcription factor 2 | transcription factor | -3.70 | 5.35 |
| Sost | Sclerostin | WNT signaling | -3.70 | 5.35 |
| Cntf | Ciliary neurotrophic factor | Cell survival | -5.00 | 7.25 |
| Tnfrsf4 | Tumor necrosis factor receptor superfamily member 4 | NF-kappa-B activation | -5.00 | 1.75 |
| Jph3 | Junctophilin-3 | cell surface ion channels | -5.26 | 7.48 |
| Myoz1 | Myozenin-1 | actin cytoskeleton | -5.26 | 2.49 |
| Muc2 | Mucin-2 | Secreted barrier glycoprotein | -6.67 | 1.78 |
| Lag3 | Lymphocyte activation gene 3 protein | immune function | -8.33 | 11.81 |
| Gfi1 | Zinc Finger Protein Gfi-1 | transcription repressor | -8.33 | 0.00 |
| Pspn | Persephin | GDNF family, protective | -10.00 | 15.00 |
| Nptx1 | Neuronal pentraxin-1 | mediates hypoxic-ischemic injury | -11.11 | 3.70 |
| Gjc2 | Gap junction gamma-2 protein | gap junction | -11.11 | 14.81 |
| Pou4f1 | POU domain, class 4, transcription factor 1 | transcription factor | -12.50 | 18.75 |
| Gfap | Glial fibrillary acidic protein | cytoskeleton | -16.67 | 19.44 |
| Mlc1 | Membrane protein MLC1 | homolog to KCNA1 (KIV1.1) | -25.00 | 12.50 |
| Reln | Reelin | Extracellular matrix serine protease-protective activities | -50.00 | 75.00 |
Table 4. Upstream regulators.
| Gene Name | Protein | Function | Fold Change Alp/WT | ||
|---|---|---|---|---|---|
| Average | Std Dev | ||||
| GATA2 | |||||
| Gata1 | Erythroid transcription factor | transcription factor | 12.00 | 8.49 | |
| Klf1 | Krueppel-like factor 1 | transcription factor | 9.50 | 1.41 | |
| Mcemp1 | Mast cell-expressed membrane protein 1 | Cell differentiation | 4.60 | 2.55 | |
| Cd177 | Cd177 antigen | activates TNF-primed cells | 4.00 | 3.64 | |
| Clec4e | C-type lectin domain family 4 member E | cell signaling | 4.00 | 1.41 | |
| Slc35d3 | Solute carrier family 35, D3 | transmembrane transport | 3.50 | 2.12 | |
| Retnlg | Resistin-like gamma | Extracellular/receptor binding | 2.71 | 0.44 | |
| Spns3 | Protein spinster homolog 3 | transporter | 2.69 | 0.65 | |
| Ngp | Neutrophilic granule protein | inhibitor of cathepsin B (CTSB) activity | -3.70 | 1.78 | |
| Hemgn | Hemogen | Cell proliferation | -5.00 | 7.00 | |
| Ear2 | V-ErbA-Related Protein 2 | transcription factor | -5.00 | 7.00 | |
| Gfi1 | Zinc Finger Protein Gfi-1 | transcription repressor | -8.33 | 0.00 | |
| Mmrn1 | Multimerin-1 | Carrier protein | -16.67 | 22.22 | |
| STAT1 | |||||
| Tnf | Tumor necrosis factor | NF-kappa-B activation, cytokine | 6.60 | 3.11 | |
| Apol6 | Apolipoprotein L6 | lipid binding | 5.00 | 1.41 | |
| Slc8a1 | Sodium/calcium exchanger 1 | calcium signaling | 3.33 | 1.41 | |
| Chil3 | Chitinase-like protein 3 | Injury response | -2.94 | 2.42 | |
| NFKBIA | |||||
| Tnf | Tumor necrosis factor | NF-kappa-B activation, cytokine | 6.60 | 3.11 | |
| Ccl7 | C-C motif chemokine 7 | chemokine | 2.79 | 0.34 | |
| Ccl3 | C-C motif chemokine 3 | chemokine | -8.33 | 4.17 | |
| FOXA2 | |||||
| Ccr5 | C-C chemokine receptor type 5 | chemokine | 2.71 | 2.14 | |
| Aldob | Fructose-bisphosphate aldolase B | gluconeogenesis | 2.07 | 1.31 | |
| Tnni1 | Troponin I, slow skeletal muscle | Muscle contraction | -2.33 | 0.54 | |
| Astn1 | Astrotactin-1 | Adhesion/cell migration | -3.03 | 0.00 | |
| Saa3 | Serum amyloid A-3 protein | inflammation | -5.00 | 3.00 | |
| Muc2 | Mucin-2 | Secreted barrier glycoprotein | -6.67 | 1.78 | |
| JUNB | |||||
| Tnf | Tumor necrosis factor | NF-kappa-B activation, cytokine | 6.60 | 3.11 | |
| Il12b | Interleukin-12 subunit beta | cytokine | 5.50 | 4.95 | |
| Ccl3 | C-C motif chemokine 3 | chemokine | -8.33 | 4.17 | |
Vascular permeability assay
8.5 wk old Alport and WT mice (n = 16/genotype) were anesthetized (Avertin (0.4 mg/g BW, IP)). Each mouse received an intra-cardiac injection (200 μl) of unconjugated Rhodamine fluorescent dye (0.04% in sterile normal saline (NS)) followed by 3 minutes incubation. At 30 and 60 sec post-injection, the lips, nose and front paws were examined under UV light. If no fluorescence was noted, the experiment was aborted. If at 3 min post-injection, the lips, nose and front paws glowed, the mouse was transcardially perfused with 5ml 0.1M cacodylate to wash out the Rhodamine dye from the vasculature. The cochleae were isolated, perilymphatically perfused with, and then immersed in cold (4°C) 4% paraformaldehyde in 0.1M cacodylate buffer. Within 20 minutes, the lateral wall (LW, Fig 1) of both cochleae representing the stria vascularis and spiral ligament was microdissected, transferred into a 0.5 ml tube and frozen in liquid nitrogen. Since the stria vascularis tended to fragment if not attached to the lateral wall, In a subset of dissections, only the spiral ligament (SL, Fig 1) was collected to determine its contribution to lateral wall diffusion. The vascular permeability of the stria vascularis was the difference between that of the whole LW and the SL as shown in Fig 1. A negative control group (n = 7) was created using LW from mice that underwent intra-cardiac injection with 0.9% sterile NS in place of Rhodamine dye followed by isolation of the LW.
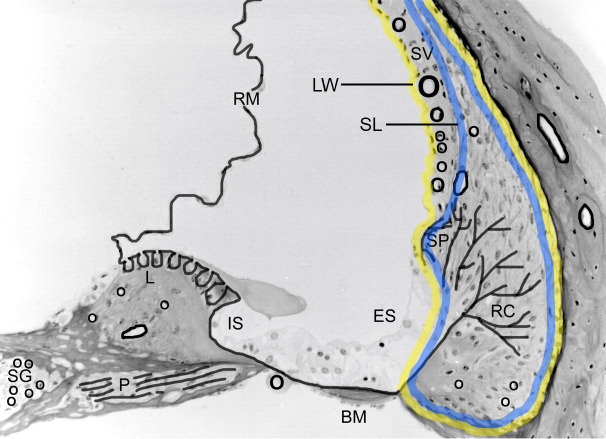
Fig 1. Mid-modiolar cross section of a mouse cochlea denoting basement membranes found in the membranous tissues.
SG = spiral ganglion; P = perineurium; L = limbus; IS = inner sulcus; BM = basilar membrane; ES = external sulcus; RC = root cells; SP = spiral prominence; SV = stria vascularis; RM = Reissner’s membrane; SL = spiral ligament; LW = lateral wall. Note as per reference for Fig 2, the lateral wall is outlined in yellow and the spiral ligament is outlined in blue.
Each sample of two LWs or two SLs was iced and micro-homogenized (100 strokes) in 30 μl of sterile phosphate buffered saline (PBS). Fluorescence (550/580) was quantified in black 384 well low-volume plates. A standard curve was constructed for the unconjugated Rhodamine fluorescent dye (y = random fluorescence units (RFU) vs. x = concentration (%w/v)). Since at low levels of fluorescence the standard curve was nonlinear, the fluorescent dye concentration in each sample was determined by solving for the quadratic equation for its positive root (x1 or x2), expressed as %w/v. The permeability of the StV was determined by subtracting the mean fluorescence of the SL samples from the of the LW samples for each genotype.
Immunofluorescence analysis
Mice were deeply anesthetized with ketamine and xylazine (300 mg/kg and 30 mg/kg, respectively, IP). The appropriate level of anesthesia was evaluated by loss of the hindquarter reflex after a pinch with thumb and index finger. If the indicated dose does not inhibit the reflex, an additional 20% of the anesthetic will be administered. Animals are euthanized by transcardial perfusion with PBS while under anesthesia. Cochleae were perfused with 4% paraformaldehyde and then decalcified overnight in 150mM EDTA on a rotator at 4°C. The samples were transferred to 15% sucrose for 1 hour and then into 30% sucrose for 2 hours. Samples were mounted in OCT and frozen at -80°C. Cochleae were sectioned at 8-μm and dipped in cold acetone. After air drying 2 hours, slides were rehydrated with 1X PBS. Cochleae sections used for collagen IV immunostaining were first denatured with acid urea (6M urea, 0.1M glycine, pH 3.5) for 1 hour at 4°C and then rinsed with 1X PBS. Slides were stained with one of the following antibodies: GFAP (Protein Tech, cat # 16825-1-AP) 1:200 in 1%BSA in 0.3% PBST (Triton X-100); NPTX1 (Protein Tech, cat# 20656-1-AP) 1:100 in 1%BSA in 0.3% PBST (Triton X-100); Reelin (ThermoFisher PA%-78413) 1:50 in 1%BSA in PBS; ICAM1 (R&D Systems Cat # AF796) 5 ug/ml in 7%non-fat dry milk in 0.3% PBST (Triton X-100) Kir4.1 1:200 in 1% BSA in 0.3% PBST (Triton X-100). For macrophage analysis in the stria, dual immunofluorescence analysis was performed using anti-desmin (pericyte marker, Abcam ab8592) and anti F4/80 (macrophage marker, GeneTex GTX26640) at 1:100 and 1:250 dilution, respectively. The experiment was performed using 4 independent animals for WT stria and 7 independent animals for Alport stria. Macrophages were counted blinded in three sections per cochlea separated by 40 μM.
Confocal microscopy
Confocal images captured using a Leica TCS SP8 MP confocal imaging system, using a 63x NA: 1.4 oil or 10x NA: 0.3 objective. Final figures were assembled using Adobe Photoshop and Illustrator software (Adobe Systems, CA).
Real-time RT-PCR analysis
cDNA was generated from the previously isolated Strial RNA (the same samples used for RNA-seq analysis), using Invitrogen SuperScript® VILO™ Master Mix (Thermo Fisher). Applied Biosystems™ TaqMan® Assays (Thermo Fisher) for GREM1 Mm00488615_s1, KCNIP1 Mm01189526_m1, CLDN 24 Mm01206808_s1, ICAM 1 Mm00516023_m1, SPINK 5 Mm00511522_m1, SPOCK 1 Mm00486393_m1 and TNF Mm0443258_m1 were run on a StepOnePlus Real-Time PCR System (Thermo Fisher). The data represent 7 independent wild-type samples and 9 independent Alport samples.
Statistical analysis
For vascular permeability studies, a Kruskal-Wallis H-test followed by Dunn’s Multiple Comparison test was used to determine whether any tissue autofluorescence contributed to the measured rhodamine fluorescence. The presence of significant differences between the rhodamine fluorescence in the wild-type and Alport spiral ligament and lateral wall samples was determined using a 2-way ANOVA with factors of genotype and tissue followed by a Holm Sidak all pairwise post hoc analysis. The negative control group was removed from the ANOVA analysis because it contained only lateral wall tissue samples and thus violated a 2-way ANOVA assumption. Finally, a t-test was conducted to determine is the WT StV fluorescence differed significantly from that of the KO StV fluorescence. Significance was set at p<0.05. The statistics and resultant data graphs were conducted using Sigma Plot 13 (SYSTAT Software, San Jose, CA). Strial macrophages were counted and the data analyzed using a two-tailed students t-test. Real time RT-PCR data was analyzed using a two-tailed students t-test.
Results
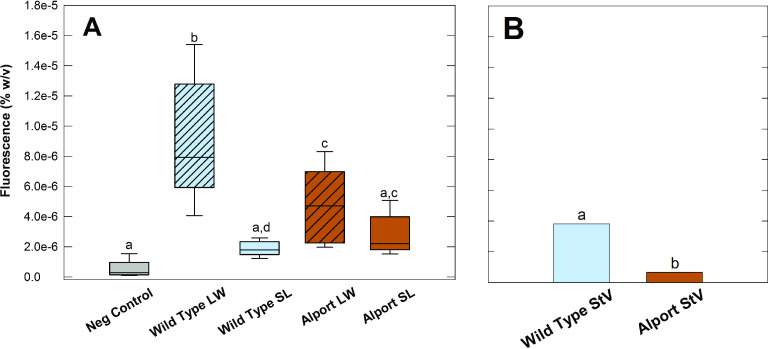
The distribution of basement membranes (black solid lines) in a mid-modiolar cross-section of the mouse cochlea are shown in Fig 1. As a reference for Fig 2, we outlined the lateral wall in yellow and the spiral ligament in blue. Previous work showed significant thickening confined to the SCBMs in Alport mice at 7 weeks of age [6]. This same study showed that basement membranes in all other cochlear regions (Fig 1) did not vary significantly in the Alport mice compared to age-matched wild-type mice. Morphometric measures of SCBMs from 9 different wild-type mice and 9 different Alport mice between 8 and 9 weeks of age showed that the SCBMs were significantly thickened (59.7 +/- 19.1 nm for wild-type versus 98.7+/-38 nm for Alport SCBMs, [8]. We surmised that the thickening of the SCBMs might compromise the permeability of the strial capillaries. To test this, Rhodamine dye was injected intracardially into 8.5 week wild-type and Alport mice. The concentration of Rhodamine dye in cochlear lateral wall tissue after a transcardial flushing of the vasculature was quantified via fluorimetry against a standard curve. The results in Fig 2A show that the SCBMs in the stria of Alport mice are indeed less permeable than those in the Alport mice, presumably owing to the thickened SCBMs. Results of the Kruskal-Wallis (H = 14, p = 0.007, df = 4) and multiple comparison test (Dunn’s, p<0.05) showed that the level of fluorescence of the negative control lateral wall (LW; Fig 1) as well as that of the spiral ligament (SL; Fig 1) tissue from both wild-type and Alport cochlea were significantly lower than that present in the lateral wall tissue of both the wild-type and Alport mice. Further analysis revealed that a significant interaction existed between the genotype and tissue (ANOVA, F(1,28) = 8.977, p = 0.006). Post hoc analysis revealed that the significance (Holm-Sidak, p<0.05) is due to the lower permeability of Rhodamine from capillaries to the surrounding lateral wall tissue in Alport versus wild-type mice. No difference (Holm-Sidak, p>0.05) was noted in the fluorescence of spiral ligament tissue in the two types of mice. Together, the lateral wall and spiral ligament data indicate that the lower vascular permeability in the Alport lateral wall is due to the accumulation of basement membrane proteins resulting in thickened strial capillaries with dysfunctional endothelial and pericyte cells. To test this conclusion, StV permeability was determined from the difference in the median level of fluorescence between LW and SL tissues for each genotype. Fig 2B shows that the permeability of the StV in the Alport mouse is significantly less (t-test, p<0.001) than that of the wild-type mice.
Fig 2.
(A). lower level of fluorescence in the Alport lateral wall tissue demonstrates a lower capillary permeability (ANOVA, F(1,28) = 8.977, p = 0.006) in 8.5 week old Alport mice compared to wild-type littermates. The dark horizontal line within each box represents the median value while the top and bottom of the box denotes the 90th and 10th percentile of the data respectively. The line denotes the standard error. The lack of a significant permeability difference between the negative control lateral wall, wild-type and Alport spiral ligament tissues (Kruskal-Wallis, H = 14, p = 0.007, df = 4) suggests that the difference seen in the permeability of the cochlear lateral wall is attributable to reduced permeability of the strial vasculature and its thickened capillary basement membranes. (B). The height of the vertical bars is the difference between the median values of LW and SL fluorescence (black horizontal lines, Fig 2A). It represents the value of the StV fluorescence. The vascular permeability in the Alport StV is significantly lower than that in the wild-type StV. LW = lateral wall, SL = spiral ligament, Same letter denotes no significant difference.
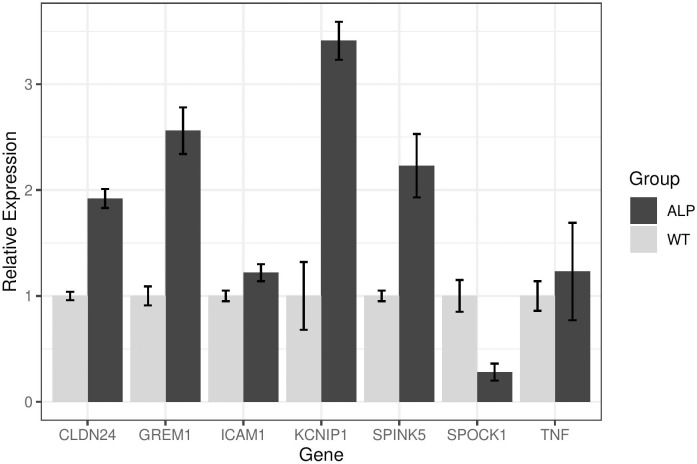
We presumed that reduced permeability might compromise strial function. To derive a clearer understanding of the health of the Alport stria vascularis we performed RNA-seq analysis using microdissected stria from 7-week-old wild-type and Alport mice. The stria vascularis was microdissected from (eight each) 7-week-old wild-type and Alport mice. The strial cDNA was sequenced and the data analyzed using Ingenuity Pathway Analysis software (QIAGEN Bioinformatics). The experiment was performed twice. Only genes that were consistently up- or down-regulated in both independent experiments by at least two-fold are presented. We broke our analysis into four categories that the literature indicates likely reflect the underlying mechanisms of strial pathology (as described in the methods): cell morphology [[10]; Table 1], Injury [[1]; Table 2], hearing/hearing loss (Table 3), and upstream regulators [[11]; Table 4]. It is notable that while the transcripts shown represent the significant differences identified from the 23000 mouse genes analyzed represent most of the genes modulated, they are not completely exhaustive. The raw data is provided in the NCBI repository to allow independent analysis by other investigators (see methods for link). For cell morphology (Table 1) most of the genes modulated regulate cell survival, cell signaling, transcriptional regulation, cell adhesion, and ion channels. For injury (Table 2), most of the genes modulated regulate pro-inflammatory cytokines, inflammation, potassium channels, cell signaling, and hypoxia/ischemia. For hearing/hearing loss (Table 3) modulated genes include transporters, intermediary metabolism, and growth factors. For upstream regulators (Table 4), which are all transcription factors, the genes modulated regulate cell signaling, adhesion/migration/differentiation, inflammation/injury response, and of course other transcription factors. There is some overlap within the categories presented in the tables as some of the genes function in multiple pathways as analyzed using the pathway-finder function of the Ingenuity software. What is notable based on the function of the genes listed in Table 2 is that the Alport stria is in an inflammatory state with considerable evidence of injury. A large percentage (approximately 80%) of these genes have never been characterized in the stria vascularis, and thus represent a novel “footprint” for strial pathology. We validated several of these genes using standard qRT-PCR. As shown in Fig 3, most of these genes validated RNA the findings observed by RNA-seq. One notable exception, TNF-α, was significantly up-regulated by RNA-seq, but did not validate by qRT-PCR. This may be due to the fact that TAQman probes only span a single exon which might miss alternatively spliced transcripts. RNA-seq has greater specificity than PCR-based methods, suggesting that the RNA-seq data is the more reliable data set [12].
Table 2. Dysregulation of genes associated with tissue injury and inflammation.
| Gene Name | Protein | Function | Fold Change Alp/WT | |
|---|---|---|---|---|
| Average | Std Dev | |||
| Kcnip1 | Kv channel-interacting protein 1 | potassium transport | 23.00 | 15.56 |
| Ccl9 | C-C motif chemokine 9 | chemokine | 10.50 | 4.60 |
| Il17f | Interleukin-17F | pro-inflammatory cytokine | 9.50 | 5.66 |
| Ros1 | Proto-oncogene tyrosine-protein kinase ROS | tyrosine kinase activity | 7.50 | 3.54 |
| Sit1 | Signaling threshold-regulating transmembrane adapter 1 | suppressor | 7.00 | 3.30 |
| Tnf | Tumor necrosis factor | NF-kappa-B activation, cytokine | 6.60 | 3.11 |
| Mmp12 | Macrophage metalloelastase | ECM protease | 6.00 | 4.53 |
| Il12b | Interleukin-12 subunit beta | cytokine | 5.50 | 4.95 |
| Grem1 | Gremlin-1 | cytokine, BMP antagonist | 4.50 | 2.12 |
| Kcnc2 | Potassium voltage-gated channel subfamily C member 2 | potassium channel | 4.00 | 2.83 |
| Has3 | Hyaluronan synthase 3 | ECM hyaluronan | 3.67 | 1.41 |
| Ipcef1 | Interactor protein for cytohesin exchange factors 1 | cell migration | 3.57 | 1.01 |
| Pvalb | Parvalbumin | calcium signaling | 3.55 | 2.70 |
| H2-DMb2 | HLA class II histocompatibility antigen, DM beta chain | endosomal membrane | 3.52 | 0.10 |
| Slc8a1 | Sodium/calcium exchanger 1 | calcium signaling | 3.33 | 1.41 |
| Tulp1 | Tubby-related protein 1 | Cell survival | 3.27 | 1.03 |
| Ms4a1 | B-lymphocyte antigen CD20 | B cell activation | 3.00 | 0.47 |
| Prkg2 | cGMP-dependent protein kinase 2 | Serine/threonine kinase | 3.00 | 2.36 |
| Spink5 | Serine protease inhibitor Kazal-type 5 | Serine protease inhibitor | 2.94 | 0.11 |
| Ccl7 | C-C motif chemokine 7 | chemokine | 2.79 | 0.34 |
| Slc6a19 | Sodium-dependent neutral amino acid transporter B(0)AT1 | transporter | 2.73 | 1.29 |
| Cd79b | B-cell antigen receptor complex-associated protein beta chain | B lymphocyte antigen receptor | 2.72 | 1.02 |
| Ccr5 | C-C chemokine receptor type 5 | chemokine | 2.71 | 2.14 |
| Gpr3 | G-protein coupled receptor 3 | G-protein coupled receptor activity. | 2.63 | 0.53 |
| Nefm | Neurofilament medium polypeptide | Neurofilament | 2.62 | 0.87 |
| Satb2 | DNA-binding protein SATB2 | transcription factor | 2.47 | 0.47 |
| Prkcg | Protein kinase C gamma type | Serine/threonine kinase | 2.42 | 0.24 |
| Cd247 | T-cell surface glycoprotein CD3 zeta chain | cell signaling | 2.41 | 1.58 |
| Mrvi1 | Protein MRVI1 | Protein complex formation | 2.31 | 1.31 |
| Grid2 | Glutamate receptor ionotropic, delta-2 | neurotransmitter | 2.19 | 0.39 |
| Otoa | Otoancorin | tectorial adhesion | 2.17 | 0.33 |
| Bach2 | Transcription regulator protein BACH2 | transcription factor, activates NFkB | 2.04 | 0.27 |
| Il27ra | Interleukin-27 receptor subunit alpha | immune response | 1.98 | 0.03 |
| Il2ra | Interleukin-2 receptor subunit alpha | immune response | 1.24 | 0.02 |
| Igf2 | Insulin-like growth factor II | WNT signaling | -2.27 | 1.50 |
| Grik2 | Glutamate receptor ionotropic, kainate 2 | neurotransmitter | -2.56 | 0.72 |
| Slc4a4 | Electrogenic sodium bicarbonate cotransporter 1 | anion exchange | -2.56 | 1.31 |
| Zap70 | Tyrosine-protein kinase ZAP-70 | immune response | -2.70 | 1.02 |
| Kcna1 | Potassium voltage-gated channel subfamily A member 1 | potassium channel | -2.94 | 1.64 |
| Chil3 | Chitinase-like protein 3 | inflammation | -2.94 | 2.42 |
| Il12a | Interleukin-12 subunit alpha | cytokine | -3.13 | 2.93 |
| Pla2g10 | Group 10 secretory phospholipase A2 | Phospholipase | -3.23 | 1.56 |
| Ptch2 | Protein patched homolog 2 | hedgehog | -3.23 | 1.14 |
| Sez6 | Seizure protein 6 | neuronal membrane signaling | -3.33 | 1.89 |
| Stap1 | Signal-transducing adaptor protein 1 | Cell signaling | -3.33 | 1.33 |
| Sost | Sclerostin | WNT signaling | -3.70 | 5.35 |
| Clec9a | C-type lectin domain family 9 member A | activation receptor | -3.70 | 5.21 |
| Ngp | Neutrophilic granule protein | inhibitor of cathepsin B (CTSB) activity | -3.70 | 1.78 |
| Kcnip2 | Kv channel-interacting protein 2 | potassium transport | -4.00 | 5.60 |
| Smtnl1 | Smoothelin-like protein 1 | Calmodulin binding protein | -4.00 | 1.92 |
| Slc2a5 | Solute carrier family 2, facilitated glucose transporter member 5 | fructose transporter | -4.55 | 6.40 |
| Cntf | Ciliary neurotrophic factor | Growth factor | -5.00 | 7.25 |
| Tnfrsf4 | Tumor necrosis factor receptor superfamily member 4 | NF-kappa-B activation | -5.00 | 1.75 |
| Slitrk3 | SLIT and NTRK-like protein 3 | Suppresses neurite outgrowth | -5.00 | 3.50 |
| Myoz1 | Myozenin-1 | actin cytoskeleton | -5.26 | 2.49 |
| Kcnc1 | Potassium voltage-gated channel subfamily C member 1 | potassium channel | -5.56 | 2.47 |
| Muc2 | Mucin-2 | Secreted barrier glycoprotein | -6.67 | 1.78 |
| Ccl3 | C-C motif chemokine 3 | chemokine | -8.33 | 4.17 |
| Cdhr1 | Cadherin-related family member 1 | cell adhesion | -9.09 | 13.22 |
| Nptx1 | Neuronal pentraxin-1 | mediates hypoxic-ischemic injury | -11.11 | 3.70 |
| Tmem178 | Transmembrane protein 178A | NF-kappa-B activation | -20.00 | 28.00 |
| Mlc1 | Membrane protein MLC1 | homolog to KCNA1 (KIV1.1) | -25.00 | 12.50 |
Table 3. Hearing loss.
| Gene Name | Protein | Function | Fold Change Alp/WT | |
|---|---|---|---|---|
| Average | Std Dev | |||
| Fgf10 | Fibroblast growth factor 10 | growth factor activity | 6.00 | 5.66 |
| Slc25a21 | Mitochondrial 2-oxodicarboxylate carrier | transporter | 5.91 | 1.67 |
| Col1a1 | Collagen alpha-1(I) chain | Extracellular matrix | 3.50 | 0.86 |
| Slc26a4 | Pendrin | Sodium-independent transporter | 2.25 | 0.06 |
| Tmc1 | Transmembrane channel-like protein 1 | probable ion channel | -2.33 | 0.27 |
| Tomt | Transmembrane O-methyltransferase homolog | mechanotransduction | -2.86 | 1.31 |
| Ocm | Oncomodulin | calcium ion-binding | -2.94 | 1.12 |
| Slc17a8 | Vesicular glutamate transporter 3 | vesicular glutamate transporter | -3.03 | 2.20 |
| Bdnf | Brain-derived neurotrophic factor | Growth factor | -4.00 | 0.00 |
| Gfi1 | Zinc Finger Protein Gfi-1 | transcription repressor | -8.33 | 0.00 |
| Hearing | ||||
| Otoa | Otoancorin | Adhesion molecule | 2.17 | 0.33 |
| Lhfpl5 | LHFPL tetraspan subfamily member 5 protein | mechanotransduction | -3.13 | 0.98 |
| Asic2 | Acid-sensing ion channel 2 | cation transport | -11.11 | 16.05 |
| Grxcr2 | Glutaredoxin domain-containing cysteine-rich protein 2 | metabolism | -14.29 | 20.41 |
Fig 3. Confirmation of RNA-seq findings using real time RT-PCR.
Strial RNA from seven wild-type mice and nine Alport mice were analyzed independently for the seven indicated transcripts by real time RT-PCR. All confirmed RNA-seq results except for Tnf-α. This may be due to taq-man probes only spanning a single exon, possibly missing alternatively spliced isoforms. Data is presented as the average with standard deviations. Data was analyzed using two-tailed students t-test. *p<0.05.
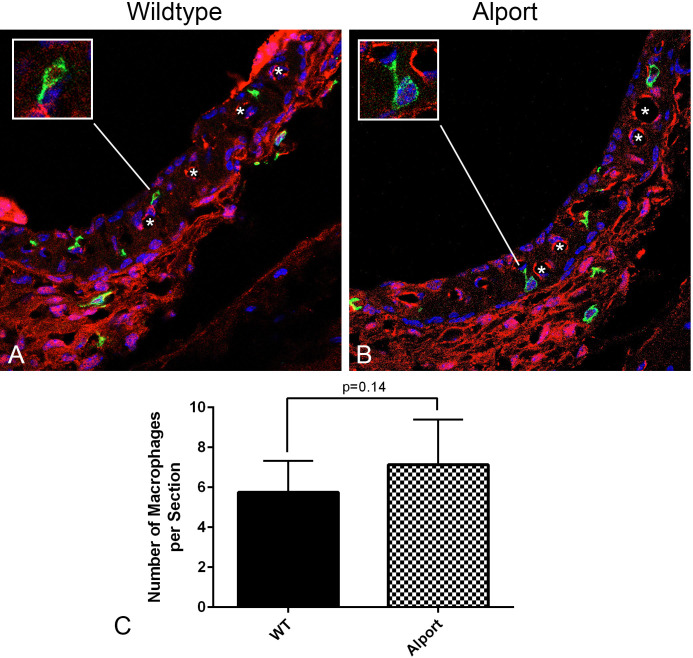
It has been previously shown that resident macrophages are activated and non-resident macrophages recruited to the lateral wall and the stria vascularis in response to noise damage, ischemia, or mitochondrial damage [13, 14]. To determine whether the inflammatory cytokines are due to activation/increased numbers of macrophages in the stria vascularis of Alport mice we performed dual immunofluorescence labeling with anti-desmin antibodies (a marker for pericytes) and anti-F4/80 9a marker for macrophages [15], antibodies. The results in Fig 4A show that the number of macrophages in the Alport and wild-type mice appear similar. To validate this observation, macrophages were quantified in mid-modiolar cross sections of the stria from eight wild-type and eight Alport mice (Fig 4B). While the numbers trended higher in the Alport stria, they did not achieve significance.
Fig 4. The number of interstitial macrophages in Alport stria versus wild-type stria (7 weeks of age) did not vary significantly, however the macrophages in Alport stria appear activated, with slightly higher numbers, larger cell bodies and numerous cell processes.
Mid-modiolar cryosections of cochleae from wild-type (A) and Alport (B) mice were immunostained using anti-desmin (pericyte marker, red) and anti-F4/80 (macrophage marker, green) antibodies. Eight wild-type and 8 Alport mice were analyzed. The data analyzed by two-tailed student’s t-test, but did not achieve significance (C).
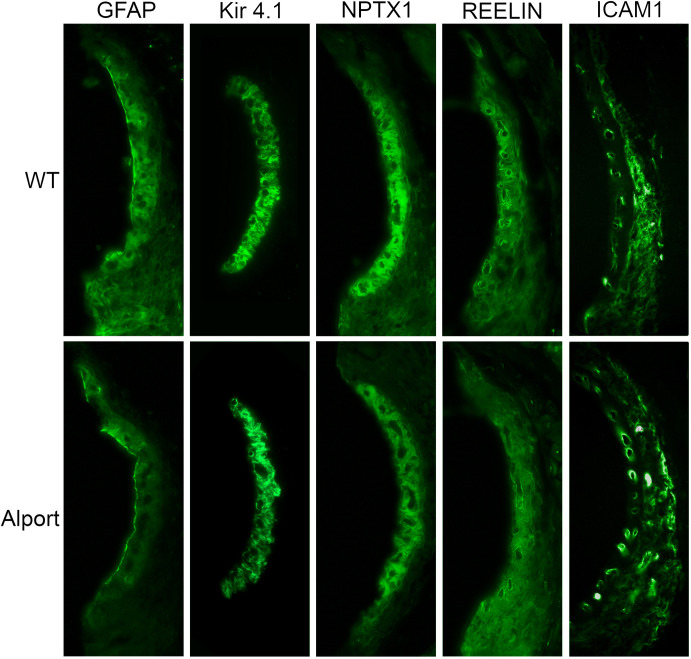
To determine whether the RNA-seq data corroborated with protein expression in the stria, we performed immunohistochemical analysis of four proteins encoded by the genes marked in bold in Tables 2 and 3. Several genes were chosen on the basis that they have never been shown to be expressed in the stria vascularis and thus may reflect novel pathogenic mechanisms. The results in Fig 5 show that a good correlation exists between protein expression and mRNA expression for the four genes/proteins. Glial fibrillary acidic protein [concentrated at the luminal surface of the marginal cells, involved in cell-cell communication; [16]], neuronal pentraxin 1 [localizing to intermediate cells, involved in acute immune response; [17]], and reelin [partially encircling strial vessels, pericyte-like localization, involved in response to tissue injury; [18]] have never been shown to be expressed in the stria, and thus represent novel genes associated with strial pathology. ICAM1, a cell adhesion molecule, has been previously shown to be expressed in strial and spiral ligament vessels [19].
Fig 5. Immunofluorescence analysis for strial expression of proteins encoded for by Gfap, Nptx1, Reelin, and Icam1 genes.
To corroborate RNA-seq data, we analyzed protein expression using mid-modiolar cross sections of 7-week-old wild-type and Alport mice. Results show down regulation in Alport compared to wild-type mice for glial fibrillary acidic protein, neuronal pentraxin 1, and reelin, and with up regulation in Alport compared to wild-type for Icam1. All are consistent with the alterations in mRNA expression. We include Kir4.1 as a control for intermediate cell staining [20]. The experiment shown is reflective of four independent experiments using independent groups of animals.
Discussion
Prior work showed that the SCBMs in Alport mice are thickened relative to age/strain-matched wild-type mice, and that the thickening is associated with an accumulation of extracellular matrix (ECM) [6–8]. As in the renal glomerulus, the mechanism underlying the SCBM thickening is mediated through activation of endothelin A receptors [8, 21]. Blocking these receptors with small molecules prevents accumulation of ECM in the SCBMs and normalizes SCBM thickness ultrastructurally [8]. A cursory look at the resting stria from Alport mice demonstrated that the tissue was in a state of oxidative/metabolic stress, much like the stria from age/strain-matched wild-type mice following noise exposure [8, 22]. Here we extend these findings to demonstrate the full spectrum of changes in gene expression in the Alport stria compared to age/strain-matched wild-type stria. To the best of our knowledge, this is the first application of RNA-seq comparing profiles in normal and diseased stria vascularis.
The results suggest that the stria vascularis is in an inflammatory state with a large number of proinflammatory cytokines and chemokines up-regulated (for example Ccr5, Ccl9, Ccl7, Il17f, and TNF-α) and a number of molecules meant to protect from inflammatory damage are down-regulated (including Ctnf, Ccl3, Il12a, and Smtnl1). A large number of genes are involved in the regulation of the pro-inflammatory NFkappaB response (including Bach2(induced), Card11 (induced), Tnf (induced), Tnfrsf4 (suppressed), and Tmem178 (suppressed)), clearly identifying activation of inflammatory responses in the strial compartment.
That strial function is impacted in Alport mice is evidenced by modulation of a number of transporters and channels that show significant changes in gene expression compared to wild-type littermates. These include Slc2a5, Slc4a4, Slc6a19, Slc8a1, Slc17a8, Slc25a21, Slc26a4, and Spns3. Potassium channels and transport mediators were also affected. For the most part these were significantly down-regulated including Kcna1, Kcnc1, Kcnip 2, Mlc1, and Kcna5. There were exceptions, however, where up-regulation was observed including Kcnc2 and Kcnip1. These channels have not been previously characterized in the stria vascularis, so the consequence of their up regulation is not clear.
Notably, a number of transcription factors are modulated that regulate genes associated with injury and inflammation (Table 4), among them STAT1, NFkappaBIA, FOXa2 and JUNB. These four transcription factors are associated with inflammatory responses and likely contribute to the inflammatory state of the Alport stria vascularis. GATA2 is a transcription factor that regulates transcriptional modulators GATA1 and Klf1, both of which are highly up-regulated in the Alport stria vascularis relative to wild-type, amplifying the transcriptional dysregulation in Alport stria.
As shown in Table 3, several genes are modulated that have been previously shown to be related to hearing loss. Brain-derived neurotrophic factor is markedly down-regulated in the Alport stria relative to wild-type stria. This growth factor has been shown to inhibit spiral ganglion degeneration and thus reduced secretion might compromise cochlear health. Slc26a4 encodes Pendrin, a well-characterized transporter required for regulation of fluid volume in the Scala media [23]. The absence of Pendrin results in deafness. Up-regulation of Pendrin in Alport stria might reflect a compensatory mechanism due to down-regulation of other transporters. FGF10 is required for expansion of the non-sensory regions of the cochlear duct during cochlear development [24]. Whether there is a functional consequence for FGF10 up-regulation (5-fold) in the Alport stria is unclear.
In a recent publication, we showed that the Alport stria was under metabolic stress resulting in elevated expression of hypoxia-related factors [8]. In the current study we provide a more comprehensive profile of strial injury and demonstrate unequivocally that the stria vascularis in the Alport mouse model is in an inflammatory state. As noted above, many of the inflammatory pathways induced in the Alport stria converge at NF-kappaB activation. NF-kappaB has long been known to play a primary role in inflammatory diseases [25]. Therefore, it is of interest to point out that NF-kappaB is induced in the lateral wall of mice subjected to acoustic overstimulation [26]. Exposure of mice to loud noise produces oxidative stress and up-regulation of genes associated with inflammation [1, 22, 27]. The Senescence Accelerated Mouse-Prone 8 (SAMP8 mouse), which shows accelerated aging, shows signs of both inflammation and oxidative stress in the stria vascularis [28]. This likely precedes degenerative changes documented for the strial capillaries in the aging mouse [29]. Collectively, these studies suggest that inflammation associated with strial pathology may be quite common and thus may reflect a more general target to protect against major causes of hearing loss such as presbycusis and noise-induced damage.
Studies of human temporal bones documented splitting of the basilar membrane in the region of the pars pectinata and cellular infilling in the tunnel of Nuel. These investigators concluded that the SNHL associated with Alport syndrome might be associated with abnormal cochlear micromechanics [30, 31]. The Merchant paper further concluded that the SCBMs were not thickened. Careful examination of the data in Merchant et al. it is clear that the SCBMs on the outside of the pericytes are indeed thickened relative to TEM images of normal SCBMs, which is what is observed in the mouse. SCBMs are bilayered, with an internal basement membrane between the endothelial cell and the pericyte and a second basement membrane lining the outer layer of the pericyte [graphically shown in [8]]. The Merchant paper was only considering the endothelial basement membranes, which were indeed of normal thickness. Early studies of human Alport organ of Corti isolated and fixed immediately following death also noted thickening of the SCBMs [32]. Importantly, Moon et al [33] noted significant hearing loss in Alport patients with normal otoaccoustic emissions, an observation that is wholly incompatible with the theory of abnormal cochlear micromechanics, which essentially rules it out. It is quite possible that the splitting of the basement membrane in the pars pectinata is either an artifact of tissue preparation, which can take up to a year for human temporal bones, or occurs long after hearing loss is established.
In summary, the RNA-seq studies presented here show that progressive thickening of the SCBMs in the Alport mouse model is associated with strial inflammation, oxidative stress, and dysregulation of ion channels and transporters. These changes likely account for the sensitivity of Alport mice to noise-induced hearing loss documented earlier [8]. It is important to remember that the Alport SCBMs have a change in the type IV collagen composition [6], which precipitates the progressive changes that culminate in reduced vascular permeability and strial inflammation. It will be of interest to determine the similarities/differences in the inflammatory response for models of presbycusis and noise-induced strial damage. If similar, they may be responsive to more generalized anti-inflammatory therapeutics, as has previously been proposed [13]. Since >80% of these genes have never been described in the stria vascularis, this work provides an important framework for validating therapies aiming to prevent Alport strial dysfunction as well as to define novel molecular pathways associated with strial dysfunction not only in Alport syndrome, but likely other disorders where SCBM thickening has been a noted feature.
Supporting information
(XLSX)
(XLSX)
(JPG)
Acknowledgments
We would like to thank John (Skip) Kennedy for his assistance in assembling this poster.
Data Availability
All relevant data are within the paper and/or Supporting Information files
Funding Statement
Funded by NIH R01 DC015385 to DC and MAG from the NIDCD.
References
- 1.Shi X., Pathophysiology of the cochlear intrastrial fluid-blood barrier (review). Hear. Res., 2016; 338: 52–63. 10.1016/j.heares.2016.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kashtan C.E., Ding J., Garosi G, Heidet L., Massella L. et al. Alport syndrome: a unified classification of genetic disorders of collagen IV α345: a position paper of the Alport Syndrome Classification Working Group. Kidney Int. 2018; 93: 1045–1051. 10.1016/j.kint.2017.12.018 [DOI] [PubMed] [Google Scholar]
- 3.Mochizuki T., Lemmink H.H., Mariyama M., Antignac C., Gubler M.C., et al. Identification of mutations in the alpha 3(IV) and alpha 4(IV) collagen genes in autosomal recessive Alport syndrome. Nat. Genet. 1994; 8: 77–81. 10.1038/ng0994-77 [DOI] [PubMed] [Google Scholar]
- 4.Lemmink H.H., Mochizuki T., van den Heuvel L.P., Schröder C.H., Barrientos A., et al. Mutations in the type IV collagen alpha 3 (COL4A3) gene in autosomal recessive Alport syndrome. Hum. Mol. Genet. 1994; 3: 1269–1273. 10.1093/hmg/3.8.1269 [DOI] [PubMed] [Google Scholar]
- 5.Barker D.F., Hostikka S.L., Zhou J., Chow L.T., Oliphant, et al. Identification of mutations in the COL4A5 collagen gene in Alport Syndrome. Science 1990; 8: 1224–1227. [DOI] [PubMed] [Google Scholar]
- 6.Cosgrove D., Samuelson G., Meehan D.T., Miller C., McGee J. et al. Ultrastructural, physiological, and molecular defects in the inner ear of a gene-knockout mouse model for autosomal Alport syndrome. Hear Res. 1998; 121: 84–98. 10.1016/s0378-5955(98)00069-0 [DOI] [PubMed] [Google Scholar]
- 7.Gratton M.A., Rao V.H., Meehan D.T., Askew C., Cosgrove D. Matrix metalloproteinase dysregulation in the stria vascularis of mice with Alport syndrome: implications for capillary basement membrane pathology. Am. J. Pathol. 2005; 166: 1465–1474. 10.1016/S0002-9440(10)62363-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meehan D.T., Delimont D., Dufek B., Zallocchi M., Phillips G. et al. Endothelin-1 Mediated Induction of Extracellular Matrix Genes in Strial Marginal Cells Underlies Strial Capillary Basement Membrane Thickening in Alport Mice. Hear. Res. 2016; 34: 100–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cosgrove D., Meehan D.T., Grunkemeyer J.A., Kornak J.M., Sayers R. et al. Collagen COL4A3 knockout: a mouse model for autosomal Alport syndrome. Genes Dev. 1996; 10: 2981–2992. 10.1101/gad.10.23.2981 [DOI] [PubMed] [Google Scholar]
- 10.Liu H., Chen L., Zhang Q., Pan N., et al. Organ of Corti and Stria vascularis: Is there interdependence for survival? Plos One 2016; 11(12): e0168953 10.1371/journal.pone.0168953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heranen A., Ikaheimo K., Virkkala J., Pirvola U. the stress response in the non-sensory cells of the cochlea under pathological conditions-possible role of mediating noise vulnerability. J. Assoc. Res. Otolaryngol. 2018; 19: 637–652. 10.1007/s10162-018-00691-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang C., Gong B., Bushel P.R., Thierry-Mieg J., et al. The concordance between RNA-seq and microarray data depends on chemical treatment and transcript abundance. Nat. Biotech. 2014; 32: 926–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujioka M., Okano H., Ogawa K. Inflammatory and immune responses in the cochlea: potential therapeutic targets for sensorineural hearing loss. Front Pharmacol. 2014; 5: 287 10.3389/fphar.2014.00287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muzushima Y., Fujimoto C., Kashio K., Yamosoba T. Macrophage recruitment, but not interleukin 1 beta activation, enhances noise-induced hearing damage. Biochem. Biophys. Res. Commun. 2017; 493: 894–900. 10.1016/j.bbrc.2017.09.124 [DOI] [PubMed] [Google Scholar]
- 15.Tournabene S.V., Sato K., Pham L., Billings P., et al. Immune cell recruitment following accoustic trauma. Hear. Res. 2006; 115–124. [DOI] [PubMed] [Google Scholar]
- 16.Sugaya-Fukasawa M., Watanabe T., Tamura M., Egashira S., & Hisatomi H. Glial fibrillary acidic protein is one of the key factors underlying neuron-like elongation in PC12 cells. Experimental and therapeutic medicine. 2011; 2(1): 85–87. 10.3892/etm.2010.162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schlimgen A.K., Hlelms J.A., Vogel H., Perin M.S. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron., 1995; 14: 519–526. 10.1016/0896-6273(95)90308-9 [DOI] [PubMed] [Google Scholar]
- 18.Micera A., Balzamino B.O., Biamonte F., Esposito G., Marino R. et al. Current progress of Reelin in development, inflammation, and tissue remodeling: from nervous to visual systems. Curr. Mol. Biol. 2016; 245: 889–903. [PubMed] [Google Scholar]
- 19.Shi X., Nutall A.L. Expression of adhesion molecular proteins in the cochlear lateral wall of normal and PARP-1 mutant mice. Hearing Res. 2007; 224: 1–14. [DOI] [PubMed] [Google Scholar]
- 20.Ando M., Takeuchi S. Immunological identification of an inward rectifier K+ channel (Kir4.1) in the intermediate cell (melanocyte) of the cochlear stria vascularis of gerbils and rats Cell Tissue Res. 1999; 298: 179–183. 10.1007/s004419900066 [DOI] [PubMed] [Google Scholar]
- 21.Dufek B., Meehan D.T., Delimont D., Cheung L., Gratton M.A., Phillips G., et al. Endothelin A receptor activation on mesangial cells initiates Alport glomerular disease. Kidney Int. 2016; 90: 300–310. 10.1016/j.kint.2016.02.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gratton M.A., Eleftheriadou A., Garcia J., Verduzco E., Martin G.K. et al. Noise-induced changes in gene expression in the cochlea of mice differing in their susceptibility to noise damage. Hear. Res., 2011; 277: 211–226. 10.1016/j.heares.2010.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hulander M, Kiernan A.E., Blomqvist S.R., Carlsson P., Samuelsson E.J. et al. Lack of pendrin expression leads to deafness and expansion of the endolymphatic compartment in inner ears of Foxi1 null mutant mice. Development. 2003; 130: 2013–2025. 10.1242/dev.00376 [DOI] [PubMed] [Google Scholar]
- 24.Urness L.D., Wang X., Shibata S., Ohyama T., Mansour S.L. Fgf10 is required for specification of non-sensory regions of the cochlear epithelium. Dev Biol. 2015; 400: 59–71. 10.1016/j.ydbio.2015.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tak P.P., Firestein G.S. NF-kappaB: a key role in inflammatory diseases. J Clin Invest. 2001; 107: 7–11 10.1172/JCI11830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto H., Omelchenko I., Shi X., Nuttall A.L. The influence of NF-kappaB signal-transduction pathways on the murine inner ear by acoustic overstimulation. J Neurosci Res. 2009; 87: 1832–1840. 10.1002/jnr.22018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wilson T., Omelchenko I., Foster S., Zhang Y., Shi X. et al. JAK2/STAT3 inhibition attenuates noise-induced hearing loss. PLoS One. 2014; 9 e108276 10.1371/journal.pone.0108276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Menardo J., Tang Y., Ladrech S., Lenoir M., Casas F. et al. Oxidative stress, inflammation, and autophagic stress as the key mechanisms of premature age-related hearing loss in SAMP8 mouse Cochlea. Antioxid. Redox Signal. 2012; 16: 263–274. 10.1089/ars.2011.4037 [DOI] [PubMed] [Google Scholar]
- 29.Carraro M, Harrison R.V. Degeneration of stria vascularis in age-related hearing loss; a corrosion cast study in a mouse model. Acta Otolaryngol. 2016; 136: 385–390. 10.3109/00016489.2015.1123291 [DOI] [PubMed] [Google Scholar]
- 30.Merchant S.N., Burgess B.J., Adams J.C., Kashtan C.E., Gregory M.C., Temporal bone histopathology in Alport syndrome. Laryngoscope. 2004; 114: 1609–1618. 10.1097/00005537-200409000-00020 [DOI] [PubMed] [Google Scholar]
- 31.Ungar O.J., Nadol J.B., Santos F. Temporal bone histopathology of x-linked inherited Alport syndrome. Laryngoscope Investig. Otolaryngol. 2018; 3: 311–314. 10.1002/lio2.177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weidauer H., Arnold W. Strukturelle Veranderungen am horogan beim Alport Syndrome. Z. Laryngol. Rhinol. Otol. 1976; 55: 6–16. [PubMed] [Google Scholar]
- 33.Moon I.S., Bang M.Y., Shim D.B., Shin S.H., Choi J.Y. Severe to profound hearing loss in patients with progressed Alport syndrome. Acta Otolaryngol. 2009; 129: 982–987. 10.1080/00016480802545588 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(JPG)
Data Availability Statement
All relevant data are within the paper and/or Supporting Information files