Abstract
EphA2 receptor kinase could become a novel target for anti-glioblastoma treatment. Doxazosin previously identified acts like the endogenous ligand of EphA2 and induces cell apoptosis. Through lead structure modification a derivative of Doxazosin possessing unique dimeric structure showed an improvement in the activity. In the current study, we expanded the dimeric scaffold by lead optimization to explore the chemical space of the conjoining moieties and a slight variation to the core structure. 27 new derivatives were synthesized and examined with EphA2 overexpressed and wild type glioblastoma cell lines for cell proliferation and EphA2 activation. Three new compounds 3d, 3e, and 7bg showed potent and selective activities against the growth of EphA2 overexpressed glioblastoma cells. Dimer 3d modification replaces the long alkyl chain with a short polyethylene glycol chain. Dimer 7bg has a relatively longer polyethylene glycol chain in comparison to compound 3d and the length is more similar to the lead compound. Whereas dimer 3e has a rigid aromatic linker exploring the chemical space. The diversity of the linkers in the active suggest additional hydrogen binding sites has a positive correlation to the activity. All three dimers showed selective activity in EphA2 overexpressed cells, indicating the activity is correlated to the EphA2 targeting effect.
Keywords: EphA2, glioblastoma, Doxazosin, agonist, dimer
Graphical Abstract
1. Introduction
Glioblastoma multiforme (GBM) is the most common and malignant primary neurological brain tumor in adults.1-3 Treatment options for GBM are still very limited, and the standard available chemotherapy agent for GBM is Temozolomide (TMZ), a DNA-alkylating reagent.4 This current treatment often fails due to resistance.5 Exploring other pathways to inhibit malignancy growth is critical in extending patient life expectancy. Erythropoietin-producing hepatocellular receptor 2 (EphA2) has shown to play an important role in cancer migration.2,3,6 It has been well documented that EphA2 is overexpressed in GBM, and the overexpression is correlated with malignant progression and poor prognosis.7,8 Studies indicate that EphA2 activation by its ligand ephrin-A1 has the ability to suppress tumor progression. In addition, the activation causes induction of apoptosis, inhibition of cell proliferation, and suppression of cell migration.9-12 In vivo studies demonstrated restoring basal levels of ephrin-A1 by systemic administration decreases tumorigenicity and invasiveness of carcinoma xenografts. 13,14 Unfortunately, EphA2 overexpression is often accompanied by loss of expression or mis-localization of ephrin-A1 in tumor tissue.3,6,15 The reduced ligand coupled with increased EphA2 expression results in frequent Akt activation, which promotes ligand-independent pro-invasive Akt-EphA2 crosstalk.3,16,17 This interaction may be in part responsible for EphA2 overexpression during tumor progression and the positive correlation of EphA2 expression and unfavorable prognosis.1,17 From a therapeutic point of view, restoring the ligand for EphA2 could recover the tumor suppressing activity of EphA2, and provide a new approach for the treatment of EphA2 overexpressed tumor. Unfortunately, the endogenous ligand of EphA2, ephrin-A1, could not effectively pass blood brain barrier (BBB), which makes this endogenous ligand less useful for the treatment of glioblastoma with overexpression of EphA2. This fact led us to hypothesize that a small molecule agonists for EphA2 can be exploited as novel cancer therapeutic.18,19
In previous studies, we identified a small molecule EphA2 agonist Doxazosin using a combination of structure-based virtual screening and cell-based assays.19 Further lead optimization of Doxazosin to activate EphA2 resulted in one particular compound with a unique dimeric structure and superior activity compared to Doxazosin (Figure 1).18 Furthermore, this dimer also has the capability to cross BBB.20 The novel symmetric structure provides us a new scaffold for further optimization.
Figure 1.
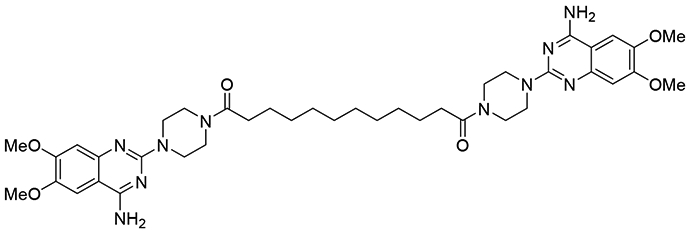
EphA2 agonist with a dimeric structure
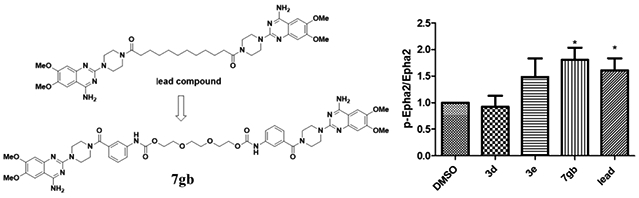
In this current study, we performed optimization of the dimeric lead compound. 27 new derivatives were synthesized and examined with glioblastoma cell lines. Three new dimers 3d, 3e, and 7bg showed similar activities compared to the lead compound (scheme 1-3). Dimer 3d exchanges the long alkyl chain with a short polyethylene glycol linker, this modification reduces the molecular weight and introduces hydrogen bonding acceptors into the structure allowing different interactions to happen inside the ligand binding domain. Dimer 7bg has a relatively longer polyethylene glycol chain compared to compound 3d, and has an additional benzamide moiety exploring the effect of the use of the polyethylene glycol chain. Dimer 3e has a rigid aromatic linker, suggesting both the flexibility and bulkiness of the compound are not critical to retain activity. All three dimers showed better activity in EphA2 overexpressed cells verse wild type glioma cells, indicating the targeting effect is through EphA2 pathways. However, only 7bg showed significant EphA2 activation, suggesting that the introduction of a benzamide moiety increases the ability to bind to the EphA2 binding domain in comparison to compound 3d which also contains a polyethylene glycol linker. Compounds 3d and 3e failed to cause EphA2 activation as effective as the lead compound indicating that the modification may lead to other targeting effect to the pathway instead of ephA2 activation.
Scheme 1.
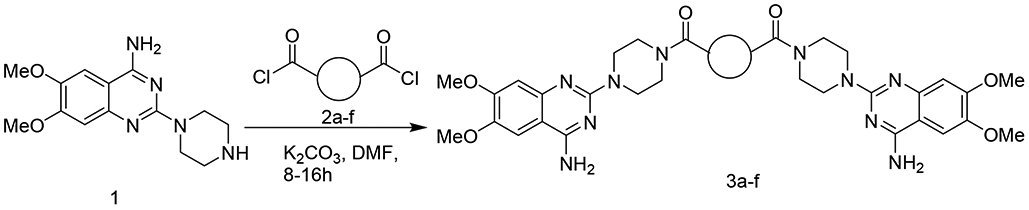
Synthesis of dimers based on 6,7-dimethoxy-2-(piperazin-1-yl) quinazolin-4-amine as the monomer
Scheme 3.
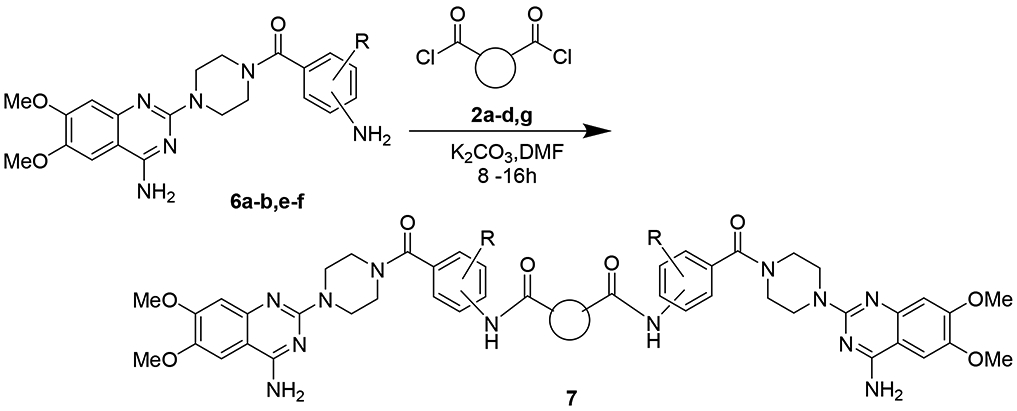
The monomeric compounds from scheme 2 introduced to the linkers from scheme 1.
2. Results and discussion
2.1. Development of an EphA2 overexpressed glioblastoma cell line
Targeting EphA2 to induce cell apoptosis provides a benefit due to lower systematic toxicity in comparison to current treatment by TMZ, a chemotherapy agent for GBM. The ideal drug candidates should selectively decrease the growth of EphA2 overexpressed cells. To examine the selectivity of the compounds, we constructed a U251 GBM cell line with overexpression of EphA2.
The western blotting results indicate that the basal EphA2 expression in U251 cells is negligible and similar to the empty vector transfected cells, whereas EphA2 transfected cells have a 5-fold increase (Figure 2). Previously the lead compound resulted in inhibition of growth based on these three cell lines wild type with an IC50 of 5.2 μM, the empty vector with an IC50 of 4.8 μM, and the EphA2 overexpressed with an IC50 of 2.1 μM. The results are consistent to our previous study that the dimeric lead compound is a selective EphA2 ligand,18 and inhibits cell growth via activation of EphA2.
Figure 2.
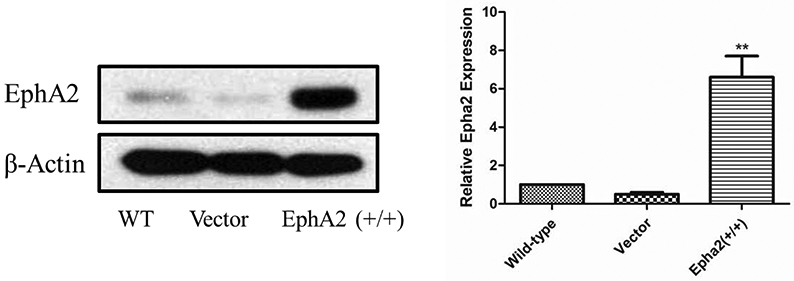
Construction of U251/EphA2 cell line based on U251 wild type cells. The intensities of the bands were quantified using ImageJ (NIH) to generate the figure. The figures are representative of 3 experiments. **p <0.001 EphA2 vs. wild type by unpaired t-test.
2.2. Lead optimization and biological evaluation
Our previous efforts resulted in a new EphA2 agonist with a dimeric structure (Figure 1).18 It significantly activated EphA2 at 2 μM in vitro, and induced EphA2 kinase internalization at 2 μM as well. In this study, a total of 27 new derivatives were synthesized using combinatorial chemistry strategy, mainly focused on the exploration of the dimeric structures.
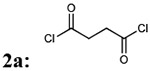
First, we focused on the modification of the linker of this lead compound (Scheme1). The alkyl linker cannot serve as either hydrogen bond acceptor or donor. To explore the ligand binding domain, theses linkers were exchanged with various structures obsessing hydrogen bonding capability. Therefore, we introduced polyethylene glycol linkers to overcome the possibility that introduction of a hydrogen bond acceptor or donor could increase the binding activity (compound 3c and 3d). To explore if length could be a contributing factor, we also generated analogs with short alkyl linkers (compounds 3a, 3b). We also wanted to explore if flexibility and bulkiness mattered in the ligand binding domain, therefore rigid aromatic linkers were used too (compounds 3e, 3f). The first set of new linkers for the dimmers are listed below (Table 1).
Table 1.
compound list of the original core with the new linkers.
| Entry | Addition of linkers: 2a-f | % yield of 3a-f | Molecular weight 3a-f |
|---|---|---|---|
| 1 | 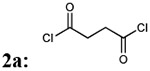 |
80% | 660.72 |
| 2 | 80% | 674.75 | |
| 3 | 57% | 676.72 | |
| 4 | 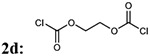 |
43% | 692.72 |
| 5 | 31% | 858.94 | |
| 6 | 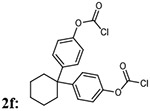 |
31% | 899.00 |
We also modified the original core of the structure by adding benzamide moieties and varying the aromatic nitro group (5a-5f). These nitro groups were reduced to amino group (7a-7f)(Scheme 2). To explore if this modification retained activity these compounds were tested with the U251 cell lines. Of these new analogs we varied the combination of nitro group with different substitutions to examine their ability to target EphA2 (Table 2).
Scheme 2.
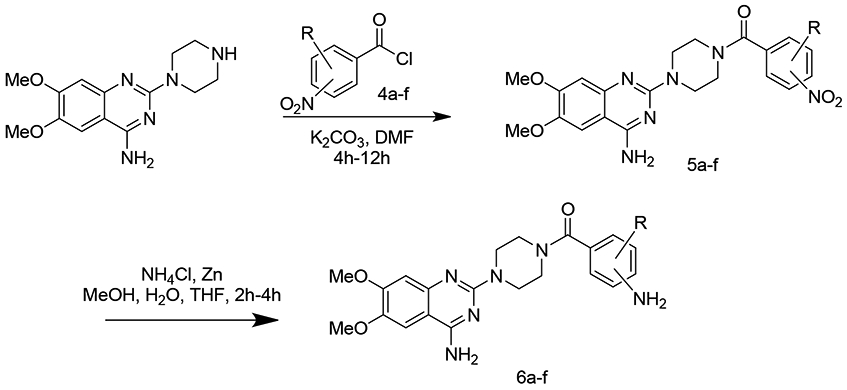
Addition of acyl chlorides to produce a small set of monomeric compounds
Table 2.
compound bearing the new aromatic core as monomers.
| Entry | Addition of acyl chlorides: 4a-f |
% yield of 5a-f |
% yield of 6a-f |
Molecular weight 5a-f |
Molecular weight 6a-f |
|---|---|---|---|---|---|
| 1 | 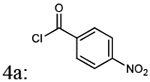 |
87% | 78% | 438.44 | 408.45 |
| 2 | 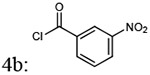 |
87% | 89% | 438.44 | 408.45 |
| 3 | 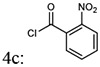 |
85% | 78% | 438.44 | 408.45 |
| 4 | 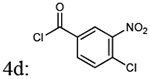 |
72% | 77% | 472.88 | 472.15 |
| 5 | 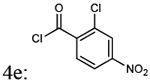 |
79% | 72% | 274.88 | 442.90 |
| 6 | 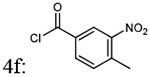 |
83% | 77% | 452.46 | 422.48 |
From the new analogs we then attempted to link the two together with the same set of linkers used beforehand (Scheme 3). Unfortunately, with the additional benzamide moiety, the formed dimers have poor solubility in various solvents making the purification not practicable. Overall, 9 new dimers were generated with this process (Table 3).
Table 3.
Dimeric compounds bearing the aromatic core with the new linkers.
| Entry | Acyl chloride from 6 | Addition of linker 2a-d,g | % yield of 7a-d, g |
Molecular weight |
|---|---|---|---|---|
| 1 | 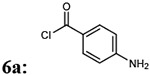 |
7ab: 70% | 912.99 | |
| 2 | 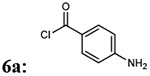 |
7ac: 70% | 914.96 | |
| 3 | 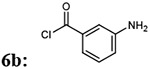 |
 |
7ba: 73% | 898.96 |
| 4 | 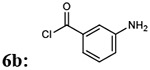 |
7bb: 30% | 912.99 | |
| 5 | 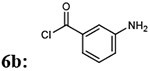 |
7bc: 23% | 914.96 | |
| 6 | 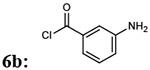 |
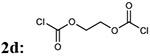 |
7bd: 70% | 930.96 |
| 7 | 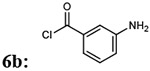 |
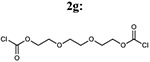 |
7bg: 70% | 1019.07 |
| 8 | 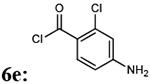 |
7ec: 43% | 983.85 | |
| 9 | 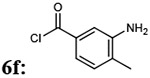 |
7fc: 23% | 943.02 |
The 27 new analogs including 15 dimers and 12 monomers show diverse substitution on the linker, and the biological results of the compounds can provide us a detailed SAR to elucidate the structural requirement for the targeting of EphA2.
To evaluate the compounds, we utilized the above developed EphA2 overexpressed U251 cells. To verify the selectivity of the compounds, we also screened the compounds with wild type U251 cells with low EphA2 expression. The selective index is calculated based on the IC50s of the two cell lines (Table 4).
Table 4.
Inhibition of the cell proliferation of U251/U251 EphA2 cells.
| Comp | U251 EphA2 overexpressed*(μM) |
U251 Wild type*(μM) |
Selectivity index wild/overexpressed** |
|---|---|---|---|
| Lead | 2.1 ± 1.05 | 5.2 ± 2.56 | 2.5 ± 2.44 |
| 3a | 6.75 ± 2.40 | 12.62 ± 6.62 | 1.9 ± 1.18 |
| 3b | 7.47 ± 3.98 | 8.07 ± 4.37 | 1.1 ± 0.82 |
| 3c | 51.86 ± 26.07 | 160 ± 80.62 | 3.1 ± 2.20 |
| 3d | 1.60 ± 0.40 | 3.05 ± 0.82 | 1.9 ± 0.70 |
| 3e | 2.45 ± 1.11 | 9.22 ± 5.56 | 3.8 ± 2.84 |
| 3f | 3.18 ± 1.44 | 7.48 ± 4.03 | 2.4 ± 1.66 |
| 5a | 12.70 ± 8.72 | 12.23 ± 8.35 | 1.0 ± .93 |
| 5b | 27.41 ± 15.05 | 24.80 ± 16.01 | 0.9 ± .77 |
| 5c | 7.41 ± 2.98 | 8.34 ± 4.42 | 1.1 ± .75 |
| 5d | 12.06 ± 5.98 | 14.73 ± 9.20 | 1.2 ± .97 |
| 5e | 26.43 ± 11.80 | 53.01 ± 29.78 | 2.0 ± 1.44 |
| 5f | 4.41 ± 2.36 | 10.77 ± 7.23 | 2.4 ± 2.10 |
| 6a | 11.14 ± 7.26 | 8.98 ± 4.61 | 0.8 ± 0.67 |
| 6b | 16.50 ± 9.16 | 8.84 ± 4.36 | 0.5 ± 0.40 |
| 6c | 14.60 ± 8.67 | 35.29 ± 21.98 | 2.4 ± 2.10 |
| 6d | 5.15 ± 3.01 | 17.91 ± 12.66 | 3.5 ± 3.19 |
| 6e | 3.00 ±1.26 | 16.49 ± 10.56 | 5.5 ± 4.21 |
| 6f | 2.70 ± 0.55 | 47.94 ± 21.13 | 17.8 ± 8.62 |
| 7ab | 27.99 ± 14.87 | 45.72 ± 34.86 | 1.6 ± 1.52 |
| 7ac | 30.44 ± 12.63 | 174.01 ± 129.21 | 5.7 ± 4.86 |
| 7ba | 41.41 ± 25.89 | 47.86 ± 26.93 | 1.2 ± 0.97 |
| 7bb | 122.30 ± 59.31 | 128.10 ± 53.59 | 1.0 ± 0.67 |
| 7bc | 9.92 ±6.20 | 8.11 ± 4.42 | 0.8 ± 0.68 |
| 7bd | 4.99 ± 2.32 | 10.12 ± 6.11 | 2.0 ± 1.55 |
| 7bg | 1.90 ± 0.55 | 7.91 ± 2.28 | 4.2 ± 1.70 |
| 7ec | 3.41 ± 1.50 | 2.59 ± 1.24 | 0.8 ± 0.49 |
| 7fc | 5.66 ± 2.94 | 11.69 ± 7.36 | 2.1 ± 1.69 |
The area shaded in gray represent the monomer compounds.
IC50 values are reported in μM. included Mean±SD.(n=4)
Selectivity index is calculated by taking the wild type average/Overexpressed average included is the error of propagation
All the compounds were examined with multiple doses, and the cells were treated with the compounds for 48 hr, and cell viability was determined after. We used the lead compound and 0.1% DMSO as the positive and negative control, respectively.
Among the first set of dimers (scheme 1 and table 1) the activity varied. Compounds 3a and 3b with reduced length of the alkyl chain resulted in decreased activity, which is consistent with our previous results and should be eliminated as an optimization direction in the future. Compounds 3d-3f all showed similar or improved activity compared to the lead, which possible is due to the introduction of hydrogen binding site or by reducing the flexibility. 3e and 3f both have a rigid and aromatic linker, and both inhibited the growth of EphA2 overexpressed cells with IC50s in the low micro mole range, suggesting that keeping the two monomers far away from each other benefit the activity. Compounds 3c and 3d have polyethylene glycol linkers which may contribute to the binding of the ligand to EphA2 via hydrogen bonding. Compound 3c lost the potency but retained the selectivity for EphA2 compared to the wild type cell lines. However, 3d has a different polyethylene glycol linker, and the potency dramatically increased and the selectivity remained. Of the first set of dimmers compound 3d showed two-fold more selectivity to the EphA2 overexpressed cells than the wild type and improved potency compared to the lead.
In the next scheme we varied the core structure by adding nitro containing benzamide moieties and then reducing the nitro group to an amino group (Scheme 2 and Table 2). For monomers 5a--6f, most of them have the IC50s above 10μM, regardless if it is the nitro group or the amino group on the benzamide moiety. Only compounds 6d, 6e and 6f showed relatively better activity, with an IC50 of 3.6, 3.0 and 2.7 μM, respectively. The position of the amino group is critical to the activity and the para position has the greatest activity of the monomers. Compound 6f had the best selectivity index among all the compounds with a value of 17.78, suggesting the reduction of the nitro group to the amine adds the possibility of improved ligand binding to EphA2 receptor. Overall, the trend in the reduction from a nitro group to an amino group benefitted the activity, suggesting that hydrogen bond donors are important for the activity on the benzamide moiety.
After synthesis of the dimers based on the amino containing benzamide, the final set of dimmers were produced (Scheme 3 and Table 3). For the 9 dimers among this group, the activities of the majority were not appealing with the exception of compound 7bg. It showed an IC50 of 1.9 μM for EphA2 overexpressed cells and 7.9 μM for wild type of cells, which results in a selective index of 4.2. Although the potency was reduced by the addition of the benzamide moiety, it is clear that introduction of hydrogen binding in the linker has a positive effect on the potency of the compounds. In most cases when the polyethylene glycol linkers are used the dimeric compounds had improved activity compared to the similar monomers. But in comparison to the original core structure, the dimers formed by monomers with benzamide moiety did not show better activity compared to dimers 3a-3f.
Considering both of the potency and selectivity, we narrow down to compounds 3d, 3e, 7bg for further mechanism investigation.
2.3. EphA2 activation
EphA2 overexpression has been identified in various different malignant cells.2,3,24,6,7,12,13,17,21-23 Decreased levels of EphA2-ligand combined with the overexpression of EphA2 in cancer cells lead to the aggressive growth of the tumor, which can be overcome in vitro by using endogenous EphA2 ligand or a small molecule ligand.12,18,19,22,25 Our lead compound could induce EphA2 phosphorylation just as the endogenous ligand ephrin-A1, and it is necessary to determine if the new analogs could cause EphA2 activation as well.18
Dimers 3e and 7bg showed better activity to induce EphA2 phosphorylation at 2μM, with only 7bg showing statistical significance compared to control (Figure 3). To our surprise, dimer 3d did not show clear EphA2 activation. Even 3d showed great activity and selectivity to EphA2 overexpressed cells. It is possible that 3d targets EphA2 with some unknow different mechanism, which needs further investigation.
Figure 3.
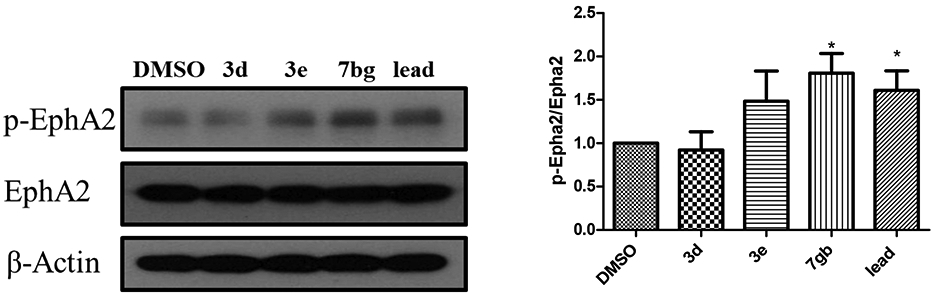
EphA2 activation of the new derivatives. U251 EphA2 cells were treated with the compounds with 2 μM (in 0.1% DMSO) for 1hr and cell lysates were subject to immunoprecipitation and immunoblot for phosphorylated EphA/B kinases (pEphA/B) and total EphA2. Treatment with 2 μM of lead was served as positive control. Treatment with 0.1% DMSO was served as vehicle control. The band intensities of the bands were quantified using ImageJ (NIH) to generate the figure. The figures are representative of 3 experiments. *p <0.05 treatment vs. DMSO by unpaired t-test.
2.4. Log P value
Since the compounds are designed to be used for the treatment of GBM, whether they can cross BBB is very critical for the potential application. Hansch and Leo found that blood-brain barrier penetration is optimal when the LogP values of the compounds are in the range of 1.5-2.7, with the mean value of 2.1. 26,27 Therefore, we measured LogP value of compounds 3d, 3e, and 7bg to predict the potential capability of these compounds to cross BBB, and the lead compound was measured as a positive control. The results are listed in Table 5. The data reported here showed that dimers 3e and 7bg showed similar LogP values compared to the lead compounds. Although the value is out of the range of the preferred LogP values indicated by Hansch, the lead compound has the ability to cross the BBB shown in our previous study. 20 We speculate that for these types of dimeric structures, may cross the BBB with an unknown mechanism instead of passive lipid penetration.
Table 5.
The measurement was repeated three times with water and 1-octanol as the two solvents, and the results are presented as Mean±SD.
| Compounds | Log P ( Mean±SD) |
|---|---|
| 3d | 1.28±0.03 |
| 3e | 0.75±0.01 |
| 7bg | 0.78±0.03 |
| Lead comp | 0.71±0.02 |
3. Conclusion
Based on the pervious structure activity relationship, the dimeric structure of Doxazosin analog showed promising ligand binding activity to EphA2. To further study this unique structure a variety of structural moieties examined if they could further improve the activity, we synthesized 27 new derivatives and 15 which retain the dimeric structure. The dimers consist of several different monomers and various linkers including alkyl, polyethylene glycol and aromatic or rigid structures. Two compounds 7bg and 3d showed great potency and selectivity toward EphA2 overexpressed GBM cells. They also stimulated EphA2 phosphorylation, which demonstrated their targeting effect to EphA2 receptor. The LogP values of the two identified dimers are not in the ideal range for their capability to cross the BBB. However, the lead compound has a similar LogP value and does cross the BBB in our previous study,20 suggesting that these types of compounds may possess a novel mechanisms to cross the BBB other than passive lipid penetration, which will need further investigation.
4. Experimental section
4.1. Chemistry
Chemicals were commercially available and used as received without further purification unless otherwise noted. Moisture sensitive reactions were carried out under a dry argon atmosphere in flame-dried glassware. Solvents were distilled before use under argon. Thin-layer chromatography was performed on precoated silica gel F254 plates (Whatman). Silica gel column chromatography was performed using silica gel 60Å(Merck, 230-400 Mesh), and ethanol/dichloromethane was used as the elution solvent. Mass spectra were obtained on the electrospray mass spectrometer at Cleveland State University MS facility Center. The molecular weight of the compounds was examined with LC-MS. All the NMR spectra were recorded on a Bruker 400 MHz in DMSO-d6. Chemical shifts (δ) for 1H NMR spectra are reported in parts per million to residual solvent protons.
Compounds 3a-3f were prepared as the following procedure.
The dimethoxyquinazolin and piperazine were dissolved in n-BuOH with 1 to 5 mole ratio, and the mixture was heated to 100°C for 24 hours, then it was cooled down to room temperature. The precipitated while solid was collected via filtration and washed with n-BuOH. After it was dried, the solid was mixed with various different linkers and K2CO3 in DMF. It was stirred until the reaction was completed. The reaction was quenched with Na2CO3 aqueous solution and stirred overnight, and the precipitated product was collected via filtration and washed with water and dried to give the corresponding compounds.
1,4-bis(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)butane-1,4-dione (3a)
Light pink solid, 80% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.44 (2H, s), 7.14 (4H, b), 6.76 (2H, s), 3.84 (6H, s), 3.81(10H, s) 3.70 (4H, s), 3.55-3.52 (4H, s), 2.63 (4H, s). 13C-NMR (400MHz, DMSO-d6) δ 170.67, 161.62, 159.10, 158.82, 154.73, 149.35, 149.26, 145.50, 145.39, 105.73, 104.20, 103.44, 56.33, 55.89, 45.33, 44.25, 43.93, 41.73, 28.15. DUIS-MS calculated for C32H40N10O6 [M + H]+: 660.31, found : 661.10
1,5-bis(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)pentane-1,5-dione (3b).
Light pink solid, 80% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (2H, s), 7.14 (4H, b), 6.75 (2H, s), 3.84 (6H, s), 3.80 (6H, s), 3.75 (4H, s), 3.70 (4H, s), 3.52 (8H, s), 2.43-2.40 (3H, t, J=7.1 Hz), 1.78 (2H, t, J= 14.0 Hz). 13C-NMR (400MHz, DMSO-d6) δ 171.09, 161.62, 159.10, 158.79, 154.73, 149.35, 149.26, 145.50, 145.39, 105.72, 104.19, 103.44, 56.33, 55.89, 45.34, 44.33, 44.18, 43.94, 41.54, 32.41, 21.09. DUIS-MS calculated for C33H42N10O6 [M + H]+: 674.33, found : 675.15
2,2'-oxybis(l-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)ethanone) (3c)
Light pink solid, 72% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (2H, s), 7.15 (4H, b), 6.76 (4H, s), 4.31 (4H, s), 3.84 (6H, s), 3.79 (6H, s), 3.75(8H,s) 3.52 (8H, s) 13C-NMR (400MHz, DMSO-d6) ) δ 167.62, 161.63, 158.76, 154.73, 149.25, 145.52, 105.74, 104.18, 103.45, 69.64, 56.33, 55.89, 44.72, 44.28, 43.87, 41.66. DUIS-MS calculated for C32H40N10O7 [M + H]+: 676.31, found : 677.20
ethane-1,2-diyl bis(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carboxylate) (3d)
Light pink solid, 43% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (2H, s), 7.14 (4H, b), 6.75 (4H, s), 4.27 (4H, s), 3.83 (6H, s), 3.79 (6H, s), 3.78(8H, s), 3.45 (8H, s) 13C-NMR (400MHz, DMSO-d6) δ 162.77, 161.63, 158.76, 155.00, 149.22, 145.52, 105.72, 104.19, 103.46, 63.67, 56.32, 55.87, 43.98, 43.80. DUIS-MS calculated for C32H40N10O8 [M + H]+: 692.30, found : 693.15
propane-2,2-diylbis(4,1-phenylene)bis(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carb oxylate) (3e)
light pink solid, 31% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.45 (2H, s), 7.24-7.18 (8H, m), 7.07-7.06 (4H, m), 6.78(2H, s) 3.84 (9H, s), 3.80 (9H, s), 3.64 (4H, m) 3.50 (4H, m), 1.66 (6H, s). 13C-NMR (400MHz, DMSO-d6) δ 161.67, 158.68, 154.77, 153.60, 149.51, 149.09, 147.45, 145.58, 127.75, 121.74. 105.66, 104.23, 103.48, 56.34, 55.91, 43.78, 42.38, 31.02. DUIS-MS calculated for C45H50N10O8 [M + H]+: 858.38, found : 859.15
cyclohexane-1,1-diylbis(4,1-phenylene)bis(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carboxylate) (3f)
light pink solid, 31% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.45 (2H, s), 7.31 (2H, s),7.28 (2H, s), 7.18(4H, b) 7.06 (2H, s), 7.04 (2H, s), 6.77 (2H, s) 3.84 (6H, s), 3.80 (13H, s), 3.63 (5H, m) 3.50 (6H, m), 2.25 (4H, s), 1.47 (6H, s). 13C-NMR (400MHz, DMSO-d6) δ 161.66, 158.64, 154.78, 153.56, 149.26, 149.04, 145.59, 145.42, 128.04, 121.91, 105.64, 104.24, 103.47, 56.35, 55.92, 45.52, 43.78, 36.80, 26.17, 22.97. DUIS-MS calculated for C48H54N10O8 [M + H]+: 898.41, found : 899.25
Compounds 5a-5f were prepared as the following procedure.
The dimethoxyquinazolin and piperazine product was mixed with various different substituted acetyl chloride and K2CO3 in THF / H2O one to one mixture. It was stirred until the reaction was completed. The reaction was quenched with Na2CO3 aqueous solution and stirred overnight, and the precipitated product was collected via filtration and washed with water and dried to give the corresponding compounds.
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(4-nitrophenyl)methanone (5a)
Light yellow solid, 87% yield for the step; 1H-NMR (400MHz, DMSO-d6) 8.30, 8.28, (1H, d, J= 7.9 Hz), 8.25-8.23 (1H, d, J= 7.9), 8.13-8.11 (1H, d, J= 8.1Hz), 7.74-7.72 (1H, d, J = 8.0 Hz), 7.47-7.45 (1H, d, J= 7.0 Hz) 7.19 (2H, b), 6.76-6.75( 1H, d, J= 5.9 Hz), 3.94 (2H, s), 3.83 (4H, s), 3.80 (3H, s), 3.73 (2H, s) 3.35 (1H, s), 3.12 ( 1H, s).13C-NMR (400MHz, DMSO-d6) δ 167.73, 167.39, 161.74, 161.65, 158.66, 158.35, 154.78, 154.73, 149.38, 149.05, 148.95, 148.29, 145.78, 145.58, 142.80, 130.77, 128.86, 124.25, 123.64, 105.70, 105.64, 104.19, 103.61, 103.48, 56.33, 55.90, 47.43, 44.22, 43.77, 43.08, 42.16, 41.36. DUIS-MS calculated for C21H22N6O5 [M + H]+: 438.17, found : 439.15
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(3-nitrophenyl)methanone (5b)
Light yellow solid, 80% yield for the step; 1H-NMR (400MHz, DMSO-d6) ) δ 8.33-8.31 (1H, d, J= 8.2 Hz), 8.27 (1H, s), 7.92-7.90 (1H, d, J= 7.6 Hz), 7.79-7.75(1H, t, J= 15.9 Hz) 7.15(2H, b), 6.75 (1H, s), 3.84 (4H, s), 3.80 (4H, s), 3.74 (3H, m), 3.40 (1H, m). 13C-NMR (400MHz, DMSO-d6) δ 167.39, 161.65, 158.78, 154.75, 149.22, 148.21, 145.58, 138.01, 133.92, 130.71, 124.72, 122.46, 105.73, 104.20, 103.50, 56.33, 55.89, 44.19, 42.23. DUIS-MS calculated for C21H22N6O5 [M + H]+: 438.17, found : 439.10
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(2-nitrophenyl)methanone (5c)
Light yellow solid, 85% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 8.23-8.212, (1H, d, J= 8.0 Hz), 7.87 (1H, t, J= 6.9 Hz), 7.72 (1H, t, J= 7.3 Hz), 7.60-7.58 (1H, d, J= 7Hz) 7.44 (1H, s), 7.17 (2H, b) 6.74 (1H, s), 3.83 (5H, s), 3.79 (4H, s), 3.72 (3H, m), 3.26 (2H, m)-NMR (400MHz, DMSO-d6) δ 166.02, 161.65, 158.77, 154.72, 149.16, 145.91, 145.59, 133.36, 132.89, 130.77, 128.62, 125.23, 105.71, 104.15, 103.51,56.31, 55.89, 46.95, 43.95, 43.65, 41.88. DUIS-MS calculated for C21H22N6O5 [M + H]+: 438.17 found : 439.10
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(4-chloro-3-nitrophenyl)methanone (5d)
Light yellow solid, 72% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 8.18, (1H, s), 7.88-7.87 (1H, s), 7.80 (1H, s), 7.44 (1H, s) 7.17 (2H, b), 6.74 (1H, s), 3.83 (6H, s), 3.79 (5H, s), 3.73-3.60 (4H, m), 3.41 (2H, m.)13C-NMR (400MHz, DMSO-d6) δ 166.45, 161.63, 158.71, 154.72, 149.12, 148.03, 145.56, 136.69, 132.76, 132.39, 126.40, 124.90, 105.66, 104.12, 103.46, 56.30, 55.89, 47.55, 44.18, 43.71, 42.32. DUIS-MS calculated for C21H21ClN6O5 [M + H]+: 472.13, found : 472.95
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(2-chloro-4-nitrophenyl)methanone (5e)
Light yellow solid, 79% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 8.42 ( 1H, s), 8.30-8.28 (1H, d, J= 7.5 Hz), 7.76-7.74 (1H, d, J= 7.8) 7.43 (1H, s), 7.17 (2H, b), 6.76-6.74 (1H, d, J= 7.0), 3.83 (4H, s), 3.79 (4H, s), 3.73 (3H, s), 3.22 (1H, s) .13C-NMR (400MHz, DMSO-d6) δ 164.61, 161.64, 161.58, 159.09, 158.68, 154.72, 154.67, 149.33, 149.13, 148.60, 145.60, 145.37, 145.31, 130.86, 129.71, 125.15, 123.39, 105.70, 104.20, 104.12, 103.50, 103.39, 56.31, 55.89, 46.62, 44.21, 44.17, 43.76, 41.81. DUIS-MS calculated for C21H21C1N6O5 [M + H]+: 472.13, found : 473.00
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(4-methyl-3-nitrophenyl)methanone (5f)
Light yellow solid, 83% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 8.05, (1H, s), 7.70 (1H, s), 7.60 (1H, s), 7.44 (1H, s) 7.16 (2H, b), 6.74 (1H, s), 3.83 (4H, s), 3.80 (4H, s), 3.74 (4H, m), 3.42 (2H, m), 2.56(3H, s).13C-NMR (400MHz, DMSO-d6) δ 167.37, 161.63, 158.76, 154.72, 149.28, 149.19, 145.55, 135.45, 134.52, 133.47, 132.12, 123.67, 105.70, 104.10, 103.48, 56.30, 55.88, 47.65, 44.19, 43.82, 42.23, 19.82. DUIS-MS calculated for C22H24N6O5 [M + H]+: 452.18, found : 453.15
Compounds 6a-6f was prepared as the following procedure.
Last step products with the nitro group were reduced by using NH4Cl 5 molar equivalence and Zn 15 molar equivalence to the nitro compounds. This was monitored by TLC in DCM/MeOH until reaction was complete it was then stopped by using an Acid base extraction. Acid was added until all zinc was dissolved then filtered to remove any undissolved zinc or other contaminates. Follow by readjusting the pH to a basic 12 by adding sodium carbonate aqueous solution. This mixture was then extracted by DCM and ethyl acetate then was dried with MgSO4 and washed with diethyl ether and hexane to give the corresponding compounds.
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(4-aminophenyl)methanone (6a)
Light yellow solid, 78% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (1H, s), 7.19-7.17 (3H, m), 6.74 (1H, s), 6.59 (1H, s) 6.56 (1H, d), 5.53 (2H, s), 3.83 (3H, s), 3.79 (3H, s), 3.76 (4H, s) 3.56 (4H, s).13C-NMR (400MHz, DMSO-d6) 170.55, 161.61, 158.82, 154.70, 150.99, 149.24, 145.48, 129.80, 122.44, 113.19, 105.70, 104.16, 103.44, 56.31, 55.88, 44.22. DUIS-MS calculated for C21H24N6O3 [M + H]+: 408.19, found : 409.15
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(3-aminophenyl)methanone (6b)
Light yellow solid, 89% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (1H, s), 7.15 (2H, b), 7.10-7.06 (1H, t, J= 7.7 Hz), 6.74 (1H, s), 6.64-6.2 (1H, d, J= 8.0 Hz), 6.59 (1H, s), 6.53-6.51 (1H, d, J= 7.4 Hz), 3.83 (3H, s), 3.79 (3H, s), 3.75 (4H, m), 3.62-3.59 (2H, t, J= 16.5 Hz), 3.42 (2H, b). 13C-NMR (400MHz, DMSO-d6) δ 170.30, 161.61, 158.80, 154.72, 149.20, 145.54, 137.13, 129.32, 115.25, 114.43, 112.63 105.71, 104.18, 103.47, 56.33, 55.89, 44.17. DUIS-MS calculated for C21H21ClN6O5 [M + H]+: 408.19, found : 409.05
(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)(2-aminophenyl)methanone (6c)
Light yellow solid, 78% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (1H, s), 7.15-7.11 (3H, m), 7.04 (1H, s), 6.74 (2H, s) 6.58 (1H, b), 5.19 (2H, s), 3.83 (4H, s), 3.79 (7H, s), 3.53 (4H, m).13C-NMR (400MHz, DMSO-d6) δ 169.28, 161.61, 158.81, 154.70, 149.22, 146.32, 145.50, 130.50, 128.29, 119.80, 116.01, 105.70, 104.14, 103.45, 56.31, 55.88, 44.20, 30.89. DUIS-MS calculated for C21H24N6O3 [M + H]+: 408.19, found : 409.15
(3-amino-4-chlorophenyl)(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)methanone (6d)
Light yellow solid, 77% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 7.44 (1H, s), 7.25 (1H, s), 7.18, (2H, s), 6.84 (1H, s), 6.74 (1H, s), 6.57 (1H, s) 5.57 (2H, b), 3.83 (3H, s), 3.79 (5H, m), 3.64 (5H, m). 13C-NMR (400MHz, DMSO-d6) δ 169.27, 161.62, 158.78, 154.69, 149.17, 145.52, 145.22, 135.92, 129.46, 118.40, 115.58, 114.29, 105.68, 104.12, 103.48, 56.30, 55.89, 47.59, 44.17, 42.17. DUIS-MS calculated for C17H23N5O3 [M + H]+: 442.15, found : 443.10
(4-amino-2-chlorophenyl)(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)methanone (6e)
Light yellow solid, 72% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (1H, s), 7.14, (2H, b), 7.01-6.99, (1H, d, J= 7.5 Hz), 6.73 (1H, s) 6.63 (1H, s), 6.55- 6.53 ( 1H, d, J= 7.5 Hz), 5.63 (2H, s) 3.83 (4H, s) 3.79 (5H, s), 3.65-3.60 (8H, m), 3.25 (2H, s), 3.17(2H, s) 13C-NMR (400MHz, DMSO-d6) δ 167.25, 161.61, 158.77, 154.70, 151.04, 149.17, 145.54, 130.48, 129.40, 122.63, 113.56, 112.90, 105.70, 104.13, 103.47, 56.32, 55.90, 49.07, 46.91, 44.34, 43.94, 41.83. DUIS-MS calculated for C19H21N5O4 [M + H]+: 442.15, found : 443.05
(3-amino-4-methylphenyl)(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazin-1-yl)methanone (6f)
Light yellow solid, 77% yield for the step; 1H-NMR (400MHz, DMSO-d6) δ 7.43 (1H, s), 7.14 (2H, b), 6.99-6.97 (1H, d, J= 7.7 Hz), 6.74 (1H, s), 6.67 (1H, s), 6.50-6.49 (1H, s, J= 3 Hz), 5.02 (2H, b) 3.83 (3H, s), 3.80 (4H, m), 3.723-3.40 (8H, m), 2.08(3H, s). 13C-NMR (400MHz, DMSO-d6) δ 170.41, 161.65, 154.82, 147.06, 145.76, 134.53, 130.16, 123.04, 1114.97, 112.95, 104.51, 103.31, 56.45, 56.10, 55.98, 44.37, 17.80. DUIS-MS calculated for C22H26N6O3 [M + H]+: 422.21, found : 423.10
Compounds 7aa-7fc were prepared as the following procedure.
The corresponding reduced benzamides 6a-6f were mixed with different dichloride linkers. They were dissolved in anhydrous DMF in a 2 to 1 ratio under argon protection. The dichloride linkers where diluted in anhydrous DMF and added slowly. It was stirred overnight and monitored by TLC after 24 hours. The reaction was stopped by reducing down the solvent and purified with PTLC in DCM/EtOH.
N1,N5-bis(4-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)glutaramide (7ab)
Light yellow solid, 70% yield for the last step; 1H-NMR (400MHz, DMSO-d6) 10.15 (2H, s), 7.71-7.69 (4H, m), 7.44-7.40 (7H, d), 7.16 (6H, m), 6.74 (3H, d), 3.83 (10H, m), 3.79 (13H, m), 3.56 (9H, m) 2.44 (4H, s) 1.93 (2H, s).13C-NMR (400MHz, DMSO-d6) δ 171.61, 169.51, 161.62, 158.78, 154.70, 154.52, 149.20, 145.51, 140.99, 130.58, 128.94, 128.60, 120.88, 118.94, 105.69, 104.15, 103.45, 56.31, 55.88, 44.17, 36.06, 21.29. DUIS-MS calculated for C47H52N12O8 [M + H]+: 912.40, found : 913.20
2,2'-oxybis(N-(4-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)acetamid e)(7ac)
Light yellow solid, 70% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 10.27 (2H, s), 7.78 (2H, s), 7.77 (2H, s), 7.47 (2H, s), 7.45 (2H, s) 7.43 (2H, s), 7.15 (4H, m) , 6.74 (2H, s), 4.33 (4H, s) 3.83(10H, s), 3.79 (11H, m) 3.61-3.56 (8H, m).13C-NMR (400MHz, DMSO-d6) δ 169.40, 168.88, 161.62, 158.77, 154.71, 149.18, 145.52, 139.99, 131.34, 128.64, 119.61, 105.68, 104.15, 103.45, 71.33, 56.31, 55.88, 44.13. DUIS-MS calculated for C46H50N12O9 [M + H]+: 914.38, found : 915.15
N1,N4-bis(3-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)succinamide (7ba)
Light yellow solid, 70% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 10.32 (2H, s), 8.55 (4H, s), 7.83(2H, s), 7.70 (2H, s) 7.64-7.62 (2H, d, J= 7.0 Hz), 7.42-7.38(2H, t, J= 14 Hz), 7.12-7.10 (2H, d, J= 7.8 Hz), 3.92 (8H, m), 3.86 (8H, s), 3.84 (7H, s) 3.76 (4H, m) 3.53 (5H, m) 2.71 (4H, s). 13C-NMR (400MHz, DMSO-d6) δ 171.18, 169.60, 161.74, 155.59, 147.08, 139.91, 129.38, 121.93, 120.57, 118.05, 105.29, 102.39, 56.68, 56.43, 45.00, 31.68. DUIS-MS calculated for C46H50N12O8 [M + H]+: 898.39, found : 899.20
N1,N5-bis(3-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)glutaramide (7bb)
Light yellow solid, 70% yield for the last step; 1H-NMR (400MHz, DMSO-d6) 10.09 (2H, s), 7.45 (2H, s), 7.64-7.62 (2H, m), 7.43 (2H, s) 7.40-7.38 (4H, m), 7.14-7.10(6H, m), 6.74 (2H, d), 3.92 (8H, m), 3.83 (9H, m), 3.79 (18H, m) 2.41 (4H, s) 1.93 (2H, s).13C-NMR (400MHz, DMSO-d6) δ 171.57, 169.44, 161.62, 158.79, 154.72, 149.18, 145.55, 139.77, 136.78, 129.33, 121.98, 120.50, 118.15, 105.71, 104.15, 103.48, 56.31, 55.89, 47.55, 44.06, 36.03, 21.28. DUIS-MS calculated for C47H52N12O8 [M + H]+: 912.40, found : 913.20
2,2'-oxybis(N-(3-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)acetamid e) (7bc)
Light yellow solid, 73% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 10.26 (2H, s), 7.82 (2H, s), 7.74-7.73 (2H, d, J= 6.7 Hz), 7.44 (4H, m) 7.15 (5H, b), 6.74 (2H, d), 4.31 (3H, s) 3.82 (8H, s), 3.79 (9H, m).13C-NMR (400MHz, DMSO-d6) δ 169.30, 168.82, 162.78, 161.62, 158.80, 154.71, 149.19, 145.55, 138.83, 129.44, 122.71, 121.16, 118.79, 105.71, 104.17, 103.49, 71.31, 56.32, 55.88, 47.64, 44.18, 42.26, 36.24, 31.25. DUIS-MS calculated for C46H50N12O9 [M + H]+: 914.38, found : 915.10
ethane-1,2-diylbis((3-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)carbamate) (7bd)
light yellow solid, 30% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 9.94 (2H, s), 7.57-7.54 (4H, m), 7.43 (3H, s), 7.37 (2H, s) 7.14 (5H, b), 7.07-7.05 (2H, d), 6.74 (3H, s) 4.37 (4H, s) 3.83 (11H, s), 3.79 (11H, m) .13C-NMR (400MHz, DMSO-d6) δ 169.38, 161.62, 158.78, 154.71, 153.81, 149.18, 145.54, 139.58, 136.91, 129.44, 121.52, 119.75, 117.30, 105.70, 104.15, 103.47, 63.27, 62.49, 56.31, 55.88, 47.56, 44.12, 42.18, 25.95. DUIS-MS calculated for C46H50N12O10 [M + H]+: 930.38, found : 931.20
(ethane-1,2-diylbis(oxy))bis(ethane-2,1-diyl)bis((3-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)phenyl)carbamate) (7bg)
Light yellow solid, 23% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 9.92 (2H, s), 7.57-7.53 (4H, m), 7.43 (2H, s), 7.37-7.36 (2H, s) 7.14 (4H, b), 7.06-7.04 (2H, d), 6.74 (2H, s) 4.22 (3H, s) 3.83 (9H, s), 3.79 (1 OH, m), 3.67 (4H, s), 3.59 (4H, s).13C-NMR (400MHz, DMSO-d6) δ 169.41, 162.77, 161.62, 158.78, 154.70, 153.96, 149.19, 145.53, 139.70, 136.90, 129.41, 121.38, 119.61, 117.15, 105.71, 104.14, 103.47, 70.18, 69.15, 64.12, 60.69, 56.31, 55.88, 47.64, 44.17, 42.24, 36.24, 31.25. DUIS-MS calculated for C50H58N12O12 [M + H]+: 1018.43, found : 1019.20
2,2'-oxybis(N-(4-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)-3-chlorophenyl) acetamide) (7ec)
Light yellow solid, 43% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 10.32 (2H, s), 7.97 (2H, s), 7.68-7.66 (2H, d, J= 8.0 Hz), 7.44 (2H, s), 7.41-7.39 (2H, s), 7.18 (5H, m), 6.74 (3H, s), 4.33 (4H, s) 3.83 (13H, s), 3.75 (11H, s), 3.72 (11H, m) 3.24 (6H, m).13C-NMR (400MHz, DMSO-d6) δ 169.09, 166.09, 161.63, 158.64, 154.74, 148.97, 145.60, 145.48, 140.35, 131.20, 129.89, 129.02, 120.15, 118.86, 105.61, 105.42, 104.27, 104.16, 103.48, 103.35, 71.13, 56.32, 55.90, 46.78, 44.31, 44.16, 43.88, 41.79. DUIS-MS calculated for C46H48C12N12O9 [M + H]+: 982.30, found : 983.10
2,2'-oxybis(N-(5-(4-(4-amino-6,7-dimethoxyquinazolin-2-yl)piperazine-1-carbonyl)-2-methylphenyl)acetamide) (7fc)
Light yellow solid, 23% yield for the last step; 1H-NMR (400MHz, DMSO-d6) δ 9.62 (2H, s), 7.60 (2H, s), 7.43 (2H, s), 7.34-7.32 (2H, d, J= 7.0 Hz), 7.21-7.19 (2H, d, J= 7.6 Hz), 7.15 (4H, b), 6.74 (2H, s), 4.33 (4H, s) 3.83 (9H, s), 3.79 (11H, s), 2.29 (6H, m) 1.36 (3H, s).13C-NMR (400MHz, DMSO-d6) δ 169.16, 168.46, 161.62, 158.77, 154.71, 149.20, 145.52, 135.90, 134.08, 133.91, 130.91, 124.73, 124.29, 105.71, 104.15, 103.46, 71.09, 56.31, 55.88, 44.16, 34.85, 30.90, 18.09. DUIS-MS calculated for C48H54N12O9 [M + H]+: 942.41, found : 943.20
4.2. LogP measurement
Compounds 3d, 3e, 7gb and lead compound were used in the determination of their log P values. This method follows the OECD test guidelines for the slow stir method, with slight modification due to the data collection method. The data was collected on a UV-Vis spectrophotometer Shimadzu UV-1280 and standard UV Quartz 10 mm cuvette was used. The chemical was purchased from VWR brand EMPLURA 1-octanol 99% purity. The Flask where equipped with Teflon coated magnetic stirrers and Coring PC420D stir plates where used. The 1-octanol and DDH2O where placed into two separate containers containing both phases to become mutual saturated by allowing them to stir for 48 h. Then the two phases where separated by a separation funnel. Compounds 3d, 3e, 7gb and lead now were dissolved in 1-octanol pre-saturated with water. These stock solutions are then centrifuged at 5,000 rpm for 5 min. The supernatant was taken to be the working solution. Take 5 mL working solution of each into new clear flask, and add identical volume water saturated with 1-octanol, then stir at room temperature for 48 h. After that, stop stirring; take two-phase solution out, centrifuge at 5,000 rpm for 5 min to separate this two-phase system. Removal of the 1-octanol phase from the solution then transferred for UV spectrophotometry to achieve absorbance at the correct wavelength. According to the procedure, concentrations were determined by UV measurement so, equation written as:
Where A0 and Ax are the initial and final absorbance values of organic layer.
HPLC-PDA analysis of the purity of the compounds.
Reversed-phase HPLC analysis of compounds were conducted on Beckman HPLC system with AutoSampler (table 6). The chromatographic separation was performed on a C18 column (2.0 mm × 150 mm, 5 μm) from Phenomenex (Torrance, CA, USA), one mobile phase was employed for isocratic elution with a flow rate of 0.2 mL/min. The injection volume was 6 μL and the PDA detector was set up 346nm
Table 6.
Purity of the compounds
| H2O/MeOH (20%/80%) | ||
|---|---|---|
| Compound | Retention time (Rt) in minutes |
Purity |
| 3a | 1.90 | 99.99% |
| 3b | 1.90 | 99.99% |
| 3c | 1.98 | 97.59% |
| 3d | 1.88 | 99.99% |
| 3e | 1.86 | 99.61% |
| 3f | 1.92 | 90.71% |
| 5a | 1.75 | 99.97% |
| 5b | 1.76 | 99.95% |
| 5c | 1.76 | 99.91% |
| 5d | 1.76 | 99.90% |
| 5e | 1.75 | 99.75% |
| 5f | 1.76 | 99.95% |
| 6a | 1.74 | 99.95% |
| 6b | 1.75 | 99.95% |
| 6c | 1.75 | 99.94% |
| 6d | 1.75 | 99.77% |
| 6e | 1.78 | 99.54% |
| 6f | 1.74 | 97.79% |
| 7ab | 1.79 | 99.29% |
| 7ac | 1.82 | 98.93% |
| 7ba | 1.86 | 98.95% |
| 7bb | 1.86 | 98.88% |
| 7bc | 1.84 | 97.08% |
| 7bd | 1.85 | 97.23% |
| 7bg | 1.83 | 98.15% |
| 7ec | 1.84 | 98.01% |
| 7fc | 1.85 | 99.44% |
4.3. Biological studies
4.3.1. Cell Culture and Antibodies.
U251 cells were obtained from ATCC (Rockville, MD). The cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 10 mg/ml L-Glutamine, 100 U/mL penicillin-streptomycin, and 0.1 mg/ml streptomycin. EphA2 overexpression cells were grown in the presence of 1 μg/ml puromycin. Cell cultures were grown at 37 °C, in a humidified atmosphere of 5% CO2 in a Thermo CO2 incubator (Grand Island, NY). Antibodies used included rabbit polyclonal antibodies against pEphA/B (Santa Cruz, Santa Cruz, CA), as well as mouse monoclonal antibody to EphA2 (clone D7, Millipore, Billerica, MA).
4.3.2. Mammalian cell viability analysis
The MTT assay was used to examine the effect of the compounds on the growth of U251 and U251 over expression EphA2 cells in four replications. 3000 cells per well were seeded with RPMI1640 medium in 96-well flat-bottomed plates for 24 h and were then exposed to various concentrations of test compounds dissolved in DMSO (final concentration ≤0.1%) in medium for 48 h. Controls received DMSO at a same concentration as that in drug-treated cells. Cells were incubated in 200 μL of 0.5 mg/mL of MTT reagent diluted in fresh media at 37°C for 3 h. Supernatants were removed from the wells, and the reduced MTT dye was solubilized with 200 μL/well DMSO. Absorbance at 570 nm was determined on a SpectraMax Plus384 spectrophotometer (Molecular Devices). Data obtained with quadruplication were normalized and fitted to a dose–response curve using GraphPad Prism v.5 (GraphPad).
4.3.3. EphA2 activation.
U251 EphA2-overexpressing cells were plated in 6-well dishes at a density of 100,000 cells/well and grown for 24 hours prior to stimulation with appropriate compounds for 30 min. 0.1% DMSO was used as vehicle control. Following treatment, cells were washed and lysed in modified RIPA Buffer (20 mM Tris-HCl pH 7.4, 20 mM NaF, 150 mM NaCl, 10% glycerol, 0.1% SDS, 0.5% DCA, 2 mM EDTA, 1% Triton X-100, 2 mM Na3VO4, and protease inhibitors, including 1 mM phenylmethylsulphonyl fluoride, and 2 μg/ml each of aprotinin and leupeptin) for 20 min, followed by immunoprecipitation. Samples were boiled 5 min and run on 4-12% Bis-Tris Plus gels (Thermo Fisher), followed by transfer to Immobilon-P PVDF membranes (Millipore, Billerica, MA). Membranes were blocked 1 hr at room temperature in 3% BSA in TBS containing 0.05% Tween-20 (TBS-T) followed by overnight incubation with pEphA/B (1:500) and EphA2 (Santa Cruz) (1:1000) primary antibodies. Membranes were washed in TBS-T and incubated with goat anti-rabbit-HRP (1:5000) secondary antibody 1 hr at room temperature, followed by washing and developing with Luminol Reagent (Santa Cruz). Band intensities were quantified using ImageJ (NIH) to generate the figure.
Supplementary Material
5. Acknowledgements
This work was supported by grants from National Institutes of Health to BW (1R01NS096956-01) and National Science Foundation Major Research Instrumentation Grants (CHE-0923398 and CHE-1126384).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Miao H, Wang B. EphA receptor signaling-Complexity and emerging themes. Semin Cell Dev Biol. 2012;23(1):16–25. doi: 10.1016/j.semcdb.2011.10.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McCarron JK, Stringer BW, Day BW, Boyd AW. Ephrin expression and function in cancer. Futur Oncol. 2010;6(1):165–176. doi: 10.2217/fon.09.146 [DOI] [PubMed] [Google Scholar]
- 3.Miao H, Li DQ, Mukherjee A, et al. EphA2 Mediates Ligand-Dependent Inhibition and Ligand-Independent Promotion of Cell Migration and Invasion via a Reciprocal Regulatory Loop with Akt. Cancer Cell. 2009;16(1):9–20. doi: 10.1016/j.ccr.2009.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Taillibert S, Kanner A, et al. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma. JAMA. 2017;318(23):2306. doi: 10.1001/jama.2017.18718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. doi: 10.1016/j.gendis.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li X, Wang L, Gu JW, et al. Up-regulation of EphA2 and down-regulation of EphrinA1 are associated with the aggressive phenotype and poor prognosis of malignant glioma. Tumour Biol. 2010;31(5):477–488. doi: 10.1007/s13277-010-0060-6 [DOI] [PubMed] [Google Scholar]
- 7.Wykosky J EphA2 as a Novel Molecular Marker and Target in Glioblastoma Multiforme. Mol Cancer Res. 2005;3(10):541–551. doi: 10.1158/1541-7786.MCR-05-0056 [DOI] [PubMed] [Google Scholar]
- 8.Liu F, Park PJ, Lai W, et al. A Genome-Wide Screen Reveals Functional Gene Clusters in the Cancer Genome and Identifies EphA2 as a Mitogen in Glioblastoma. Cancer Res. 2006;66(22):10815–10823. doi: 10.1158/0008-5472.CAN-06-1408 [DOI] [PubMed] [Google Scholar]
- 9.Miao H, Burnett E, Kinch M, Simon E, Wang B. Activation of EphA2 kinase suppresses integrin function and causes focal-adhesion-kinase dephosphorylation. Nat Cell Biol. 2000;2(2):62–69. doi: 10.1038/35000008 [DOI] [PubMed] [Google Scholar]
- 10.Carles-Kinch K, Kilpatrick KE, Stewart JC, Kinch MS. Antibody targeting of the EphA2 tyrosine kinase inhibits malignant cell behavior. Cancer Res. 2002;62(10):2840–2847. doi:12019162 [PubMed] [Google Scholar]
- 11.Noblitt LW, Bangari DS, Shukla S, et al. Decreased tumorigenic potential of EphA2-overexpressing breast cancer cells following treatment with adenoviral vectors that express EphrinA1. Cancer Gene Ther. 2004;11(11):757–766. doi: 10.1038/sj.cgt.7700761 [DOI] [PubMed] [Google Scholar]
- 12.Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Ligation of EphA2 by Ephrin A1-Fc inhibits pancreatic adenocarcinoma cellular invasiveness. Biochem Biophys Res Commun. 2004;320(4):1096–1102. doi: 10.1016/j.bbrc.2004.06.054 [DOI] [PubMed] [Google Scholar]
- 13.Guo H, Miao H, Gerber L, et al. Disruption of EphA2 receptor tyrosine kinase leads to increased susceptibility to carcinogenesis in mouse skin. Cancer Res. 2006;66(14):7050–7058. doi: 10.1158/0008-5472.CAN-06-0004 [DOI] [PubMed] [Google Scholar]
- 14.Jackson D, Gooya J, Mao S, et al. A human antibody-drug conjugate targeting EphA2 inhibits tumor growth in vivo. Cancer Res. 2008;68(22):9367–9374. doi: 10.1158/0008-5472.CAN-08-1933 [DOI] [PubMed] [Google Scholar]
- 15.Pasquale EB. Eph-Ephrin Bidirectional Signaling in Physiology and Disease. Cell. 2008;133(1):38–52. doi: 10.1016/j.cell.2008.03.011 [DOI] [PubMed] [Google Scholar]
- 16.Astin JW, Batson J, Kadir S, et al. Competition amongst Eph receptors regulates contact inhibition of locomotion and invasiveness in prostate cancer cells. Nat Cell Biol. 2010;12(12):1194–1204. doi: 10.1038/ncb2122 [DOI] [PubMed] [Google Scholar]
- 17.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10(3):165–180. doi: 10.1038/nrc2806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petty A, Idippily N, Bobba V, et al. Design and synthesis of small molecule agonists of EphA2 receptor. Eur J Med Chem. 2018;143:1261–1276. doi: 10.1016/j.ejmech.2017.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Petty A, Myshkin E, Qin H, et al. A Small Molecule Agonist of EphA2 Receptor Tyrosine Kinase Inhibits Tumor Cell Migration In Vitro and Prostate Cancer Metastasis In Vivo. Ling MT, ed. PLoS One. 2012;7(8):e42120. doi: 10.1371/journal.pone.0042120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhong B, Li Y, Idippily N, Petty A, Su B, Wang B. A quantitative LC-MS/MS method for determination of a small molecule agonist of EphA2 in mouse plasma and brain tissue. Biomed Chromatogr. December 2018:e4461. doi: 10.1002/bmc.4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barquilla A, Lamberto I, Noberini R, Heynen-Genel S, Brill LM, Pasquale EB. Protein kinase A can block EphA2 receptor-mediated cell repulsion by increasing EphA2 S897 phosphorylation. Mol Biol Cell. 2016;27(17):2757–2770. doi: 10.1091/mbc.E16-01-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JW, Han HD, Shahzad MMK, et al. EphA2 immunoconjugate as molecularly targeted chemotherapy for ovarian carcinoma. J Natl Cancer Inst. 2009;101(17):1193–1205. doi: 10.1093/jnci/djp231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B Cancer cells exploit the Eph-ephrin system to promote invasion and metastasis: tales of unwitting partners. Sci Signal. 2011;4(175):pe28. doi: 10.1126/scisignal.2002153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miao H, Gale NW, Guo H, et al. EphA2 promotes infiltrative invasion of glioma stem cells in vivo through cross-talk with Akt and regulates stem cell properties. Oncogene. 2015;34(5):558–567. doi: 10.1038/onc.2013.590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Duggineni S, Mitra S, Lamberto I, et al. Design and synthesis of potent bivalent peptide agonists targeting the EphA2 receptor. ACS Med Chem Lett. 2013;4(3):344–348. doi: 10.1021/ml3004523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mertsch K, Maas J. Blood-Brain Barrier Penetration and Drug Development from an Industrial Point of View. Curr Med Chem Nerv Syst Agents. 2002;2(3):187–201. doi: 10.2174/1568015023358067 [DOI] [Google Scholar]
- 27.Leo A, Hansch C, Elkins D. Partition coefficients and their uses. Chem Rev. 1971;71(6):525–616. doi: 10.1021/cr60274a001 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


